Abstract
Cardiac glycosides are plant-derived molecules that have shown antiproliferative properties against cancer cells, though the mechanism of action is not completely understood. We show that one cardiac glycoside, convallatoxin, presents antiproliferative effects against colorectal cancer cells in culture and that the resulting cell death is independent of the p53 tumor suppressor. Our data suggest that convallatoxin may be useful in the treatment of cancers that harbor inactivating mutations in the p53 signaling pathway.
Keywords: Cancer, Apoptosis, Cell cycle, Convallatoxin, p53
1. Introduction
It is becoming more evident that cancer can be defined as a metabolic disease [1]. One type, colorectal cancer, is on the rise with current statistics showing it as the second most common cancer in women and the third most prevalent cancer in men [2], [3]. These cancers are characterized by mutations in a number of oncogenes and tumor suppressors, with KRAS and p53, respectively, being the most commonly mutated [4]. In cancers that maintain intact p53 signaling, there is some promise in using small molecules to reactivate this pathway to inhibit cancer cell growth [5]. However, given the high mutational rate of p53 in colorectal cancers, it is imperative that research continue to explore and identify molecules that are effective at stopping the growth of colorectal cancers that lack functional p53.
Cardiac glycosides are plant-derived molecules that have traditionally been used to treat symptoms of cardiac congestion and arrhythmias [6], with one mechanism being inhibition of the Na+/K+ transporter [7], [8]. In addition to their functional effect on cardiac physiology, a number of cardiac glycosides have been reported to have anti-proliferative properties [9], [10], [11]. One of these cardiac glycosides, convallatoxin, exhibits antiproliferative properties against a number of cancer types [11], [12], [13]. It has previously been suggested that convallatoxin exposure presents a cytotoxic effect on colorectal cancer cells in culture [14], however the mechanism of action and whether this antiproliferative effect is dependent on p53 has not yet been explored.
In this study, we hypothesized that convallatoxin would be an effective antiproliferative agent against colorectal cancer cells in vitro, and that this effect would be dependent on the activity of the p53 tumor suppressor. We present data showing that convallatoxin inhibits the growth and initiates an apoptotic pathway in HCT116 colorectal cancer cells. Most importantly, we show that this effect is independent of p53, suggesting that convallatoxin may be a useful agent to treat colorectal cancers harboring inactivating mutations in the p53 pathway.
2. Methods
2.1. Cell lines and culture conditions
HCT116+/+ (#CCL-247; p53-positive) and WI-38 (#CCL-75) cells were purchased from ATCC. HCT116−/− (p53-null) cells were kindly provided by Dr. Jennifer Pietenpol (Vanderbilt University). All cells were cultured in monolayer in Dulbecco's Modified Eagle Medium (DMEM; Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco), and 1% Penicillin/Streptamycin (Gibco).
2.2. Cell proliferation and viability assays
Convallatoxin (CNT) was purchased from Sigma-Aldrich (#C9140), suspended in sterile water, and stored at 4 °C. For proliferation assays, cells were plated at 2.5 × 104 cells/well in 12-well plates and exposed to different concentrations of CNT. 24 h after treatment, cells were trypsinized and counted with a TC10 Automated Cell counter (BioRad) and cell numbers were normalized to control-treated wells. For viability assays, HCT116 cells (1 × 104) and WI-38 cells (2 × 103) were seeded in triplicate in 96-well plates and incubated for 24 h. Medium was then replaced with either control medium or medium containing 2-fold dilutions of CNT (starting concentration = 500 nM) and incubated for 24 h. Viability was determined by the addition of alamarBlue for 4 h and reading at 570 nm. 600 nm readings were used to detect background signal and data were normalized by determining the ratio of 570 nm/600 nm.
2.3. Mitotic and apoptotic analyses
To analyze mitotic and/or apoptotic cells, HCT116 cells (1 × 104) were grown on glass coverslips and exposed to CNT (40 nM) for 24 h. Following exposure, cells were stained with Hoechst 33,342 (Molecular Probes) per the manufacturer's protocol. Mitotic cells were visualized by the presence of chromosome condensation and metaphase/anaphase characteristics, while apoptotic cells were visualized by the presence of chromosomal and nuclear blebbing. A minimum of five random images were taken from each slide and all cells on each image were counted and characterized as interphase, mitotic, or apoptotic. CNT-treated cell counts were normalized by comparing numbers to control-treated cells.
2.4. Gene expression analysis
Total RNA was isolated using the SurePrep™ TrueTotal™ RNA Purification Kit (Fisher Scientific). cDNA was produced using TaqMan Reverse Transcription Reagents (Applied Biosystems) according to the manufacturer's protocol. 500 ng of total RNA was used in the reaction. SYBR Green reagents (BIO-Rad) were used to carry out real-time PCR using the QuantStudio3 qPCR machine (Thermo-Fisher). Primer sequences are described in Table 1. Data from select genes were normalized to Actin expression and analyzed using the ΔΔCt method for relative quantification.
Table 1.
Primer sequences used in this study.
| Gene symbol | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| ACTB | CATGTACGTTGCTATCCAGGC | CTCCTTAATGTCACGCACGAT |
| CDKN1A | TGTCCGTCAGAACCCATGC | AAAGTCGAAGTTCCATCGCTC |
| SFN | TTGACGACAAGAAGCGCATCAT | GTAGTGGAAGACGGAAAAGTTCA |
| PUMA | GACCTCAACGCACAGTACGAG | AGGAGTCCCATGATGAGATTGT |
| NOXA | ACCAAGCCGGATTTGCGATT | ACTTGCACTTGTTCCTCGTGG |
| BCL2 | GGTGGGGTCATGTGTGTGG | CGGTTCAGGTACTCAGTCATCC |
| BCL2L1 | GAGCTGGTGGTTGACTTTCTC | TCCATCTCCGATTCAGTCCCT |
3. Results
3.1. Convallatoxin inhibits the growth of HCT116 cells in culture
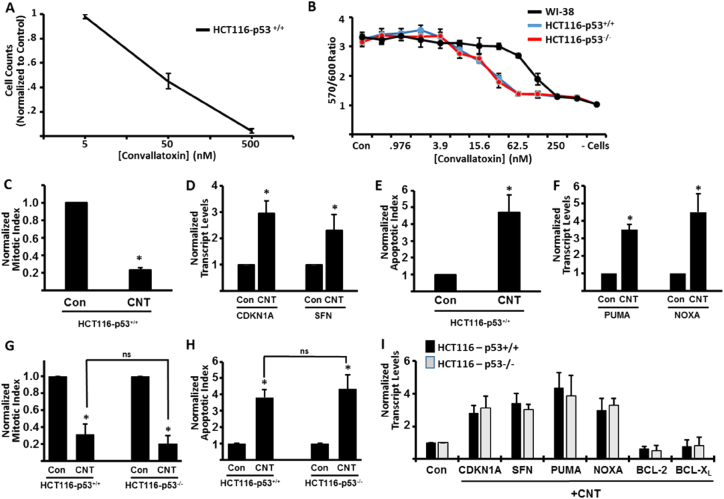
Convallatoxin has previously been shown to be an effective antiproliferative agent against some cancer types, but its effectiveness against colorectal cancer is currently unknown. To test the efficacy of convallatoxin (CNT) as an antiproliferative agent against colorectal cancer, HCT116+/+ (harboring two functional p53 alleles) cells were exposed to increasing concentrations of CNT and then counted after 24 h. While low doses were ineffective at halting cellular growth, we found that high doses (500 nM) of CNT nearly completely blocked cell growth, with the LD50 concentration being approximately 50 nM (Fig. 1A). To confirm these results via a different methodology, HCT116+/+ cells were exposed to serially diluted concentrations of CNT for 24 h and then assayed for viability using alamarBlue reagent. Consistent with Fig. 1A, CNT was effective in reducing the viability of HCT116+/+ cells (Fig. 1B). It should also be noted that, in this experiment, CNT was less effective at reducing the viability of normal, diploid, WI-38 cells, suggesting that the colorectal cancer cells being used are more sensitive to the cytotoxic effects of CNT than normal cells in culture (Fig. 1B). To determine the mechanism driving the CNT-induced reduction in viability, HCT116+/+ cells were exposed to CNT for 24 h and stained with Hoechst to visualize nuclear components. This staining procedure allows for easy analysis of mitotic cells as well as cells that have initiated an apoptotic cell death pathway [15]. Fluorescent microscopy analyses confirmed that CNT exposure resulted in a decreased mitotic index relative to control-treated cells (Fig. 1C). qPCR analysis supported our findings, as we observed an increased expression of CDKN1A (p21) and Stratifin (SFN), genes known to be upregulated during cell cycle arrest (Fig. 1D). In addition to the decreased mitotic index following CNT exposure, we also observed a significant increase in apoptotic cells (Fig. 1E), indicating that CNT is effective at inducing cell death in these cells. The increased apoptotic index was supported by increased transcript levels of PUMA and NOXA (Fig. 1F), both of which are expressed following the initiation of apoptosis [16].
Fig. 1.
Effects of CNT exposure on HCT116 cells. Control or CNT-treated HCT116+/+ cells were grown for 24 h. Following exposure, cell counts were assayed and normalized to control treated cells (A). Similarly, the indicated cells were grown for 24 h in the presence of control media or the indicated concentrations of CNT, and viability determined via alamarBlue reduction (B). For this assay, (− cells) indicates the reduction of alamarBlue in the absence of any plated cells. HCT116 +/+ cells were exposed to CNT (50 nM) for 24 h, stained with Hoechst and counted to determine the mitotic (C) and apoptotic (E) indices. Real-time PCR was used to determine the expression of cell cycle regulatory genes (D) and apoptotic genes (F) after 24 h of CNT treatment. Hoechst analysis (G–H) and real-time PCR analysis (I) was performed in HCT116+/+ and HCT116−/− cells after 24 h of CNT exposure. All data are normalized to control-treated cells and presented ± standard deviation. (ns) = not significant. *p < 0.05.
3.2. Convallatoxin inhibits cancer cell proliferation in a p53-independent manner
Of the genes analyzed by real-time PCR, all have been shown to be direct transcriptional targets of the p53 tumor suppressor [16]. In addition, HCT116+/+ cells maintain an intact p53 signaling pathway [17]. As a result, we hypothesized that the functional effects of CNT may require the activity of p53. Previously, an isogenic HCT116 cell line was generated by deleting both alleles of the p53 gene [18], so we exposed HCT116−/− cells to serially diluted concentrations of CNT for 24 h and assayed for viability via alamarBlue reduction. We found that HCT116−/− cells responded nearly identically to HCT116+/+ cells, suggesting that the CNT-mediated reduction in viability did not require functional p53 activity (Fig. 1B). We subsequently exposed HCT116−/− cells to CNT for 24 h and analyzed the resulting cells via Hoechst staining. Interestingly, we observed a decreased mitotic index and an increased apoptotic index that was not significantly different than HCT116+/+ cells (Fig. 1G–H), suggesting that p53 is not required for the anti-proliferative effects of CNT on colorectal cancer cells. In support of these data, we observed increased transcript levels of cell cycle inhibitory/apoptotic genes (CDKN1A, SFN, PUMA, NOXA), along with a slight decrease in expression of anti-apoptotic genes (BCL-2) that was also not dependent upon p53, suggesting that the anticancer effects of CNT do not require the p53 tumor suppressor.
4. Discussion
The incidence of colorectal cancer is high [2], [3] and there is always a need for novel therapeutic options for this disease. Given the strong anti-growth properties of p53, it is not surprising that previous reports have centered on the ability to reactivate this gene in cancers that maintain wild-type p53 [19]. While this is a promising approach, a significant number of colorectal cancers harbor a p53 gene that is often mutated or genomically deleted [4], making these treatment options difficult to implement. We show that convallatoxin (CNT), a cardiac glycoside, has antiproliferative and apoptotis-inducing properties towards colorectal cancer cells. Additionally, the effects observed are independent of p53. Our data suggest that convallatoxin may be useful for blocking the proliferation of colorectal cancers that have lost functional p53 due to mutational events or genomic deletion.
We found it interesting that, even in the absence of p53, CNT exposure induces the expression of PUMA and NOXA, both of which are validated p53 target genes. It should be noted that PUMA and NOXA are regulated by a number of transcription factors in addition to p53. For example, previous reports show that PUMA is a transcriptional target of p73, FoxO3a, C/EBP homologous protein (CHOP), and the E2F1 transcription factors [20], [21], [22], while NOXA can be transcriptionally controlled by additional proteins such as p73 and E2F1 [23], [24], among others. Further research is necessary to determine the specific proteins and/or pathways that are regulating the expression of these pro-apoptotic genes in response to CNT. We also analyzed the expression of antiapoptotic genes (Bcl-2/Bcl-XL) following CNT treatment. While we saw negligible changes in Bcl-XL expression, we did observe a reproducible decrease in Bcl-2 expression following exposure to CNT. It should also be noted that this decrease in Bcl-2 occurred regardless of p53 status, again emphasizing that the effects of CNT do not require functional p53.
Cardiac glycosides have previously been shown to function by selectively antagonizing the Na+ K+ transporter [7], [8]. It is possible that this is a potential mechanism driving the antiproliferative properties of CNT on HCT116 cells in our study, as previous reports have suggested that colorectal cancers upregulate specific isoforms of this transporter [25]. In addition to this potential mechanism of action, previous research suggests that cardiac glycosides maintain cytotoxic properties against cells that are independent of their ability to abrogate sodium and potassium transport [14], [26]. Cardiac glycosides have been shown to modulate calcium-dependent caspase activation [8], increase the production of reactive oxygen species [27], and negatively affect the function of topoisomerase enzymes [28]. Given these previously reported functions of cardiac glycosides, along with data from our group (Fig. 1B) and others [10] suggesting that cancer-derived cells are more sensitive to their effects, we believe that research should continue to examine the many mechanisms behind the use of CNT as a potential anti-proliferative compound. Further, most interesting is the observation that CNT induces an apoptotic cascade as observed by apoptosis-induced membrane blebbing, and that this cell death pathway is activated in the absence of p53. While the absence of p53 is known to grant cells the ability to proliferate at higher rates, it has also been suggested that the lack of functional p53 can sensitize cancer cells to chemotherapeutic agents that modify chromatin and/or induce high amounts of DNA damage [29]. While there is no current evidence that CNT specifically acts on chromatin or is able to generate genotoxic stress, further research is required to identify the specific mechanism(s) that are driving CNT-mediated cellular death. Collectively, our data suggests that CNT should be further explored as a potential therapeutic for colorectal cancers, in particular those that have inactivated the p53 tumor suppressor signaling pathway.
Funding support
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- 1.Seyfried T.N., Flores R.E., Poff A.M., D'Agostino D.P. Cancer as a metabolic disease: implications for novel therapeutics. Carcinogenesis. 2014;35:515–527. doi: 10.1093/carcin/bgt480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tariq K., Ghias K. Colorectal cancer carcinogenesis: a review of mechanisms. Cancer Biol. Med. 2016;13:120–135. doi: 10.28092/j.issn.2095-3941.2015.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 4.Yan W.F., Wu G., Sun P.C., Qiu D. P53 mutations occur more commonly than KRAS mutations in colorectal adenoma. Int. J. Clin. Exp. Med. 2015;8:1370–1375. [PMC free article] [PubMed] [Google Scholar]
- 5.Shen H., Maki C.G. Pharmacologic activation of p53 by small-molecule MDM2 antagonists. Curr. Pharm. Des. 2011;17:560–568. doi: 10.2174/138161211795222603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calderon-Montano J.M., Burgos-Moron E., Orta M.L., Maldonado-Navas D., Garcia-Dominguez I., Lopez-Lazaro M. Evaluating the cancer therapeutic potential of cardiac glycosides. Biomed. Res. Int. 2014;2014:794930. doi: 10.1155/2014/794930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rahimtoola S.H., Tak T. The use of digitalis in heart failure. Curr. Probl. Cardiol. 1996;21:781–853. doi: 10.1016/s0146-2806(96)80001-6. [DOI] [PubMed] [Google Scholar]
- 8.Schoner W., Scheiner-Bobis G. Endogenous and exogenous cardiac glycosides: their roles in hypertension, salt metabolism, and cell growth. Am. J. Physiol. Cell. Physiol. 2007;293:C509–36. doi: 10.1152/ajpcell.00098.2007. [DOI] [PubMed] [Google Scholar]
- 9.Hallbook H., Felth J., Eriksson A., Fryknas M., Bohlin L., Larsson R., Gullbo J. Ex vivo activity of cardiac glycosides in acute leukaemia. PLoS One. 2011;6 doi: 10.1371/journal.pone.0015718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haux J. Digitoxin is a potential anticancer agent for several types of cancer. Med. Hypotheses. 1999;53:543–548. doi: 10.1054/mehy.1999.0985. [DOI] [PubMed] [Google Scholar]
- 11.Kaushik V., Azad N., Yakisich J.S., Iyer A.K. Antitumor effects of naturally occurring cardiac glycosides convallatoxin and peruvoside on human ER + and triple-negative breast cancers. Cell. Death Discov. 2017;3:17009. doi: 10.1038/cddiscovery.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang S.Y., Kim N.H., Cho Y.S., Lee H., Kwon H.J. Convallatoxin, a dual inducer of autophagy and apoptosis, inhibits angiogenesis in vitro and in vivo. PLoS One. 2014;9 doi: 10.1371/journal.pone.0091094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaushik V., Yakisich J.S., Azad N., Kulkarni Y., Venkatadri R., Wright C., Rojanasakul Y., Iyer A.K. Anti-tumor effects of cardiac glycosides on human lung cancer cells and lung tumorspheres. J. Cell. Physiol. 2016 doi: 10.1002/jcp.25611. [DOI] [PubMed] [Google Scholar]
- 14.Felth J., Rickardson L., Rosen J., Wickstrom M., Fryknas M., Lindskog M., Bohlin L., Gullbo J. Cytotoxic effects of cardiac glycosides in colon cancer cells, alone and in combination with standard chemotherapeutic drugs. J. Nat. Prod. 2009;72:1969–1974. doi: 10.1021/np900210m. [DOI] [PubMed] [Google Scholar]
- 15.Frey T. Nucleic acid dyes for detection of apoptosis in live cells. Cytometry. 1995;21:265–274. doi: 10.1002/cyto.990210307. [DOI] [PubMed] [Google Scholar]
- 16.Schuler M., Green D.R. Mechanisms of p53-dependent apoptosis. Biochem. Soc. Trans. 2001;29:684–688. doi: 10.1042/0300-5127:0290684. [DOI] [PubMed] [Google Scholar]
- 17.Take Y., Kumano M., Teraoka H., Nishimura S., Okuyama A. DNA-dependent protein kinase inhibitor (OK-1035) suppresses p21 expression in HCT116 cells containing wild-type p53 induced by adriamycin. Biochem. Biophys. Res. Commun. 1996;221:207–212. doi: 10.1006/bbrc.1996.0575. [DOI] [PubMed] [Google Scholar]
- 18.Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J.P., Sedivy J.M., Kinzler K.W., Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 19.Selivanova G. Wild type p53 reactivation: from lab bench to clinic. FEBS Lett. 2014;588:2628–2638. doi: 10.1016/j.febslet.2014.03.049. [DOI] [PubMed] [Google Scholar]
- 20.Melino G., Bernassola F., Ranalli M., Yee K., Zong W.X., Corazzari M., Knight R.A., Green D.R., Thompson C., Vousden K.H. p73 induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J. Biol. Chem. 2004;279:8076–8083. doi: 10.1074/jbc.M307469200. [DOI] [PubMed] [Google Scholar]
- 21.You H., Pellegrini M., Tsuchihara K., Yamamoto K., Hacker G., Erlacher M., Villunger A., Mak T.W. FOXO3a-dependent regulation of Puma in response to cytokine/growth factor withdrawal. J. Exp. Med. 2006;203:1657–1663. doi: 10.1084/jem.20060353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Futami T., Miyagishi M., Taira K. Identification of a network involved in thapsigargin-induced apoptosis using a library of small interfering RNA expression vectors. J. Biol. Chem. 2005;280:826–831. doi: 10.1074/jbc.M409948200. [DOI] [PubMed] [Google Scholar]
- 23.Flinterman M., Guelen L., Ezzati-Nik S., Killick R., Melino G., Tominaga K., Mymryk J.S., Gaken J., Tavassoli M. E1A activates transcription of p73 and Noxa to induce apoptosis. J. Biol. Chem. 2005;280:5945–5959. doi: 10.1074/jbc.M406661200. [DOI] [PubMed] [Google Scholar]
- 24.Hershko T., Ginsberg D. Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J. Biol. Chem. 2004;279:8627–8634. doi: 10.1074/jbc.M312866200. [DOI] [PubMed] [Google Scholar]
- 25.Sakai H., Suzuki T., Maeda M., Takahashi Y., Horikawa N., Minamimura T., Tsukada K., Takeguchi N. Up-regulation of Na(+),K(+)-ATPase alpha 3-isoform and down-regulation of the alpha1-isoform in human colorectal cancer. FEBS Lett. 2004;563:151–154. doi: 10.1016/S0014-5793(04)00292-3. [DOI] [PubMed] [Google Scholar]
- 26.Paula S., Tabet M.R., Ball W.J., Jr. Interactions between cardiac glycosides and sodium/potassium-ATPase: three-dimensional structure-activity relationship models for ligand binding to the E2-Pi form of the enzyme versus activity inhibition. Biochemistry. 2005;44:498–510. doi: 10.1021/bi048680w. [DOI] [PubMed] [Google Scholar]
- 27.Newman R.A., Yang P., Hittelman W.N., Lu T., Ho D.H., Ni D., Chan D., Vijjeswarapu M., Cartwright C., Dixon S., Felix E., Addington C. Oleandrin-mediated oxidative stress in human melanoma cells. J. Exp. Ther. Oncol. 2006;5:167–181. [PubMed] [Google Scholar]
- 28.Bielawski K., Winnicka K., Bielawska A. Inhibition of DNA topoisomerases I and II, and growth inhibition of breast cancer MCF-7 cells by ouabain, digoxin and proscillaridin A. Biol. Pharm. Bull. 2006;29:1493–1497. doi: 10.1248/bpb.29.1493. [DOI] [PubMed] [Google Scholar]
- 29.Nieto M., Samper E., Fraga M.F., Gonzalez de Buitrago G., Esteller M., Serrano M. The absence of p53 is critical for the induction of apoptosis by 5-aza-2′-deoxycytidine. Oncogene. 2004;23:735–743. doi: 10.1038/sj.onc.1207175. [DOI] [PubMed] [Google Scholar]