Abstract
Squamous cell carcinoma of the esophagus (SCC) is the most common form of esophageal cancer in the world and is typically diagnosed at an advanced stage when successful treatment is challenging. Understanding the mutational profile of this cancer may identify new treatment strategies. Because somatic retrotransposition has been shown in tumors of the gastrointestinal system, we focused on LINE-1 (L1) mobilization as a source of genetic instability in this cancer. We hypothesized that retrotransposition is ongoing in SCC patients. The expression of L1 encoded proteins is necessary for retrotransposition to occur; therefore, we evaluated the expression of L1 open reading frame 1 protein (ORF1p). Using immunohistochemistry, we detected ORF1p expression in all four SCC cases evaluated. Using L1-seq, we identified and validated 74 somatic insertions in eight tumors of the nine evaluated. Of these, 12 insertions appeared to be somatic, not genetically inherited, and sub-clonal (i.e., present in less than one copy per genome equivalent) in the adjacent normal esophagus while clonal in the tumor. Our results indicate that L1 retrotransposition is active in squamous cell carcinoma of the esophagus and that insertion events are present in histologically normal esophagus that expand clonally in the subsequent tumor.
Keywords: LINE-1, L1, retrotransposition, sub-clonal, squamous cell carcinoma, esophagus
INTRODUCTION
Esophageal squamous cell carcinoma (SCC) is the most common esophageal cancer in the world and its incidence differs across various geographic areas; it rarely manifests in North America, but it is the major histologic type of esophageal cancer in parts of Eastern Asia (Abedi-Ardekani and Hainaut, 2014; Enzinger and Mayer, 2003; Ohashi, et al., 2015). SCC develops from the cells comprising the squamous esophageal mucosa and its main risk factors include combined alcohol and tobacco use, consumption of scalding beverages, and a diet low in fresh fruits and vegetables (Abedi-Ardekani and Hainaut, 2014; Kamangar, et al., 2007). This aggressive cancer is especially common in rural, mountainous areas with little access to resources and minimal dietary diversity such as Northern Iran, central China, parts of South-East Africa, and South America (Islami, et al., 2009; Kamangar, et al., 2007; Lambert and Hainaut, 2007; Tran, et al., 2005). Unfortunately, by the time SCC is diagnosed, greater than half of the patients have inoperable tumors or obvious metastases (Enzinger and Mayer, 2003). Even if the tumor is removable, the prognosis for most patients is still very poor; therefore, better methods for early detection and treatment are necessary (Baba, et al., 2014).
Recently many groups, including our own, have evaluated a mutation generating mechanism known as retrotransposition and its role in epithelial cell cancer development(Doucet-O’Hare, et al., 2015; Hancks and Kazazian, 2012; Helman, et al., 2014; Iskow, et al., 2010, Lee, et al., 2012; Rodic, et al., 2015; Shukla, et al., 2013; Solyom, et al., 2012). During retrotransposition, a sequence of DNA mobilizes through reverse transcription of an RNA intermediate, a so-called “copy and paste” mechanism. L1 elements are the only autonomous retrotransposons in the human genome. These elements mobilize by promoting their own transcription followed by the translation of the two open reading frames which code for proteins necessary for the element’s reintegration into the genome (Scott, et al., 1987). The movement of all other active retro-elements in the human genome [Alu and SINE–VNTR–Alu (SVA)] is dependent on the activity of L1 elements (Esnault, et al., 2000; Wei, et al., 2001). Cellular mRNAs can also be inserted into the genome by L1, becoming processed pseudogenes. Because retrotransposons are potentially mutagenic when inserting into new sites in the genome, host cells inhibit their activity by suppressing transcription as well as L1 RNA translation and L1 protein functions (Arjan-Odedra S, 2012; Bogerd, et al., 2006; Chen, et al., 2006; Chow, et al., 2010; Goodier, et al., 2012; Heras, et al., 2013; Levin and Moran, 2011; Stenglein and Harris, 2006). L1 expression levels are inversely correlated with methylation of its promoter in the 5’ UTR of the element; numerous epigenetic modifiers contribute to establishing and maintaining the methylation status of L1 elements (Ecco, et al., 2016; Imbeault and Trono, 2014; Irahara, et al., 2010; Jacobs, et al., 2014; Muotri, et al., 2010; Rowe and Trono, 2011; Shigaki, et al., 2013; Turelli, et al., 2004; Yoder, et al., 1997). A known feature of SCC is global hypomethylation of L1 elements throughout the genome; furthermore, the less methylation present, the worse the prognosis for the patient (Baba, et al., 2014; Iwagami S, 2013). Additionally, we have observed expression of ORF1p encoded by L1 in normal squamous epithelium of the esophagus indicating that L1 is potentially active in the relevant normal tissue (Doucet-O’Hare, et al., 2015).
We hypothesized that L1 is active in normal squamous epithelium and that resulting L1 insertions in the genome may contribute to esophageal squamous cell tumor development or expand in these tumors as so-called passenger mutations. In cancer cells, we expected that L1 hypomethylation and other effects may create an increasingly hospitable environment for continued L1 expression and mobilization. To test these hypothesis, we evaluated L1expression and mobilization in individuals with esophageal squamous cell carcinoma using L1-seq, a high-throughput L1 targeted next-generation sequencing method (Ewing and Kazazian, 2010). We observed L1 activity in SCC and in normal esophagus, and demonstrated that sub-clonal insertions are present in normal squamous epithelium at a higher rate than observed in our previous studies (Doucet-O’Hare, et al., 2015; Ewing, et al., 2015).
MATERIALS AND METHODS
Tissue microdissection and processing
We obtained both tumor and matched normal samples from the Johns Hopkins Hospital Surgical Pathology frozen tissue bank in two groups. For the first group (n=5), genomic DNA was extracted directly from frozen samples; for the second group (n=4) we micro-dissected the tissue. For microdissection, each fresh frozen tissue sample was embedded in optimum cutting temperature (O.C.T.) compound. Following embedding, tissues were sliced in a cryostat, mounted onto glass slides, and stained with hematoxylin. Following staining, a gastrointestinal pathologist, Dr. Robert A. Anders, reviewed each piece of normal and cancerous tissue to assess whether or not these were contaminated with other cells. No malignant cells were seen in any normal sample used for gDNA extraction. Micro-dissected tumor samples were also checked for necrotic tissue which was removed in two cases.
The samples were prepared for DNA extraction by slicing with a cryostat and cutting sections of tissue between 10 μm and 30 μm (Doucet and Kazazian, 2016). Between different samples, the blade of the cryostat was cleaned with 100% ethanol, and normal and tumor samples were cut on different days. Different blades were used for the normal and the tumor samples to ensure no cross contamination during this portion of the procedure. Furthermore, several eppendorf tubes of tissue for each sample were stored at -80°C so that DNA could be reisolated separately for repeat validation experiments; all reported results were consistent in these replicates.
Immunohistochemistry (IHC)
We performed our immunohistochemistry (IHC) experiments using the EnVision System-HRP (catalog K4006; Dako) according the manufacturer’s protocol. We performed the primary antibody incubation with a mouse monoclonal ORF1p (1.25 mg/mL) at a 1: 3,000 dilution for 40 minutes at room temperature. We performed secondary antibody incubation as per the manufacturer’s protocol. The monoclonal antibody used detects amino acids 35-44 of the ORF1 protein, and is the same antibody we used previously (Doucet-O’Hare, et al., 2015). A second antibody (JH74) that detects amino acids 137-337 was used at a 1 :2,000 dilution with an overnight incubation at 4 °C to validate the results (Doucet-O’Hare, et al., 2015). Results from IHC are presented in Figure 1.
Figure 1. ORF1p expression in normal esophagus and squamous cell carcinoma.
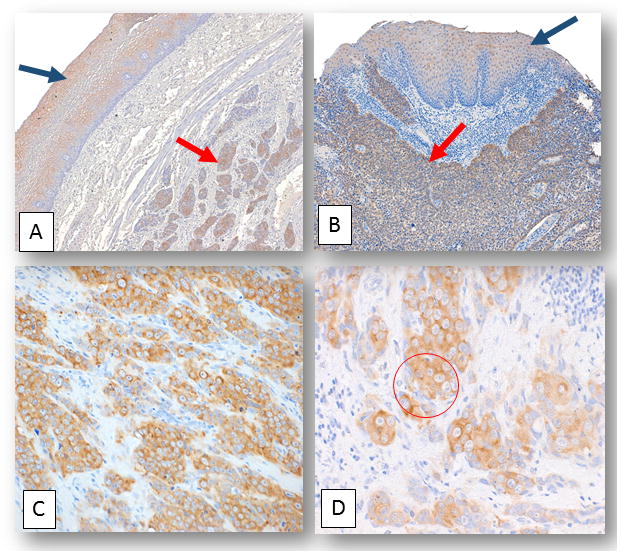
A) and B) Representative photomicrographs depicting LINE-1 ORF1p expression in normal esophageal tissue (black arrows) and in esophageal squamous cell carcinoma cases (red arrows): A) and B) Normal esophageal tissue adjacent to squamous cell carcinoma from two distinct individuals stained with LINE1 ORF1p (final magnification x100). C) and D) are photomicrographs showing a squamous cell carcinoma case where peri-nuclear staining patterns manifest for ORF1p. C) Final magnification x100. D) The same case as C) with a zoomed-in image of ORF1p peri-nuclear staining accentuation (within red circle). Final magnification x160.
L1-seq
Using DNeasy (Qiagen), we isolated DNA from thinly shaved sections of fresh-frozen tissue embedded in OTC freezing media Four pairs of patient samples were micro-dissected (including samples: 20, 21, 22, 23, and 24 N and T). The microdissection removed all normal tissue from the tumor, and all tumor tissue from the normal, and additionally removed areas where necrosis was evident. Equal amounts of genomic DNA from each individual were pooled by group (either normal or tumor) in the same manner previously described (Doucet-O’Hare, et al., 2015). L1-seq uses a hemi-specific PCR and eight degenerate primers to selectively amplify human-specific active L1 elements in the human genome (Ewing and Kazazian, 2010).
Following the PCR phase of L1-seq, we excised products between 200 and 500 nucleotides from a 1% agarose gel and purified these with the Qiagen gel purification kit. We analyzed libraries on the Bioanalyzer 2100 and combined the products in equimolar ratios from the eight different degenerate primer reactions. We sent the libraries for next-generation sequencing on the Illumina HiSeq 2500, aligned resulting reads with Bowtie2, and sorted the reads based on the presence or absence of L1 sequence. We also segregated and excluded all previously published polymorphic L1 insertions and reference insertions from our list of putative somatic insertions using the L1-seq pipeline established by Adam Ewing (Ewing and Kazazian, 2010). Overall, our bioinformatics analysis was identical to previous studies (Doucet-O’Hare, et al., 2015; Ewing and Kazazian, 2010).
Stringency filter
Initially, we randomly selected insertions for PCR validation and Sanger sequencing. Thereafter, we determined that the yield of positive insertions would be improved by filtering the insertions and selecting those most likely to validate. We focused on predicted somatic insertions with a map score of 0.5 or greater, several unique reads, and maximum peak widths of 90 nucleotides or more. The map score originates from the UCSC genome browser map-ability tracks (Kent, et al., 2002) and corresponds to how unique a given sequence is in the genome. A score of 1 indicates that a sequence maps unambiguously in the genome, while a score of 0.5 indicates a sequence maps to two locations in the genome. Unique reads are defined as reads which span different sequences within the genome; our sequencing data are comprised of a series of different overlapping reads. The more unique reads correspond to a predicted somatic insertion, the more likely we are able to validate that insertion. Unique reads are stacked and the distance of nucleotides covered by all the reads is referred to as the maximum width of the peak. Using the map score, the number of unique reads and confining our validations to peaks that span at least 90 nucleotides greatly improves our ability to validate and catalog the somatic insertions predicted by L1-seq.
We use the number of known reference and known non-reference insertions detected in each L1-seq library to evaluate assay performance. Because we sequenced two sets of samples separately, the number of known non-reference and known reference insertions must be considered separately for each group. Therefore, the two groups of samples were handled in parallel and separated throughout our study with regard to insertion coverage, detection, and validation (Table 1).
Table 1.
L1-seq detected reference, non-reference, and somatic insertions in SCC patients
| Disease | Group | Number of Individuals | SCC patients | Known reference | Known non-reference | Reads required | Map score |
|---|---|---|---|---|---|---|---|
| SCC/EAC | 1 | 5 | 4 | 1005 | 350 | 10 | 0.3 |
| SCC | 2 | 5 | 5 | 640 | 253 | 10 | 0.3 |
| Disease | Group | Number of Individuals | SCC patients | Patients with insertions | Tumor-only | NE sub-clonal/SCC clonal | Normal only |
| SCC/EAC | 1 | 5 | 4 | 4 | 18 | 0 | 0 |
| SCC | 2 | 5 | 5 | 4 | 44 | 12 | 0 |
In the table, the number of known reference, known non-reference, and somatic L1 insertions are listed. The number of known reference and known non-reference insertions detected with L1-seq were used to estimate the level of sensitivity across the individuals assayed. We expect ~900 known reference insertions and ~200 known non-reference insertions to be detected for full sensitivity. The table also includes the reads required to identify a known insertion as well as the map score cut-off for quality control purposes. In the second part of the table, the number of SCC patients in each cohort can be found and the number of individuals with insertions. Normal esophagus is abbreviated NE, squamous cell carcinoma is abbreviated at SCC, and esophageal adenocarcinoma is abbreviated EAC in the table. Normal only and tumor only indicate the insertions were detected in only the normal or only the tumor, respectively. Known non reference insertions refer to insertions which have previously been reported in the literature, but are not present in the hg19. Likewise, known reference insertions are those which are included in the reference genome. Finally, the NE sub-clonal insertions are the potentially sub-clonal insertions detected in the tumor tissue with a conventional PCR and the normal tissue with a nested PCR. For a description of map score, please see Materials and Methods.
Insertion Validation
Somatic insertions are validated with site specific PCR (Fig. 2 and 3). If the samples are not barcoded, all samples in a pool were evaluated for the presence or absence of a predicted insertion. A volume of 2μL of 12.5 ng/μL DNA was used per reaction. For each validation, the FS and L1SP1A2 primer reaction was performed on all samples in the pool (Doucet and Kazazian, 2016). When comparing tumor and normal DNA, validation PCR was performed on both samples. For the FS/L1 primer (filled site PCR) we use the following in the master mix (1x): 12.5 μL of Promega GoTaq green (2X) master mix, 0.8 μL of FS primer 20 μM, 1.6 μL of L1 primer 20 μM, 2 μL of genomic DNA (12.5 ng/μL), 8.2 μL of DNase free water. For the FS/ES primers (empty site PCR) we use the following in the master mix (1X): 12.5 μL of Promega GoTaq green (2X) master mix, 1 μL of FS primer 20 μM, 1 μL of ES primer 20 μM, 2 μL of genomic DNA (12.5 ng/μL), 9.5 μL of DNase free H2O. We use the following parameters for the PCR: (1) 95°C for 2 minutes, (2) 95°C for 30 seconds , (3) 57°C for 30 seconds, (4) 72°C for 1 minute and 30 seconds, (5) Go to step 2 (29x), (6) 72°C for 5 minutes, (8) 4°C hold. Next, we run the PCR products on a 1.5% TAE gel to resolve them. We excise fragments which are unique to only one of the samples and isolate the DNA to send for Sanger sequencing. If no clear filled site band is uniquely present in one of the samples tested, a nested PCR is carried out.
Figure 2. L1 structure and L1-seq validation scheme.
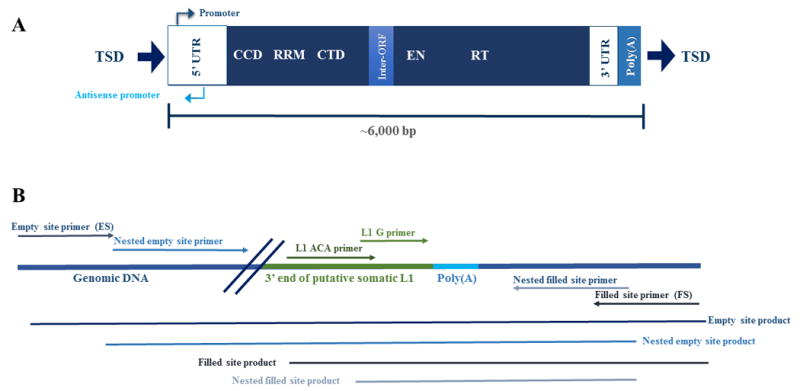
A) LINE-1 structure of a human specific active element in the human genome. Full-length L1 elements are approximately 6,000 nucleotides in length and have a 5’ untranslated region, containing both a promoter and an anti-sense promoter. L1 encodes two open reading frames for proteins on which it relies for its mobilization in the cell (Feng, et al., 1996; Mathias, et al., 1991; Moran, et al., 1996; Scott, et al., 1987). The first open reading frame encodes ORF1p, a protein with RNA chaperoning activity that includes a coiled-coil domain (CCD), an RNA recognition motif (RRM), and a carboxy-terminal domain (CTD) (Martin, 2010; Martin and Bushman, 2001). The second open reading encodes ORF2p, possesses an endonuclease domain (EN) and a reverse transcriptase domain (RT) (Fanning, 1987; Feng, et al., 1996; Mathias, et al., 1991; Moran, et al., 1996). The element also has a poly(A) tail between the 3’ untranslated region and the target site duplication (TSD). Both the poly(A) tail and TSDs are hallmarks of the target-primed reverse transcription, the process by which L1 elements mobilize (Ostertag and Kazazian, 2001). B) The PCR validation method for predicted somatic insertions utilizes two unique areas of sequence in the 3’ end of the L1 element. The sequence ‘ACA’ is incorporated into the L1 ACA primer and is positioned about 90 nucleotides from the poly(A) tail. The ‘ACA’ nucleotides (Boissinot, et al., 2000; Skowronski, et al., 1988) distinguish the human specific transcriptionally active L1 element from other L1s in the genome and allow for a specific PCR product to amplify. The L1 G primer has a ‘G’ nucleotide which is also unique to the human specific active L1 element and the nucleotide is approximately ten nucleotides preceding the poly(A) tail (Ewing and Kazazian, 2010). To validate an insertion, pictured adjacent to a poly-A tail, the L1 primers are used in conjunction with filled and empty site primers to amplify the 3’ end of the somatic insertion. The nested empty site and nested filled site primers are flanked by the empty and filled site primers and are used to verify that insertions predicted in tumor are truly absent from normal tissue. Additionally, nested primers are utilized when amplifying low copy number somatic insertions because they are able to detect when one cell out of a thousand has a copy of an insertion. The size of the expected products and relative locations of the primers are pictured in the figure.
Figure 3. Examples of sub-clonal insertions in normal esophagus.
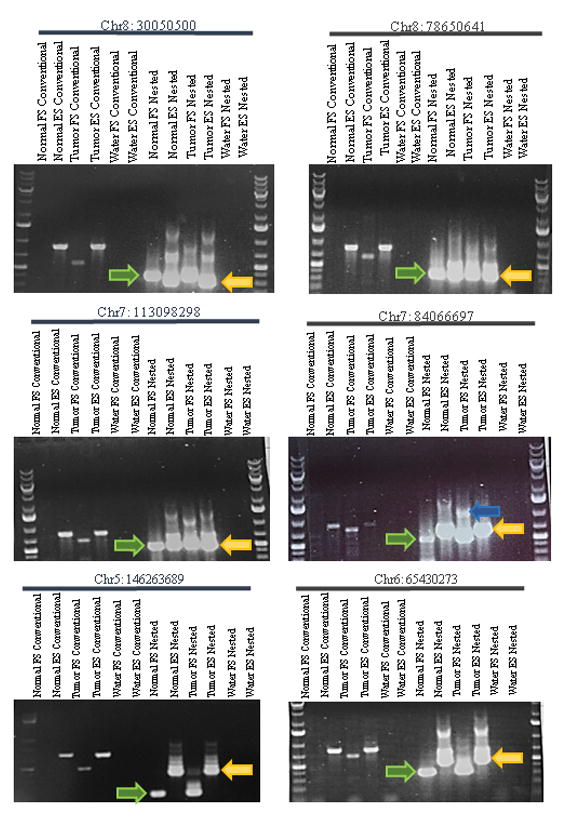
Conventional PCRs done on gDNA isolated from normal esophagus and esophageal squamous cell carcinoma alongside the corresponding nested PCRs. The nested PCRs use the product from the conventional PCRs and an internal pair of primers, positioned nearer to the predicted breakpoint of the somatic insertion. For the nested PCRs, the correct band size is denoted by a green (right-pointing) arrow for the filled site and a yellow (left-pointing) arrow for the empty site. These PCRs showcase events which presumably occur in the normal esophageal tissue and are expanded clonally in the adjacent tumors. FS and ES indicate filled-site and empty site primers, respectively. The upper empty site band of the insertion labeled Chr7: 84066697 in the tumor sample was sequenced and contained the full L1 insertion and the flanking sequence (blue arrow). The remaining upper bands in the nested empty site reactions were all sequenced and found to be non-specific amplification products.
For the nested FS PCR, use the following in the master mix (1x): 12.5 μL of Promega GoTaq green (2X) master mix, 0.8 μL of FS nested primer 20 μM, 1.6 μL of L1”G” primer (located 3’ of the L1SP1A2 primer) 20 μM, 2 μL of genomic DNA (12.5 ng/μL), 8.2 μL of DNase free water. For the FS/ES nested primers (nested empty site PCR) we use the following in the master mix (1X): 12.5 μL of Promega GoTaq green (2X) master mix, 1 μL of FS nested primer 20 μM, 1 μL of ES nested primer 20 μM, 2 μL of previous PCR product, 8.5 μL of DNase free H2O. We use the following parameters for the PCR: (1) 95°C for 2 minutes, (2) 95°C for 30 seconds , (3) 57°C for 30 seconds, (4) 72°C for 1 minute and 30 seconds, (5) Go to step 2 (29x), (6) 72°C for 5 minutes, (8) 4°C hold. Products are run on a 1.5% TAE gel to resolve them. We excise fragments which are unique to only one of the samples and isolate the DNA to send for Sanger sequencing. Additional details can be found in the molecular methods chapter on L1seq (Doucet and Kazazian, 2016). For the insertions which did not validate, one of three scenarios occurred: (i.) we did not detect a filled site band in any sample tested; (ii.) we did not obtain useable sequence for the filled site band excised for a given prediction, or (iii.) the filled site band occurred in both normal and cancer tissues only with a nested PCR. The insertions for which we did not find a filled site band are likely false positives in L1-seq. None of the insertions appeared to be private germ-line insertions following PCR validation. Our overall rate of insertion validation is similar to our previously reported experience using this technique (Doucet-O’Hare, et al., 2015; Solyom, et al., 2012).
RESULTS
L1 Protein Expression in Normal Esophagus (NE) and Squamous Cell Carcinoma of the esophagus (SCC)
To evaluate L1 activity in SCC and in the normal esophagus of SCC patients, we assayed L1 protein expression in both normal and tumor samples (Fig. 1). LINE-1 protein expression (e.g., ORF1p) is necessary for L1 mobilization (Moran, et al., 1996), but it is not sufficient to ensure somatic insertions will occur, even in cancer (Doucet-O’Hare, et al., 2015). Even without evidence of somatically acquired L1 insertions, we previously observed ORF1p expression in all esophageal adenocarcinoma samples evaluated (Doucet-O’Hare, et al., 2015). Furthermore, we observed weaker ORF1p expression in the normal squamous epithelial tissues of many patients coinciding with insertions that were present in normal tissue originally and expanded in the adjacent malignant lesion (Doucet-O’Hare, et al., 2015).
Since ORF1p is expressed in many epithelial cancers (Rodic, et al., 2014), we hypothesized that ORF1p would be expressed in SCC. Additionally, we expected to observe weaker ORF1p expression in the normal squamous epithelium (Doucet-O’Hare, et al., 2015). We obtained formalin-fixed paraffin-embedded tissue samples for four of the nine patients, and using a monoclonal ORF1p antibody (Rodic, et al., 2014) we confirmed ORF1p expression in all samples evaluated. Three of the four cases evaluated with immunohistochemistry (IHC) showed robust ORF1p staining in the malignant tissue while, in one case the staining was much weaker (Fig. 1). The weakest level of ORF1p expression in a tumor sample was comparable to the level of expression in the normal esophagus of the other samples. Intriguingly, the SCC case in which ORF1p expression was low was the only case in which no somatic insertions were confirmed (Supp. Fig. S1). We also observed low-level ORF1p expression in normal squamous epithelium of all four patients evaluated (Fig. 1).
ORF1p expression in epithelial cancers typically appears in a diffuse to speckled distribution in the cytoplasm of the cancer cells. Although ORF1p was present in all four samples evaluated, the distribution pattern differed among samples and across regions of the same sample. In some samples, we observed ORF1p expression predominantly in a diffuse pattern in the cytoplasm of the cells, while in three samples, we observed an accentuated perinuclear pattern oftentimes with aggregates of the protein localizing near the nuclear periphery (Fig. 1). We confirmed the perinuclear staining pattern with a second primary antibody targeting a different portion of ORF1p (Doucet-O’Hare, et al., 2015). The significance of these different cellular distributions is unknown.
Somatic L1 Insertions in Esophageal Squamous Cell Carcinoma
After observing ORF1p expression in all SCC cases evaluated, we characterized the potential mutations caused by L1 activity in esophageal SCC by studying matched fresh-frozen normal and cancer tissues. Because SCC is a relatively rare cancer in the United States, we only acquired nine paired samples for our study (Zhang, et al., 2012). We received the samples in two groups, the first group consisted of four individuals with SCC and one individual with esophageal adenocarcinoma (EAC), and the second group consisted of five individuals with SCC (Table 1). The latter samples were micro-dissected to eliminate cross-contamination between tumor and normal tissue. Each of the two groups was prepared into L1-seq libraries separately and then next-generation sequenced and analyzed. L1-seq is a high throughput technique which enriches for the human-specific sub-family of L1 elements using specific PCR primers (Ewing and Kazazian, 2010). After sequencing, data were subjected to a computational pipeline, designed by Ewing and colleagues, that analyzes sequencing data and identifies potential somatic insertions present in tissue (Ewing and Kazazian, 2010). By comparing somatic insertions detected by L1-seq in ESCC to the database of non-reference L1 insertions maintained by Ewing, we identified putative unique insertions in the tumor samples and selected them for PCR and sequencing validation. Non-reference insertions are insertions previously detected in the human population but absent from the UCSC human genome browser.
As in our previous studies, we defined somatic insertions as those which were not inherited from a previous generation and are therefore present in only a subset of cells within a tissue (Doucet-O’Hare, et al., 2015). We hypothesized that some somatic insertions may exist in a sub-clonal population of normal esophageal cells. Sub-clonal insertions could be amplified when cells are selectively amplified during tumor initiation and progression; therefore we evaluated every putative insertion with conventional PCR and nested PCR in both normal and tumor DNA (Fig. 2) (Doucet-O’Hare, et al., 2015).
In order to select putative insertions for validation with PCR and Sanger sequencing, we filtered our results by selecting insertions with 3 unique reads or more, greater than a 90 base-pair nucleotide window, and a map score, a value which represents the accuracy of the alignment, of 0.5 or greater (Ewing and Kazazian, 2010). After filtering, in the first group of samples, we observed 100 potential tumor-only insertions and tested 36 of them. We selected a subset of filtered insertions for validation and using PCR and Sanger sequencing we were able to validate 18 insertions distributed among the four individuals with SCC.
After filtering results for the second group of samples, we found 133 potential insertions unique to the tumor, 82 of which were tested. Again we selected a subset of filtered insertions and validated 56 insertions distributed among four of the five individuals with 12 of the insertions appearing to be sub-clonal in the adjacent normal esophagus and clonal in the tumor (Fig. 3). Interestingly, all four patients with confirmed somatic insertions in this second group of micro-dissected samples harbored sub-clonal insertions in the normal tissue.
We previously detected somatic insertions in normal tissue with nested PCR for approximately 5% of somatic insertions (Doucet-O’Hare, et al., 2015; Ewing, et al., 2015). In esophageal squamous cell carcinoma samples, we observed that ~16% (12/74 insertions) were present in the normal esophagus with nested PCR. The sensitivity of the conventional PCR method for somatic insertion confirmation is approximately one in ten, i.e., if one in ten cells has the insertion present it is detected at a low level by conventional PCR (Ewing, et al., 2015). Using nested PCR, we consistently detect an insertion present in one out of a thousand cells (Ewing, et al., 2015). Our results are in line with the known sensitivity of L1-seq as demonstrated in previous work (Ewing, et al., 2015). Thus, we found sub-clonal insertions in matched normal epithelial tissue at a greater frequency in SCC samples than in the EAC, gastric, pancreatic, and colon cancers previously studied (Doucet-O’Hare, et al., 2015; Solyom, et al., 2012). Somatic insertions could be selectively amplified and, if they disrupt a tumor suppressor or induce an oncogene, may actively participate in carcinogenesis. If, retrotransposition is an ongoing process in the normal esophagus of some or all individuals, it would be an active source of mutations which could begin or contribute to tumorigenesis.
We speculate that the sub-clonal insertions in the normal esophagus could have occurred in a single cell which eventually gave rise to a lineage that underwent malignant transformation. Unfortunately we lack a third tissue type from the patient to test the possibility that the insertion originated in early embryonic development (Kano, et al., 2009); therefore, we cannot conclude that the sub-clonal insertions occurred in the normal adult esophagus. In the process of tumorigenesis, cells with insertions may have been selectively amplified, making the insertions in the tumor cells easily detectable by conventional PCR (Fig. 4) (Doucet-O’Hare, et al., 2015; Goodier, 2014). Furthermore, the incidence of sub-clonal insertions in the normal tissue does not appear to be a rare phenomenon in SCC as it is present in four out of the eight patients with confirmed somatic insertions (Table 2). In addition to the 12 sub-clonal insertions in the normal tissue, we confirmed that 33 of the previously mentioned tumor specific insertions were likely sub-clonal somatic insertions because they were only detectable with nested PCR. These insertions presumably occurred after initiation of tumor development.
Figure 4. Acquisition, Detection, and Validation of sub-clonal and clonal somatic insertions in L1-seq.
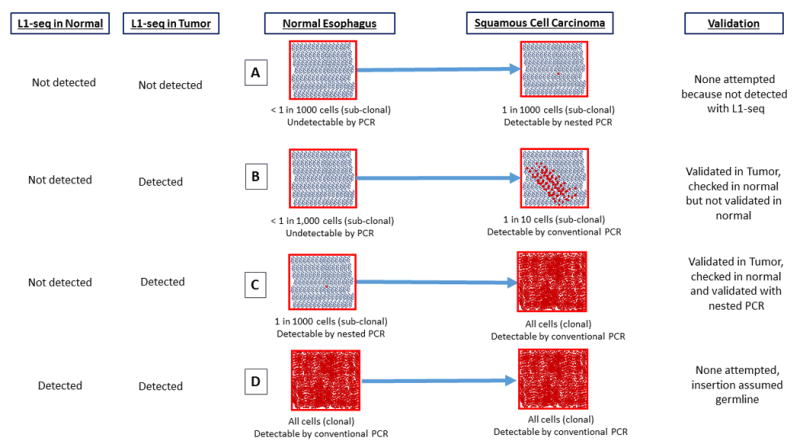
This diagram details the sensitivity of L1-seq with regard to detecting somatic insertions at differing levels of clonality in a tissue and different scenarios by which a somatic insertion could become amplified in a tumor. (A) An insertion present at a very low frequency (less than one in a thousand cells) in the normal esophagus and at an undetectable level in the tumor when evaluated by L1-seq. (B) An insertion at an undetectable level in the normal esophagus and a sub-clonal but detectable level in the tumor. (C) An insertion at undetectable level for L1-seq in the normal esophagus which is clonal in the tumor. (D) An insertion which is clonal in both the normal esophagus and the tumor. This insertion is not tested by PCR because it is presumed to be germline.
Table 2.
Validated somatic insertions in SCC
| Esophageal Squamous Cell Cancer Validated Insertions | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Insertion number |
Predicted breakpoint |
Group | Sample | Patient Age | 5’ end validated |
Inversion | TSD sequence | Insertion size (w/o poly A) |
Exact Genomic Breakpoint |
Subclonal in NE |
Likely clonal in SCC |
Predicted Endo site |
poly(A) length |
Gene(s) | Stand of insertion (relative to hg19) |
| 1 | chr1:74225746 | 2 | 20T | 79 | Yes | No | 300 bp (TATCTCCAGCTTACC | 120 bp | chr1:74225531 | Yes | AGAT/AA | 80 bp | “-” | ||
| 2 | chr1:91054960 | 2 | 20T | 79 | 50 bp | “+” | |||||||||
| 3 | chr1:110159225 | 2 | 20T | 79 | Yes | 100 bp | “+” | ||||||||
| 4 | chr1:188390840 | 2 | 21T | 71 | 60 bp | “+” | |||||||||
| 5 | chr2:7391894 | 2 | 23T | 76 | Yes | Yes | 40 bp | “-” | |||||||
| 6 | chr3:106586376 | 2 | 20T | 79 | 100 bp | LINC00882 | “-” | ||||||||
| 7 | chr3:158778615 | 2 | 21T | 71 | Yes | Yes | 15 bp (CAGAATATGCATTTT | 1859 bp | chr3:158778722 | Yes | TTCT/GA | 150 bp | “-” | ||
| 8 | chr3:173244207 | 2 | 21T | 71 | Yes | 75 bp | NLGN1 | “+” | |||||||
| 9 | chr4:21015215 | 2 | 21T | 71 | yes | Yes | 11 bp (TAAAGCTTCTTT) | 1229 bp | chr4:21015270 | Yes | CTTT/AT | 75 bp | KCNIP4 | “-” | |
| 10 | chr4:21295696 | 2 | 20T | 79 | Yes | No | 16 bp (AAAAACTGTTAATA | 520 bp | chr4:21295716 | TATG/AA | 40 bp | KCNIP4 | “+” | ||
| 11 | chr4:33062493 | 2 | 20T | 79 | 100 bp | “-” | |||||||||
| 12 | chr4:34773929 | 2 | 23T | 76 | Yes | 45 bp | “-” | ||||||||
| 13 | chr4:100470748 | 2 | 20T | 79 | 50 bp | TRMT1 | “-” | ||||||||
| 14 | chr4:122022647 | 2 | 20T | 79 | Yes | No | 5 bp (GTTTT) | 1110 bp | chr4:122022858 | AAAA/CA | 50 bp | “-” | |||
| 15 | chr5:18343101 | 2 | 20T | 79 | Yes | 100 bp | “-” | ||||||||
| 16 | chr5:22972054 | 2 | 21T | 71 | Yes | 50 bp | “+” | ||||||||
| 17 | chr5:27118145 | 2 | 23T | 76 | Yes | No | 376 bp (GTATATCTCCAAATT | 121 bp | chr5:27117892 | Yes | TATA/CA | 150 bp | “-” | ||
| 18 | chr5:63570580 | 2 | 21T | 71 | Yes | No | 389 bp deletion (TGACTCCTA | 630 bp | chr5:63571024 | Yes | Endo indep | 50 bp | RNF180 | “-” | |
| 19 | chr5:146263689 | 2 | 21T | 71 | Yes | No | 12 bp (AAACATTTACCA) | 1039 bp | chr5:146263701 | Yes | Yes | ATTT/AC | 50 bp | PPP2R2B | “+” |
| 20 | chr6:54007315 | 2 | 20T | 79 | Yes | Yes | 12 bp (TCTTTGATTTTT) | 258 bp | chr6:54007426 | AAAG/AC | 80 bp | MLIP | “-” | ||
| 21 | chr6:65430273 | 2 | 20T | 79 | Yes | Yes | 100 bp | EYS | “-” | ||||||
| 22 | chr6:93175977 | 2 | 20T | 79 | Yes | “+” | |||||||||
| 23 | chr6:149512094 | 2 | 21T | 71 | “+” | ||||||||||
| 24 | chr6:167049997 | 2 | 21T | 71 | 130 bp | RPS6KA2 | “-” | ||||||||
| 25 | chr7:78377347 | 2 | 20T | 79 | Yes | No | 20 bp (AAAAAAAAAAAAA | 773 bp | chr7:78377476 | Yes | TTTT/TT | 50 bp | MAGI2 | “-” | |
| 26 | chr7:84066697 | 2 | 22T | 59 | Yes | No | 1 bp deletion | 354 bp | chr7:84066755 | Yes | Yes | Endo indep | 55 bp | “-” | |
| 27 | chr7:113098298 | 2 | 22T | 59 | Yes | No | None | 1189 bp | chr7:113098416 | Yes | Yes | GTTA/TA | 100 bp | “-” | |
| 28 | chr7:144876298 | 2 | 21T | 71 | Yes | No | 11 bp (ACTGCTCTCTT) | 361 bp | chr7: 144876634 | GCAG/TG | 50 bp | “-” | |||
| 29 | chr8:62731519 | 2 | 20T | 79 | Yes | No | 5 bp (TTTTA) | 568 bp | chr8:62731576 | TAAA/AA | 50 bp | “-” | |||
| 30 | chr8:63452731 | 2 | 23T | 76 | Yes | Yes | 5 bp | NKAIN3 | “-” | ||||||
| 31 | chr8:78650641 | 2 | 23T | 76 | Yes | Yes | 70 bp | “+” | |||||||
| 32 | chr8:96814193 | 2 | 20T | 79 | Yes | No | 2 bp (TC) | 737 bp | chr8:96814250 | Yes | Yes | CTTG/AA | 50 bp | C8orf37-AS1, LOC100616530 | “-” |
| 33 | chr8:105742587 | 2 | 21T | 71 | 50 bp | “+” | |||||||||
| 34 | chr8:130050500 | 2 | 23T | 76 | Yes | Yes | 50 bp | “+” | |||||||
| 35 | chr8:137876889 | 2 | 21T | 71 | 12 | “-” | |||||||||
| 36 | chr8:137876889 | 2 | 21T | 71 | 25 bp | “+” | |||||||||
| 37 | chr9:27521686 | 2 | 21T | 71 | MOB3B | “+” | |||||||||
| 38 | chr9:30914267 | 2 | 21T | 71 | Yes | Yes | 14 bp (AAGAAGGCATAGAA | 1305 bp | chr9:82494253 | Yes | GATC/AA | 20 bp | “+” | ||
| 39 | chr9:82494252 | 2 | 21T | 71 | Yes | No | 12 bp (AAAAAAAATCAA) | 282 bp | chr9:30914249 | ATCA/AG | 100 bp | LINC01507 | “+” | ||
| 40 | chr10:11187849 | 2 | 22T | 59 | Yes | 100 bp | CELF2 | “+” | |||||||
| 41 | chr10:55846139 | 2 | 23T | 76 | Yes | No | 3 bp deletion | 175 bp | chr10:55846282 | Yes | Yes | Endo indep | 40 bp | PCDH15 | “-” |
| 42 | chr10:115267083 | 2 | 23T | 76 | 55 bp | “+” | |||||||||
| 43 | chr11:23140915 | 2 | 20T | 79 | Yes | No | 14 bp (AAAAAACATAAACA | 1043 bp | chr11:23140929 | ACAT/AA | 35 bp | “+” | |||
| 44 | chr11:43634948 | 2 | 23T | 76 | “+” | ||||||||||
| 45 | chr11:78577470 | 2 | 20T | 79 | Yes | Yes | 109 bp deletion | 346 bp | chr11:78577397 | Yes | Endo indep | 60 bp | TENM4 | “+” | |
| 46 | chr11:95617289 | 2 | 23T | 76 | Yes | Yes | 70 bp | MTMR2 | “-” | ||||||
| 47 | chr12:72582208 | 2 | 21T | 71 | Yes | Yes | 130 bp | “+” | |||||||
| 48 | chr13:90364217 | 2 | 20T | 79 | “-” | ||||||||||
| 49 | chr14:21603456 | 2 | 20T | 79 | Yes | 50 bp | “+” | ||||||||
| 50 | chr14:95721343 | 2 | 20T | 79 | 50 bp | CLMN | “+” | ||||||||
| 51 | chr15:55401145 | 2 | 20T | 79 | 50 bp | “-” | |||||||||
| 52 | chr16:32621143 | 2 | 23T | 76 | “-” | ||||||||||
| 53 | chr18:7914950 | 2 | 21T | 71 | 100 bp | PTPRM | “+” | ||||||||
| 54 | chr19:31676750 | 2 | 23T | 76 | 80 bp | “-” | |||||||||
| 55 | chr20:12193544 | 2 | 21T | 71 | 40 bp | “+” | |||||||||
| 56 | chr20:31767412 | 2 | 20T | 79 | 20 bp | BPIFA2 | “+” | ||||||||
| 57 | chr1:169592527 | 1 | 3T | 51 | Yes | No | 3 bp (TAT) | 290 bp | chr1:169592905 | Yes | SELP | “-” | |||
| 58 | chr2:62784947 | 1 | 3T | 51 | Yes | 50 bp | “-” | ||||||||
| 59 | chr2:118147901 | 1 | 1T | 76 | Yes | “+” | |||||||||
| 60 | chr2:192054832 | 1 | 1T | 76 | Yes | GATT/AA | “-” | ||||||||
| 61 | chr3:108723582 | 1 | 8T | 78 | Yes | 20 bp | MORC1 | “+” | |||||||
| 62 | chr4:178589680 | 1 | 2T | 67 | Yes | 74 bp | AK094945 | “-” | |||||||
| 63 | chr5:8875319 | 1 | 3T | 51 | Yes | BC032891 | “+” | ||||||||
| 64 | chr5:6447510 | 1 | 8T | 78 | Yes | “-” | |||||||||
| 65 | chr6:5599305 | 1 | 3T | 51 | CAGT/AT | 83 bp | FARS2 | “-” | |||||||
| 66 | chr7:69594334 | 1 | 8T | 78 | Yes | No | 2 bp (TA) | 365 bp | chr11:122340070 | Yes | AUTS2 | “+” | |||
| 67 | chr7:18316210 | 1 | 8T | 78 | 10 bp | HDAC9 | “-” | ||||||||
| 68 | chr7:71682380 | 1 | 8T | 78 | Yes | No | 6 bp (AGAAAA) | 528 bp | chr7:71682386 | 30 bp | CALN1 | “+” | |||
| 69 | chr9:8381705 | 1 | 1T | 76 | 20 bp | PTPRD | “+” | ||||||||
| 70 | chr9:81288257 | 1 | 8T | 78 | 40 bp | “+” | |||||||||
| 71 | chr11:122339699 | 1 | 3T | 51 | Yes | 35 bp | “-” | ||||||||
| 72 | chr14:81244336 | 1 | 3T | 51 | Yes | TTAT/AG | CEP128 | “-” | |||||||
| 73 | chr16:20172684 | 1 | 3T | 51 | Yes | 70 bp | “-” | ||||||||
| 74 | chrX:144755675 | 1 | 3T | 51 | Yes | 43 bp | “-” | ||||||||
In this table all of the validated somatic insertions’ information is listed, including: the group/ sample in which they were detected, the patient’s age, whether or not the 5’ end of the insertion was validated (including all of the related information such as the target-site duplication (TSD) and endonuclease cleavage site, etc.), if the insertion was amplified with a conventional PCR, whether or not the insertion was sub-clonal in normal esophagus, the gene into which the insertion occurred, and finally the strand, with respect to the hg19 reference assembly, into which the insertion occurred. All genomic positions listed in the table are from hg19.
Characterization of Tumor Specific Insertions
After we observed evidence of L1 activity in eight out of nine cases of SCC evaluated, we studied the characteristics of the confirmed somatic insertions more thoroughly. To identify the precise base-pair at which the insertions occurred, we performed PCRs to amplify the 5’ end of the insertions. We successfully amplified and sequenced 23 out of 74 attempted somatic L1 5’ ends (Table 2). For 51 of the somatic insertions, we were only able to validate the 3’ end of the insertion with site-specific PCR and Sanger sequencing. For a subset of validated insertions, we identified endonuclease cleavage sites and target site duplications (TSDs) which are both hallmarks of the process of retrotransposition (Table 2). Out of the 20 putative endonuclease cleavage sites identified in our study, 15/20 were similar (differed by 3 base-pairs or less) to the canonical endonuclease cleavage site in target-primed reverse transcription reactions (Jurka, 1997). Four of the aforementioned twenty validated insertions are potentially endonuclease independent (Morrish, et al., 2007), because they lack canonical TSDs and obvious endonuclease cleavage sites. All of the potentially endonuclease independent insertions had genomic deletions at the site of insertion integration (Gilbert, et al., 2002). The remaining 4 insertions had non-canonical endonuclease cleavage sites which differed by more than three nucleotides from the expected cleavage site of 5’ TTTT/AA 3’ (Jurka, 1997). For 18/72 of the confirmed insertions, we identified TSDs ranging from two base pairs in length to 376 base pairs in our samples with a median size of 12 base-pairs. The insertion sizes varied from 120 to 1,859 base pairs with 11 of the insertions under 500 base pairs (Table 2). Interestingly, five of the insertions had 5’ end inversions, a finding consistent with previous studies of germ-line insertions and L1 insertions in cancer (Doucet-O’Hare, et al., 2015; Hancks and Kazazian, 2012; Helman, et al., 2014; Lee, et al., 2012; Rodic, et al., 2015; Shukla, et al., 2013; Solyom, et al., 2012). In contrast to others’ work, we did not detect any 3’ transductions (Tubio, et al., 2014); but L1-seq rarely detects such events.
Of the insertions validated in SCC patients, 41 of the 3’ ends amplified with a single PCR in tumor, while the remaining 3’ ends of 33 insertions amplified in tumor only following a nested PCR (Fig. 2). The tumor only insertions which could only be confirmed with nested PCR may have been acquired in the tumor suggesting that in the tumor retrotransposition was ongoing. Additionally, in 12 out of 41 instances where the 3’ end validated with a conventional PCR in tumor, the insertion also validated with nested PCR in the normal epithelium. The large proportion of sub-clonal insertions validated in the normal tissue indicates that L1 elements are active either in embryonic tissues giving rise to the normal esophagus or in the normal esophagus itself of some individuals (Kano, et al., 2009). The intrinsic activity of LINE-1 in the normal tissue of individuals may contribute to cancer development through the generation of additional mutations which could undergo positive selection during carcinogenesis (Fig. 4).
None of the somatic insertions evaluated occurred in exons; however, 32 insertions did occur into the introns of 32 different genes. The genes C8orf37-AS1 and LOC100616530 share an intron into which one of the insertions occurred. Another gene, KCNIP4, had two different validated insertions in an intron approximately 280 kb apart. Although KCNIP4 is approximately 1.2 Mb in size, the insertions occurred in two different individuals, meaning that this gene was recurrently subjected to L1 insertions (Supp. Fig. S2). The insertions in KCNIP4, a potassium voltage-gated channel interacting protein, are in opposite orientations relative to the gene (Supp. Fig. S2). We previously observed a statistically significant correlation between age and insertion occurrence in colon cancer (Solyom, et al., 2012). In contrast, the correlation between age and insertion occurrence in SCC was not statistically significant (P = 0.1025) (Supp. Fig. S1).
Of the 32 genes with validated somatic insertions, 22 occurred into genes that have previously been associated with cancer (Supp. Table S1) (see Supp. References). Our findings reveal a statistically significant enrichment of validated somatic insertions into genes previously associated with cancer (P< 1 × 10-10) (40-82). We considered the probability that a somatic insertion would hit more of the cancer-associated genes than would be expected due to chance alone. After accounting for gene size, we observed a significant enrichment of somatic insertions into cancer associated genes using a chi squared test (P< 1 × 10-10) (Supp. References). Four of the 22 genes are considered “known cancer genes” by the network of cancer genes (An, et al., 2016), and the remaining 18 genes are candidate cancer genes which have been associated with cancer in the literature (Supp. Table S1). Interestingly, out of the 22 genes associated with cancer (Supp. References), variation in 16 of the genes have also been associated with smoking (Supp. References) (Supp. Table S1). Smoking is one of the main risk factors for SCC; therefore, it is notable that 16 of the cancer-associated genes into which L1 elements inserted are also associated with smoking (Supp. References) (Supp. Table S1).
DISCUSSION
Expanding our understanding of mutational mechanisms in cancer may lead to earlier diagnosis and better treatment options. The prevalence of squamous cell carcinoma throughout the world and its deadly nature necessitate its study. In order to detect, diagnose, and treat SCC effectively, we need a thorough evaluation of mutations acquired in the normal squamous tissue of the esophagus which transitions to cancer. Cellular processes often become dysregulated during carcinogenesis and may provide a favorable environment for the activity of retrotransposons.
In parallel with evaluating cancer samples for newly acquired genomic insertions, we looked for L1 protein expression differences between the normal squamous epithelium and the tumor. We observed higher ORF1p expression in SCC tumor samples possessing a larger number of confirmed somatic insertions. Because we were only able to acquire tissue for four of the nine SCC patients studied, our results are not statistically significant. Previously, we hypothesized that higher ORF1p expression would correlate with a larger number of somatic insertions in patients; nevertheless, this did not hold true for EAC and Barrett’s esophagus patients. Interestingly, in the present study, multiple patterns of ORF1p staining were apparent in three individuals, including a diffuse cytoplasmic pattern and a perinuclear pattern with accentuation and protein aggregates near the nuclear periphery in some foci within the tumor (Fig. 1). While we do not understand the significance of these ORF1 protein expression patterns in SCC, we can conclude that the protein is present in all samples evaluated, and that there are differences in level and pattern of expression between patient samples.
Our group and others have firmly established that L1 somatic insertions occur frequently in epithelial cancers and presented evidence that insertions may sometimes contribute to cancer development (Doucet-O’Hare, et al., 2015; Ewing, et al., 2015; Helman, et al., 2014; Iskow, et al., 2010; Lee, et al., 2012; Miki, et al., 1992; Rodic, et al., 2015; Shukla, et al., 2013; Solyom, et al., 2012; Tubio, et al., 2014). In this study, we demonstrate retrotransposition is an active process in patients with SCC. We observed 62 somatic insertions absent from normal tissue and present in tumor samples. Moreover, we found 12 insertions that amplified easily with a conventional PCR in tumor and were also present in the matched normal tissue using nested PCR (Fig. 2, 3). In the patients with sub-clonal insertions in normal tissue, 37 of the insertions validated in tumor appeared to be clonal. The low frequency of sub-clonal insertions in the normal tissue (<1/3) coupled with the microdissection of the tissues strongly supports that these insertions were present in the normal tissue and were propagated in the resulting lesion. However, it is possible that the insertions present in normal esophagus occurred much earlier than the adult esophagus, namely in early embryonic development (Kano, et al., 2009).
Faulkner and colleagues first detected somatic insertions into normal hippocampus and we also detected a somatic insertion in normal colon, two somatic insertions in normal stomach, and two somatic insertions in normal esophagus (Baillie, et al., 2011; Doucet-O’Hare, et al., 2015; Ewing, et al., 2015). Aside from the aforementioned examples, observations of somatic insertions in normal tissues are relatively uncommon (Evrony, et al., 2012). Surprisingly, we detected twelve instances of somatic insertions in normal tissue with nested PCR suggesting the insertions are in only a few cells. With conventional PCR, the same insertions were observed only in tumor DNA, demonstrating that a larger number of cells contain the insertions in the tumor. Because the frequency of sub-clonal insertions in normal esophagus of SCC patients is much higher than we have previously observed in cancer patients, it appears that either the esophagus itself or embryonic cells giving rise to the esophagus are permissive for L1 somatic insertions (Doucet-O’Hare, et al., 2015; Ewing, et al., 2015; Kano, et al., 2009).
Our data are consistent with two different models explaining the role of retrotransposition in cancer. First, it is possible, as previously suggested (Doucet-O’Hare, et al., 2015; Ewing, et al., 2015; Goodier, 2014), that some somatic L1 insertions are acquired in the normal tissue and subsequently expand in the cancer (Fig. 4). In this case, we speculate that the clonal nature of a tumor facilitates the expansion of somatic insertions that occur over time, making the insertions easier to detect. A second scenario consistent with our data is that many insertions are acquired during or after carcinogenesis due to L1 hypomethylation and consequentially increased L1 ORF1p expression; the increased expression of ORF1p suggests retrotransposition may be increased in the tumor as compared to the normal tissue (Fig. 4). Understanding the role of somatic insertions in cancer relies heavily on having an accurate estimation of retrotransposon activity in normal cells. In light of these data, we must establish the underlying rate of retrotransposition in any normal tissue subsequently evaluated in a disease state in order to interpret the data obtained. In future studies, single-cell sequencing should reveal the occurrence of somatic L1 insertions in normal cells and give insight with regard to its frequency.
Supplementary Material
Acknowledgments
We thank Dr. Norman J Barker for assistance with photomicrographs and Dr. Jeffrey Han for the use of the JH74 rabbit monoclonal ORF1p antibody. The work was funded by the following: National Institute of General Medical Science (NIGMS) Grant R01 GM 099875 (to H.H.K.), the Sol Goldman award (to H.H.K.), the Burroughs Wellcome Fund Career Award for Biomedical Scientists (to K.H.B.), National Institutes of Health (NIH) Grant R01CA163705 (to K.H.B.), and NIGMS P50 grant GM107632 (to K.H.B. and H.H.K.). We also thank the Johns Hopkins Conte Digestive Diseases Basic and Translational Research Core Center for technical assistance.
References
- Abedi-Ardekani B, Hainaut P. Cancers of the upper gastro-intestinal tract: a review of somatic mutation distributions. Arch Iran Med. 2014;17(4):286–92. [PubMed] [Google Scholar]
- An O, Dall’Olio GM, Mourikis TP, Ciccarelli FD. NCG 5.0: updates of a manually curated repository of cancer genes and associated properties from cancer mutational screenings. Nucleic Acids Res. 2016;44(D1):D992–9. doi: 10.1093/nar/gkv1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjan-Odedra SSC, Sherer NM, Wolinsky SM, Malim MH. Endogenous MOV10 inhibits the retrotransposition of endogenous retroelements but not the replication of exogenous retroviruses. Retrovirology. 2012;9(53) doi: 10.1186/1742-4690-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba Y, Murata A, Watanabe M, Baba H. Clinical implications of the LINE-1 methylation levels in patients with gastrointestinal cancer. Surg Today. 2014;44(10):1807–16. doi: 10.1007/s00595-013-0763-6. [DOI] [PubMed] [Google Scholar]
- Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479(7374):534–7. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O’Shea KS, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103(23):8780–5. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, Chevret P, Furano AV. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol. 2000;17(6):915–28. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Current Biology. 2006;16(5):480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, Attreed M, Avner P, Wutz A, Barillot E, et al. LINE-1 Activity in Facultative Heterochromatin Formation during X Chromosome Inactivation. Cell. 2010;141(6):956–969. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- Doucet-O’Hare TT, Rodic N, Sharma R, Darbari I, Abril G, Choi JA, Young Ahn J, Cheng Y, Anders RA, Burns KH, et al. LINE-1 expression and retrotransposition in Barrett’s esophagus and esophageal carcinoma. Proc Natl Acad Sci U S A. 2015;112(35):E4894–900. doi: 10.1073/pnas.1502474112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet TT, Kazazian HH., Jr Long Interspersed Element Sequencing (L1-Seq): A Method to Identify Somatic LINE-1 Insertions in the Human Genome. Methods Mol Biol. 2016;1400:79–93. doi: 10.1007/978-1-4939-3372-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecco G, Rowe HM, Trono D. A Large-Scale Functional Screen to Identify Epigenetic Repressors of Retrotransposon Expression. Methods Mol Biol. 2016;1400:403–17. doi: 10.1007/978-1-4939-3372-3_25. [DOI] [PubMed] [Google Scholar]
- Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349(23):2241–52. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nature Genetics. 2000;24(4):363–7. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, Parker JJ, Atabay KD, Gilmore EC, Poduri A. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151(3):483–96. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing AD, Gacita A, Wood LD, Ma F, Xing D, Kim MS, Manda SS, Abril G, Pereira G, Makohon-Moore A, et al. Widespread somatic L1 retrotransposition occurs early during gastrointestinal cancer evolution. Genome Res. 2015;25(10):1536–45. doi: 10.1101/gr.196238.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing AD, Kazazian HH., Jr High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 2010;20(9):1262–70. doi: 10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning TaS M. The LINE-1 DNA sequences in four mammalian orders predict proteins that conserve homologies to retrovirus proteins. Nucleic Acids Research. 1987;15(5) doi: 10.1093/nar/15.5.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87(5):905–16. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110(3):315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Goodier JL. Retrotransposition in tumors and brains. Mobile DNA. 2014;5 doi: 10.1186/1759-8753-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Cheung LE, Kazazian HH., Jr MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet. 2012;8(10):e1002941. doi: 10.1371/journal.pgen.1002941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH. Active human retrotransposons: variation and disease. Current Opinion in Genetics & Development. 2012;22(3):191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helman E, Lawrence MS, Stewart C, Sougnez C, Getz G, Meyerson M. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 2014;24(7):1053–63. doi: 10.1101/gr.163659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heras SR, Macias S, Plass M, Fernandez N, Cano D, Eyras E, Garcia-Perez JL, Caceres JF. The Microprocessor controls the activity of mammalian retrotransposons. Nat Struct Mol Biol. 2013;20(10):1173–81. doi: 10.1038/nsmb.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbeault M, Trono D. As time goes by: KRABs evolve to KAP endogenous retroelements. Dev Cell. 2014;31(3):257–8. doi: 10.1016/j.devcel.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irahara N, Nosho K, Baba Y, Shima K, Lindeman NI, Hazra A, Schernhammer ES, Hunter DJ, Fuchs CS, Ogino S. Precision of pyrosequencing assay to measure LINE-1 methylation in colon cancer, normal colonic mucosa, and peripheral blood cells. J Mol Diagn. 2010;12(2):177–83. doi: 10.2353/jmoldx.2010.090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islami F, Kamangar F, Nasrollahzadeh D, Moller H, Boffetta P, Malekzadeh R. Oesophageal cancer in Golestan Province, a high-incidence area in northern Iran - a review. Eur J Cancer. 2009;45(18):3156–65. doi: 10.1016/j.ejca.2009.09.018. [DOI] [PubMed] [Google Scholar]
- Iwagami SBY, Watanabe M, Shigaki H, Miyake K, Ishimoto T, Iwatsuki M, Sakamaki K, Ohashi Y, Baba H. LINE-1 hypomethylation is associated with a poor prognosis among patients with curatively resected esophageal squamous cell carcinoma. Annals of Surgery. 2013;257(3):449–455. doi: 10.1097/SLA.0b013e31826d8602. [DOI] [PubMed] [Google Scholar]
- Jacobs FM, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014;516(7530):242–5. doi: 10.1038/nature13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci U S A. 1997;94(5):1872–7. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamangar F, Malekzadeh R, Dawsey SM, Saidi F. Esophageal cancer in Northeastern Iran: a review. Arch Iran Med. 2007;10(1):70–82. [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert R, Hainaut P. The multidisciplinary management of gastrointestinal cancer. Epidemiology of oesophagogastric cancer. Best Pract Res Clin Gastroenterol. 2007;21(6):921–45. doi: 10.1016/j.bpg.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, 3rd, Lohr JG, Harris CC, Ding L, Wilson RK, et al. Landscape of somatic retrotransposition in human cancers. Science. 2012;337(6097):967–71. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. 2011;12(9):615–27. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL. The ORF1 protein encoded by LINE-1: structure and function during L1 retrotransposition. J Biomed Biotechnol. 2006;2006(1):45621. doi: 10.1155/JBB/2006/45621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL. Nucleic acid chaperone properties of ORF1p from the non-LTR retrotransposon, LINE-1. RNA Biol. 2010;7(6):706–11. doi: 10.4161/rna.7.6.13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol Cell Biol. 2001;21(2):467–75. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254(5039):1808–10. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, Vogelstein B, Nakamura Y. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52(3):643–5. [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87(5):917–27. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Morrish TA, Garcia-Perez JL, Stamato TD, Taccioli GE, Sekiguchi J, Moran JV. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446(7132):208–212. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nature Genetics. 2002;31(2):159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468(7322):443–6. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi S, Miyamoto S, Kikuchi O, Goto T, Amanuma Y, Muto M. Recent Advances From Basic and Clinical Studies of Esophageal Squamous Cell Carcinoma. Gastroenterology. 2015;149(7):1700–15. doi: 10.1053/j.gastro.2015.08.054. [DOI] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–38. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- Rodic N, Sharma R, Sharma R, Zampella J, Dai L, Taylor MS, Hruban RH, Iacobuzio-Donahue CA, Maitra A, Torbenson MS, et al. Long interspersed element-1 protein expression is a hallmark of many human cancers. Am J Pathol. 2014;184(5):1280–6. doi: 10.1016/j.ajpath.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodic N, Steranka JP, Makohon-Moore A, Moyer A, Shen P, Sharma R, Kohutek ZA, Huang CR, Ahn D, Mita P, et al. Retrotransposon insertions in the clonal evolution of pancreatic ductal adenocarcinoma. Nature Medicine. 2015;21(9):1060. doi: 10.1038/nm.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Trono D. Dynamic control of endogenous retroviruses during development. Virology. 2011;411(2):273–87. doi: 10.1016/j.virol.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Scott AF, Schmeckpeper BJ, Abdelrazik M, Comey CT, O’Hara B, Rossiter JP, Cooley T, Heath P, Smith KD, Margolet L. Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics. 1987;1(2):113–25. doi: 10.1016/0888-7543(87)90003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigaki H, Baba Y, Watanabe M, Murata A, Iwagami S, Miyake K, Ishimoto T, Iwatsuki M, Baba H. LINE-1 hypomethylation in gastric cancer, detected by bisulfite pyrosequencing, is associated with poor prognosis. Gastric Cancer. 2013;16(4):480–7. doi: 10.1007/s10120-012-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla R, Upton KR, Munoz-Lopez M, Gerhardt DJ, Fisher ME, Nguyen T, Brennan PM, Baillie JK, Collino A, Ghisletti S, et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell. 2013;153(1):101–11. doi: 10.1016/j.cell.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronski J, Fanning TG, Singer MF. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol Cell Biol. 1988;8(4):1385–97. doi: 10.1128/mcb.8.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solyom S, Ewing AD, Rahrmann EP, Doucet T, Nelson HH, Burns MB, Harris RS, Sigmon DF, Casella A, Erlanger B, et al. Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 2012;22(12):2328–38. doi: 10.1101/gr.145235.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J Biol Chem. 2006;281(25):16837–41. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- Tran GD, Sun XD, Abnet CC, Fan JH, Dawsey SM, Dong ZW, Mark SD, Qiao YL, Taylor PR. Prospective study of risk factors for esophageal and gastric cancers in the Linxian general population trial cohort in China. Int J Cancer. 2005;113(3):456–63. doi: 10.1002/ijc.20616. [DOI] [PubMed] [Google Scholar]
- Tubio JM, Li Y, Ju YS, Martincorena I, Cooke SL, Tojo M, Gundem G, Pipinikas CP, Zamora J, Raine K, et al. Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science. 2014;345(6196):1251343. doi: 10.1126/science.1251343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli P, Vianin S, Trono D. The innate antiretroviral factor APOBEC3G does not affect human LINE-1 retrotransposition in a cell culture assay. J Biol Chem. 2004;279(42):43371–3. doi: 10.1074/jbc.C400334200. [DOI] [PubMed] [Google Scholar]
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21(4):1429–39. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13(8):335–40. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- Zhang HZ, Jin GF, Shen HB. Epidemiologic differences in esophageal cancer between Asian and Western populations. Chinese Journal of Cancer. 2012;31(6):281–286. doi: 10.5732/cjc.011.10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


