Abstract
Background
This study retrospectively evaluated the capability of computed-tomography (CT) based radiomic features to predict EGFR mutation status in surgically-resected peripheral lung adenocarcinomas in an Asian cohort of patients.
Materials and Methods
298 patients with surgically resected peripheral lung adenocarcinomas were investigated in this institutional review board-approved retrospective study with waived consent. 219 quantitative 3D features were extracted from segmented volumes of each tumor, and 59 of these which were considered as independent features were included in the analysis. Clinical and pathological information were obtained from the institutional database.
Results
Mutant EGFR was significantly associated with female gender (p=0.0005); never smoker status (p<0.0001), lepidic predominant adenocarcinomas (p=0.017), and low or intermediate pathologic grade (p=0.0002). Statistically significant differences were found in 11 radiomic features between EGFR mutant and wild type groups on univariate analysis. Mutant EGFR status could be predicted by a set of five radiomic features that fall in three broad groups: CT attenuation energy, tumor main direction and texture defined by wavelets and Laws (AUC 0.647). Multiple logistic regression model showed that adding radiomic features to a clinical model resulted in a significant improvement of predicting power, as the AUC increased from 0.667 to 0.709 (p<0.0001).
Conclusions
CT based radiomic features of peripheral lung adenocarcinomas can capture useful information regarding tumor phenotype, and the model we built can be useful to predict the presence of EGFR mutations in peripheral lung adenocarcinoma in Asian patients when mutational profiling is not available or possible.
Keywords: epidermal growth factor receptor, peripheral lung adenocarcinoma, radiomic features, Tomography, X-ray computed, Logistic Models
Introduction
Lung cancer accounts for 13% of new cancers diagnosed world-wide and is the leading cause of cancer related deaths 1. 85% of all lung cancers are non-small cell lung carcinoma (NSCLC), with adenocarcinoma being the most common histologic subtype 2. Over the last decade, molecular translational research advances have heralded major breakthroughs in the understanding, diagnosis and management of lung cancer, particularly the development of new target-based therapies directed against key signaling pathways involved in lung cancer growth and malignant progression 3, 4. Small-molecule tyrosine kinase inhibitors (TKIs) that target the epidermal growth factor receptor (EGFR) were the first targeted drugs to enter clinical use for the treatment of NSCLC. Patients with EGFR mutations have a higher response rate to EGFR-TKIs (60%-80%) than those with EGFR–wild type or unknown mutation status (10%-20%) 5. Randomized trials have clearly shown that treatments with targeted TKIs, such as erlotinib, gefitinib, or afatinib are associated with longer progression-free survival (PFS) and higher objective radiographic response rates than standard first-line chemotherapy in patients with mutated EGFR lung cancer 6-9. However, if gefitinib is administered in the case of non-EGFR mutated lung cancer, the patient will experience a shorter PFS compared to platinum-based chemotherapy 3, highlighting the importance of identifying this genetically unique subset of patients.
Cumulative epidemiology studies have identified several clinicopathological factors such as female, non-smokers, adenocarcinoma histology, and East Asian origin that associated with high prevalence of EGFR mutation 10, 11. Unfortunately, there are no reliable clinical characteristics that allow for accurate prediction of EGFR mutation status. For some patients, biopsy samples may be the only tumor materials available for testing EGFR mutation status and they are often composed of variable ratios of tumor to normal cells,12. Thus, mutant DNA alleles present at extremely low concentrations become difficult to detect, leading to a false negative result. Further, due to intratumoral heterogeneity, the portion of the tumor tested for EGFR mutation may also result as negative but may be truly positive 13-15. In a recent study 16, two sequencing-based mutation detection approaches (dideoxy- and pyrosequencing) were validated against parallel sequencing in a clinical setting, and the results showed that dideoxy sequencing missed four responders and pyrosequencing missed two responders; meanwhile, precise quantification of mutant alleles revealed a low correlation of histopathological estimates of tumor content and frequency of mutant alleles, indicating that sequencing technologies with inferior sensitivity may fail to detect clinically relevant oncogene mutations in cancer patients. Therefore, receiving a negative mutation analysis result, one must consider whether the cell sample was truly representative for the EGFR mutation status of the lung tumor.
There have been several reports regarding the relationship between Computed- Tomography (CT) features and EGFR mutation status in NSCLC 17-20; however, the findings were not consistent with each other. According to a study conducted by Zhou et al. 18, there were no differences in morphological CT features between EGFR mutation and wild-type tumors. In contrast, Rizzo et al. 17reported that EGFR mutation was significantly associated with air bronchogram, pleural retraction, small lesion size, and absence of fibrosis. Recent technological advances in medical imaging allow high throughput extraction of quantitative imaging features. Radiomics is the process of converting images to mineable data through computational approaches. These data can be used to develop decision support systems to accurately estimate patient risk and improve individualized treatment 21, 22. Several studies have showed that such features extracted from CT images of lung cancers can be useful to distinguish radiation-induced fibrosis from tumor recurrence 23, differentiate the presence of K-ras mutation from pan-wild type NSCLC 24, provide independent predictive indicator of response to first-line chemotherapy 25, and identify patients with locally advanced lung adenocarcinoma at risk of developing distant metastasis 26. CT imaging is routinely used in lung cancer, and we thus hypothesize that if CT-based radiomic features associated with EGFR mutation status can be determined, they could provide a useful clinical predictor in patients with unresectable lung cancer or those wherein biopsy is unable to be performed. Imaging-based risk models may also provide additional information for clinicians on whether re-biopsy is needed for those patients with negative EGFR mutation result. Therefore, in this retrospective study, we performed a radiomic analysis to identify image biomarkers of harboring EGFR mutation in peripheral lung adenocarcinomas in a Chinese cohort of patients.
Materials and Methods
This retrospective study was approved by the institutional review board. Informed consent was waived.
Study Population
A consecutive search of the surgical database at our institution between December 2012 and March 2014 identified 397 patients with primary lung adenocarcinoma who fulfilled the following inclusion criteria: (a) pathology reports with diagnosis of lung adenocarcinoma; (b) preoperative thin-section CT images at Picture Archiving and Communication System (PACS) and the location of the lesion was peripheral (tumor involving subsegmental bronchus or smaller airway); (c) available test results for EGFR mutation status; (d) available clinical data. Thereafter, 99 patients were excluded due to the following reasons: receiving preoperative treatment, such as radiotherapy or chemotherapy; the duration between CT examination and subsequence surgery exceeded one month; and cases with lung cancer that is difficult to contour the tumor margin on CT images.
Clinical and pathological data collected for analysis included gender, age at diagnosis, smoking status, pathologic TNM stage, and histologic lung adenocarcinoma subtypes. Smoking status was categorized into two groups, never smokers and smokers which included former or current smokers. Tumors were staged pathologically according to the seventh edition of the American Joint Committee on Cancer Staging Manual 27. Tumors were diagnosed as adenocarcinoma and then categorized according to the 2011 IASLC/ATS/ERS classification system 28.
CT Examination
Chest CT examinations were conducted using Somatom Sensation 64 (Siemens Medical Solutions, Forchheim, Germany), Light speed 16 (GE Medical Systems, Milwaukee, WI), or Discovery CT750 HD scanner (GE Medical Systems, Milwaukee, WI). The parameters used were as follows: 120 kVp with tube current adjusted automatically, pitch was 0.969, reconstruction thickness was 1.5 mm, and reconstruction interval was 1.5 mm for the 64-detector scanner; tube voltage was 120 kVp, tube current was 150-200 mA, pitch was 0.969; reconstruction thickness was 1.25 mm, and reconstruction interval was 1.25 mm for the 16-detector scanner and Discovery CT750 HD scanner.
Detection of EGFR mutations
For the gene mutation analysis, tumor specimens were obtained by surgical resection. We performed EGFR mutation analyses of four tyrosine kinase domain (exons 18-21), which are frequently mutated in lung cancer, as previously described 29. EGFR mutations were determined by amplification refractory mutation system (ARMS) real-time technology using Human EGFR Gene Mutations Detection Kit (Beijing ACCB Biotech Ltd).
Tumor segmentation
We used Definiens Developer XD© (Munich, Germany) as the image analysis platform to perform tumor segmentation and feature extraction. A lung tumor analysis (LuTA) tool within Definiens Cognition Network Technology was used. Lesions were volumetrically segmented using semi-automatic approach by two radiologists with more than 6 and 3 years of experience in CT imaging of thoracic malignancies (Figure 1). The semi-automatic segmentation workflow which contained the following four steps named pre-processing, semi-automated correction of the pulmonary boundary, click and grow, and manual refinement and generation of lesion statistics were described in detail in previous studies 30-32. Here, the single click ensemble segmentation (SCES) algorithm 31 which is an advanced version of the previous click & grow algorithm and reduces sensitivity towards the location of the initial seed-point was used. The SCES makes use of the original algorithm by choosing different seed points automatically within a specified area of the lesion and performing region growing with each generated seed point. Each of the two radiologists reviewed the segmented images in consensus, and any discrepancies were resolved by discussion until consensus was reached.
Fig. 1.


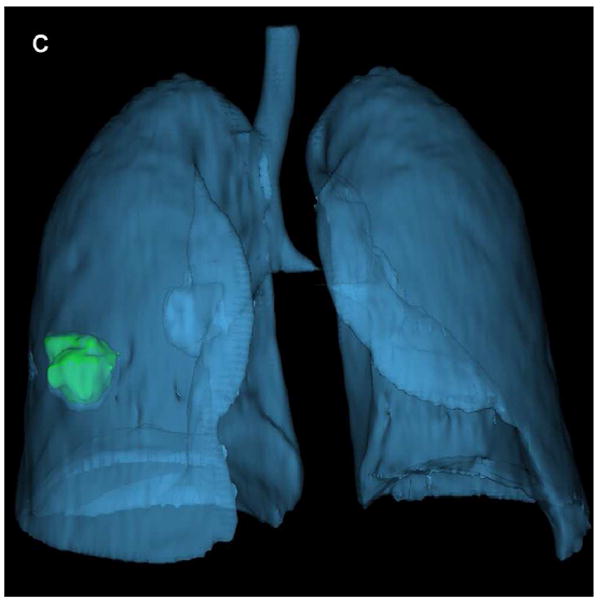
Representative CT image with tumor segmentation using semi-automated algorithm for peripheral adenocarcinoma: (a) A lobulated lung tumor in the right middle lobe was chosen for segmentation. (b) One radiologist segmented the boundary of the tumor which was shown in green outline. (c) 3D view of the lung and segmented tumor.
Radiomic feature extraction
We extracted a total of 219 features from each of the 3D objects. These features were divided into eight categories, including tumor size, shape, location, air space, pixel intensity histogram, run length & co-occurrence, laws texture, and wavelets. A description of all features is provided (Supplementary Table 1), and detailed description of texture features could be found in the previous study 33. Our features cover a variety of descriptors from size, location, attachment of the lesion of interest, to CT pixel distribution and texture as appearing on CT image. We characterize texture in observed CT images using Laws feature descriptor and in the decomposed domain by using wavelets. Texture feature are known carry information that are not always typically observed by human eye. Metrics described by Wavelet transformation, Laws features are considered to describe subtle characteristics in the image and have been shown to be useful in image classifications 34.
Statistical Analysis
All statistical analyses were performed using SAS software (version 9.4, Cary NC) and MATLAB (Natick, MA). Power transformation was considered to apply parametric analytical tools. The correlation between features was investigated in order to address collinearity issue. The highly correlated features (correlation > 0.9) were regarded as dependent features which were not considered in this analysis. Thus, 59 of 219 features were considered as mutually independent features for prediction of EGFR mutation. Fisher’s exact test and the Kruskal-Wallis test were used for categorical and continuous variables between two groups, respectively. Multiple logistic regression analysis was performed. The final model was selected using the backward elimination method. Variables with p-value of < 0.25 in univariate model were entered in the initial model, and then a variable with p-value of > 0.15 was eliminated at each step. The elimination procedure was terminated when the p-value of all variables in the model is < 0.15. The accuracy and error of the predictive models (i.e., the area under the curve (AUC) and 95% confidence interval (CI)) were estimated by the Bootstrap method with 1000 Bootstrap samples. Receiver operating characteristic (ROC) curves for each model were constructed and the AUC was calculated with EGFR mutation status determined by amplification refractory mutation system-PCR as outcome. Further, various predictive models were developed by the support vector machine (SVM) and the principal component analysis (PCA) and were compared with the logistic regression model. In this study, the regression model was selected as the AUC of the model which was higher than any other models (results were omitted). A two-sided p-value of < 0.05 was regarded as statistically significant.
Results
The patients’ demographic and clinicopathological data are presented in Table 1. All the enrolled 298 patients were surgically treated: lobectomy in 280 patients, pneumonectomy in 4, wedge resection in 6, and segmentectomy in 8 patients. Overall, there were 126 men and 172 women with a median age of 60 years (range: 30 – 80 years). The pathologic stage distribution was as follows: IA in 117 patients (39.26%), IB in 63 patients (21.14%), IIA in 22 patients (7.38%), IIB in 6 patients (2.01%), IIIA in 72 patients (24.16%), IIIB in 2 patients (0.67%), and IV in 16 patients (5.37%). Most of the tumors were early stage (stage I or II) (69.80%). All cases were lung adenocarcinomas and the most common histologic subtype among invasive adenocarcinomas was acinar predominant subtype (42.95%), followed by lepidic predominant subtype (23.83%). EGFR mutation results (Supplementary Table 2) were satisfactorily demonstrated in all patents, with 137 (45.97%) patients out of the full cohort of 298 cases were identified as EGFR mutant and 161 (54.03%) as EGFR wild-type.
Table 1.
Clinicopathological characteristics of patients
| Variables | Overall dataset (n=298) | EGFR mutant (n=137) | EGFR wild-type (n=161) | p-value | |
|---|---|---|---|---|---|
| Median Age (range) | 60 (30 – 80) | 59 (37 – 80) | 60 (30 – 79) | 0.51 | |
| Gender | Male | 126 | 43 (31.39%) | 83 (51.55%) | 0.001 |
| Female | 172 | 94 (68.61%) | 78 (48.45%) | ||
| Smoking status | Smokers | 136 | 43 (31.39%) | 93 (57.76%) | <0.0001 |
| Never smokers | 162 | 94 (68.61%) | 68 (42.24%) | ||
| Pathologic grade† | High | 74 | 20 (14.60%) | 54 (33.75%) | 0.0002 |
| Low/intermediate | 223 | 117 (85.40%) | 106 (66.25%) | ||
| Histologic subtype* | Lepidic predominant adenocarcinomas | 74 | 43 (31.39%) | 31 (19.25%) | 0.022 |
| others | 224 | 94 (68.61%) | 130 (80.75%) | ||
| Stage | I or II | 208 | 93 (67.88%) | 115 (71.43%) | 0.53 |
| III or IV | 90 | 44 (32.12%) | 46 (28.57%) | ||
Adenocarcinoma in situ (AIS) and minimally invasive adenocarcinoma (MIA) were classified as low grade; lepidic, acinar, and papillary as intermediate grade; micropapillary, solid and invasive mucinous adenocarcinoma as high grade. One case with enteric was eliminated in this analysis.
Histologic subtype was categorized as lepidic predominant adenocarcinomas (AIS, MIA, and lepidic predominant invasive adenocarcinoma) and other subtypes of dominant histologic findings (acinar, papillary, micropapillary, and solid predominant as well as variants of invasive adenocarcinoma).
There were significant differences in gender, smoking status, pathologic grade, and histologic subtype between EGFR wile-type group and EGFR mutant group (Table 1). Concerning gender, there were significantly more female patients with mutant, compared to wild-type EGFR in lung adenocarcinomas (OR=2.33, 95% CI: 1.45 – 3.74, p=0.001). Smokers with mutant EGFR were significant lower than smokers with wild-type (OR=2.99, 95% CI: 1.85 – 4.82, p<0.0001). EGFR mutations were also significantly more frequent in patients with low or intermediate pathologic grade (OR=2.98, 95% CI: 1.67 – 5.30, p=0.0002), and patients with lepidic-predominant adenocarcinomas (OR=1.92, 95% CI: 1.13 – 3.27, p=0.022). There were no differences in stage distribution or median age between EGFR mutant and wild-type groups (p=0.53; p=0.51). Univariate analyses revealed that four clinical factors may be associated with EGFR mutation status in peripheral lung adenocarcinoma: gender, smoking status, histologic subtype, and pathologic grade were all significant predictors of harboring and EGFR mutation (Table 2).
Table 2.
Univariate analysis for clinicopathological factors that predict EGFR mutation status
| Clinical Features | Odds Ratio | p-value | |||
|---|---|---|---|---|---|
| Point | 95% CI | ||||
| Lower | Upper | ||||
| Stage | I or II | reference | |||
| III or IV | 1.18 | 0.72 | 1.94 | 0.51 | |
| Gender | Female | reference | |||
| Male | 0.43 | 0.27 | 0.69 | 0.0005 | |
| Histologic subtype | Others | reference | |||
| Lepidic predominant adenocarcinomas | 1.92 | 1.13 | 3.27 | 0.017 | |
| Pathologic grade | Low or Intermediate | reference | |||
| High | 0.34 | 0.19 | 0.60 | 0.0002 | |
| Smoking status | Never smokers | reference | |||
| Smokers | 0.34 | 0.21 | 0.54 | <0.0001 | |
| Age (per 5 years old increase) | 0.97 | 0.85 | 1.11 | 0.68 | |
CI – confidence interval
We then investigated the association of radiomic features with EGFR mutation status. Giving that this analysis produced far more features that were considered dependent, only a prioritized subset of features were selected for further analysis to avoid over fitting. Prioritization methods are described in ref. 29. Fifty-nine independent features were selected finally (Supplementary Table 3). Among these 59 independent features, 11 were independent predictors of harboring EGFR mutation (Table 3, Supplementary Table 4) on univariate analysis, including one feature describing tumor shape (F26), two features describing tumor location (F17, F27), one feature describing airspace (F19), two pixel intensity histogram based features (F185, F186), two runlength & co-occurrence based features (F47, F51), two laws texture features (F90, F111), and one wavelet texture feature (F190).
Table 3.
Univariate analysis for Radiomic features that predict EGFR mutation status
| Radiomic Features | Odds Ratio | p-value | |||
|---|---|---|---|---|---|
| Point | 95% CI | ||||
| Lower | Upper | ||||
| F19 | 10a_3D_Relative_Volume_AirSpaces | 1.16 | 1.04 | 1.3 | 0.007 |
| F186 | Histogram ENERGY Layer 1 | 0.49 | 0.29 | 0.82 | 0.007 |
| F90 | 3D Laws features L5 L5 L5 Layer 1 | 0.82 | 0.7 | 0.96 | 0.015 |
| F51 | AvgLRE | 0.11 | 0.02 | 0.72 | 0.021 |
| F47 | AvgCoocurrence-mean | 0.98 | 0.97 | 1 | 0.025 |
| F190 | 3D Wavelet decomposition. P2 L2 C9 Layer 1 | 1.26 | 1.03 | 1.55 | 0.025 |
| F185 | Histogram SD Layer 1 | 1.88 | 1.07 | 3.31 | 0.027 |
| F26 | Elliptic Fit | 0.11 | 0.02 | 0.79 | 0.028 |
| F17 | 9f-3D-Min-Dist-COG-to-Border | 0.59 | 0.36 | 0.96 | 0.032 |
| F111 | 3D Laws features R5 E5 R5 Layer 1 | 0.71 | 0.51 | 0.99 | 0.041 |
| F27 | Main direction | 1.21 | 1 | 1.45 | 0.048 |
With multiple logistic regression analyses, clinical features of smoking status and pathologic grade proved to be independent predictors of EGFR mutation, and the AUC of ROC was 0.667 (95% CI: 0.604 - 0.721). model produced from radiomic features alone showed moderate predictive power (AUC=0.647; 95% CI: 0.576 - 0.701) for identifying EGFR mutant status. There was a significant difference between AUCs of the logistic regression model incorporating only clinical features, and that incorporating only radiomic features (p<0.0001). When clinical and radiomic features were combined, the AUC was increased to 0.709 (95% CI: 0.654 - 0.766) (Table 4). The model generated with combined clinical and radiomic features was superior to both the model generated with clinical features alone (p<0.0001) and the model created with radiomic features alone (p<0.0001) (Figure 2).
Table 4.
Multiple logistic regression analysis of clinicopathological parameters and radiomic features predicting the presence of EGFR mutation in peripheral lung adenocarcinomas
| Model | Features | p-value | Odds Ratio | AUC | |||||
|---|---|---|---|---|---|---|---|---|---|
| Point | 95% CI | Point | 95% CI | ||||||
| Lower | Upper | Lower | Upper | ||||||
| Clinical features | Pathologic grade (ref= Lower or Intermediate) | High | 0.002 | 0.39 | 0.22 | 0.70 | 0.667 | 0.604 | 0.721 |
| Smoking status (ref = Never smokers) | Yes | <.0001 | 0.38 | 0.23 | 0.61 | ||||
| Radiomic features | Main direction (F27) | 0.115 | 1.17 | 0.96 | 1.42 | 0.647 | 0.576 | 0.701 | |
| 3D Laws features L5 L5 S5 Layer 1 (F92) | 0.010 | 0.52 | 0.31 | 0.85 | |||||
| Histogram ENERGY Layer 1 (F186) | 0.025 | 0.50 | 0.27 | 0.92 | |||||
| 3D Wavelet decomposition. P2 L2 C9 Layer 1 (F190) | 0.048 | 1.29 | 1.00 | 1.67 | |||||
| 3D Wavelet decomposition. P1 L2 C5 Layer 1 (F216) | 0.137 | 1.16 | 0.96 | 1.40 | |||||
| Clinical + Radiomic features | Pathologic grade (ref= Lower or Intermediate) | High | 0.016 | 0.47 | 0.25 | 0.87 | 0.709 | 0.654 | 0.766 |
| Smoking status (ref = Never smokers) | Yes | 0.001 | 0.41 | 0.25 | 0.67 | ||||
| Main direction (F27) | 0.20 | 1.14 | 0.93 | 1.40 | |||||
| 3D Laws features L5 L5 S5 Layer 1 (F92) | 0.056 | 0.60 | 0.36 | 1.01 | |||||
| Histogram ENERGY Layer 1 (F186) | 0.11 | 0.60 | 0.32 | 1.13 | |||||
| 3D Wavelet decomposition. P2 L2 C9 Layer 1 (F190) | 0.13 | 1.22 | 0.94 | 1.59 | |||||
| 3D Wavelet decomposition. P1 L2 C5 Layer 1 (F216) | 0.24 | 1.13 | 0.92 | 1.38 | |||||
AUC – area under curve
Fig. 2.

Receiver operating characteristic (ROC) curves for the prediction of EGFR mutation using logistic regression model that included clinical factors alone (green line), a model that use radiomic features alone (red line), and a model that combined clinical factors and radiomic features (blue line). The highest area under the curve (AUC) was achieved for the combination of clinical factors and radiomic features (AUC=0.709).
Discussion
There are various methods to detect EGFR mutations, such as direct sequencing of polymerase chain reaction (PCR)-amplified genomic DNA, high-resolution melting analysis, fragment analysis, restriction fragment length polymorphism and the amplification refractory mutation system 35; however, these molecular methods are generally costly, and sometimes in a low percentage of tumor cells, we would have most likely missed the mutations and re-biopsy had to be recommended. In this study, we sought to apply radiomic features to peripheral lung adenocarcinomas to determine if we could noninvasively discriminate EGFR-mutant from EGFR- wild type case on a routine practice without adding additional cost.
We found that 137 of the tumors harbored EGFR mutation, which corresponded to 45.97% of the 298 tumor samples, this is in keeping with previous reports on Asian patients 36; in addition, exon 19 and exon 21, the most common mutation types of the EGFR gene, showed nearly the same percentage in these patients (46.72%, 48.91% respectively). Dual mutations were detected in one patient (0.73%) in this study, confirming that the existence of both EGFR TKI resistant mutation (exon 20) and sensitive mutation (exon 21) is rare. Several studies indicated that EGFR mutation was strongly related with never smokers, female, adenocarcinoma, and pathologic stage 29, 36. Consistent with most of these findings, the rate of EGFR mutation in female and never smoker patients with lung adenocarcinomas was considerably higher compared to male and smokers in this study. Although EGFR mutation was detected more frequently in early stage patients (67.88%) compared to advanced stage (32.12%), the difference was not significant. The results of the multiple logistic regression analysis revealed that pathologic grade and smoking status were independent predictors for EGFR mutation and the AUC was 0.667. Besides clinical information, more detailed factors will likely be required to identify those at high probability of harboring EGFR mutations.
CT imaging is used widely in oncologic practice for lung tumor characterization. Usually, we interpret the image based on visual assessment; there are features, however, within each image that may not be perceived by the naked eye and requires computer-aided techniques. Previous authors have demonstrated the potential of quantitative CT based texture analysis in differentiation of K-ras mutation from pan-wildtype NSCLC 24. To the best of our knowledge, only one study 37that published recently has explored the association between CT gray-level texture features and EGFR mutations in a relatively small sample size (25 patients with EGFR mutation and 20 patients with EGFR wild-type). In this study, we present comprehensive radiomic analysis using semi-automatic segmentation in 298 peripheral lung adenocarcinomas. 219 radiomic features were extracted to assess the ability to predict EGFR mutation status. Considering that using too many features in the classification algorithm can lead to over-fitting, in which noise or irrelevant features may exert undue influence on classification decisions, only those features which are not associated with other features were selected for further analysis. With this approach, we found that 11 radiomic features from seven different feature categories were significantly associated with EGFR mutations. Texture features, Wavelet features, Laws features, and along with pixel statistical have seen a resurgent use in medical images, especially CT and MRI images 24, 38. In our study we formed sets of five one dimensional Laws filters, each designed to describe different structures in the image (E: Edges, S: Spots, R: Ripple, W: Waves, L: Lowpass). The wavelet transformation was limited to two levels (L) of decomposition on each of the nine faces (C) on the 3D tumor, with two types of metrics (P), namely energy and entropy. Our statistical model finds Laws features L5L5S5 and Wavelet P2L2C9 & P1L2C5 to be one of the five predictors of EGFR mutation status. Several previous studies 17, 39showed that EGFR mutation was associated with small tumor size. In our analysis, size based features including longest diameter and short axis were not significant predictor for EGFR mutation. We believe that this difference can be explain in part by the fact that most clinically assessed tumor diameters are manually drawn on the central slice of CT images and limited to one dimension of the tumor, while whole tumor volume was taken into account in the size based radiomic features.
With logistic regression analysis, we identified that radiomic features could be served as an imaging surrogate for EGFR mutation, although the AUC of radiomic features alone was not as high as the model created with clinical features, they provide complementary information, as the combination results in an improved ROC curve (AUC, 0.709), and we were able to significantly increase the predictive performance of EGFR mutation. These findings suggested that the combination of radiomic data and demographic information in a system model is more effective.
There are some limitations in this study. One limitation is that this study is retrospective and limited to only Eastern Asian populations; care should be taken before generalizing our findings to other populations. Second, radiomic features were derived from semi-automatic segmentation by radiologists, which can be influence by observers’ subjective trend. However, the results of automatic boundary extraction method were not satisfactory for all the lesions, particularly in the case of tumors with GGO components, as their margins are usually unclear form the adjacent normal lung parenchyma. Furthermore, since atelectasis is common in patients with central lung cancers, and differentiation between them is rather difficult as both appear as solid density on CT, our analysis did not include central lung cancers. A prospective multi-Institutional study with a large patient cohort would be required to confirm our observations.
Conclusions
In summary, this study revealed associations between CT based radiomic features and EGFR mutation status in peripheral lung adenocarcinomas, and therefore non-invasive radiomic phenotype analysis has the potential to improve the differentiation of EGFR mutate from wild type when used in addition to clinical predictors.
Supplementary Material
Clinical Practice Points.
The presence of activating EGFR mutations has been shown to be prognostic for a more favorable outcome to TKI therapy in lung adenocarcinomas.
Several reports have described the relationship between mutation status of EGFR and traditional radiological features. Radiomic-based approach allows high throughput extraction of quantitative parameters from CT images which beyond what is visually perceived by the human eyes. Thus, we hypothesized that radiomic analysis of routinely performed preoperative CT images could provide imaging biomarkers for EGFR mutations.
In this study we demonstrated that eleven CT based radiomic features have significant association with EGFR mutations. Adding radiomic features to clinical model could improve the predicting power of EGFR mutations, thus helping in formulating a better clinical decision without adding additional cost.
Acknowledgments
We thank Zhongli Zhan (Department of Pathology, Tianjin Medical University Cancer Institute and Hospital) for doing histologic evaluation and EGFR mutation analysis.
This work was supported by the National Cancer Institute (grants U01 CA143062), Tianjin Science and Technology Major Project (No. 12ZCDZSY1550), and Public science and technology research funds projects of NHFPC of the P.R. China (No. 201402013). This work has been supported in part by the Biostatistics Core Facility at the Moffitt Cancer Center & Research Institute, a National Cancer Institute designated Comprehensive Cancer Center (5P30CA076292-16).
Abbreviations
- NSCLCs
non-small cell lung cancers
- TKIs
tyrosine kinase inhibitors
- EGFR
epidermal growth factor receptor
- PFS
progression-free survival
- CT
computed tomography
- PACS
picture archiving and communication system
- IASLC/ATS/ERS
International Association for the Study of Lung Cancer, American Thoracic Society, and European Respiratory Society
- ARMS
amplification refractory mutation system
- SCES
single click ensemble segmentation
- CI
confidence interval
- ROC
receiver operating characteristic
- AUC
area under the curve
- PCR
polymerase chain reaction
- AIS
adenocarcinoma in situ
- MIA
minimally invasive adenocarcinoma
Footnotes
Conflict of Interest
RJG is a consultant and shareholder in HealthMyne, Inc. and oncology-specific PACS system. No other authors of this manuscript have relationships with any companies, whose products or services may be related to the subject matter of the article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Halpenny DF, Riely GJ, Hayes S, et al. Are there imaging characteristics associated with lung adenocarcinomas harboring ALK rearrangements? Lung Cancer. 2014;86:190–194. doi: 10.1016/j.lungcan.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganeshan B, Panayiotou E, Burnand K, Dizdarevic S, Miles K. Tumour heterogeneity in non-small cell lung carcinoma assessed by CT texture analysis: a potential marker of survival. European radiology. 2012;22:796–802. doi: 10.1007/s00330-011-2319-8. [DOI] [PubMed] [Google Scholar]
- 3.Mok TS, Wu Y-L, Thongprasert S, et al. Gefitinib or carboplatin–paclitaxel in pulmonary adenocarcinoma. New England Journal of Medicine. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 4.Jackman DM, Miller VA, Cioffredi L-A, et al. Impact of epidermal growth factor receptor and KRAS mutations on clinical outcomes in previously untreated non–small cell lung cancer patients: results of an online tumor registry of clinical trials. Clinical Cancer Research. 2009;15:5267–5273. doi: 10.1158/1078-0432.CCR-09-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riely GJ, Pao W, Pham D, et al. Clinical course of patients with non–small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clinical Cancer Research. 2006;12:839–844. doi: 10.1158/1078-0432.CCR-05-1846. [DOI] [PubMed] [Google Scholar]
- 6.Yang JC-H, Schuler MH, Yamamoto N, et al. LUX-Lung 3: A randomized, open-label, phase III study of afatinib versus pemetrexed and cisplatin as first-line treatment for patients with advanced adenocarcinoma of the lung harboring EGFR-activating mutations. J Clin Oncol. 2012;30 LBA7500. [Google Scholar]
- 7.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non–small-cell lung cancer with mutated EGFR. New England Journal of Medicine. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi K, Inoue A, Maemondo M, et al. First-line gefitinib versus first-line chemotherapy by carboplatin (CBDCA) plus paclitaxel (TXL) in non-small cell lung cancer (NSCLC) patients (pts) with EGFR mutations: a phase III study (002) by North East Japan Gefitinib Study Group. ASCO Annual Meeting Proceedings. 2009;27:8016. [Google Scholar]
- 9.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. The lancet oncology. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 10.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 11.Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer biological and clinical implications. Cancer research. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 12.Shahi RB, De Brakeleer S, De Greve J, Geers C, In’t Veld P, Teugels E. Detection of EGFR-TK domain-activating mutations in NSCLC with generic PCR-based methods. Applied immunohistochemistry & molecular morphology : AIMM / official publication of the Society for Applied Immunohistochemistry. 2015;23:163–171. doi: 10.1097/PDM.0000000000000035. [DOI] [PubMed] [Google Scholar]
- 13.Tomonaga N, Nakamura Y, Yamaguchi H, et al. Analysis of intratumor heterogeneity of EGFR mutations in mixed type lung adenocarcinoma. Clinical lung cancer. 2013;14:521–526. doi: 10.1016/j.cllc.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Bai H, Wang Z, Wang Y, et al. Detection and clinical significance of intratumoral EGFR mutational heterogeneity in Chinese patients with advanced non-small cell lung cancer. PLoS One. 2013;8:e54170. doi: 10.1371/journal.pone.0054170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taniguchi K, Okami J, Kodama K, Higashiyama M, Kato K. Intratumor heterogeneity of epidermal growth factor receptor mutations in lung cancer and its correlation to the response to gefitinib. Cancer science. 2008;99:929–935. doi: 10.1111/j.1349-7006.2008.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Querings S, Altmüller J, Ansén S, et al. Benchmarking of mutation diagnostics in clinical lung cancer specimens. PloS one. 2011;6:e19601. doi: 10.1371/journal.pone.0019601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rizzo S, Petrella F, Buscarino V, et al. CT Radiogenomic Characterization of EGFR, K-RAS, and ALK Mutations in Non-Small Cell Lung Cancer. Eur Radiol. 2016;26:32–42. doi: 10.1007/s00330-015-3814-0. [DOI] [PubMed] [Google Scholar]
- 18.Zhou JY, Zheng J, Yu ZF, et al. Comparative analysis of clinicoradiologic characteristics of lung adenocarcinomas with ALK rearrangements or EGFR mutations. Eur Radiol. 2015;25:1257–1266. doi: 10.1007/s00330-014-3516-z. [DOI] [PubMed] [Google Scholar]
- 19.Lee Y, Lee HJ, Kim YT, et al. Imaging characteristics of stage I non-small cell lung cancer on CT and FDG-PET: relationship with epidermal growth factor receptor protein expression status and survival. Korean J Radiol. 2013;14:375–383. doi: 10.3348/kjr.2013.14.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee HJ, Kim YT, Kang CH, et al. Epidermal growth factor receptor mutation in lung adenocarcinomas: relationship with CT characteristics and histologic subtypes. Radiology. 2013;268:254–264. doi: 10.1148/radiol.13112553. [DOI] [PubMed] [Google Scholar]
- 21.Kumar V, Gu Y, Basu S, et al. Radiomics: the process and the challenges. Magnetic resonance imaging. 2012;30:1234–1248. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parmar C, Velazquez ER, Leijenaar R, et al. Robust radiomics feature quantification using semiautomatic volumetric segmentation. PloS one. 2014;9:e102107. doi: 10.1371/journal.pone.0102107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mattonen SA, Palma DA, Haasbeek CJ, Senan S, Ward AD. Early prediction of tumor recurrence based on CT texture changes after stereotactic ablative radiotherapy (SABR) for lung cancer. Medical physics. 2014;41:033502. doi: 10.1118/1.4866219. [DOI] [PubMed] [Google Scholar]
- 24.Weiss GJ, Ganeshan B, Miles KA, et al. Noninvasive Image Texture Analysis Differentiates K-ras Mutation from Pan-Wildtype NSCLC and Is Prognostic. PloS one. 2014;9:e100244. doi: 10.1371/journal.pone.0100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravanelli M, Farina D, Morassi M, et al. Texture analysis of advanced non-small cell lung cancer (NSCLC) on contrast-enhanced computed tomography: prediction of the response to the first-line chemotherapy. European radiology. 2013;23:3450–3455. doi: 10.1007/s00330-013-2965-0. [DOI] [PubMed] [Google Scholar]
- 26.Coroller TP, Grossmann P, Hou Y, et al. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology. 2015;114:345–350. doi: 10.1016/j.radonc.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Annals of surgical oncology. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 28.Travis WD, Brambilla E, Noguchi M, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society: international multidisciplinary classification of lung adenocarcinoma: executive summary. Proceedings of the American Thoracic Society. 2011;8:381–385. doi: 10.1513/pats.201107-042ST. [DOI] [PubMed] [Google Scholar]
- 29.Locatelli-Sanchez M, Couraud S, Arpin D, Riou R, Bringuier P-P, Souquet P-J. Routine EGFR molecular analysis in non-small-cell lung cancer patients is feasible: exons 18-21 sequencing results of 753 patients and subsequent clinical outcomes. Lung. 2013;191:491–499. doi: 10.1007/s00408-013-9482-4. [DOI] [PubMed] [Google Scholar]
- 30.Balagurunathan Y, Kumar V, Gu Y, et al. Test–Retest Reproducibility Analysis of Lung CT Image Features. Journal of digital imaging. 2014;27:805–823. doi: 10.1007/s10278-014-9716-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu Y, Kumar V, Hall LO, et al. Automated delineation of lung tumors from CT images using a single click ensemble segmentation approach. Pattern recognition. 2013;46:692–702. doi: 10.1016/j.patcog.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Velazquez ER, Aerts HJ, Gu Y, et al. A semiautomatic CT-based ensemble segmentation of lung tumors: Comparison with oncologists’ delineations and with the surgical specimen. Radiotherapy and Oncology. 2012;105:167–173. doi: 10.1016/j.radonc.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balagurunathan Y, Gu Y, Wang H, et al. Reproducibility and Prognosis of Quantitative Features Extracted from CT Images. Translational oncology. 2014;7:72–87. doi: 10.1593/tlo.13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Randen T, Husoy JH. Filtering for texture classification: A comparative study. Pattern Analysis and Machine Intelligence, IEEE Transactions on. 1999;21:291–310. [Google Scholar]
- 35.da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annual Review of Pathology: Mechanisms of Disease. 2011;6:49–69. doi: 10.1146/annurev-pathol-011110-130206. [DOI] [PubMed] [Google Scholar]
- 36.Liu WS, Zhao LJ, Pang QS, Yuan ZY, Li B, Wang P. Prognostic value of epidermal growth factor receptor mutations in resected lung adenocarcinomas. Med Oncol. 2014;31:771. doi: 10.1007/s12032-013-0771-9. [DOI] [PubMed] [Google Scholar]
- 37.Ozkan E, West A, Dedelow JA, et al. CT Gray-Level Texture Analysis as a Quantitative Imaging Biomarker of Epidermal Growth Factor Receptor Mutation Status in Adenocarcinoma of the Lung. AJR Am J Roentgenol. 2015;205:1016–1025. doi: 10.2214/AJR.14.14147. [DOI] [PubMed] [Google Scholar]
- 38.Zhao B, J LP, Moskowitz CS, Guo P, Ginsberg MS, Lefkowitz RA, Qin Y, Riely GJ, Kris MG, Schwartz LH. Evaluating variability in tumor measurements from same-day repeat CT scans of patients with non-small cell lung cancer. Radiology. 2009;252:263–272. doi: 10.1148/radiol.2522081593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu J-S, Huang M-S, Chen C-Y, et al. Correlation between EGFR mutation status and computed tomography features in patients with advanced pulmonary adenocarcinoma. Journal of thoracic imaging. 2014;29:357–363. doi: 10.1097/RTI.0000000000000116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


