Abstract
The problem of the propagation of conformational changes over long distances or through a closely packed protein is shown to fit a model of a ligand-induced conformational change between two protein states selected by evolution. Moreover, the kinetics of the pathway between these states is also selected so that the energy of ligand binding and the speed of the transition between conformational states are physiologically appropriate. The crystallographic data of a wild-type aspartate receptor that has negative cooperativity and a mutant that has no cooperativity but has native transmembrane signaling are shown to support this model.
The aspartate receptor of chemotaxis receives a stimulus on the outside of a cell that produces a response on the inside of the cell that, as for most receptors, is an enormous amplification over the signal it receives. This has been shown to be achieved by a receptor that transmits a 1-Å conformational change >100 Å across the membrane from the ligand-binding site to a cytoplasmic activation site (1). Because allosteric proteins in general propagate conformational charges over considerable distances, and substrates at active sites generate analogous changes (2–5), how these conformational changes are generated and transmitted is of major interest for understanding the regulatory, kinetic, and recognition properties of proteins.
Because the aspartate receptor is an example of a change that travels a long distance and because its crystallography has already revealed important features of its structure, we have examined the wild type and the mutant S68A as clues to the elucidation of the transmission problem.
The aspartate receptor of Escherichia coli is a dimer that shows negative cooperativity (6), a phenomenon that offers the advantage that one can crystallize the receptor when only one aspartate is bound (7, 8). To ensure that we could dissect the conformational effects causing the propagation of the excitation from the conformational effects causing the negative cooperativity, we compared the crystal structures of the wild type to a mutant, S68A, that retained the transmembrane propagations but had no cooperativity.
Experimental
E. coli XL1 Blue pMK 155 (residues 35–180) cells were used for overproduction of the ligand-binding domain of the S68A (replacement of serine-68 by alanine by using directed mutagenesis) aspartate receptor. The receptor protein was purified as described (8). Crystals with and without bound aspartate have I41 symmetry and contain one protein molecule in the asymmetric unit. The unit-cell dimensions of the apo crystals are a = b = 65.0 Å and c = 68.7 Å, and those for the crystals with ligand aspartates are a = b = 63.8 Å and c = 70.3 Å. The apo S68A crystals were grown by mixing 2 μl S68A protein (15–20 mg/ml in a buffer containing 10 mM Tris⋅HCl, pH 8.0) with 2 μl of a reservoir solution containing 12% (vol/vol) polyethylene glycol (PEG) 6000 and 1.5 M NaCl. Similarly, crystals containing ligand aspartates were grown by mixing 2 μl of protein (15–20 mg/ml in a buffer containing 10 mM Tris⋅HCl, pH 8.0, and 25 mM aspartic acid) with 2 μl of a reservoir solution containing 0.5 M sodium acetate, 0.2 M NaMes, pH 6.5, and 0.05 M CdSO4. Diffraction data were collected at the Stanford Synchrotron Radiation Laboratory (Stanford, CA) beamline 7–1 at 100 K after freezing the crystals in cryoprotectants consisting of 20% PEG 400, 12% PEG 600, and 1.5 M NaCl for the apo and 20% PEG 400, 0.5 M sodium acetate, 0.2 M NaMes (pH 6.5) and 0.05 M CdSO4 for the complex crystals. X-ray data from both crystals were refined to a resolution of 1.9 Å. The data were processed and integrated by denzo, and scaled by using scalepack. The crystal structures were determined by molecular replacement, and refined by using x-plor with free R factors of 0.27 (apo) and 0.29 (complex), respectively.
Results
The crystallographic data for the S68A protein with and without bound aspartate are summarized in Table 1.
Table 1.
Crystallographic data
| Diffraction data | S68A apo | S68A complex |
|---|---|---|
| Resolution (Å) | 1.9 | 1.9 |
| Unique reflections | 12,078 | 16,213 |
| Completeness, % | 97 | 99 |
| Rmerge, % | 9.7 | 4.4 |
| Rfree, % | 27 | 29 |
The binding of aspartate, the initiating impulse for the conformation change, is seen by the crystallography to induce formation of a number of new hydrogen bonds and breaking of a number of bonds that existed in the apoprotein as described in Table 2. The bonds formed when aspartate is bound (designated as AspS to distinguish substrate from amino acid residues of the wild-type aspartate receptor) are Arg-64⋅⋅⋅AspS, Tyr-149⋅⋅⋅AspS, Gln-152⋅⋅⋅AspS, Thr-154⋅⋅⋅AspS, Arg-69′⋅⋅⋅AspS, and Arg-73′⋅⋅⋅AspS (Table 2). One drastic change is the swing of Ser-68′ in the second (unoccupied) site, resulting in a formation of a new hydrogen bond with Tyr-149′. The distance between Arg-64 and Gln-155 becomes less and that between Arg-64 and Gln-158 becomes longer.
Table 2.
Bond changes when aspartate binds to the wild-type aspartate receptor
| Bond Forming/strengthening | Bond breaking/weakening |
|---|---|
| Arg-64⋅⋅⋅Gln-155 | Arg-64⋅⋅⋅Gln-158 |
| Arg-64⋅⋅⋅AspS | |
| Tyr-149⋅⋅⋅AspS | |
| Gln-152⋅⋅⋅AspS | |
| Thr-154⋅⋅⋅AspS | |
| Arg-69′⋅⋅⋅AspS | |
| Arg-73′⋅⋅⋅AspS |
The structural change within a subunit of the S68A receptor protein depicted by the relative motion between helices α1 and α4 is shown in Fig. 1b, and it seems to be similar to the motion for the wild type in Fig. 1a. The conformational change triggered by the presence of aspartate involves a downward shift of helix α4 with respect to α1 by ≈1.3 Å. For the wild-type periplasmic domain, helix α4 shifts downward with a magnitude of about 1.5 Å. Coupled to this downward shift, helix α4 also tilts by 5° in the wild-type protein. The tilt at helix α4, however, is more subtle in the case of the S68A mutant, where it is about 2° in magnitude (Table 3).
Figure 1.
(a) The backbone atoms in the α4 helix, shown at the left, compared with the backbone atoms of the α1 helix, shown at the right, for the wild-type protein. The red line is the apoprotein and the yellow line is the protein with aspartate bound. (b) The same comparison as a, but for the S68A receptor.
Table 3.
Differences from wild type when aspartate binds to the S68A receptor
| Structural feature | Wild type | S68A |
|---|---|---|
| Downward shift at helix α4, Å | 1.5 | 1.3 |
| Tilt at helix α4 relative to helix α1, degrees | 5 | 2 |
| Inter-subunit angle, degrees | 4 | 0 |
| New hydrogen bond formation | Ser-68′ to Tyr-149′ in the unoccupied site | Both sites have aspartate |
Conformational changes through helix α4 are accompanied by a piston-like motion in the helix interface. The main chain of helix α4 remains comparatively rigid and the side chains undergo small angular adjustments. This motion is also associated with a rearrangement of H-bond strengths between the side chains of helices α1 and α4. For example, the H-bond distance between Arg-64 and Gln-155 is 3.51 Å, and that between Arg-64 and Gln-158 is 2.85 Å in the apo crystal structure. Upon aspartate binding, these distances become 2.75 Å and 3.10 Å, respectively.
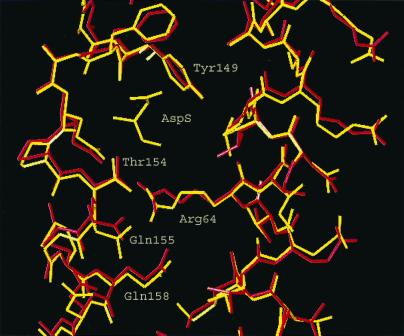
In Fig. 2 are shown the side chains of the residues near Thr-154 at the position of the ligand binding to the receptor, showing a similar 1-Å shift downwards of the side chains. Further down in the protein it can be seen that the side chains relative to each other and to the backbones are far enough apart so that there are no van der Waal's repulsions or steric hindrances to a 1 Å shift in the protein.
Figure 2.
Side chains in the helices α1 and α4 that shift relative to each other on binding aspartate.
The mutant that lacks cooperativity and the wild type that has strong negative cooperativity have similar movements of α4 helix downward in both cases. The main difference is that the Ala-68 that replaces Ser-68 has no role in the interactions with Tyr-149. Because Ser-68 has been shown to play a major role in the cooperative interactions in the structure (9), it seems clear that this Ser-68 is a big actor in the cooperativity of the protein. It is intriguing that the negatively cooperative protein has, if anything, a greater movement downwards of helix α4 than the noncooperative protein, indicating that the cooperativity does not “bleed off” energy from the excitation mechanism but, if anything, evolution has selected it for both the negative cooperativity and the propagation of the signal.
Discussion
To interpret these observations in the necessary thermodynamic and
kinetic background, the alternative pathways for an induced
conformational change are shown in Fig. 3
(a glossary for the various terms and their symbols and relationships
is included in the Fig. 3 legend). The figure shows three possible
pathways for the conformational change: (i) an isomerization
of the protein to its final conformation, followed by binding of ligand
( →
→
 →
→
 );
(ii) a binding of ligand to the unchanged protein, followed
by an isomerization of protein bonds to the final conformation
(
);
(ii) a binding of ligand to the unchanged protein, followed
by an isomerization of protein bonds to the final conformation
( →
→
 →
→
 ); or
(iii) an initial isomerization to a conformation
intermediate between the initial and final conformation, followed by
binding of ligand and further isomerization to the final conformation
(
); or
(iii) an initial isomerization to a conformation
intermediate between the initial and final conformation, followed by
binding of ligand and further isomerization to the final conformation
( →
→
 →
→
 →
→
 ). The overall
thermodynamics of binding and isomerization must be the same in all
three pathways
(KtAB⋅KSB
=
KtAC⋅KSC⋅KtCB
=
KSA⋅K′tAB)
and must equal the observed binding constant to the receptor
Kobs, which in this case is
105 M. Which pathway is actually chosen by the
protein is, of course, a matter of kinetics. Thermodynamics and
crystallography can focus on logical suggestions, but kinetics will
decide the actual path.
). The overall
thermodynamics of binding and isomerization must be the same in all
three pathways
(KtAB⋅KSB
=
KtAC⋅KSC⋅KtCB
=
KSA⋅K′tAB)
and must equal the observed binding constant to the receptor
Kobs, which in this case is
105 M. Which pathway is actually chosen by the
protein is, of course, a matter of kinetics. Thermodynamics and
crystallography can focus on logical suggestions, but kinetics will
decide the actual path.
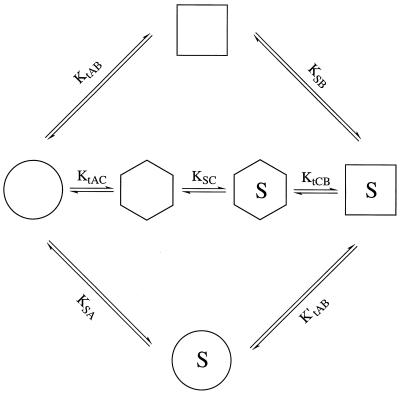
Figure 3.
The alternative pathways for the ligand-induced change. The extremes
represent pathways in which (i) the apoprotein
isomerizes to conformation B (=
 ), which then
binds substrate
), which then
binds substrate
 (top);
(ii) the ligand binds to the apoprotein conformation that
then isomerizes to form the final bound-ligand conformation
(
(top);
(ii) the ligand binds to the apoprotein conformation that
then isomerizes to form the final bound-ligand conformation
( →
→
 →
→
 ) (bottom);
or (iii) the protein isomerizes to a conformation somewhere
in between
) (bottom);
or (iii) the protein isomerizes to a conformation somewhere
in between
 and
and
 and then
binds to the ligand and reaches the final state
(
and then
binds to the ligand and reaches the final state
( →
→
 →
→
 →
→
 )
(middle). Glossary: A =
)
(middle). Glossary: A =
 , the
conformation of the protein in the absence of ligand. B
=
, the
conformation of the protein in the absence of ligand. B
=
 , the
conformation of the protein in the presence of ligand. C =
, the
conformation of the protein in the presence of ligand. C =
 , an
intermediate conformation containing elements of A and B. AS =
, an
intermediate conformation containing elements of A and B. AS =
 , BS =
, BS =
 , CS =
, CS =
 , the ligand
bound to the protein in the A, B, and C conformations, respectively.
, the ligand
bound to the protein in the A, B, and C conformations, respectively.
Kobs = the observed affinity constant of the receptor = KtAB⋅KSB.
KtAB = [B]/[A].
KtAC = [C]/[A].
KtCB = [B]/[C].
In the present case it seems likely that Pro153, which breaks the helix
so that residues 151 and 152 are no longer helical, makes the active
site more accessible to aspartate, the binding of which initiates the
shift in bonds described above, suggesting that the kinetic pathway is
 →
→
 →
→
 . In the case
of hemoglobin, the movement of histidine F8 away from contact with the
Fe of the heme is needed to allow oxygen to bind to the Fe of the heme,
which then initiates the cascade described by Perutz (4) for hemoglobin
(suggesting the pathway
. In the case
of hemoglobin, the movement of histidine F8 away from contact with the
Fe of the heme is needed to allow oxygen to bind to the Fe of the heme,
which then initiates the cascade described by Perutz (4) for hemoglobin
(suggesting the pathway
 →
→
 →
→
 ).
).
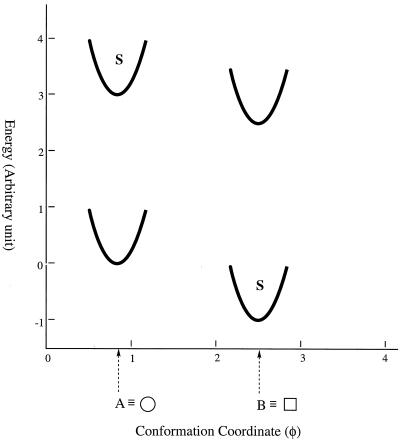
The thermodynamic observations of an allosteric protein can be illustrated in another way as shown in Fig. 4, where an arbitrary parameter on the abscissa, φ, stands for the conformation of the protein (a function describing all the angles and distances of the atoms in the protein), and a parameter E on the ordinate stands for the energy of that conformation (and any binding affinity calculation). Here A, designated as in the glossary, is the conformation of the apoprotein in its native state, and B is the conformation of the final liganded state.
Figure 4.
The energetics of the conformation changes when a ligand binds to a protein. Conformational states are shown as potential wells depicting the changing energetics of small displacement from the most stable conformation of that well. The conformation A is the most stable conformation of the apoprotein, but B exists at a higher energy state, its amount depending on the kinetics and KtAB. The presence of a ligand will stabilize conformation B but will destabilize the A conformation because S has very little attraction to the apo conformation.
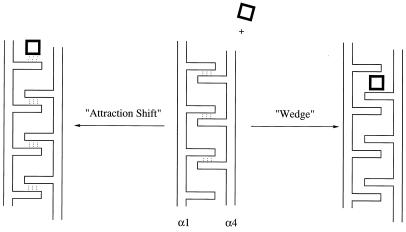
In hypothesizing the kinetic mechanisms that applied to the aspartate receptor, one that we considered initially (10) we will call a “wedge mechanism” (shown in Fig. 5) in which the ligand inserts itself between relatively rigid side chains extending from a relatively rigid helix, causing a downward shift (because of steric repulsion between the helix and the ligand). But the current crystallographic data suggest that the “attraction shift” mechanism of Fig. 5 is more accurate. In this mechanism, the new ligand binds to the receptor, causing a shift in the previous bonds of the apoprotein to a new alignment of hydrogen bonds as summarized in Table 2, causing a downward shift of one helix relative to the other.
Figure 5.
Possible mechanisms for ligand-induced changes in the aspartate receptor. On the right is a wedge mechanism in which a ligand is attracted into a position between rigid side chains and deflects one helix downwards relative to the other. On the left is an attraction shift mechanism in which the binding of a ligand attracts hydrogen bonds, leading to a shifting of hydrogen bonds and causing the downward shift of helix α4 relative to helix α1.
As one proceeds along the protein chain, it is noted that there is a tilt between helices α1 and α4 that becomes more pronounced (5° shift) when aspartate is bound (Fig. 4). That shift separates the chains enough so that there will be very little resistance to the downward shift of helix α4 relative to helix α1 or the other subunit. These conclusions agree with the crystallographic facts and, like the thermodynamics, are completely accurate for the final states. However, the kinetic mechanism by which the final states are achieved is only a hypothesis.
In connecting the lessons learned from the aspartate receptor and their generalization to propagation of conformational changes in general, several principles stand out.
Principle 1.
The ligand-induced shift occurs between two conformations that are thermodynamically close enough so that the binding of one ligand tilts the equilibrium from conformation A to conformation B. The ligand binding constant, 105 M, is an overall observed constant made up of KtAB⋅KSB; therefore KSB must be big enough to overwhelm an unfavorable KtAB so that the observed constant, in this case 105 M, is in a physiologically appropriate range. The overall mechanism seems to be composed theoretically of two factors, an energetically unfavorable conformational transition (e.g., KtAB ≈ 10−2) followed by a very favorable binding to the final conformation (e.g., KSB ≈ 107); thus the binding constant observed is less than that for a template type protein because the binding must induce the new conformation that produces the transmembrane signal.
Principle 2.
Because in the case of a receptor the conformational change must travel
long distances, and in the case of a tightly packed globular protein
conformational changes produce friction, both conformations and
kinetics must be “evolutionized,” i.e., optimized by mutations
over evolutionary time. Thus evolution selects for (i) an
initial conformation that attracts a ligand or can rapidly convert to a
conformation,  ,
that attracts a ligand, and (ii) a kinetic process that goes
rapidly enough to be physiologically relevant.
,
that attracts a ligand, and (ii) a kinetic process that goes
rapidly enough to be physiologically relevant.
Principle 3.
Some conformation B =
 in the
initial apoprotein is, of course, a theoretical possibility, but it
would in most cases be present only in a small concentration
(KtAB ≤ 10−2 or
ΔG ≈ 2,700 cal; 1 cal = 4.18 J). The more
in the
initial apoprotein is, of course, a theoretical possibility, but it
would in most cases be present only in a small concentration
(KtAB ≤ 10−2 or
ΔG ≈ 2,700 cal; 1 cal = 4.18 J). The more
 = B
conformation that is present initially, the smaller will be the
physiological effects of induced changes (i.e., the lower the
activation, inhibition, or cooperativity induced by the ligand). It is
erroneously assumed by some that if there is a small amount of the
final conformation present initially, that inevitably means that the
= B
conformation that is present initially, the smaller will be the
physiological effects of induced changes (i.e., the lower the
activation, inhibition, or cooperativity induced by the ligand). It is
erroneously assumed by some that if there is a small amount of the
final conformation present initially, that inevitably means that the
 →
→
 →
→
 pathway is
selected. Every protein must conceptually have some concentration of a
different conformation, the amount being determined by the equilibrium
constant, KtAB,
KtAC, etc. (every compound or element
has a vapor pressure indicating the existence of some gas phase, but
that is not an indication that chemical reactions, e.g., Cu with HCl,
go through the gas phase). If the rearrangement of
pathway is
selected. Every protein must conceptually have some concentration of a
different conformation, the amount being determined by the equilibrium
constant, KtAB,
KtAC, etc. (every compound or element
has a vapor pressure indicating the existence of some gas phase, but
that is not an indication that chemical reactions, e.g., Cu with HCl,
go through the gas phase). If the rearrangement of
 ⇌
⇌
 is slow or
the association
is slow or
the association
 + S ⇌
+ S ⇌
 is slow, then
is slow, then
 →
→
 →
→
 may not be
the favored pathway. In each case, the induced fit theory postulates
that the ligand induces a new shape of the protein but does not specify
the pathway.
may not be
the favored pathway. In each case, the induced fit theory postulates
that the ligand induces a new shape of the protein but does not specify
the pathway.
Principle 4.
The forgoing discussion of conformational changes covers the general situation of the protein but does not discuss the subunit interactions that would need to be considered for quantitative analysis of a multisubunit protein; the manner in which such analysis could be done has already been published (11).
Principle 5.
The observation that the enormous amplification of the response of the aspartate receptor is generated by a 1-Å conformation change may be surprising to some, but on careful reflection it is an excellent compromise between the multiple facets of protein structure and the needs of biological responses. Fortunately, enzymes are known to be highly specific and therefore for them to detect a 1-Å shift in a conformation is not too demanding a chore. If that is so, the smaller the conformational change the better, because it requires the least thermodynamic energy and is more likely to be easy kinetically. In the case of the piston model of the aspartate receptor, that nomenclature was chosen to distinguish it from models such as scissors, see-saw rotation, etc. because an α-helix has many of the characteristics of a piston, i.e., it is not compressible and not stretchable. Such a model would fit into the “shear” category of Gerstein, Lesk, and Chothia's description of conformation changes in induced-fit proteins (2). Gerstein, Lesk, and Chothia's categorization of observed conformational changes in proteins is highly relevant because their excellent analysis demonstrated that many conformational changes occurred as fixed domain movements. The movement of a fixed domain in a tightly packed protein will be subject to the same thermodynamic requirements as discussed here and therefore would be expected: (i) to also be “evolutionized” so that the kinetic and thermodynamic requirements could be achieved, and (ii) to generally be in small movements so as to adjust the thermodynamic requirements of the change to the finite limits of the binding energy of the ligand to the protein. Thus, propagation of conformational changes over long distances in proteins (or short distances in tightly packed proteins) is likely to require a model (such as that described above) in which the ligand changes the equilibrium between two thermodynamic minima by a mechanism that has used evolution to optimize thermodynamic and time constraints. The original conformational change generated by the binding of ligand (either wedge or attraction shift or some other) probably was not the one we see today but was improved by evolution to optimize propagation and to bring the thermodynamics into the range of binding energy of small molecules and the kinetics into biologically relevant times.
Acknowledgments
We are grateful for the valuable advice on crystallography from Prof. Tom Alber and the members of his laboratory, and for the critical reading of the manuscript by Profs. Robert Glaeser, Jack Kirsch, and Susan Marqusee. We are also appreciative of the financial support from National Institutes of Health Grant DK09765 and a National Institutes of Health Postdoctoral Fellowship to E.W.Y.
Abbreviations
- PEG
polyethylene glycol
- AspS
substrate aspartate
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1JMW).
References
- 1.Otteman K M, Xiao W, Shin Y-K, Koshland D E., Jr Science. 1999;285:1751–1753. doi: 10.1126/science.285.5434.1751. [DOI] [PubMed] [Google Scholar]
- 2.Gerstein M, Lesk A M, Chothia G. Biochemistry. 1994;33:6739–6749. doi: 10.1021/bi00188a001. [DOI] [PubMed] [Google Scholar]
- 3.Johnson L N, Noble M E M, Owen D J. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 4.Perutz M F. Nature (London) 1970;228:726–739. doi: 10.1038/228726a0. [DOI] [PubMed] [Google Scholar]
- 5.Nissen P, Hansen J, Ban N, Moore P B, Steitz T A. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 6.Biemann H-P, Koshland D E., Jr Biochemistry. 1994;33:629–634. doi: 10.1021/bi00169a002. [DOI] [PubMed] [Google Scholar]
- 7.Scott W G, Milligan D L, Milburn M V, Privé G G, Yeh J, Koshland D E, Jr, Kim S H. J Mol Biol. 1993;232:555–573. doi: 10.1006/jmbi.1993.1411. [DOI] [PubMed] [Google Scholar]
- 8.Yeh J I, Biemann H P, Prive G G, Pandit J, Koshland D E, Jr, Kim S H. J Mol Biol. 1996;262:186–201. doi: 10.1006/jmbi.1996.0507. [DOI] [PubMed] [Google Scholar]
- 9.Kolodziej A F, Tan T, Koshland D E., Jr Biochemistry. 1996;35:14782–14792. doi: 10.1021/bi961481v. [DOI] [PubMed] [Google Scholar]
- 10.Lynch B A, Koshland D E., Jr FEBS Lett. 1992;307:3–9. doi: 10.1016/0014-5793(92)80891-j. [DOI] [PubMed] [Google Scholar]
- 11.Koshland D E, Jr, Nemethy G, Filmer D. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]