Abstract
In recent years, protein engineers have succeeded in tuning the excitation spectra of natural fluorescent proteins from green wavelengths into orange and red wavelengths, resulting in the creation of a series of fluorescent proteins with emission in the far-red portions of the optical spectrum. These results have arisen from the synergistic combination of structural knowledge of fluorescent proteins, chemical intuition, and high-throughput screening methods. Here we review structural features found in autocatalytic far-red fluorescent proteins, and discuss how they add to our understanding of the biophysical mechanisms of wavelength tuning in biological chromophores.
Introduction
Red is the color of blood, and I will seek it.
– Conrad Aiken
Fluorescent proteins (FPs), originally identified as biological curiosities in sea creatures such as jellyfish [1], coral [2] and anemone [3], have become central tools of cell biology research. FPs, as genetically encoded fluorescent labels, are used as real-time protein tags, gene expression reporters, and cell lineage tracers [4]. FPs have also served as surprisingly adaptable templates for engineering chemical sensors for pH and ions [5], membrane voltage potential [6, 7], and metabolites [8]. A broad palette of engineered FPs has enabled Forster resonance energy transfer experiments to characterize protein-protein interactions and various biochemical phenomena associated with protein conformational changes [9, 10]. FPs are fundamental tools powering exciting new fields such as optogenetics and super-resolution microscopy [11, 12].
Two motivations drive efforts to develop FPs with excitability beyond 600 nm. First, excitation beyond 600 nm would facilitate non-invasive imaging of cells in vascularized tissues. Hemoglobin is the primary absorber of light in mammalian tissue, with broad absorbance up to wavelengths to 600 nm [13], thus use of excitation light beyond 600 nm should enable better detection of fluorescently labelled cells through tissue. The second reason is to provide an additional channel for multi-wavelength imaging. The availability of a FP that can be efficiently excited at 633 nm would allow the use of this common and less phototoxic laser wavelength for imaging multiple events in living samples.
A note on nomenclature may be useful. Some orange FPs have historically been called red FPs (RFPs). In this review, we will use “red” to describe FPs with average (not peak) emission above 620 nm, a commonly accepted orange/red boundary [14]. As there is no standard definition of “far red”, we will use a 650 nm average emission as a cutoff to be consistent with existing far-RFP naming. This differs from the plant literature, where 650–700 nm light is designated red and 700–750 nm light is designated far red [15].
Recent engineering efforts have successfully produced a number of FPs with excitation maxima above 600 nm and average emission in the far-red. Here we review structural and biophysical features of red-shifting of both excitation and emission in RFPs and far-RFPs of the GFP superfamily.
Evolutionary relationships of RFPs and far-RFPs
Far-red FPs from coral
Autocatalytic far-red FPs are derived by mutagenesis of tetrameric orange or red FPs. DsRed from Discosoma sp. coral, was the first orange-red FP discovered. DsRed is the natural parent of some of the most commonly used RFPs (Table 1). DsRed, which emits in the orange (peak excitation/emission 558/583 nm) [2], was engineered into the dimeric orange dTomato [16•], with similar spectra and brightness, and into the monomeric mRFP1 [17•], with true red emission but with brightness reduced to 22% of dsRed (Table 1).
Table 1.
Characteristics of selected far-red fluorescent proteins
| Fluorescent protein | Quarternary structure | Ex. peaka | Em. peaka | Extinction coefficientb | Quantum yieldc | Peak brightnessd | PDB number |
|---|---|---|---|---|---|---|---|
| DsRed | tetramer | 558 | 583 | 75 | 0.79 | 59 | 1GGX |
| E2-Crimson | tetramer | 611 | 646 | 59 | 0.12 | 7.0 | |
| dTomato | dimer | 554 | 581 | 69 | 0.69 | 48 | |
| mRFP1 | monomer | 584 | 607 | 50 | 0.25 | 13 | |
| mCherry | monomer | 587 | 610 | 72 | 0.22 | 16 | 2H5Q |
| mPlum | monomer | 590 | 649 | 41 | 0.10 | 4.1 | 2QLG |
| AQ143 | tetramer | 595 | 655 | 90 | 0.04 | 3.6 | 4OHS |
| mRaspberry | monomer | 598 | 625 | 86 | 0.15 | 13 | |
| mGrape3 | monomer | 608 | 646 | 40e | 0.03 | 1.2 | |
|
| |||||||
| eqFP578 | dimer | 552 | 578 | 102 | 0.54 | 55 | 3PIB |
| Katushka2 | dimer | 588 | 633 | 67 | 0.44 | 29 | 3PJ7 |
| eqFP650 | dimer | 592 | 650 | 65 | 0.24 | 16 | 4EDO |
| eqFP670 | dimer | 605 | 670 | 70 | 0.06 | 4.2 | 4EDS |
| TagRFP | monomer | 555 | 584 | 98 | 0.41 | 40 | 3M22 |
| mKate | monomer | 585 | 635 | 42 | 0.30 | 13 | 3BXB |
| mKate2 | monomer | 586 | 630 | 50 | 0.36 | 18 | |
| TagRFP675 | monomer | 598 | 675 | 56 | 0.08 | 4.5 | 4KGF |
| mNeptune2.5 | monomer | 599 | 643 | 95 | 0.28 | 27 | |
| mNeptune2.5 | monomer | 599 | 643 | 95 | 0.28 | 27 | |
| mNeptune | monomer | 600 | 650 | 67 | 0.20 | 13 | 3IP2 |
| mCardinal | monomer | 604 | 659 | 87 | 0.19 | 17 | 4OJ0, 4OQW |
| TagRFP657 | monomer | 611 | 657 | 34 | 0.10 | 3.4 | |
Excitation and emission maxima in nm.
Maximum extinction coefficient per chromophore in mM−1cm−1 measured by the alkali denaturation method.
Quantum yield of fluorescence.
Calculated as peak extinction coefficient per chain in mM−1cm−1 multiplied by quantum yield.
After photoactivation by 470 nm light. See text for references. FPs are grouped into DsRed and eqFP578 clades. Within each clade, they are ordered first by quarternary structure then by excitation peak.
From mRFP1, further red-shifted RFPs were engineered. mCherry, perhaps the most used RFP, shows slightly redder spectra and slightly improved brightness [16•]. mRaspberry and mPlum feature more dramatically red-shifted spectra with mPlum being the first FP with average emission in the far-red [18•]. Raspberry impressively is as bright as its parent, but, the red-shifted emission of mPlum came at the cost of reduced brightness. mGrape3, in which a π-π interaction was introduced at the chromophore for the purpose of red-shifting excitation (discussed below), exhibited a red-shifted excitation peak at 608 nm, but only transiently after illumination by 470 nm light [19••]. mGrape3 is also rather dim, retaining only 2% of the brightness of dsRed (Table 1).
The same π-π interaction as in mGrape3 was also introduced in a DsRed variant, creating the tetrameric E2-Crimson [20•], which was brighter than mGrape3, retaining 12% of the brightness of dsRed (Table 1). The dimness of mGrape relative to E2-Crimson mirrors the dimness of mRFP1 relative to DsRed, and in both cases are primarily due to decreased quantum yield. This suggests a role for tetramerization in stiffening the chromophore pocket and thereby improving quantum yield.
Far-red FPs from anemone corals
An orange FP from the bubble-tip anemone Entacmaea quadricolor, eqFP578 [21•], has served as a highly successful scaffold for engineering red and far-red FPs (Table 1). The dimeric eqFP578 was red-shifted to create the dimeric RFP Katushka [22•], which was further evolved into the dimeric far-RFPs eqFP650 and eqFP670 [23] (Table 1). Meanwhile eqFP578 was also monomerized to create TagRFP, which was evolved into monomeric RFPs mKate [22•] and mKate2 [24]. mKate then served as the parent for the far-RFPs mNeptune [19••], mNeptune2.5 [25••], TagRFP657 [26], and TagRFP675[27]. Finally, mNeptune was further red-shifted to create the bright mCardinal [25••] and a series of mNeptune variants with significantly redder emissions but lower brightness similar to TagRFP675 [28] (Table 1). Thus eqFP578 has been exceptionally productive as a scaffold for the engineering of far-red FPs.
Interestingly, eqFP578-derived RFPs and far-RFPs generally show brighter fluorescence than their DsRed-derived counterparts at the same excitation wavelengths. For example, mKate is brighter than mCherry, and mNeptune is brighter than mRaspberry (Table 1). The trend to higher brightness of eqFP derivatives has extended to even redder excitation wavelengths, so that the eqFP578-derived mCardinal and TagRFP657 are both many times brighter than the dsRed-derived mGrape3 (Table 1). Unfortunately, the trend to decreasing quantum yields with redder excitation wavelengths also persists in the monomeric series TagRFP, mKate2, mNeptune2.5, mCardinal, and TagRFP657 (Table 1), suggesting that red-shifting usually comes at the expense of brightness.
Applications
Excitation wavelengths beyond 600 nm have been shown to be useful for whole-body imaging in mammals. When expressed in the liver, a deeply-situated vascularized organ, Neptune excited slightly off-peak at 600–620 nm produced more signal than mKate excited on-peak at 570–590 nm, despite the higher intrinsic brightness of mKate [19••]. Beyond 600 nm, intrinsic brightness matters more; Neptune excited at 600–620 nm was brighter than a phytochrome-based infrared fluorescent protein excited on-peak, due to the 2-fold higher brightness of Neptune [19••]. mCardinal also revealed differentiating myocytes in leg muscle while a co-expressed GFP could not, when both were excited at their peak absorbances [25••].
Researchers have taken advantage of the in vivo contrast provided by red-excitable far-RFPs to improve non-invasive imaging of cells in animals [25••, 29, 30••]. Here, fluorescence offers a faster and more reproducible method for assessing cell number than luciferase reporters, as long as signal can be detected above autofluorescent background. In a comparison of E2-Crimson and firefly luciferase, E2-Crimson signal was found to better correlate with tumor weight, although luciferase could detect 5-fold fewer cells [30••]. The relative transparency of tissue to red excitation and far-red emission light also makes far-red FPs better sutied for fluorescence tomography [31].
Furthermore, the ability of far-RFPs to be excited by common 633- and 635-nm laser lines, albeit off-peak, can been useful. For example, mCardinal provides an imaging channel beyond orange FPs such as dsRed [25••]. When exciting to the right of the absorption peak, even small red-shifts of a few nm can improve effective brightness substantially. Far-red FPs also were at one point uniquely useful for imaging in the first commercial STED microscope, which featured only a single 635-nm excitation laser and a 670-nm depletion laser [26, 32], but newer STED microscopes now work well with GFP and YFP [33, 34].
Structures and mechanisms
Atomic structures have played an essential role throughout the engineering of red-shifted FPs. For example, the very first example of wavelength red-shifting, YFP, was created rationally by the introduction of Tyr in place of Thr203 in GFP (homologous to 197 in RFPs) to create a π-π stack with the chromophore [35••]. This was enabled directly by knowledge of the crystal structure of GFP, which revealed the location of the Thr203 side chain above the chromophore and the existence of enough space for the larger Tyr side chain. Indeed the creation of YFP was included together with the first description of the GFP three-dimensional structure. The dsRed crystal structure also revealed the structure of the dsRed chromophore, including the existence of an acylimine group that extended the π-conjugated system [36]. This in turn allowed prediction of shifts in electron density away from the phenolate oxygen to the acylimine oxygen in the excited state [37, 38], providing a further rational avenue for red-shifting. Further structural studies have subsequently been useful in two ways: First, to confirm mechanisms of red-shifting in mutants obtained through semi-rational mutagenesis of chromophore-interacting residues, and second to reveal new mechanisms for more subtle wavelength changes acquired incidentally or from screens of randomly generated mutants. These contributions of structural knowledge are often intermixed in the same studies, and so we will simply discuss structural studies by wavelength order, presenting individual findings as they arise.
A conserved chromophore covalent structure
RFPs and far-RFPs are structurally similar to jellyfish GFP, comprising an eleven-stranded beta-barrel with the chromophore embedded in an alpha helix running through the center, like a candle in a lantern [39]. The beta barrel limits movement of the chromophore, preventing excited state energies from dissipating due to vibrational interactions. Crystal structures of the RFPs and far-RFPs described here reveal chromophores of the DsRed type [36]. This chromophore contains the same p-hydroxybenzylideneimidazolinone (HBDI) group as GFP but also has an additional double bond between the backbone nitrogen and alpha carbon of Gln66 [36, 40](Figure 1A). Together with the backbone carbonyl of amino acid 65, this double bond forms an acylimine group that extends π conjugation by four atoms. This allows more delocalized electron density, decreasing the energy gap between the highest occupied molecular orbital (HOMO) and the lowest unoccupied molecular orbital (LUMO), which represent the ground and excited states for the most significant electron transition.
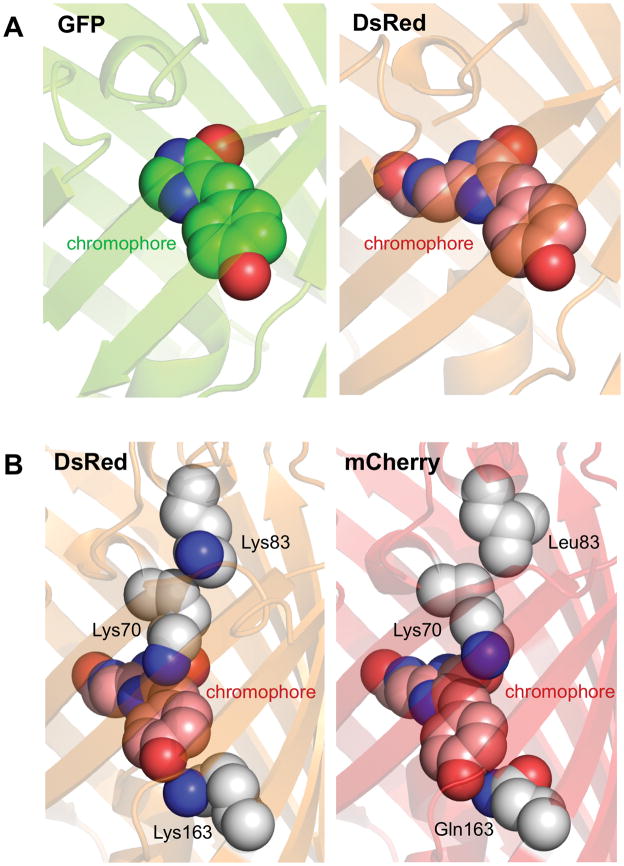
Figure 1.
Basic structural biology of RFPs. (A) Structures of GFP and DsRed chromophores. The GFP chromophore (left), shown with carbon in green, nitrogen blue, and oxygen red, is a π-conjugated system consisting of a phenolate ring (front) connected to a imidazolinone ring (rear) by a methylene bridge. The DsRed chromophore (right), shown with carbon in pink, consists of the same groups formed from cyclization of a Gln-Tyr-Gly sequence, but with conjugation extended through an acylimine group formed by additional oxidation of the backbone N-Cα bond of the Gln residue. (B) Altered electronic interactions of the chromophore with its environment in mCherry. In DsRed (left), the positive charges of Lys70 and Lys163 stabilize the ground state, where electron density preferentially resides in the phenol ring. In mCherry (right), movement of Lys70 away from the chromophore and mutation of Lys163 to Gln reduces this effect. Movement of Lys70 in mCherry is due to loss of electrostatic repulsion by Leu83. Side chains and alpha carbons of these important amino acids are shown a spheres, with carbon in white, nitrogen blue, and oxygen red.
The residues in the barrel near the chromophore exert steric and electronic effects on the chromophore that modulate the excitation and emission spectra and the brightness of the chromophore. Extensive theoretical studies have investigated the physical chemical mechanisms for wavelength shifting and tuning of GFP [41] and RFPs [37, 38, 42, 43]. As the RFP chromophore is extended relative to GFP, some of the mechanisms of wavelength tuning that have evolved were not previously observed in GFP-family proteins.
Altered electronic interactions with the chromophore rings in mCherry
The crystal structure of mCherry shows two features that may account for the substantial red-shift from its progenitor, DsRed [44]. First, Lys163 (pKa 8.0 by PROPKA 3.0) has been substituted with Gln. This side chain occupies a position underneath the phenol ring of the chromophore (when N- and C-termini are oriented up), and Lys163 was observed in a conformation indicating hydrogen bonding with the chromophore phenolate [36, 40]. The chromophore is deprotonated within the protein based on its distinct absorbance within the protein compared to in solution [45], and thus the Lys163 is believed to be protonated and capable of hydrogen bond donation to the chromophore [36, 40]. It thus is expected to stabilize electron density on the phnolate in DsRed (Figure 1B). As electron density in the phenol ring decreases in the excited state [38], this is expected to preferentially stabilize the ground state, accentuating the energy gap between the ground and excited states. Replacement of cationic Lys163 in mCherry with the neutral and less acidic Gln would be expected to result in a weaker hydrogen bond in which the distance betwen the donor proton and the phenolate oxygen is increased. This would lead to destabilization of the ground state to produce the observed red-shift in excitation spectrum.
A second change is mutation of Lys83 to Leu. A similar Lys83Met mutation was shown to red-shift DsRed [46•]. The DsRed Lys83Met crystal structure reveals movement of a conserved Lys70 toward Met83 and away from the phenol ring. As Lys83 only interacts with other canonical amino acids (as opposed to the chromophore), it is suitable to attempt to predict its pKa using PROPKA3 [47], which yields values of 7.67–7.93. Thus Lys83 is likely to be protonated at neutral pH, and the movement of Lys70 toward Met83 may be due to loss of electrostatic repulsion. A similar movement of Lys70 was also observed in mCherry (Figure 1B). Interestingly, quantum mechanics/molecular mechanics modeling suggests that the bond between the methylene group and the phenol ring has higher electron density in the ground versus the excited state [38, 41]. Thus Lys70 may also contribute to stabilization of the ground state, and its movement away from the phenol group would also be expected to cause a red-shift in excitation.
Chromophore isomerization and altered electronic distributions in mKate
The crystal structure of mKate reveals multiple structural features responsible for red-shifting from its progenitor, TagRFP [48•]. The mKate chromophore is in a cis configuration like DsRed (Figure 2) but unlike its precursor, TagRFP, which is in the trans configuration [49, 50]. The chromophore phenolate group of TagRFP makes two hydrogen bonds to Asn143 and Ser158. The chromophore of mKate in the cis conformation is only able to make a hydrogen bond to Ser143. The loss of a hydrogen bond likely destabilizes electron distribution on the phenolate in the ground state, causing a red-shift.
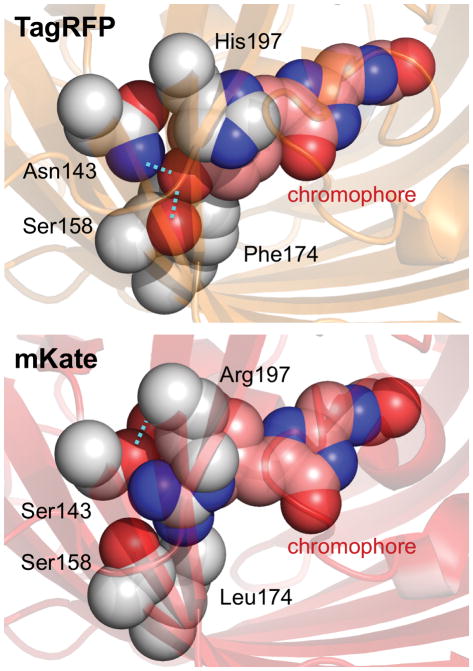
Figure 2.
Chromophore isomerization, reduced hydrogen bonding, and altered electrostatic interactions in mKate. The chromophore rings in TagRFP are in a trans conformation, where the phenolate oxygen makes two hydrogen bonds (cyan dotted lines) and the His197 engages in a cation-π interaction with the phenolate ring. In mKate, mutations Asn143Ser, Phe174Leu, and His197Arg cause the chromophore to assume a cis conformation, losing one hydrogen bond and the cation-π interaction.
A second red-shifting mechanism in mKate may be the loss of a positively charged side chain above the chromophore. His197 of TagRFP is directly above the chromophore. While its protonation state has not been directly measured, quantum mechanical calculations of asFP595, which also has a His197 above a dsRed-type chromophore, suggests that His197 can be cationic and stabilize the anionic phenolate in a cation-π interaction [51]. As with Lys163 in DsRed, this would be expected to increase the energy required to transition an electron to the excited state [52]. In mKate, replacement of His197 by Arg shifts the positive charge from directly above the phenolate ring to its side (Figure 2), removing the potential cation-π interaction.
Increased chromophore planarity in Neptune and eqFP670
The structure of Neptune revealed two major novel structural features that explain its large red-shift of both excitation and emission maxima: increased chromophore planarity and a hydrogen bond to the acylimine oxygen [19]. We will discuss increased coplanarity first. In Neptune, the side chain of Arg197, instead of extending across to the side of the phenolate ring, is parallel above the two rings of the chromophore (Figure 3A). This change may be due to the mutation Ser158Cys, which extends a larger side chain into the space previously occupied by Arg197. With Arg197 now extended along the chromophore axis, the two chromophore rings are able to become more coplanar, which is expected to promote electron delocalization and thereby reduce the energy of the excited state [37]. A Cys158Ser reversion produces a 7 nm blue-shift in excitation, confirming the importance of this mutation.
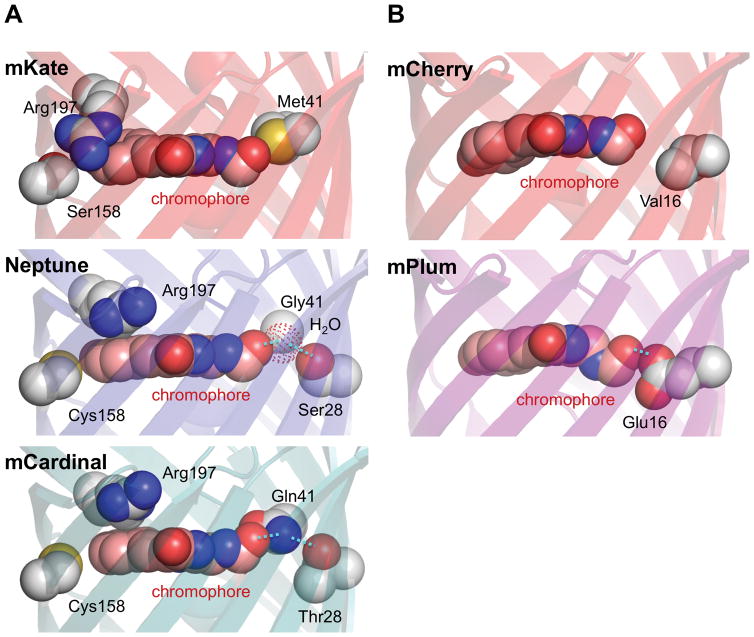
Figure 3.
Structural basis of red-shifted excitation in far-RFPs. (A) Increased chromophore planarity and a new hydrogen bond in Neptune and mCardinal. In mKate, the Arg197 side chain extends across and to the side of the phenolate ring of the chromophore, which is not coplanar with the imidazolinone ring. In Neptune, Cys158 pushes Arg197 to a new position above the chromophore, allowing for a more coplanar arrangement of the two chromophore rings. In addition, mutation Met41Gly creates a new cavity for a water molecule (red dotted sphere) to reside in near the acylimine. The white sphere labeled Gly41 is the Gly41 alpha carbon. Hydrogen bonding between the water molecule and the acylimine oxygen stabilizes the excited state and leads to an excitation red-shift. In mCardinal, the role of the water molecule is replaced by Gln41, which is held in position by Thr28. (B) Excited-state energetic relaxations near the acylimine in the excited state of mPlum. mPlum contains a new hydrogen bond from Glu16 to the acylimine oxygen of the chromophore. Conformational changes to the chromophore or to Glu16 occur in the excited state, causing a large Stokes shift.
A correlation between chromophore planarity and excitation red-shifting was also seen in the structures of eqFP650 and eqFP670 [53]. The two rings of the eqFP650 chromophore deviate from coplanarity by 18° due to the above-described extension of Arg197 to the side of the chromophore, which could contribute to its bluer excitation than Neptune (592 vs 600 nm). In eqFP670, a Ser158Asn mutation fills the space occupied by Arg197 in eqFP650, resulting in positioning of Arg197 above the chromophore and improved coplanarity.
Hydrogen bonding to the acylimine oxygen in Neptune, eqFP670, and mCardinal
The second novel feature found in Neptune is a hydrogen bond to the acylimine oxygen. A cavity created by a Met41Gly mutation contains a water molecule that forms a hydrogen bond with the chromophore acylimine oxygen (Figure 3A) [19]. This is expected to preferentially stabilize the excited state, which is more electron-rich at the acylimine oxygen than the ground state, thereby lowering the energetic gap between ground and excited states. Gly41Met reversion causes a 5-nm blue-shift to the excitation spectrum, supporting the importance of the water molecule. Mutations of Met41 to small side chains also contribute to red-shifting in E2-Crimson, TagRFP657, and eqFP670, demonstrating the generalizability of this mechanism [23, 26, 54]. The structure of eqFP670 also revealed a water molecule in the cavity positioned to hydrogen-bond with the acylimine oxygen [53].
In mCardinal, an amino acid side chain serves as the hydrogen bond donor to the acylimine oxygen. mCardinal was evolved from mNeptune by targeted mutagenesis of Gly41 and nearby positions to possible hydrogen bond donors in an effort to create an additional excitation red-shift. Indeed, a 5-nm red-shift was obtained in mCardinal by Ser28Thr and Gly41Gln mutations. The crystal structure of mCardinal confirmed that the Gln41 side chain donates a hydrogen bond to the acylimine oxygen (Figure 3A) [25••]. Gln41 also engages in a hydrogen bond with Thr28, which could stabilize Gln41 in position to interact with the acylimine. Thus the crystal structure confirmed the design hypothesis.
Emission red-shifting by excited-state-specific acylimine interactions
The mPlum chromophore acylimine also engages in a hydrogen bond with an amino acid side chain, but in a way that specifically red-shifts only the emission spectrum. mPlum demonstrates a large Stokes shift of 59 nm compared to 23 nm for its parent mCherry. A Glu16 residue is necessary for this large Stokes shift, as a Glu16Gln mutation reduces the Stokes shift to 40 nm without having any effect on the excitation peak. The crystal structure of mPlum reveals a small interatomic distance between a terminal Glu16 oxygen and the acylimine oxygen that can only be explained by a hydrogen bond [44]. As the acylimine oxygen lacks a proton, Glu16 is resumably protonated (Figure 3B). Time-resolved emission spectroscopy revealed energetic relaxation of the chromophore during the excited state with kinetics consistent with rotation of Glu16 [44]. This rotation of Glu16 could further stabilize the excited state and reduce the emitted photon energy [44], explaining the large Stokes shift in mPlum and the lack of a significant excitation red-shift compared to mCherry.
A similar mechanism of Stokes shift extension likely exists in the tetrameric AQ143 as well as TagRFP675 and redder-emitting mNeptune variants such as mNeptune684. Like mPlum, AQ143 has Glu at position 16 and exhibits an emission peak near 650 nm while its excitation spectrum is similar to mCherry. Its crystal structure, while lacking electron density in the chromophore rings, confirmed Glu16 is in a hydrogen-bonding interaction with the acylimine oxygen. TagRFP675 and mNeptune684 have a Gln at position 41, similar to mCardinal. However they have longer Stokes shifts with redder emission spectra than mCardinal. The crystal structure of TagRFP675 reveals hydrogen bonding between Gln41 and the acylimine oxygen [27]. In TagRFP675, the plane of the amide group of Gln41 is oriented more perpendicular to the plane of the chromophore than in mCardinal, resulting in a slightly longer hydrogen bond. If rotation of Gln41 occurs in the excited state to shorten the bond, as hypothesized for Glu16 in mPlum, this could explain the large Stokes shift and reduced quantum yield (due to additional paths for non-radiative decay). TagRFP675 and mNeptune684 have the smaller Ser28 instead of the Thr28 in mCardinal, which may facilitate excited-state movements of Gln41. In mCardinal, the more restricted orientation of Thr28 may affix Gln41 in a stronger hydrogen-bonding orientation that does not change.
Increased environment polarizability
mGrape3, E2-Crimson, and TagRFP657 all have a tyrosine at position 197 (DsRed numbering), and are currently the most red-shifted FPs in terms of excitation, with peaks at 608–611 nm. Crystal structures have not been obtained for these proteins, but they probably utilize the same red-shifting mechanism as YFP. To create YFP, a tyrosine was rationally introduced at position 203 (homologous to 197 of DsRed) in GFP S65T, leading to red-shifts of excitation and emission spectra by about 20 nm. Tyrosine engages in a π-π stacking interaction with the phenolate group of the chromophore [35••]. The highly polarizable tyrosine is expected to red-shift the chromophore excitation. This is because the excited state of a π-conjugated fluorophore usually has a larger dipole moment than the ground state, but if it forms within a polarizable solvent, the dipole moment can be counteracted by corresponding charge shifts in the solvent [55]. The tyrosine in effect acts as a polarizable solvent, reducing the energetic gap between ground and excited states.
Increased environment polarizability may also contribute to red-shifting in eqFP670. Pletnev et al. found that Asn143 and Asn 158 were both required for the full excitation red-shift to 605 nm of eqFP670 [53]. Asn 158 improves chromophore planarity as discussed above. Asn143 is dually hydrogen-bonded, to the chromophore and to Asn 158, which then engages a hydrogen-bond network of a water molecule, Thr176, and Arg92. Reverting Asn143 to Ser causes a dramatic excitation blue-shift. Taken alone, this could be interpreted as Ser143 creating a stronger hydrogen bond with the chromophore that preferentially stabilizes the chromophore ground state. However, introducing Asn143 into Katushka, which has Ser at both 143 and 158, causes a blue-shift, suggesting that Asn143 is not causing red-shifting through reduced stabilization of the ground state. It was instead suggested that the hydrogen bond network comprising Asn143, Asn158, water, Thr176, and Arg92 was important for the red-shifted excitation, perhaps by creating a polarizable electronic environment.
Summary and future directions
The structural studies discussed above have revealed in detail several mechanisms of red-shifting in far-RFPs. The mutational history of these proteins conveniently allows the contribution of specific residues in wavelength tuning to be unambiguously assigned. This understanding has in turn allowed the rational targeting of specific sites for mutagenesis to derive even more red-shifted FPs. An example is the repeated mutagenesis at positions 41 and 158, identified as important in the Neptune structure, in the engineering of further derivatives. Indeed, most red-shifting efforts involved a mixture of directed and random mutagenesis[16•, 19••, 20•, 22•, 23, 26], with only one example each of completely random mutagenesis [18•] and purely directed mutagenesis [25••]. Rather, researchers have taken advantage of structural and chemical knowledge to formulate informed hypotheses on the protein regions to target by mutagenesis.
Approaches combinining structural knowledge and computational prediction have become increasingly useful for protein engineering, with demonstrated successes in altering physical stability, protein-protein interactions, or enzymatic functions. A rigorous test of our understanding of far-red FPs would be the ability to engineer new capabilities by entirely non-random methods, ie., computational engineering. Computation was used to design libraries for red-shifting of mCherry by predicting substitutions that would fulfill one of three red-shifting mechanisms known from structural studies: 1) hydrogen bonding to the acylimine oxygen, 2) π-π stacking to stabilize the excited state, and 3) tighter packing around the phenolate to destabilize the ground state [56]. This allowed creation of a library of 5,000 clones from which red-shifted mutants were isolated. Crystal structures revealed that the first two of the three hypotheses were fulfilled, by Thr16 making a hydrogen bond to the acylimine oxygen and Tyr197 stacking over the phenolate. However, these two red-shifting mechanisms had already been established in previous engineering efforts, some of which deliberately targeted these sites without computational assistance [18•, 19••, 20•], whereas the more novel hypothesis of ground-state destabilization was not validated. Furthermore, both excitation wavelength and brightness of the identified mutants fell short of those of existing far-red FPs. The physical bases for desired properties such as brightness, quick maturation, and photostability are probably still not well understood enough for deterministic computer modeling. Future computational attempts at capturing multiple mechanisms for red shifting and brightness, incorporating quantum mechanical models of chromophore ground and excited states [42, 43], will likely lead to improved results.
Further improvements in brightness and red-shifting may be possible with different families of FPs than the two (DsRed and eqFP578) that have been most extensively evolved. For example, eqFP611 [3] has given rise to the monomeric RFPs mRuby1, mRuby2, and mRuby3 [57–59]. mRuby3 is currently the brightest RFP and represents an unexplored starting point for the development of far-red FPs.
What are the limits to red-shifting in far-RFPs? The DsRed chromophore is constrained by space and needs be encoded within a turn of an alpha helix. Strain is still present along the acylimine group even in the most red-shifted FPs such as mCardinal. Interestingly, strain is completely released in the photoconverted state of PSmOrange, where a photochemical reaction requiring three photons leads to cleavage of the backbone immediately N-terminal to the chromophore [60••]. Indeed PSmOrange has a dramatically red-shifted excitation peak of 634 nm, which is a perfect match for the common 633- and 635-nm lasers. It would thus be extremely desirable to create a RFP that could undergo spontaneous cleavage of the backbone near the chromophore.
We expect that further improvements in protein structure predictions, quantum mechanics/molecular mechanics predictions of absorbance spectra, and computational power will eventually enable more rational design of small and still effective libraries within reasonable computation times. Regardless, atomic-level structural analysis will certainly continue to play an essential role in understanding the optical characteristics of far-RFPs for the foreseeable future.
Acknowledgments
We gratefully acknowledge research funding from the Rita Allen Foundation (M.Z.L.), NIH Pioneer Award 1DP1OD017209 (M.Z.L.), and NIH BRAIN Initiative grant 1U01NS090600 (M.Z.L.).
References
- 1.Shimomura O, Johnson FH, Saiga Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J Cell Comp Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- 2.Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 3.Wiedenmann J, Schenk A, Rocker C, Girod A, Spindler KD, Nienhaus GU. A far-red fluorescent protein with fast maturation and reduced oligomerization tendency from Entacmaea quadricolor (Anthozoa, Actinaria) Proc Natl Acad Sci USA. 2002;99:11646–11651. doi: 10.1073/pnas.182157199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sugiyama M, Sakaue-Sawano A, Iimura T, Fukami K, Kitaguchi T, Kawakami K, Okamoto H, Higashijima S, Miyawaki A. Illuminating cell-cycle progression in the developing zebrafish embryo. Proc Natl Acad Sci USA. 2009;106:20812–20817. doi: 10.1073/pnas.0906464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter KP, Young AM, Palmer AE. Fluorescent Sensors for Measuring Metal Ions in Living Systems. Chemical Reviews. 2014;114:4564–4601. doi: 10.1021/cr400546e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.St-Pierre F, Marshall JD, Yang Y, Gong Y, Schnitzer MJ, Lin MZ. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nature Neuroscience. 2014;17:884–889. doi: 10.1038/nn.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.St-Pierre F, Chavarha M, Lin MZ. Designs and sensing mechanisms of genetically encoded fluorescent voltage indicators. Current Opinion in Chemical Biology. 2015;27:31–38. doi: 10.1016/j.cbpa.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moussa R, Baierl A, Steffen V, Kubitzki T, Wiechert W, Pohl M. An evaluation of genetically encoded FRET-based biosensors for quantitative metabolite analyses in vivo. J Biotechnol. 2014;191:250–259. doi: 10.1016/j.jbiotec.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Sato M, Umezawa Y. Genetically Encoded Fluorescent Indicators to Visualize Protein Phosphorylation in Living Cells. Methods in Molecular Biology. 2016;1360:149–156. doi: 10.1007/978-1-4939-3073-9_11. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, Ni Q, Newman RH, editors. Fluorescent Protein-Based Biosensors: Methods and Protocols (Methods in Molecular Biology) Humana Press; 2013. [Google Scholar]
- 11.Shcherbakova DM, Sengupta P, Lippincott-Schwartz J, Verkhusha VV. Photocontrollable Fluorescent Proteins for Superresolution Imaging. Annual review of biophysics. 2014;43:303–329. doi: 10.1146/annurev-biophys-051013-022836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou XX, Chung HK, Lam AJ, Lin MZ. Optical control of protein activity by fluorescent protein domains. Science (New York, NY) 2012;338:810–814. doi: 10.1126/science.1226854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weissleder R. A clearer vision for in vivo imaging. Nat Biotechnol. 2001;19:316–317. doi: 10.1038/86684. [DOI] [PubMed] [Google Scholar]
- 14.Bruno TJ, Svoronos PDN, editors. CRC Handbook of Fundamental Spectroscopic Correlation Charts. CRC Press; 2005. [Google Scholar]
- 15.Sharrock RA. The phytochrome red/far-red photoreceptor superfamily. Genome Biol. 2008;9:230. doi: 10.1186/gb-2008-9-8-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. Describes the engineering of a series of fluorescent proteins derived from mRFP1 with various excitation and emission spectra, demonstrating the spectral flexibility of the DsRed chromophore. [DOI] [PubMed] [Google Scholar]
- 17•.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. Describes the engineering of dimeric and monomeric forms of DsRed, serving as precedent for later monomerization of other tetrameric fluorescent protein species. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18•.Wang L, Jackson WC, Steinbach PA, Tsien RY. Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc Natl Acad Sci USA. 2004;101:16745–16749. doi: 10.1073/pnas.0407752101. Describes the engineering of mRaspberry and mPlum, the DsRed derivatives with the most red-shifted excitation and emission spectra. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19••.Lin MZ, McKeown MR, Ng HL, Aguilera TA, Shaner NC, Campbell RE, Adams SR, Gross LA, Ma W, Alber T, Tsien RY. Autofluorescent proteins with excitation in the optical window for intravital imaging in mammals. Chem Biol. 2009;16:1169–1179. doi: 10.1016/j.chembiol.2009.10.009. Describes the engineering of mNeptune, the first monomeric fluorescent protein with peak excitation at 600 nm or above. Also describes the structural basis for red-shifted excitation, uncovering novel mechanisms for hydrogen bond formation with the acylimine group of the chromophore and for enhancement of chromophore planarity. Also demonstrates the beneficial effect of exciting with red light during non-invasive imaging in mouse liver. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Strack RL, Hein B, Bhattacharyya D, Hell SW, Keenan RJ, Glick BS. A rapidly maturing far-red derivative of DsRed-Express2 for whole-cell labeling. Biochemistry. 2009;48:8279–8281. doi: 10.1021/bi900870u. Describes the engineering of the first fluorescent protein with peak excitation above 600 nm and a successful example of mechanism-informed design. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21•.Merzlyak EM, Goedhart J, Shcherbo D, Bulina ME, Shcheglov AS, Fradkov AF, Gaintzeva A, Lukyanov KA, Lukyanov S, Gadella TW, Chudakov DM. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. Describes the isolation of eqFP578 and its engineering into TagRFP which will eventually serve as the basis for monomeric red-excitable far-RFPs. [DOI] [PubMed] [Google Scholar]
- 22•.Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, Lukyanov KA, Bogdanova EA, Zaraisky AG, Lukyanov S, Chudakov DM. Bright far-red fluorescent protein for whole-body imaging. Nat Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. Describes the engineering of mKate, a bright RFP that will serve as the basis for the engineering of multiple far-RFPs. [DOI] [PubMed] [Google Scholar]
- 23.Shcherbo D, Shemiakina II, Ryabova AV, Luker KE, Schmidt BT, Souslova EA, Gorodnicheva TV, Strukova L, Shidlovskiy KM, Britanova OV, et al. Near-infrared fluorescent proteins. Nat Methods. 2010;7:827–829. doi: 10.1038/nmeth.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shcherbo D, Murphy CS, Ermakova GV, Solovieva EA, Chepurnykh TV, Shcheglov AS, Verkhusha VV, Pletnev VZ, Hazelwood KL, Roche PM, et al. Far-red fluorescent tags for protein imaging in living tissues. Biochem J. 2009;418:567–574. doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Chu J, Haynes RD, Corbel SY, Li P, González-González E, Burg JS, Ataie NJ, Lam AJ, Cranfill PJ, Baird MA, et al. Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein. Nature Methods. 2014;11:572–578. doi: 10.1038/nmeth.2888. Describes the engineering of mCardinal, a far-RFP with enhanced brightness and high excitability at 633 nm, describes its structure, uses it for longitudinal imaging of stem cell differentiation in muscle over months, and uses it for orthogonal imaging with orange FPs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morozova KS, Piatkevich KD, Gould TJ, Zhang J, Bewersdorf J, Verkhusha VV. Far-red fluorescent protein excitable with red lasers for flow cytometry and superresolution STED nanoscopy. Biophys J. 2010;99:L13–5. doi: 10.1016/j.bpj.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piatkevich KD, Malashkevich VN, Morozova KS, Nemkovich NA, Almo SC, Verkhusha VV. Extended Stokes shift in fluorescent proteins: chromophore-protein interactions in a near-infrared TagRFP675 variant. Scientific Reports. 2013;3:1847. doi: 10.1038/srep01847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z, Zhang Z, Bi L, Cui Z, Deng J, Wang D, Zhang X-E. Mutagenesis of mNeptune Red-Shifts Emission Spectrum to 681–685 nm. PLOS ONE. 2016;11:e0148749. doi: 10.1371/journal.pone.0148749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christensen J, Vonwil D, Shastri VP. Non-Invasive In Vivo Imaging and Quantification of Tumor Growth and Metastasis in Rats Using Cells Expressing Far-Red Fluorescence Protein. PloS One. 2015;10:e0132725. doi: 10.1371/journal.pone.0132725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30••.Levin RA, Felsen CN, Yang J, Lin JY, Whitney MA, Nguyen QT, Tsien RY. An optimized triple modality reporter for quantitative in vivo tumor imaging and therapy evaluation. PloS One. 2014;9:e97415. doi: 10.1371/journal.pone.0097415. Demonstrates intravital fluorescent protein signals are better correlated with tumor weight than luciferase signals, using E2-Crimson. Also determined fluorescence required more cells than bioluminescence for detection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu M, Schmitner N, Sandrian MG, Zabihian B, Hermann B, Salvenmoser W, Meyer D, Drexler W. In vivo three dimensional dual wavelength photoacoustic tomography imaging of the far red fluorescent protein E2-Crimson expressed in adult zebrafish. Biomed Opt Express. 2013;4:1846–1855. doi: 10.1364/BOE.4.001846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hense A, Prunsche B, Gao P, Ishitsuka Y, Nienhaus K, Nienhaus GU. Monomeric Garnet, a far-red fluorescent protein for live-cell STED imaging. Sci Rep. 2015;5:18006. doi: 10.1038/srep18006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hein B, Willig KI, Hell SW. Stimulated emission depletion (STED) nanoscopy of a fluorescent protein-labeled organelle inside a living cell. Proc Natl Acad Sci U S A. 2008;105:14271–14276. doi: 10.1073/pnas.0807705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willig KI, Kellner RR, Medda R, Hein B, Jakobs S, Hell SW. Nanoscale resolution in GFP-based microscopy. Nat Methods. 2006;3:721–723. doi: 10.1038/nmeth922. [DOI] [PubMed] [Google Scholar]
- 35••.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. Describes the atomic structure of GFP. Also describes the creation of YFP by the rational introduction of the polarizable tyrosine side chain above the chromophore, a red-shifting strategy later used in several far-red FPs. [DOI] [PubMed] [Google Scholar]
- 36.Wall MA, Socolich M, Ranganathan R. The structural basis for red fluorescence in the tetrameric GFP homolog DsRed. Nat Struct Biol. 2000;7:1133–1138. doi: 10.1038/81992. [DOI] [PubMed] [Google Scholar]
- 37.Bravaya KB, Subach OM, Korovina N, Verkhusha VV, Krylov AI. Insight into the Common Mechanism of the Chromophore Formation in the Red Fluorescent Proteins: The Elusive Blue Intermediate Revealed. J Am Chem Soc. 2012;134:2807–2814. doi: 10.1021/ja2114568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasegawa JY, Ise T, Fujimoto KJ, Kikuchi A, Fukumura E, Miyawaki A, Shiro Y. Excited states of fluorescent proteins, mKO and DsRed: chromophore-protein electrostatic interaction behind the color variations. J Phys Chem B. 2010;114:2971–2979. doi: 10.1021/jp9099573. [DOI] [PubMed] [Google Scholar]
- 39.Remington SJ. Green fluorescent protein: A perspective. Protein Sci. 2011;20:1509–1519. doi: 10.1002/pro.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ. Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2. 0-A resolution. Proc Natl Acad Sci U S A. 2001;98:462–467. doi: 10.1073/pnas.98.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bravaya KB, Grigorenko BL, Nemukhin AV, Krylov AI. Quantum Chemistry Behind Bioimaging: Insights from Ab Initio Studies of Fluorescent Proteins and Their Chromophores. Acc Chem Res. 2012;45:265–275. doi: 10.1021/ar2001556. [DOI] [PubMed] [Google Scholar]
- 42.Nemukhin AV, Grigorenko BL, Savitsky AP. Computer Modeling of the Structure and Spectra of Fluorescent Proteins. Acta Naturae. 2009;1:33–43. [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez-Garcia E, Doerr M, Thiel W. QM/MM study of the absorption spectra of DsRed.M1 chromophores. J Comput Chem. 2010;31:1603–1612. doi: 10.1002/jcc.21443. [DOI] [PubMed] [Google Scholar]
- 44.Shu X, Wang L, Colip L, Kallio K, Remington SJ. Unique interactions between the chromophore and glutamate 16 lead to far-red emission in a red fluorescent protein. Protein Sci. 2009;18:460–466. doi: 10.1002/pro.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY. The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci U S A. 2000;97:11990–11995. doi: 10.1073/pnas.97.22.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46•.Shu X, Shaner NC, Yarbrough CA, Tsien RY, Remington SJ. Novel chromophores and buried charges control color in mFruits. Biochemistry. 2006;45:9639–9647. doi: 10.1021/bi060773l. Describes the atomic structure of mCherry, revealing the basis for its red-shift compared to the parental dsRed. [DOI] [PubMed] [Google Scholar]
- 47.Olsson MH, Søndergaard CR, Rostkowski M, Jensen JH. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J Chem Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 48•.Pletnev S, Shcherbo D, Chudakov DM, Pletneva N, Merzlyak EM, Wlodawer A, Dauter Z, Pletnev V. A crystallographic study of bright far-red fluorescent protein mKate reveals pH-induced cis-trans isomerization of the chromophore. J Biol Chem. 2008;283:28980–28987. doi: 10.1074/jbc.M800599200. Describes the atomic structure of mKate, revealing the basis for its red-shift compared to the parental TagRFP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subach OM, Malashkevich VN, Zencheck WD, Morozova KS, Piatkevich KD, Almo SC, Verkhusha VV. Structural Characterization of Acylimine-Containing Blue and Red Chromophores in mTagBFP and TagRFP Fluorescent Proteins. Chem Biol. 2010;17:333–341. doi: 10.1016/j.chembiol.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petersen J, Wilmann PG, Beddoe T, Oakley AJ, Devenish RJ, Prescott M, Rossjohn J. The 2. 0-A crystal structure of eqFP611, a far red fluorescent protein from the sea anemone Entacmaea quadricolor. J Biol Chem. 2003;278:44626–44631. doi: 10.1074/jbc.M307896200. [DOI] [PubMed] [Google Scholar]
- 51.Schäfer LV, Groenhof G, Klingen AR, Ullmann GM, Boggio-Pasqua M, Robb MA, Grubmüller H. Photoswitching of the fluorescent protein asFP595: mechanism, proton pathways, and absorption spectra. Angew Chem Int Ed Engl. 2007;46:530–536. doi: 10.1002/anie.200602315. [DOI] [PubMed] [Google Scholar]
- 52.Ai HW, Henderson JN, Remington SJ, Campbell RE. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem J. 2006;400:531–540. doi: 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pletnev S, Pletneva NV, Souslova EA, Chudakov DM, Lukyanov S, Wlodawer A, Dauter Z, Pletnev V. Structural basis for bathochromic shift of fluorescence in far-red fluorescent proteins eqFP650 and eqFP670. Acta Crystallographica Section D, Biological Crystallography. 2012;68:1088–1097. doi: 10.1107/S0907444912020598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strack RL, Strongin DE, Bhattacharyya D, Tao W, Berman A, Broxmeyer HE, Keenan RJ, Glick BS. A noncytotoxic DsRed variant for whole-cell labeling. Nat Methods. 2008;5:955–957. doi: 10.1038/nmeth.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wachter RM, Elsliger MA, Kallio K, Hanson GT, Remington SJ. Structural basis of spectral shifts in the yellow-emission variants of green fluorescent protein. Structure. 1998;6:1267–1277. doi: 10.1016/s0969-2126(98)00127-0. [DOI] [PubMed] [Google Scholar]
- 56.Chica RA, Moore MM, Allen BD, Mayo SL. Generation of longer emission wavelength red fluorescent proteins using computationally designed libraries. Proc Natl Acad Sci U S A. 2010;107:20257–20262. doi: 10.1073/pnas.1013910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bajar BT, Wang ES, Lam AJ, Kim BB, Jacobs CL, Howe ES, Davidson MW, Lin MZ, Chu J. Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Scientific Reports. 2016;6:20889. doi: 10.1038/srep20889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kredel S, Oswald F, Nienhaus K, Deuschle K, Rocker C, Wolff M, Heilker R, Nienhaus GU, Wiedenmann J. mRuby, a bright monomeric red fluorescent protein for labeling of subcellular structures. PLoS One. 2009;4:e4391. doi: 10.1371/journal.pone.0004391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lam AJ, St-Pierre F, Gong Y, Marshall JD, Cranfill PJ, Baird MA, McKeown MR, Wiedenmann J, Davidson MW, Schnitzer MJ, et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nature Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60••.Subach OM, Patterson GH, Ting L-M, Wang Y, Condeelis JS, Verkhusha VV. A photoswitchable orange-to-far-red fluorescent protein, PSmOrange. Nature Methods. 2011;8:771–777. doi: 10.1038/nmeth.1664. Describes the photoconversion of an orange fluorescent protein into a far-RFP with excitation maximum at 634 nm due to photoinduced backbone cleavage, demonstrating a DsRed-like chromophore has the potential for further red-shifting in excitation maximum. [DOI] [PMC free article] [PubMed] [Google Scholar]