Abstract
An invariant substructure that forms two interlocked pairs of neighboring β-strands occurs in essentially all known sandwich-like proteins. Eight conserved positions in these strands were recently shown to act as structural determinants. To test whether the residues at these invariant positions are conserved for mechanistic (i.e., part of folding nucleus) or energetic (i.e., governing native-state stability) reasons, we characterized the folding behavior of eight point-mutated variants of the sandwich-like protein Pseudomonas aeruginosa apo-azurin. We find a simple relationship among the conserved positions: half of the residues form native-like interactions in the folding transition state, whereas the others do not participate in the folding nucleus but govern high native-state stability. Thus, evolutionary preservation of these specific positions gives both mechanistic and energetic advantages to members of the sandwich-like protein family.
Keywords: protein folding, folding transition state
Sandwich-like proteins are a group of very diverse proteins comprising 69 superfamilies in 38 protein folds (1). The general architecture of these proteins involves β-strands that form two main β-sheets that pack face-to-face. A recent structural and sequence analysis of the arrangements of strands within the sandwich sheets revealed a rigorously defined constraint on the supersecondary structure that holds true for 94% of all known sandwich-like structures (1). This invariant substructure consists of two interlocked pairs of neighboring β-strands. Within these four strands, eight hydrophobic positions (two in each strand) were found to have fixed structural roles at the interface between the β-sheets forming the common geometrical core. The hydrophobic residues at these eight positions are conserved across all sandwich-like proteins (1). The identification of a distinct set of structural determinants in a group of such diverse proteins as the sandwich-like proteins may shed light on how this architecture is controlled by the primary structure. Explicitly, are the eight residues invariant across all sandwich-like proteins preferentially conserved to direct the folding reaction (i.e., participating in the folding nucleus) or are they selected to stabilize the final structure? Here, we answer this question by using Pseudomonas aeruginosa azurin as our model system.
P. aeruginosa azurin is a small (128 residues) single-domain protein with a β-barrel structure composed of eight β-strands, which belongs to the sandwich-like protein family (2). In vivo, a redox-active copper is coordinated to the protein, allowing for electron-transfer activity (2). The copper in azurin can be eliminated, creating apo-azurin, without change of the overall structure (3). Apo-azurin is an excellent model system because equilibrium- and kinetic-folding processes for apo-azurin are two-state reactions (4-8). Moreover, the stability of the apoprotein is rather high, and several mutants with native-like structure have been created (2, 6, 9, 10). Apo-azurin folds via a ≈60% native-like transition state with a speed that is in good agreement with the predicted value based on the native-state topology (4, 5, 11). Here, we report the folding behavior of eight single-point-mutated apo-azurin variants in which the residues at the positions acting as structural determinants across sandwich-like proteins have been exchanged individually for alanine. We find that the conserved positions play mutually exclusive roles in defining the folding mechanism and protein stability: half of the residues participate in the folding nucleus with little affect on native-state stability, whereas the other half governs high native-state stability without participating in the folding transition state.
Materials and Methods
Construction of Variants. Upon structurally based sequence alignment of P. aeruginosa azurin with other sandwich-like proteins, we identified the eight interlocked-pair residues as Val-31 and Leu-33 in strand three, Trp-48 and Leu-50 in strand four, Val-95 and Phe-97 in strand six, and Tyr-108 and Phe-110 in strand seven. Based on this identification, we generated eight single-point-mutated variants of azurin with target residues replaced by alanines: Val-31–Ala, Leu-33–Ala, Trp-48–Ala, Leu-50–Ala, Val-95–Ala, Phe-97–Ala, Tyr-108–Ala, and Phe-110–Ala. A pUC18 vector containing the WT P. aeruginosa azurin gene was used as template to create the variants via QuikChange (Stratagene) site-directed mutagenesis as in ref. 12. Variants were expressed in Escherichia coli BL21PlysS cells, and purification was performed by using an established procedure (6). Apoprotein was prepared by extensive cyanide dialysis and verified by copper-titration monitoring absorption at 630 nm (6).
Equilibrium Unfolding. Guanidine hydrochloride (GuHCl)-induced equilibrium unfolding was performed in 100 mM phosphate, pH 7.0, 25°C by using fluorescence (excitation at 285 nm; emission monitored at 308 nm) and far-UV CD detection (200–300 nm). Samples were incubated for 2 h before measurements. The equilibrium-unfolding reactions were reversible without any display of protein concentration dependence (in the 5–50 μM protein range). For Trp-48–Ala apo-azurin, tyrosine fluorescence was used in combination with far-UV CD. The equilibrium-unfolding curves for WT and mutant apo-azurins were analyzed by using a two-state model (13, 14). For each apo-azurin variant, far-UV CD- and fluorescence-detected transitions overlapped. The ΔGU(H2O) values listed in Table 1 are average values from fits to both CD and fluorescence curves. Errors (standard deviations) are derived from multiple experiments.
Table 1. Thermodynamic stability, folding speed, and φ values for WT and point-mutated apo-azurin variants (pH 7, 25°C).
| Apo-azurin variant | Mutated β-strand | ΔGU(H2O), kJ/mol | ΔΔGU(H2O), kJ/mol | kF(H2O), s-1 | ΔΔG‡(H2O), kJ/mol | φ† |
|---|---|---|---|---|---|---|
| WT | — | 25.0 ± 0.7 | — | 160 ± 20 | — | — |
| Val-31—Ala | 3 | 20.0 ± 0.4 | -5.0 ± 0.7 | 23 ± 3 | -4.72 ± 0.3 | 0.93 ± 0.14 |
| Leu-33—Ala | 3 | 21.0 ± 0.3 | -4.0 ± 0.7 | 35 ± 2 | -3.70 ± 0.3 | 0.91 ± 0.18 |
| Trp-48—Ala | 4 | 4.0 ± 0.3 | -21.0 ± 0.7 | 22 ± 6 | -4.83 ± 0.7 | 0.23 ± 0.03§ |
| Leu-50—Ala | 4 | 18.0 ± 0.2 | -7.0 ± 0.7 | 5 ± 1 | -7.3 ± 0.3 | 1.04 ± 0.11 |
| Val-95—Ala | 6 | 22.5 ± 0.2 | -2.5 ± 0.7 | 78 ± 3 | -1.75 ± 0.3 | 0.63 ± 0.21¶ |
| Phe-97—Ala | 6 | 15.0 ± 0.7 | -10.0 ± 0.7 | 40 ± 6 | -3.38 ± 0.3 | 0.35 ± 0.04 |
| Tyr-108—Ala | 7 | 12.5 ± 0.2 | -12.5 ± 0.7 | 23 ± 3 | -4.72 ± 0.3 | 0.37 ± 0.03 |
| Phe-110—Ala | 7 | 11.0 ± 0.9 | -14.0 ± 0.9 | 54 ± 5 | -2.64 ± 0.3 | 0.19 ± 0.02 |
The φ values are calculated as φ = ΔΔG‡(H2O)/ΔΔGU(H2O), where ΔΔGU(H2O) is calculated as ΔGU(mutant,H2O)-ΔGU(WT,H2O) and ΔΔG‡(H2O) as RTln[kf(mutant,H2O)/kf(WT,H2O)].
Standard deviations (σ) for the φ values were calculated as in ref. 19: σφ = |φ|*√[(σΔΔG‡/ΔΔG‡)2+(σΔΔGU/ΔΔGU))2].
Not used in analysis because this value may contain large errors; ΔGU(H2O) is only 4.0 kJ/mol and the refolding rate constant in water had to be estimated, assuming a two-state reaction, using the equilibrium stability and the unfolding speed.
Not used in analysis because ΔGU(mutant,H2O)-ΔGU(WT,H2O) is only 2.5 kJ/mol and the φ-value estimation may be unreliable.
Folding Dynamics. Time-resolved folding and unfolding was probed by fluorescence (excitation at 285 nm; emission monitored at 308 nm) and far-UV CD (at 220 nm) by using an Applied Photophysics (Surrey, U.K.) Pi-Star stopped-flow mixer. Both detection modes gave identical kinetic traces in all cases. Apoazurin variants were mixed in a 1:10 ratio with appropriate GuHCl/buffer solutions. Six kinetic traces were averaged and fit to monophasic decay equations. There were no missing amplitudes (within the 2- to 3-ms dead time) in either fluorescence or far-UV CD kinetic traces, and no protein concentration dependence was found (in the 5–50 μM protein range tested). Unfolding and refolding rate constants at different GuHCl concentrations were fit by assuming standard linear dependence of lnkF and lnkU on GuHCl concentration (15). Because of the low stability of Trp-48–Ala apo-azurin, its kF(H2O) was derived from the equilibrium stability, ΔGU(H2O), and the extrapolated unfolding rate constant, kU(H2O). For all other variants, equilibrium [i.e., ΔGU(H2O)] and kinetically derived {i.e., RTln [kF(H2O)/kU(H2O)]} stabilities agreed within 10%. Errors (standard deviations) in rate constants were derived from multiple experiments.
Results and Discussion
To probe the role in folding of the eight structural determinants found across most sandwich-like proteins, we exchanged each of the conserved residues individually with alanine by using P. aeruginosa azurin as our model. All eight apo-azurin variants (Val-31–Ala, Leu-33–Ala, Trp-48–Ala, Leu-50–Ala, Val-95–Ala, Phe-97–Ala, Tyr-108–Ala, and Phe-110–Ala) adopted folded structures identical to WT azurin as deduced from their far-UV CD and tryptophan (Trp-48) emission (except Trp-48–Ala) spectra, as well as their ability to bind copper (data not shown).
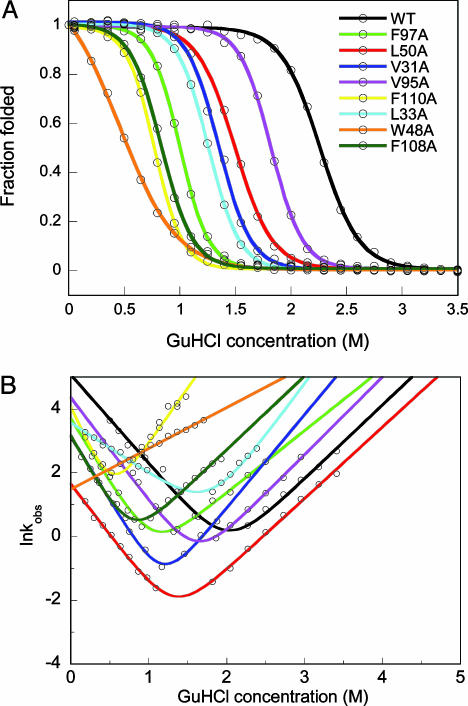
The thermodynamic stability of each apo-variant was tested by GuHCl titrations monitored by far-UV CD and fluorescence. Like WT apo-azurin (4, 6, 8, 10, 11), the variants unfold in single, reversible transitions (Fig. 1A); curves derived from CD and fluorescence data overlap for each protein, which is indicative of two-state equilibrium-unfolding processes. The thermodynamic stability is lower for all of the variants as compared with the stability of WT apo-azurin (Table 1): the most dramatic reduction in stability is found for Trp-48–Ala apo-azurin. In accord, the unique fluorescence of Trp-48 is believed to arise because of rigid encapsulation of this side chain (16). Folding and unfolding kinetics of the variants were probed by fluorescence and far-UV CD detection methods using stopped-flow mixing (Fig. 1B). In support of two-state reactions, all kinetic traces are single-exponential decays, without any missing amplitudes; CD and fluorescence data overlap at each condition. Moreover, no protein concentration dependence in either unfolding or refolding phases was observed. For all variants, the folding speed in water is slower than that of WT apo-azurin (Table 1).
Fig. 1.
Equilibrium and kinetic folding data. (A) GuHCl-induced equilibrium unfolding of WT and apo-azurin variants. For each protein, far-UV CD- and fluorescence-detected transitions overlap (fluorescence data shown). Solid curves are two-state fits to each set of data (see Table 1). (B) Semilogarithmic graph of lnkobs versus GuHCl for WT and point-mutated apo-azurins. For each protein at each solvent condition, kinetic folding/unfolding traces monitored by far-UV CD and fluorescence are identical (fluorescence data shown). Solid lines are fits to two-state kinetic equations. Because of low stability of Trp-48–Ala apo-azurin, only kU values were measured (these data are fitted to a line, the unfolding arm). Folding rate constants are reported in Table 1.
We used φ-value analysis (17, 18) to obtain information about the involvement of the mutated residues in azurin's folding transition state. By measuring the change in protein stability, ΔΔGU(H2O), and the change in folding barrier [i.e., folding speed; ΔΔG‡(H2O)] caused by mutation of a particular residue, the φ value for that residue can be calculated as φ = ΔΔG‡/ΔΔGU (see definitions in Table 1 legend). A φ value of 0 indicates that the site of mutation is unfolded in the transition state, whereas a φ value of 1 indicates that the site of mutation forms native-like interactions in the transition-state structure. Fractional φ values may be interpreted as possessing different degrees of structure in the folding nucleus. The φ values derived for the eight mutated positions in azurin are given in Table 1. Concern regarding φ values involving low ΔΔGU(H2O) values was recently raised by Sanchez and Keifhaber (19), suggesting a cutoff at 7 kJ/mol in ΔΔGU(H2O) for reliable φ values. In a response, Fersht and Sato (20) demonstrated that φ values using ΔΔGU(H2O) as low as 2.5 kJ/mol can be accurately determined in many cases; this finding was reinforced by molecular dynamics simulations of reversible peptide folding (21). Cutoffs in ΔΔGU(H2O) that have generally been used in φ-value literature range from 0.8 to 2.9 kJ/mol (20, 22, 23). Because ΔΔGU(H2O) is only 2.5 kJ/mol for the Val-95–Ala apo-azurin variant, we exclude its φ value from the analysis (Table 1). In addition, we note that the stability of Trp-48–Ala apo-azurin is only 4.0 kJ/mol (i.e., it is only folded to ≈85% in buffer), and the refolding rate constant in water had to be estimated by using the equilibrium stability and the unfolding speed, assuming a two-state reaction. Therefore, the φ value derived for this position may include propagated errors and is not used in our analysis.
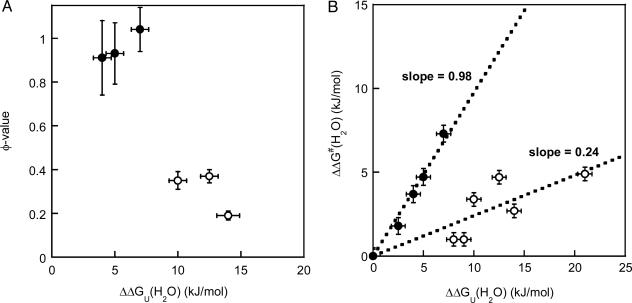
Among the six variants for which reliable φ values could be calculated, we find a surprisingly simple correlation between effect on thermodynamic stability and involvement in the folding transition state. The residues fall into two classes, those with high φ values (φ >0.9; Val-31–Ala, Val-33–Ala, and Leu-50–Ala) and those with relatively low φ values (φ <0.4; Phe-97–Ala, Tyr-108–Ala, and Phe-110–Ala). Moreover, the residues with high φ values do not contribute much to the overall stability [ΔΔGU(H2O) of 4–7 kJ/mol for Val-31–Ala, Val-33–Ala, and Leu-50–Ala], whereas the residues with low φ values contribute significantly to the thermodynamic stability [ΔΔGU(H2O) of 10–14 kJ/mol for Phe-97–Ala, Tyr-108–Ala, and Phe-110–Ala] (Fig. 2A). The two groups of residues can also be illustrated by the use of a Brønsted plot (Fig. 2B) (24). Here, we also include the Trp-48–Ala and Val-31–Ala data and earlier data on two other azurin mutants (6). It has been shown for some two-state proteins (for example, CI-2) that if all mutants fall on the same line, this correlation reflects a uniform transition state that is an expanded version of the native state (25). This is not the case for azurin: the data appear separated into two discrete lines, implying that one region folds early (slope = φav = 0.98) and other parts do not form native-like interactions until after the folding transition state (slope = φav = 0.24).
Fig. 2.
Residue-specific transition-state analysis. (A) φ values as a function of protein stability effect [ΔΔGU(H2O); with respect to WT apo-azurin] for six apo-azurin variants (data taken from Table 1). The two classes of variants, as discussed in the text, are shown by open (low φ) and filled (high φ) circles. Standard deviations are shown in both dimensions. (B) Brønsted analysis of azurin mutations. Plot shows ΔΔG‡(H2O) versus ΔΔGU(H2O) (see Table 1 for definitions), with standard deviations in both dimensions, for the eight positions studied here and His-117–Gly and His-46–Gly apo-azurin variants (6). The separation of the data points into two discrete sets illustrates that the folding nucleus is not uniform but divided into (at least) two regions. The two groups are represented by filled (slope = 0.98, R = 0.99) and open circles (slope = 0.24, R = 0.84).
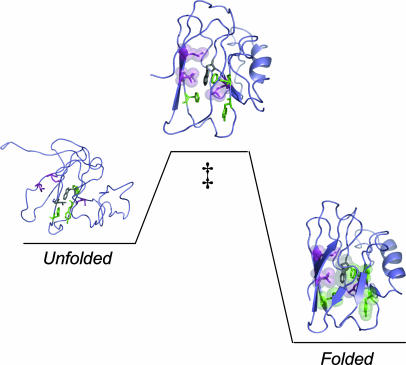
The three residues with φ values close to 1 are highlighted in the folding scheme shown in Fig. 3. Val-31 and Leu-50 are within 4–5 Å of each other in the same layer of the β-sandwich (26), and Leu-33 packs with Val-31. If these residues form native-like interactions in the folding transition state, they bring parts of strands three and four together. The low φ values of the other three residues suggest that strand seven and part of strand six are not well defined in the folding transition state. Previous work showed that position 46 (in loop between strands three and four) and 117 (in loop between strands seven and eight) are not involved in apo-azurin's folding nucleus (i.e., φ < 0.2 for these two positions) (6, 11). In a recent study of a Cys-112–Ser variant of azurin, Karlsson and coworkers (27) concluded that Val-31, Leu-33, and Leu-50 are important in the folding transition state (27). Their finding is in agreement with our observations on WT apo-azurin, although their absolute φ values are lower and folding is not two-state for the Cys-112–Ser variant (27). Taken together, azurin's folding transition state is localized with at least one region of near-complete structure.
Fig. 3.
Illustration of apo-azurin going from unfolded to folded via a high-energy transition state (‡). The eight residues constituting the structural determinants across sandwich-like proteins are shown in stick format. The three residues forming native-like interactions in the transition state (high φ) are shown in magenta (space-filled in transition state) and the three residues involved in governing high native-state stability (low φ) are shown in green. The remaining two residues, with unreliable φ values, are shown in gray (prepared by using Protein Data Bank entry 1AZU and pymol).
Among other proteins with sandwich-like structures, only two fibronectin type III (fnIII) domains have been extensively characterized with respect to their folding nuclei (23, 28). The third fnIII domain of human tenascin (TNfnIII), which folds by a two-state kinetic mechanism like apo-azurin, was shown to have a localized folding transition state dominated by four core residues, one in each of the β-strands B, C, E, and F (23). Upon alignment of TNfnIII's sequence with other sandwich-like proteins in ref. 1, we discovered that these strands are responsible for forming the interlocked pair and the four high-φ positions are in fact among the eight structural determinants found in all sandwich-like proteins. Three of the remaining four conserved positions were tested, and they all had low φ values (23). Two of the high-φ positions and two low-φ positions are identical between the azurin and TNfnIII, whereas high and low φ values are switched at two positions. The remaining two positions could not be compared because the φ value for position 95 in azurin was unreliable and position 22 in TNfnIII was not tested. Taken together, the distribution between high and low φ values among the structural determinants is the same, but only half of the positions have identical properties, in these two proteins. Clearly, studies of other sandwich-like proteins are of high importance for making general conclusions.
If evolution preserves kinetically important residues or not has been a subject of debate in the literature (29–31). In several experimental studies little or no correlation between sequence conservation and participation in the folding transition state was found. The only exception is CheY, where the residues in the folding nucleus appear significantly more conserved than other residues (29). Here, we demonstrate that residues involved in the folding transition state are among conserved structural determinants in the sandwich-like proteins P. aeruginosa azurin and TNfnIII. Thus, during the evolution of sandwich-like proteins, clearly some residues have been preserved for folding mechanistic reasons. To determine whether the residues in the folding transition state of azurin are preferentially conserved will require additional studies of many nonconserved residues. In the case of TNfnIII, an extensive study of 32 different positions was made (23); because the four positions with the highest φ values are among the conserved interlocked-pair residues, the folding transition state in TNfnIII appears to be preferentially conserved.
In summary, using P. aeruginosa apo-azurin as our model, we have shown that six of eight conserved positions acting as structural determinants across sandwich-like proteins (1) play mutually exclusive roles in defining azurin's folding mechanism and structural stability. Half of these residues form native-like interactions in the folding transition state [i.e., φ ≈1 with small effect on ΔGU(H2O)], whereas the other half governs high native-state stability [i.e., large effect on ΔGU(H2O) with φ ≈0.24)]. We propose that the evolutionary conservation of these specific positions gives a combination of mechanistic and energetic advantages to most members of the sequence-diverse, but structurally similar, sandwich-like protein family.
Acknowledgments
We thank Jessica Marks and Tom Guu for initial work. This work was supported by National Institutes of Health Grant GM059663 and Robert A. Welch Foundation Grant C-1588 (to P.W.-S.). C.J.W. is supported by the Houston Area Molecular Biophysics Program (Grant GM08280).
Author contributions: C.J.W. and P.W.-S. designed research; C.J.W. performed research; C.J.W. and P.W.-S. analyzed data; and P.W.-S. wrote the paper.
Abbreviations: GuHCl, guanidine hydrochloride; fnIII, fibronectin type III; TNfnIII, fnIII domain of human tenascin.
References
- 1.Kister, A. E., Finkelstein, A. V. & Gelfand, I. M. (2002) Proc. Natl. Acad. Sci. USA 99, 14137–14141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adman, E. T. (1991) Adv. Protein Chem. 42, 145–197. [DOI] [PubMed] [Google Scholar]
- 3.Nar, H., Messerschmidt, A., Huber, R., van de Kamp, M. & Canters, G. W. (1992) FEBS Lett. 306, 119–124. [DOI] [PubMed] [Google Scholar]
- 4.Pozdnyakova, I., Guidry, J. & Wittung-Stafshede, P. (2001) Arch. Biochem. Biophys. 390, 146–148. [DOI] [PubMed] [Google Scholar]
- 5.Pozdnyakova, I. & Wittung-Stafshede, P. (2001) Biochemistry 40, 13728–13733. [DOI] [PubMed] [Google Scholar]
- 6.Pozdnyakova, I., Guidry, J. & Wittung-Stafshede, P. (2002) Biophys. J. 82, 2645–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pozdnyakova, I. & Wittung-Stafshede, P. (2003) Biochim. Biophys. Acta 1651, 1–4. [DOI] [PubMed] [Google Scholar]
- 8.Fuentes, L., Oyola, J., Fernandez, M. & Quinones, E. (2004) Biophys. J. 87, 1873–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guzzi, R., Sportelli, L., La Rosa, C., Milardi, D., Grasso, D., Verbeet, M. P. & Canters, G. W. (1999) Biophys. J. 77, 1052–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mei, G., Di Venere, A., Campeggi, F. M., Gilardi, G., Rosato, N., De Matteis, F. & Finazzi-Agro, A. (1999) Eur. J. Biochem. 265, 619–626. [DOI] [PubMed] [Google Scholar]
- 11.Pozdnyakova, I. & Wittung-Stafshede, P. (2001) J. Am. Chem. Soc. 123, 10135–10136. [DOI] [PubMed] [Google Scholar]
- 12.Marks, J., Pozdnyakova, I., Guidry, J. & Wittung-Stafshede, P. (2004) J. Biol. Inorg. Chem. 9, 281–288. [DOI] [PubMed] [Google Scholar]
- 13.Fersht, A. (1999) Structure and Mechanism in Protein Science (Freeman, New York).
- 14.Pace, C. N. & Shaw, K. L. (2000) Proteins 4, Suppl., 1–7. [DOI] [PubMed] [Google Scholar]
- 15.Fersht, A. R. (1997) Curr. Opin. Struct. Biol. 7, 3–9. [DOI] [PubMed] [Google Scholar]
- 16.Gilardi, G., Mei, G., Rosato, N., Canters, G. W. & Finazzi-Agro, A. (1994) Biochemistry 33, 1425–1432. [DOI] [PubMed] [Google Scholar]
- 17.Matouschek, A., Kellis, J. T., Jr., Serrano, L., Bycroft, M. & Fersht, A. R. (1990) Nature 346, 440–445. [DOI] [PubMed] [Google Scholar]
- 18.Matouschek, A. & Fersht, A. R. (1991) Methods Enzymol. 202, 82–112. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez, I. E. & Kiefhaber, T. (2003) J. Mol. Biol. 334, 1077–1085. [DOI] [PubMed] [Google Scholar]
- 20.Fersht, A. R. & Sato, S. (2004) Proc. Natl. Acad. Sci. USA 101, 7976–7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Settanni, G., Rao, F. & Caflisch, A. (2005) Proc. Natl. Acad. Sci. USA 102, 628–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riddle, D. S., Grantcharova, V. P., Santiago, J. V., Alm, E., Ruczinski, I. & Baker, D. (1999) Nat. Struct. Biol. 6, 1016–1024. [DOI] [PubMed] [Google Scholar]
- 23.Hamill, S. J., Steward, A. & Clarke, J. (2000) J. Mol. Biol. 297, 165–178. [DOI] [PubMed] [Google Scholar]
- 24.Fersht, A. R., Itzhaki, L. S., elMasry, N. F., Matthews, J. M. & Otzen, D. E. (1994) Proc. Natl. Acad. Sci. USA 91, 10426–10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Itzhaki, L. S., Otzen, D. E. & Fersht, A. R. (1995) J. Mol. Biol. 254, 260–288. [DOI] [PubMed] [Google Scholar]
- 26.Nar, H., Messerschmidt, A., Huber, R., van de Kamp, M. & Canters, G. W. (1991) J. Mol. Biol. 221, 765–772. [DOI] [PubMed] [Google Scholar]
- 27.Engman, K. C., Sandberg, A., Leckner, J. & Karlsson, B. G. (2004) Protein Sci. 13, 2706–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cota, E., Steward, A., Fowler, S. B. & Clarke, J. (2001) J. Mol. Biol. 305, 1185–1194. [DOI] [PubMed] [Google Scholar]
- 29.Larson, S. M., Ruczinski, I., Davidson, A. R., Baker, D. & Plaxco, K. W. (2002) J. Mol. Biol. 316, 225–233. [DOI] [PubMed] [Google Scholar]
- 30.Mirny, L. & Shakhnovich, E. (2001) J. Mol. Biol. 308, 123–129. [DOI] [PubMed] [Google Scholar]
- 31.Tseng, Y. Y. & Liang, J. (2004) J. Mol. Biol. 335, 869–880. [DOI] [PubMed] [Google Scholar]