Abstract
A robust method for simultaneous visualization of all four cell cycle phases in living cells would be highly desirable. We developed an intensiometric reporter of the S/G2 transition and engineered a far-red fluorescent protein, mMaroon1, to visualize chromatin condensation in mitosis. We combined these new reporters with the previously described Fucci system to create Fucci4, a set of four orthogonal fluorescent indicators that together resolve all cell cycle phases.
Proper regulation of the cell division cycle is essential for multicellular life, including for normal development1,2, stem cell renewal3, and tissue maintenance4. In addition, cell cycle abnormalities in cancer contribute to tumor growth and further genetic damage5. As any cell cycle phase can be regulated or perturbed, methods to report all four phases in living cells would be useful. In the widely used Fucci reporter system, G1-phase cells are labeled with one fluorescent protein (FP) fused to Cdt1(30-120), while cells in S, G2, or M are labeled with another FP fused to Geminin(1-110)6,7. Fucci has also been modified to add a G0/G1 transition reporter8. However, while Fucci and its variants reveal whether cells are within one of the proliferative phases of the cell cycle (S, G2, or M), they do not report which of these three phases cells are in.
Additional channels for live-cell imaging would be desirable for reporting multiple events such as cell cycle phases. For three-color time-lapse imaging, cyan, yellow, and red fluorescent proteins (CFPs, YFPs, and RFPs) are often used8,9, avoiding the phototoxic excitation wavelengths required for blue fluorescent proteins (BFPs)10. Some large-Stokes shift RFPs, those excited exclusively by blue light and not by the yellow-orange excitation wavelengths for standard RFPs, could constitute a fourth channel, but their photobleaching rates are not well suited for time-lapse imaging11. We instead explored the possibility of creating four channels by further red-shifting of far-red fluorescent proteins (far-RFPs) to enable orthogonal imaging with CFP, a green fluorescent protein (GFP), and an orange fluorescent protein (OFP). We performed multi-site structure-guided mutagenesis of the mNeptune2 far-RFP12, discovering a T60P and M160W double mutant with peak excitation at 610 nm that we named Maroon0.1 (Supplementary Table 1 and Fig. 1a). These mutations face each other beneath the chromophore and were mutually dependent for chromophore maturation. Structural modeling suggests π-π stacking between the chromophore and Trp-160 (Supplementary Fig. 1b). While π-π stacking is a well known mechanism for excitation red-shifting in fluorescent proteins13, it has not been previously observed with a Trp side-chain or with position 160. We performed further structure-guided mutagenesis while selecting for monomericity and brightness, in both bacteria and mammalian cells, and while maintaining red-shifted absorbance (Supplementary Fig. 2, Supplementary Table 1, Supplementary Note). The final protein, mMaroon1, contains 26 mutations (Supplementary Fig. 3a).
Figure 1.
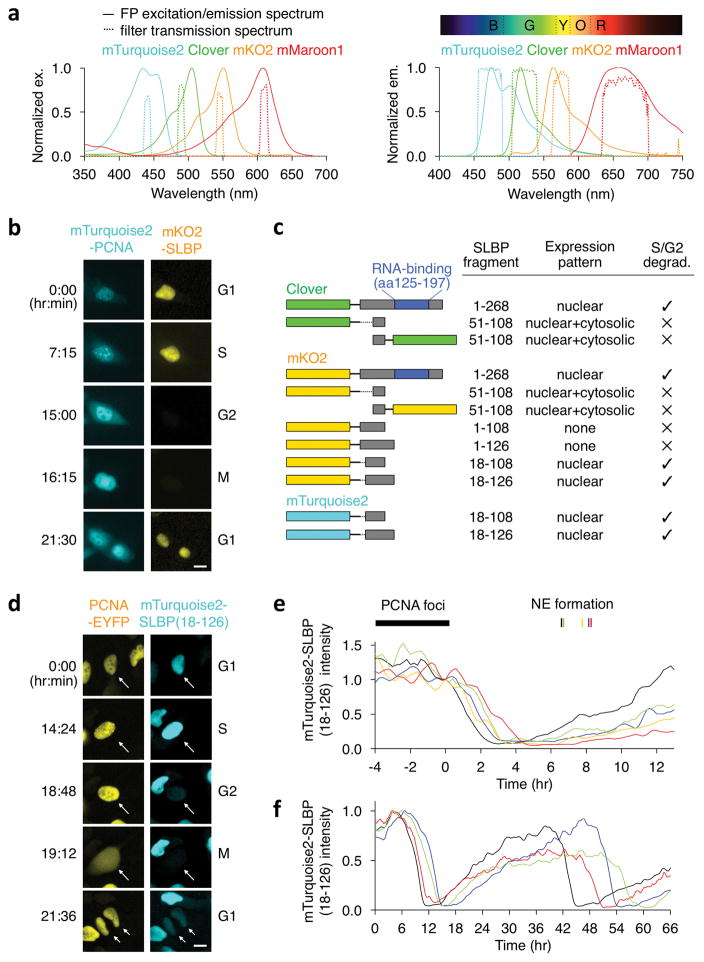
Development of a four-color imaging method and a S/G2 transition reporter. (a) mTurquoise2, Clover, mKO2, and mMaroon1 can be imaged orthogonally with appropriate excitation (left) and emission (right) filters. Excitation filters are: 440/10-nm for CFP, 490/10-nm for GFP, 545/10-nm for OFP, and 610/10-nm for far-RFP. Emission filters are: 472/30-nm for CFP, 525/30-nm for GFP, 575/25-nm for OFP, and 665/65-nm for far-RFP. The spectrum above the emission chart indicates definitions for blue (B), green (G), yellow (Y), orange (O), and red (R) in the CRC Handbook of Fundamental Spectroscopic Correlation Charts. (b) A mKO2-SLBP fusion is degraded after the end of S phase, as marked by disappearance of PCNA foci in HeLa cells. Scale bar, 10 μm. (c) FP fusions to SLBP fragments tested as candidate S/G2 transition reporters. (d) mTurquoise2-SLBP(18-126) is degraded after the end of S phase, as marked by disappearance of EYFP-PCNA foci. Arrows point to a doubly transfected cell and its daughter cells. Scale bar, 10μm. (e) Time-courses of mTurquoise2-SLBP(18-126) fluorescence during and after S phase. For each cell, fluorescence intensity was normalized to the intensity at PCNA foci disappearance. Time of nuclear envelope (NE) formation is indicated for each trace. (f) mTurquoise2-SLBP(18-126) fluorescence is cyclical through multiple cell divisions.
We performed further characterization of mMaroon1. Fluorescence excitation and emission peaked at 609 nm and 657 nm, respectively (Supplementary Table 1). mMaroon1 matured completely into a single orange-absorbing species (Supplementary Fig. 3b,c). In contrast, the only monomeric far-RFP with more red-shifted excitation, TagRFP657, matures into mixed violet-, green-, and orange-absorbing species (Supplementary Fig. 3c). Consequently, mMaroon1 is 3-fold brighter than TagRFP657 (Table 1). mMaroon1 exhibited a pKa of 6.2 and brightness-normalized photostability similar to enhanced GFP (Supplementary Fig. 3d,e). A variety of mMaroon1 fusions were correctly targeted in cells (Supplementary Fig. 4). In the standardized organized smooth endoplasmic reticulum (OSER) monomericity assay14, mMaroon1 produced a score of 46.3%, similar to mCardinal (Supplementary Table 2). This degree of monomericity surpasses Venus and Citrine YFPs14, which can label histones in cultured cells and transgenic mice without apparent deleterious effects15,16.
Table 1.
Properties of monomeric far-red fluorescent proteins with peak excitation ≥ 600 nm
| mNeptune1h | mCardinalh | TagRFP657h | mMaroon1 | |
|---|---|---|---|---|
| Excitation peak (nm) | 600 | 604 | 611 | 609 |
| Emission peak (nm) | 651 | 659 | 659 | 657 |
| Peak ε (mM−1 cm−1)a | 75 | 87 | 29 | 80 |
| Total φb | 0.23 | 0.19 | 0.10 | 0.11 |
| Maximum brightnessc | 17 | 17 | 2.9 | 8.8 |
| Short-λ emission boundary (nm)d | 564 | 566 | 577 | 588 |
| Maturation efficiency (%)e | 0.48 | 0.60 | 0.66 | 0.78 |
| Maturation half-time (min)f | 28 | 27 | 88 | 20 |
| pKa | 5.4 | 5.3 | 5.0 | 6.2 |
| Photostability (s)g | 160 | 730 | 110 | 178 |
ε = extinction coefficient.
φ = quantum yield.
Calculated as the product of ε at peak excitation and φ in units of mM−1 cm−1.
Wavelength beyond which 99.9% of total emission occurs.
Functional chromophore concentration divided by total protein concentration.
Time for fluorescence to obtain half-maximal value after exposure to oxygen.
Predicted time for fluorescence to photobleach by 50% under arc lamp illumination with excitation intensity adjusted to produce 1000 emission photons per molecule per s.
Data from ref. 12.
The red-shifted emission of mMaroon1 should facilitate orthogonal imaging of four FPs. mMaroon1 emission begins at a longer wavelength than in other monomeric far-RFPs such as mCardinal (Supplementary Fig. 5a, Table 1), allowing OFP detection up to ~590 nm without cross-detection of mMaroon1 (Fig. 1a, Supplementary Fig. 5b). Meanwhile, wavelengths above 600 nm can excite mMaroon1 without exciting OFPs (Fig. 1a). We selected mTurquoise2 CFP, Clover GFP, and either mKO2 or mOrange2 OFP as fluorophores that can be imaged orthogonally from each other and mMaroon1 with properly selected filters (Fig. 1a). These proteins are bright and photostable17, and Clover is better separated from mTurquoise2 than other GFPs due to its sharper and more red-shifted spectra18. To confirm orthogonality, we expressed mTurquoise2, Clover, mKO2, or mMaroon1 fused to a nuclear protein and imaged cells in cyan, green, orange, and far-red channels. Each fluorophore was only detected in the expected channel without bleed-through into others (Supplementary Fig. 5c). Finally, mTurquoise2, Clover, mKO2, and mMaroon1 fused to sequences targeting distinct subcellular locations were imaged together in the same cell. Each channel revealed some signals unique to that channel, indicating orthogonality (Supplementary Fig. 6). Thus, simultaneous four-channel imaging is possible with a CFP, Clover, an OFP, and mMaroon1.
We next considered how to create an intensiometric reporter for the S/G2 transition not reported by existing Fucci systems. We hypothesized that sequences from human stem-loop binding protein (SLBP), a RNA-binding protein that is degraded following S phase19, could confer degradation to a fused FP. We first performed time-lapse imaging of an mKO2-SLBP fusion protein together with mTurquoise-tagged proliferating cell nuclear antigen (PCNA), using dissolution of nuclear PCNA foci to report the S/G2 transition20. In HeLa cells, mKO2 was indeed degraded to undetectable levels following PCNA foci disappearance (Fig. 1b). We next attampted to dissociate SLBP degradation from RNA-binding. Amino acids (aa) 51-108 of SLBP outside the RNA-binding domain had been found to mediate degradation of a heterologous protein after S phase21. Unexpectedly, this fragment did not induce degradation of mKO2 or Clover when fused to either terminus (Fig. 1c). We then tested other segments encompassing the known degradation signals21 and discovered aa 18-107 or aa 18-126 were sufficient to degrade mKO2 or mTurquoise2 after S phase (Fig. 1c–e), with aa 18-126 fusions producing slightly higher signals. The fusion with mTurquoise2, mTurquoise2-SLBP(18-126) (Supplementary Fig. 7a), was expressed cyclically through multiple divisions, indicating that degradation was sufficiently active to prevent long-term accumulation (Fig. 1f, Supplementary Fig. 7b). Stable expression of mTurquoise2-SLBP(18-126) did not discernibly alter phase distributions as assessed by DNA content (Supplementary Fig. 8a) or the percentage of cells in S phase as assessed by thymidine analogue incorporation (Supplementary Fig. 8b). Cell cycle phase durations were also not changed in cells stably expressing mTurquoise2-SLBP(18-126) compared to parental cells (Supplementary Fig. 8c). These results demonstrate that mTurquoise2-SLBP(18-126) can serve as an intensiometric reporter of the S/G2 transition.
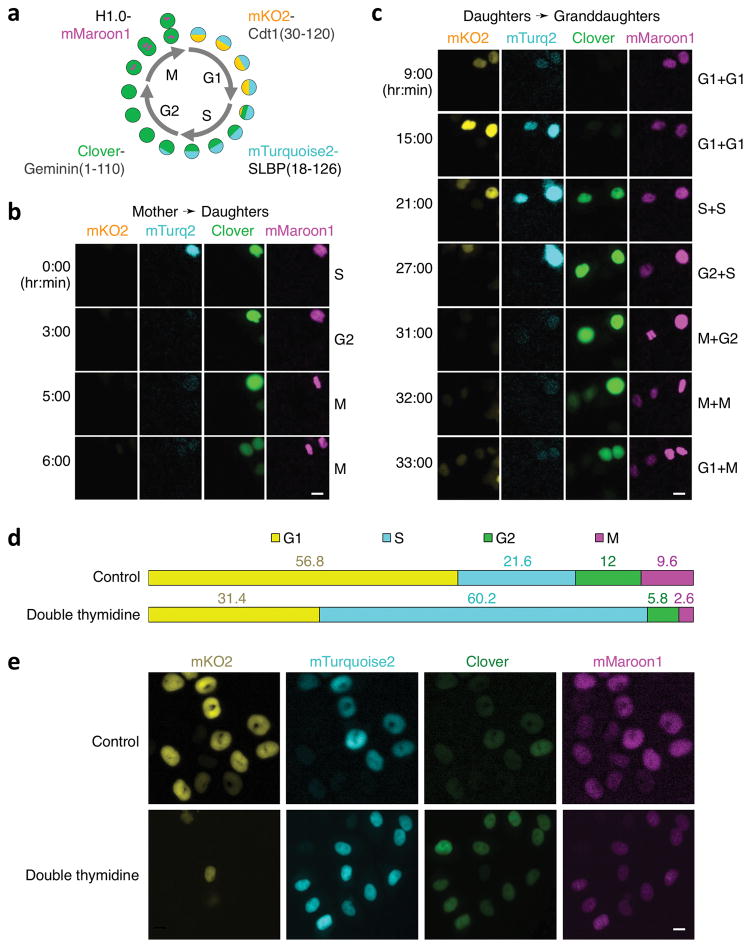
To report the G2/M transition, we fused mMaroon1 to histone H1.0, a linker histone responsible for higher-order chromatin structures (Supplementary Fig. 9a). Chromatin condensation, alignment, and separation during M phase were well visualized with H1.0-mMaroon1 (Supplementary Fig. 9b). We then combined mTurquoise2-SLBP(18-126), H1.0-Maroon1, Clover-Geminin(1-110), and mKO2-Cdt1(30-120) to create a four-phase cell cycle reporter system, which we named Fucci4 (Fig. 2a). Time-lapse imaging of HeLa cells revealed that each transition in the cell cycle could be discerned from the combination of the four reporters (Fig. 2b,c, Supplementary Fig. 10). As with the original Fucci system, the G1/S transition was marked by appearance of Clover-Geminin(1-110), while mTurquoise2-SLBP(18-126) persisted. mTurquoise2-SLBP(18-126) fluorescence loss then marked the S/G2 transition, while Clover-Geminin(1-110) persisted. Chromosome condensation in the mMaroon1 channel then marked the G2/M transition. Finally, loss of Clover-Geminin(1-110) and reappearance of mKO2-Cdt(30-120) and mTurquoise2-SLBP(18-126) marked the beginning of G1 phase. This pattern of fluorescence expression could be visualized across multiple generations (Fig. 2b,c, Supplementary Video 1). We also observed similar patterns in human U2OS and mouse NIH3T3 cells (Supplementary Fig. 11, Supplementary Video 2). Thus, Fucci4 is able to report all four cell cycle phase transitions in live cells.
Figure 2.
Fucci4, a reporter system for visualizing all four cell cycle stages. (a) Diagram of indicators mTurquoise2-SLBP(18-126), Clover-Geminin(1-110), mKO2-Cdt1(30–120), and H1.0-mMaroon1. (b,c) Tracking of cell cycle stages in HeLa cells via time-lapse imaging of Fucci4, including a mother cell dividing into two daughter cells (b) and daughter cells dividing into granddaughter cells (c). Deduced cell cycle phase is indicated to the right. Scale bar, 10 μm. (d) With Fucci4, S phase can be identified by high SLBP expression and low Cdt1 expression, revealing S-phase arrest by double-thymidine block. n = 125 and 226 cells for control and double thymidine treatment, respectively. (e) Representative field of cells treated by double-thymidine block. Scale bar, 10 μm.
Finally, we tested whether Fucci4 could visualize S-phase arrest, something not possible with existing two-color Fucci systems in which S, G2, and M phases all feature the presence of the Geminin(1-110) reporter and absence of the Cdt1(30-120) reporter. We applied a double thymidine block to HeLa cells expressing Fucci4, then quantified cells in each phase. We observed a shift of cells to an S phase profile expressing mTurquoise2-SLBP(18-126) without mKO2-Cdt(30-120) and depletion from all other phases, consistent with previously observed effects of double-thymidine block in HeLa cells22 (Fig. 2d,e). Thus, Fucci4 can be used to detect S-phase cell cycle arrest.
We also tested mTurquoise2-PCNA instead of mTurquoise2-SLBP(18-126) in an alternative four-phase indicator system. In time-lapse imaging, this system was indeed able to report all four phases, with mTurquoise2-PCNA foci dissappearance indicating the S/G2 transition (Supplementary Fig. 12a–c, Supplementary Video 3). In snapshot imaging, double-thymidine block reduced the percentage of cells with a G1-like profile and increased the percentage of cells with mTurquoise2-PCNA foci, as expected (Supplementary Fig. 12d,e). However, the percentage of cells exhibiting a G2-like profile of high Clover-Geminin(1-110) without mTurquoise2-PCNA puncta also increased, inconsistent with the G2 depletion expected from S-phase arrest22. A potential explanation is that PCNA foci increase in size gradually23 and can escape detection in early S phase, as previously noted24. Such cells would also express Clover-Geminin, appearing to have a G2 profile. Thus, a PCNA-based reporter may be unreliable in reporting S phase block. Other disadvantages of PCNA reporters are that they can cause G1 shortening20 and PCNA foci cannot be assessed by flow cytometry in living cells.
To summarize, we have engineered a far-RFP, mMaroon1, to facilitate orthogonal imaging with three other FPs. We also created the first intensiometric reporter of the S-G2 transition, mTurquoise2-SLBP(18-126). We combined these two new techniques to create Fucci4, a reporter system that discriminates all four cell cycle stages.
Compared to previous far-RFPs, mMaroon1 shows reduced emission at orange wavelengths. This allows mMaroon1 and OFPs to be imaged orthogonally without spectral unmixing, creating four channels when combined with a CFP and Clover GFP. By excluding BFP, this four-color imaging system avoids the autofluorescence and phototoxicity caused by violet excitation light. Furthermore, CFPs, Clover, OFPs, and mMaroon1 are excited well by common laser lines at 440, 488, 559 or 561, and 633 nm, respectively. Thus, four-color fluorescence imaging utilizing mMaroon1 should be widely applicable. In addition, a fifth channel using biliverdin-dependent infrared fluorescent proteins should be possible if excitation wavelengths of ~680 nm are available25.
Compared to earlier cell cycle reporters, Fucci4 supplies additional essential information. Fucci4 can reveal the distributions of all four cell cycle phases in snapshot imaging, as well as the durations of these phases in time-lapse imaging. Fucci4 also contains some degree of optical redundancy that could be useful. The H1.0-mMaroon1 marker tracks daughter cells during cytokinesis before mTurquoise2-SLBP(18-126) or mKO2-Cdt1(1-120) become visible. In addition, in the original Fucci, mKO2-Cdt1(1-120) was used to track cell movements through G1, even if G1 was also reported by the absence of GFP-Geminin(1-110). In Fucci4, mTurquoise2-SLBP(18-126) or H1.0-mMaroon1 can perform this tracking function. We retained mKO2-Cdt1(1-120) in Fucci4 to preserve the option of direct single-channel detection of cells in G1, but the orange channel could be reassigned to other uses if desired.
Fucci4 has potentially many applications in development, physiology, and cancer. Fucci4 should enable finer analysis of how developmental signals, extracellular stimuli, or genetic changes alter the cell cycle. Fucci4 should also facilitate research into molecular mechanisms regulating specific phase transitions. Finally, Fucci4 should facilitate screening for drugs that affect a particular cell cycle phase.
METHODS
Cloning
All cloning and plasmid construction were accomplished using standard molecular biology techniques, including PCR extension, overlap PCR, and In-Fusion cloning (Clontech). Complete plasmid sequences are available upon request.
Mutagenesis and library screening
Mutations for specific residues were introduced by overlap-extension PCR. Mutants were expressed and screened in a constitutive bacterial expression vector, pNCS. Plasmids were transformed via heat shock into chemically competent XL10-Gold bacteria (Agilent), and bacteria were plated on LB agar plates with ampicillin (100 μg mL−1) for selective growth, incubated at 37 °C for 16–20 hours, and grown an additional 20–24 hours at room temperature. For each round of mutagenesis, the number of colonies screened was 10-fold the expected library diversity to ensure full coverage. Colonies and bacterial patches were screened for transmitted color by eye and for fluorescence in a dark enclosure with a KL2500 fiber-optic light source (Leica), 610/20 nm excitation and 645/30 nm emission filters (Chroma), and a ST-8300M cooled CCD camera controlled with CCDOps software (Santa Barbara Instrument Group) on a MacBook Pro running OS 10.6.8 (Apple). Promising mutants were selected for spectral characterization.
Structural modeling
As an alternative to homology-based structural prediction programs, which do not model the atomic structure of the chromophores of fluorescent proteins, a model of the three-dimensional structure of mMaroon1 including chromophore atoms was generated by threading the amino acid sequence of Maroon0.1 through an electron density map generated by X-ray diffraction of low-resolution crystals of Maroon0.1. Briefly, Maroon0.1 crystals were grown in 17% PEG3350, 0.1 M MgCl2, 0.1 M Sodium citrate, pH 5.3. X-ray diffraction data were collected on Beamline BL17A at the High Energy Accelerator Research Organization (KEK) Photon Factory, Tsukuba, Japan, and processed in the program HKL2000. MOLREP was used to create structural models of the electron densities using the Neptune structure (PDB file 3IP2) as the search model. Model refinement was performed manually in the programs Coot and Refmac.
Fluorescent protein characterization
For FP purification, XL10-Gold bacteria were transformed with plasmid pNCS expressing each FP with an N-terminal polyhistidine tag, and plated on LB agar plates with ampicillin (100 μg mL−1). An individual colony was used to inoculate a 2-mL culture of LB with ampicillin, which was grown at 37 °C with shaking for 8 hr. The culture was then diluted into 200 mL of LB with ampicillin, which was grown at 37 °C with shaking overnight. Bacterial were pelleted, then lysed in B-PER II (Pierce). Proteins were captured with HisPur Cobalt Resin (Pierce), washed in phosphate-buffered saline (PBS) with 25 mM imidazole pH 7.4, eluted in PBS with 250 mM imidazole pH 7.4, and desalted into PBS pH 7.4 using Econo-Pac 10DG gravity flow chromatography columns (Bio-Rad), all at room temperature. pH titrations were performed using a series of buffers (33 mM HOAc/NaOAc 100 mM NaCl for pH 3.0–5.5; 33 mM NaH2PO4/Na2HPO4 100 mM NaCl for pH 6–8.0; 33 mM glycine 100 mM NaCl for pH 8.5–9.0). Mature fraction of a purified protein sample was calculated as the concentration of chromophore divided by the concentration of total protein. Concentration of functional chromophore was calculated using Beer’s law, absorbance of base-denatured protein at 440 nm measured in a 1-cm cuvette, and an extinction coefficient at 440 nm of 44 mM−1 cm−1 (ref. 26). Concentration of total protein was calculated using Beer’s law, absorbance at 280 nm measured in a 1-cm cuvette, and an extinction coefficient at 280 nm of 30.4 mM−1 cm−1 as determined by the method of Gill and von Hippel27 using the program ProtParam. Absorbance, excitation, and emission spectra were acquired with a Safire2 microplate reader (TECAN). Quantum yields were determined using cresyl violet in methanol as a standard (φ = 0.54). For native polyacrylamide gel electrophoresis (PAGE), purified proteins at 5 μM concentration were loaded onto 4–16% Bis-Tris polyacrylamide gels (Life Technologies) and electrophoresed in 1× NativePAGE Running Buffer (Life Technologies) with dark blue cathode buffer (with 0.02% Coomassie G-250) at 150 V for 75–90 min. Gels were stained in 0.02% Coomassie R-250 in 30% methanol and 10% acetic acid. In vitro photobleaching measurements were performed on purified protein in PBS in mineral oil droplets using an IX81 inverted microscope with a 40× 0.90-numerical aperture (NA) UPlanSApo objective (Olympus), an X-Cite 120-W metal halide lamp (Lumen Dynamics), a 615/30 nm excitation filter (Omega Optical), and an Orca ER camera (Hamamatsu), controlled by Micro-Manager software28. Images were acquired every 1 s under continuous illumination. Times were normalized to produce photon output rates of 1000 per molecule per s as previously described25, using the 72-s half-life of mKate as reference.
Cell culture and lentivirus production
HeLa cells (ATCC), HEK293A cells (Life Technologies), NIH3T3 cells (ATCC), and U2OS cells (ATCC) were used for protein localization and cell culture studies to allow comparisons with published data obtained in these lines. Cell lines were tested for mycoplasma contamination. Cells were maintained in high-glucose Dulbecco’s Modified Eagle Medium (DMEM, HyClone) with 5% fetal bovine serum (FBS, Gemini Bio) and 2 mM glutamine (Life Technologies) at 37 °C with 5% CO2. To package lentivirus, HEK293T cells at ~70% confluency were transfected with psPAX2, pMD2.G, and pLL3.7 plasmids using the calcium phosphate method. Two days post-transfection, viral supernatant was filtered with a 0.45 μm polyethersulfone filter before using to infect target cells.
Brightness comparison of Maroon variants in mammalian cells
mTurquoise2-P2A-Maroon fusions were expressed using Lipofectamine 2000 (Life Technologies) in HEK293A cells grown in the high-glucose DMEM with 10% FBS and 2 mM glutamine at 37 °C with 5% CO2. Two days post-transfection, cells were transferred to a transparent-bottom 96-well plate in PBS and fluorescence spectra were obtained on the Safire2 microplate reader. Relative brightness was calculated from peak Maroon variant emission divided by peak mTurquoise2 emission.
Imaging of fusion proteins
mMaroon1 was fused with sequences from human calnexin (NM_001746.3), Lifeact (HQ993061), mouse mannosidase II (NM_008549.2), human lamin B1 (NM_005573.2), human pyruvate dehydrogenase (PDHA, NM_000284), chicken paxillin (NM_204984.1), human calnexin (NM_001746.3), mouse histone H1.0 (NM_008197.3), and mouse mannosidase II (NM_008549.2), all gifts of M. Davidson (Florida State University). HeLa and U2OS cells were cultured as above, plated onto glass bottom plates (In Vitro Scientific), and imaged in Live Cell Imaging Solution (Life Technologies) on the IX81 inverted microscope with a 60× 1.42-NA PlanApoN oil objective (Olympus), the X-Cite 120-W metal halide lamp, the 615/30 nm excitation filter, a 665/40 nm emission filter (Chroma), and the Orca ER camera, controlled by Micro-Manager software.
Four-color imaging
HEK293T or NIH3T3 cells in LabTek 8-chambered coverglasses (Nunc) were transfected with Lipofectamine 2000. 24 hr later, they were imaged in Live Cell Imaging Solution on an Axiovert 200M inverted microscope with a 20× 0.75-NA Plan-Apochromat objective (Zeiss), an X-Cite 120-W metal halide lamp, and an Orca ER camera, controlled by Micromanager software. The following excitation (ex) filters and emission (em) filters were used: cyan, ex 440/10 nm (Chroma), em 472/30 nm (Semrock); green, ex 490/10 nm (Chroma), em 525/30 nm (Semrock); orange, ex 545/10 nm (Omega), em 575/25 nm (Chroma); far-red, ex 610/10 nm (Omega), em 665/65 nm (Chroma). Background was subtracted using the background subtraction tool in the ImageJ program29. For confocal four-color imaging, U2OS cells were infected with two lentiviruses with encephalomyocarditis virus (EMCV) internal ribosome entry sites (IRESes) for bicistronic expression, one co-expressing PDHA-10aa-mKO2 and Calnexin-14aa-Clover, and another co-expressing H1.0-10aa-mMaroon1 and mannosidaseII-10aa-mTurquoise2. Cells were imaged in Live Cell Imaging Solution in LabTek 8-chambered coverglasses on an IX81 inverted microscope with a 40× 1.15-NA UApo340 water objective and a FV1000 confocal scanner, controlled by Fluoview v3.1b software (Olympus). The following excitation lasers and emission acousto-optic tunable filter settings were used: cyan, ex 440 nm, em 460–500 nm; green, ex 488 nm, em 500–550 nm; orange, ex 559 nm, em 570–600 nm; far-red, ex 635 nm, em 650–750 nm.
Imaging of SLBP and PCNA time-courses
A pLL3.7 plasmid expressing mTurquoise2-SLBP(18-126) was used to package VSVG-pseudotyped lentivirus, which was then used to infect HeLa[EYFP-PCNA] cells (a gift of J. Ferrell, Stanford University) at a multiplicity of infection of 1. HeLa[EYFP-PCNA,mTurquoise2-SLBP(18-126)] cells were purified by fluorescence-activated cell sorting, then imaged in parallel with parental cells at 37 °C and under 5% CO2 in an ImageXpress IXMicro system (Axon Instruments) with a 20× 0.75-NA objective (Olympus) at 12-min time intervals. PCNA puncta disappearance was manually scored, and mTurquoise2-SLBP(18-126) intensity was quantified in ImageJ.
Cell cycle distribution analysis
For DNA content and 5-ethynyl-2′-deoxyuridine (EdU) analyses, mTurquoise2-SLBP(18-126), consisting of mTurquoise2 fused at its C-terminus to SLBP aa 18-126, was cloned into a pcDNA5/FRT expression vector (Life Technologies). This plasmid and pOG44 expressing flp recombinase were cotransfected into HeLa Flp-In TRex cells using Lipofectamine 2000. These cells permit stable incorporation of transgenes at a single defined chromosomal location and have been used extensively for cell cycle studies30. Transfected cells were selected for incorporation at the FRT locus in 0.1 mg/mL hygromycin (Life Technologies) for 3 weeks. For DNA content analysis, parental or mTurquoise2-SLBP(18-126)-expressing cells were seeded at 5% density on 10-cm plates and grown for 2 days, then trypsinized, pelleted, and resuspended in PBS in 0.1 μM SyTox Green for 30 min. Cells were then analyzed on a LSR II flow cytometer with a 488-nm laser line (BD). For EdU incorporation analysis, cells were seeded at 5% density on 10-cm plates and grown for 2 days. EdU was added to the growth media to a final concentration of 10 μM for 2 h, then cells were trypsinized, pelleted, and resuspended in PBS. Cells were then fixed with 4% paraformaldehyde and washed with PBS. Incorporated EdU was reacted with Alexa Fluor 555 in an alkyne-azide Huisgen cycloaddition according to supplier instructions (Click-It EdU, Life Technologies). Cells were analyzed for Alexa Fluor 555 content on the LSR II flow cytometer with a 532-nm laser line. For cell cycle phase duration analysis, time-lapse microscopy of HeLa[EYFP-PCNA] and HeLa[EYFP-PCNA, mTurquoise2-SLBP(18-126)] cells was performed in adjacent wells at 37 °C under 5% CO2 in the ImageXpress IXMicro system with the 20× 0.75-NA objective at 12-min time intervals. For cells that completed an entire cycle, times of phase transitions were determined by visualizing EYFP-PCNA, with nuclear envelope formation marking M/G1, foci formation marking G1/S, foci dissolution marking S/G2, and nuclear envelope breakdown marking G2/M.
Statistics
In cell cycle phase duration analysis, sample size of 14 cells was pre-determined to detect a 10% difference with 95% power given variance measured in preliminary experiments. Times were scored and statistics were performed blinded to the identity of the wells. Welch’s t-test was performed for comparisons within each cell cycle phase without decreasing the alpha level for multiple comparisons to be more sensitive for detecting a difference in any cell cycle phase.
Time-lapse and snapshot imaging of Fucci4
Two bicistronic lentiviruses containing the EMCV IRES were used to infect cells. One co-expressed Clover fused to human Geminin aa 1-110 (Life Technologies) with a 10-aa linker (Clover-Geminin(1-110)) and mKO2 fused to human Cdt1 aa 30-120 (Life Technologies) with a 10-aa linker (mKO2-Cdt1(30-120)). The other virus co-expressed either mTurquoise2 fused to PCNA (J. Ferrell, Stanford University) with a 23-aa linker (mTurquoise2-PCNA) or mTurquoise2-SLBP(18-126) and H1.0-mMaroon1. Cells were cultured in LabTek 8-chamber slides, infected by packaged viruses, and imaged using the above four-color epifluorescence imaging protocol, except an environmental control system (Live Cell Instrument) was used to incubate the cells at 37 °C under 5% CO2 and images were acquired every 15 min. For double thymidine block, cells were infected with the two lentiviruses expressing Fucci4 indicators in LabTek 8-chambered coverglasses. Cells were incubated in 2 mM thymidine (Chem-Impex) for 18 hr, fresh media for 9 hr, and 2 mM thymidine again for 15 hr, then imaged using the above four-color epifluorescence imaging protocol.
Data availability
Nucleotide sequences are available at GenBank for mMaroon1 (#####), H1.0-mMaroon1 (#####), Clover-Geminin(1-110) (#####), mKO2-Cdt(30-120) (#####), mTurquoise2-SLBP(18-126) (#####), and mKO2-SLBP(18-126) (#####). Mammalian expression plasmid for mMaroon1 (#####) and lentiviral packaging plasmids for mTurquoise2-SLBP(18-126)-IRES-H1.0-mMaroon1 (#####), Clover-Geminin(1-110)-IRES-mKO2-Cdt1(30-120) (#####), Clover-Gem(1-110) (#####), mKO2-SLBP(18-126) (#####), and H1.0-mMaroon1 (#####) are available at Addgene.
Supplementary Material
Acknowledgments
We thank A. Moskaleva and J. Ferrell (Stanford University) for plasmids, cell lines, and advice on cell cycle experiments, M. Davidson for plasmids, A. Straight for HeLa-Flp-In-TREX cells, and other members of the Lin laboratory for assistance with experiments. This work was supported by a Siebel Scholar Award (A.J.L.), a Stanford Graduate Fellowship (X.X.Z.), a Howard Hughes Medical Institute International Student Research Fellowship (X.X.Z.), NIH Molecular Biophysics Predoctoral Research Training Grant 5T32GM008294 (A.L.Y.), Stanford Research Experience for Undergraduates grants (K.K. and J.J.T.), NIH grant R01GM118377 (T.M.), NIH grant P50GM107615 to the Stanford Center for Systems Biology (M.C., T.M., M.Z.L.), a Burroughs Wellcome Foundation Career Award for Medical Scientists (M.Z.L.), a Rita Allen Foundation Scholar Award (M.Z.L.), and Pioneer Award 5DP1GM111003 (M.Z.L.).
References
- 1.Artus J, Cohen-Tannoudji M. Cell cycle regulation during early mouse embryogenesis. Mol Cell Endocrinol. 2008;282:78–86. doi: 10.1016/j.mce.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 2.Dong X, et al. Control of G1 in the developing Drosophila eye: rca1 regulates Cyclin A. Genes Dev. 1997;11:94–105. doi: 10.1101/gad.11.1.94. [DOI] [PubMed] [Google Scholar]
- 3.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 4.Pruitt SC, Freeland A, Kudla A. Cell cycle heterogeneity in the small intestinal crypt and maintenance of genome integrity. Stem Cells. 2010;28:1250–1259. doi: 10.1002/stem.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 6.Sakaue-Sawano A, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 7.Abe T, et al. Visualization of cell cycle in mouse embryos with Fucci2 reporter directed by Rosa26 promoter. Development. 2013;140:237–246. doi: 10.1242/dev.084111. [DOI] [PubMed] [Google Scholar]
- 8.Oki T, et al. A novel cell-cycle-indicator, mVenus-p27K-, identifies quiescent cells and visualizes G0-G1 transition. Sci Rep. 2014;4:4012. doi: 10.1038/srep04012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higuchi-Sanabria R, et al. Characterization of Fluorescent Proteins for Three- and Four-Color Live-Cell Imaging in S. cerevisiae. PLoS One. 2016;11:e0146120. doi: 10.1371/journal.pone.0146120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge J, et al. Standard fluorescent imaging of live cells is highly genotoxic. Cytometry A. 2013;83:552–560. doi: 10.1002/cyto.a.22291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shcherbakova DM, Hink MA, Joosen L, Gadella TW, Verkhusha VV. An orange fluorescent protein with a large Stokes shift for single-excitation multicolor FCCS and FRET imaging. J Am Chem Soc. 2012;134:7913–7923. doi: 10.1021/ja3018972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu J, et al. Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein. Nat Methods. 2014;11:572–578. doi: 10.1038/nmeth.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng HL, Lin MZ. Structure-guided wavelength tuning in far-red fluorescent proteins. Curr Opin Struct Biol. 2016;39:124–133. doi: 10.1016/j.sbi.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cranfill PJ, et al. Quantitative assessment of fluorescent proteins. Nat Methods. 2016;13:557–562. doi: 10.1038/nmeth.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freyer L, et al. A loss-of-function and H2B-Venus transcriptional reporter allele for Gata6 in mice. BMC Dev Biol. 2015;15:38. doi: 10.1186/s12861-015-0086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Megason SG. In toto imaging of embryogenesis with confocal time-lapse microscopy. Methods Mol Biol. 2009;546:317–332. doi: 10.1007/978-1-60327-977-2_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bajar TB, Wang SE, Zhang S, Lin ZM, Chu J. A guide to fluorescent protein FRET pairs. Sensors. 2016;16 doi: 10.3390/s16091488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lam AJ, et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitfield ML, et al. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol Cell Biol. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn AT, Jones JT, Meyer T. Quantitative analysis of cell cycle phase durations and PC12 differentiation using fluorescent biosensors. Cell Cycle. 2009;8:1044–1052. doi: 10.4161/cc.8.7.8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koseoglu MM, Graves LM, Marzluff WF. Phosphorylation of threonine 61 by cyclin a/Cdk1 triggers degradation of stem-loop binding protein at the end of S phase. Mol Cell Biol. 2008;28:4469–4479. doi: 10.1128/MCB.01416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao GD, McKenna WG, Yen TJ. Detection of repair activity during the DNA damage-induced G2 delay in human cancer cells. Oncogene. 2001;20:3486–3496. doi: 10.1038/sj.onc.1204445. [DOI] [PubMed] [Google Scholar]
- 23.Leonhardt H, et al. Dynamics of DNA replication factories in living cells. J Cell Biol. 2000;149:271–280. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schönenberger F, Deutzmann A, Ferrando-May E, Merhof D. Discrimination of cell cycle phases in PCNA-immunolabeled cells. BMC Bioinformatics. 2015;16:180. doi: 10.1186/s12859-015-0618-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin MZ, et al. Autofluorescent proteins with excitation in the optical window for intravital imaging in mammals. Chem Biol. 2009;16:1169–1179. doi: 10.1016/j.chembiol.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chalfie M, Kain SR. Green Fluorescent Protein: Properties, Applications and Protocols (Methods of Biochemical Analysis) New York: Wiley-Liss; 2005. [Google Scholar]
- 27.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 28.Edelstein AD, et al. Advanced methods of microscope control using uManager software. J Biol Methods. 2014;1:e10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tighe A, Staples O, Taylor S. Mps1 kinase activity restrains anaphase during an unperturbed mitosis and targets Mad2 to kinetochores. J Cell Biol. 2008;181:893–901. doi: 10.1083/jcb.200712028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Nucleotide sequences are available at GenBank for mMaroon1 (#####), H1.0-mMaroon1 (#####), Clover-Geminin(1-110) (#####), mKO2-Cdt(30-120) (#####), mTurquoise2-SLBP(18-126) (#####), and mKO2-SLBP(18-126) (#####). Mammalian expression plasmid for mMaroon1 (#####) and lentiviral packaging plasmids for mTurquoise2-SLBP(18-126)-IRES-H1.0-mMaroon1 (#####), Clover-Geminin(1-110)-IRES-mKO2-Cdt1(30-120) (#####), Clover-Gem(1-110) (#####), mKO2-SLBP(18-126) (#####), and H1.0-mMaroon1 (#####) are available at Addgene.