Abstract
Cutaneous melanoma (CM) is the most lethal skin cancer. The Wnt pathway has an impact on development, invasion and metastasis of CM, thus likely affecting CM prognosis. Using data from a published genome-wide association study (GWAS) from The University of Texas M.D. Anderson Cancer Center, we assessed the associations of 19,830 common single-nucleotide polymorphisms (SNPs) in 151 Wnt pathway autosomal genes with CM-specific survival (CMSS) and then validated significant SNPs in another GWAS from Harvard University. In the single-locus analysis, 1,855 SNPs were significantly associated with CMSS at P < 0.05, of which 547 SNPs were still considered noteworthy after the correction by the false positive report probability. In the replication, two SNPs remained significantly associated with CMSS after multiple comparison correction. By performing functional prediction and stepwise selection, we identified two independent SNPs (i.e., WNT2B rs1175649 G>T and BTRC rs61873997 G>A) that showed a predictive role in CMSS, with an effect-allele-attributed hazards ratio [adjHR of 1.99 (95% confidence interval (CI) = 1.41-2.81, P = 8.10E-05) and 0.61 (0.46-0.80, 3.12E-04), respectively]. Collectively, these variants in the Wnt pathway genes may be biomarkers for outcomes of CM patients, if validated by larger studies.
Keywords: cutaneous melanoma, genome-wide association study, single-nucleotide polymorphism, Wnt pathway, cutaneous melanoma-specific survival
Introduction
Cutaneous melanoma (CM) is one of the most aggressive forms of skin cancers, ranking the fifth most common cancer among males and the seventh among females in the United States. In 2016, it is estimated that there will be 76,380 new cases of CM and that 10,130 CM patients will die in the United States (Siegel et al., 2016). Although the incidence rate of CM has been increasing, the mortality rate has remained steady over the past decade (Simard et al., 2012). While the increase of CM incidence may partially be attributed to more effective clinical screening (Jemal et al., 2011; Purdue et al., 2008), there has been little improvement in the ability to accurately assess the prognosis of CM patient. The overall 5-year survival rate of CM patients varies substantially, from 97% for localized CM to 65.8% and 15.2% for regional and distant metastasis, respectively (Balch et al., 2009).
Current prognostic tools mainly include clinicopathological variables, such as tumor stage, Breslow thickness, and mitotic rate (Balch et al., 2009). However, there is a wide range of survival rates among CM patients with similar clinical characteristics, even for the same tumor stage (Schramm and Mann, 2011), which possibly results from the heterogeneity of CM and the weak specificity of those clinicopathological variables used to evaluate the prognosis of CM patients. For example, 30% of CM deaths were found to be attributable to thin lesions that were thought to have a good prognosis (Jemal et al., 2011). Therefore, the development of potential biomarkers with more prognostic specificity is necessary, which can facilitate the individuality of clinical assessment. Genetic variants, such as single-nucleotide polymorphisms (SNPs), have been associated with individual variation in susceptibility to and clinical outcome of cancer (Dong et al., 2013; Yang et al., 2011). Recently, genome-wide association studies (GWASs) have been used to identify genetic variants to be associated with clinical outcomes of pancreatic adenocarcinoma patients with a high statistical power (Wu et al., 2014).
WNTs are a group of glycoproteins that regulate multiple biological processes, including the proliferation, survival, migration and polarity of cells, the specification of cell fate, and the self-renewal of stem cells (Clevers and Nusse, 2012; Gomez-Orte et al., 2013). Aberrant activity of this pathway plays a role in a number of diseases, such as fibrosis (Chilosi et al., 2003), neurodegeneration (Berwick and Harvey, 2012; Inestrosa et al., 2012), metabolic disease (Schinner, 2009), and many types of cancer (de La Coste et al., 1998; Palacios and Gamallo, 1998; Zurawel et al., 1998).
The Wnt signaling pathway, which is required for melanocyte survival and differentiation (Hari et al., 2002; Rabbani et al., 2011), is deregulated in at least 1/3 human melanomas (Rimm et al., 1999). The pathway appears to have important roles in melanomagenesis, during both tumor initiation (Chien et al., 2009; Delmas et al., 2007) and progression (Damsky et al., 2011; Gallagher et al., 2013; Sinnberg et al., 2011). Notably, it has reported that one SNP in WNT3 is involved in CM predisposition (Ibarrola-Villava et al., 2015). However, the impact of gene variants in the Wnt pathway on CM disease progression has not been systematically tested. Here, we hypothesize that genetic variants in the Wnt pathway genes modulate clinical outcome of CM patients. To test this hypothesis, we used genotyping data of SNPs in Wnt pathway genes extracted from a previously published GWAS of CM from The University of Texas MD Anderson Cancer Center (MDACC) (Amos et al., 2011), assessed their associations with the survival of CM patients and then validated the SNPs of interest in another GWAS dataset from Harvard University.
Results
Patient characteristics
The final analysis included 858 patients from MDACC and 409 patients from Harvard University (Supplementary Table S1 online). All patients with CM were non-Hispanic white. The MDACC study included 858 CM patients, whose complete information about clinical variables, risk factor data, and GWAS data was described elsewhere (Li et al., 2013). The ages at diagnosis of these patients were between 17 and 94 years (mean age ± standard deviation, 52.4 ± 14.4 years), and there were more men (496, 57.8%) than women (362, 42.2%). Of these patients, 17.4% (149) had been diagnosed with regional/distant metastasis (stage III/IV). The median thickness of localized tumors for the MDACC cohort was 1.1 cm. The patients had a median follow-up time of 81.1 months, during which 133 (15.5%) had died for any reason at the last follow-up. Among these deaths, 95 were caused by CM. Univariate analysis indicated that age, sex, regional/distant metastasis, Breslow thickness, ulceration, and mitotic rate were significant predictors of CM-specific survival (CMSS). In the Harvard study, only age, sex, survival outcome and genotype data were available for analysis. Patients had an age range between 34 to 87 years at diagnosis (61.1 ± 10.8 years), of whom 66.3% (271) were women. The patients had a relatively longer median follow-up time (179.0 months), compared with 175.3 months for MDACC patients. During the follow-up, there were 48 (11.5%) patients died of CM, and only age was significantly associated with CMSS in univariate analysis of the Harvard dataset.
Associations between SNPs in the Wnt pathway genes and CMSS
For the illustrative purpose, Figure 1a provides the flowchart of study design performed in the present study. In the discovery using the MDACC study, we first performed multivariate Cox proportional hazards regression analyses to assess associations of 19,830 common SNPs (3,151 genotyped and 16,679 imputed SNPs) of the Wnt pathway genes with CMSS with adjustments for age, sex, regional/distant metastasis, Breslow thickness, ulceration and mitotic rate. As shown in Figure 1b, 1,855 SNPs were individually significantly associated with CMSS at P < 0.05 in an additive model, of which 547 SNPs were still considered noteworthy after the correction for the false positive report probability (FPRP). Among these, three SNPs were validated and remained statistically significantly associated with CMSS at P ≤ 0.05 in the Harvard study, including rs1175649 in WNT2B (Wnt family member 2B), rs61873997 and rs4919545 in BTRC (beta-transducin repeat containing E3 ubiquitin protein ligase). Then, we performed FPRP for multiple test correction of the three replicated SNPs in the Harvard dataset and found that rs1175649 and rs61873997 were still considered noteworthy after the correction (Supplementary Table S2). In the subsequent meta-analysis of these two studies, both of the two SNPs showed significant associations with CMSS (Table 1). None of the effects of the two SNPs were significantly heterogeneous among the studies (All Phet > 0.05). Using the TCGA dataset of 306 melanoma cases, we performed the validation analysis for the effects of two tagSNPs (WNT2B rs1175649 and BTRC rs61873997) on survival of melanoma patients, in which the follow-up information, genotype data, age, and sex were available. Although neither of SNPs was statistically significantly associated with OS of melanoma patients, the BTRC rs61873997 A allele (HR = 0.81, 95% CI = 0.62–1.05, P = 0.109) appeared to be associated with a protective effect on survival of melanoma patients, which is consistent with what we observed in both MDACC and Harvard datasets. Because the patients from the TCGA dataset had a relatively shorter median follow-up time (59.8 months), a smaller sample size (306 patients) and unavailable data of DSS information, additional validation is warranted. However, we did not find any evidence for an effect of WNT2B rs1175649 and/or BTRC rs61873997 on survival of non-melanoma skin cancer or other solid tumors.
Figure 1. Screening for SNPs in the Wnt pathway genes.
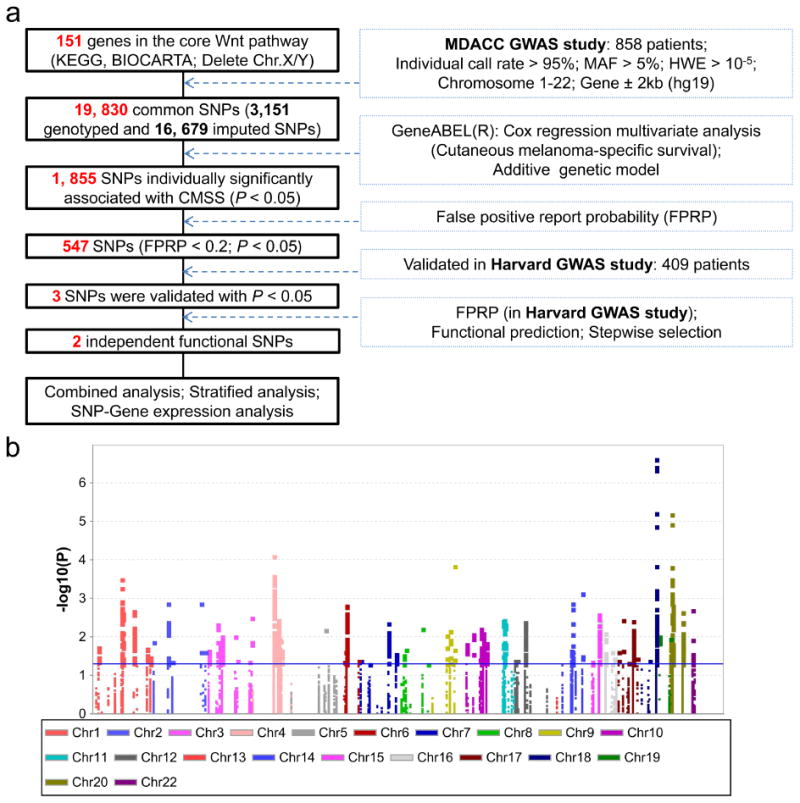
(a) Research workflow. MDACC, The University of Texas M.D. Anderson Cancer Center; GWAS, genome-wide association study; SNP, single-nucleotide polymorphism; HWE, Hardy Weinberg equilibrium; MAF, minor allele frequency. (b) Manhattan plot of associations between 19,830 SNPs and CMSS. There are 1,855 SNPs with P < 0.05. The blue horizontal line indicates P = 0.05.
Table 1. Combined analysis of two validated SNPs using two published melanoma GWAS datasets.
| SNP | Allele1 | MAF | Gene | MDACC (n=858) | Harvard (n=409) | Combined analysis | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||||
| HR (95% CI)2 | P2 | HR (95% CI)3 | P3 | Phet | I2 | HR (95% CI)4 | P4 | ||||
| rs1175649 | G/T | T: 0.11 | WNT2B | 1.97 (1.26-3.09) | 0.003 | 2.03 (1.19-3.46) | 0.010 | 0.933 | 0 | 1.99 (1.41-2.81) | 8.10E-05 |
| rs61873997 | G/A | A: 0.34 | BTRC | 0.62 (0.44-0.87) | 0.006 | 0.58 (0.37-0.92) | 0.021 | 0.818 | 0 | 0.61 (0.46-0.80) | 3.12E-04 |
Abbreviations: SNP, single-nucleotide polymorphism; GWAS, genome-wide association studies; MAF, minor allele frequency; MDACC, M.D. Anderson Cancer Center; HR: hazards ratio; CI: confidence interval; Phet: P-value for heterogeneity by Cochrane's Q test;
Reference allele/effect allele;
Adjusted for age, sex, Breslow's tumor thickness, regional/distant metastasis, ulceration of tumor and tumor cell mitotic rate;
Adjusted for age and sex;
Meta-analysis in a fixed-effects model.
Genetic variants in Wnt pathway genes as independent death predictors
Based on the results of in silico functional prediction using SNPinfo (http://snpinfo.niehs.nih.gov/snpinfo/snpfunc.htm), RegulomeDB (http://www.regulomedb.org/) and F-SNP (http://compbio.cs.queensu.ca/F-SNP, Supplementary Table S3 online), we identified the two SNPs (i.e., WNT2B rs1175649 G>T and BTRC rs61873997 G>A) as tagSNPs. Initial stepwise Cox proportional hazards regression analyses suggested these two tagSNPs as independent predictors for CMSS of CM patients (Supplementary Table S4 online). For each of the two significant SNPs, we further performed multivariate Cox proportional hazards regression analyses to evaluate their effects on death risk with adjustment for other clinicopathological covariates (i.e., age, sex, Breslow tumor thickness, regional/distant metastasis, ulceration of tumor and tumor cell mitotic rate) for the MDACC dataset but only age and sex for the Harvard dataset. In the MDACC study, the risk effect of the WNT2B rs1175649 T allele (adjHR = 1.97; 95% CI = 1.26–3.09, P = 0.003) and protective effect of the BTRC rs61873997 A allele (adjHR = 0.62; 95% CI = 0.44–0.87, P = 0.006) on CM survival were statistically significant in the trend test. In the validation using the Harvard study, similar results were obtained for the WNT2B rs1175649 T allele (adjHR = 2.03; 95% CI = 1.19-3.46, P = 0.010) and for the BTRC rs61873997 A allele (adjHR = 0.58; 95% CI = 0.37–0.92, P = 0.021) (Table 2). To further visualize the HR effects, we present Kaplan–Meier survival curves of the associations with CMSS for risk genotypes of WNT2B rs1175649 and BTRC rs61873997 in Figure 2a-2d. The regional association results from the MDACC GWAS dataset were plotted for WNT2B and BTRC, including the 250 kb regions flanking the neighborhoods of these two genes (Supplementary Figure S1 online).
Table 2. Associations between tagging SNPs in WNT2B and BTRC of the Wnt pathway and CMSS of CM patients in the MDACC Study and the Harvard study.
| Genotype | MDACC study | Harvard study | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| All | Death (%) | HR (95% CI)1 | P1 | All | Death (%) | HR (95% CI)2 | P2 | |
| WNT2B | ||||||||
| rs1175649 | ||||||||
| GG | 682 | 68 (10.0) | 1.0 0 | 321 | 32 (10.0) | 1.0 0 | ||
| GT | 171 | 27 (15.8) | 2.11 (1.32-3.38) | 0.002 | 85 | 14 (16.5) | 1.76 (0.94-3.29) | 0.079 |
| TT | 5 | 0 (0.0) | -- | -- | 3 | 2 (66.7) | 7.02 (1.62-30.30) | 0.009 |
| Trend test | 1.97 (1.26-3.09) | 0.003 | 2.03 (1.19-3.46) | 0.010 | ||||
| GT+TT vs GG | 2.08 (1.30-3.33) | 0.002 | 1.93 (1.06-3.53) | 0.031 | ||||
| BTRC | ||||||||
| rs61873997 | ||||||||
| GG | 367 | 52 (14.2) | 1.0 0 | 171 | 29 (17.0) | 1.0 0 | ||
| GA | 391 | 37 (9.5) | 0.60 (0.39-0.93) | 0.022 | 182 | 14 (7.7) | 0.44 (0.23-0.83) | 0.011 |
| AA | 100 | 6 (6.0) | 0.45 (0.19-1.07) | 0.070 | 56 | 5 (8.9) | 0.50 (0.19-1.29) | 0.152 |
| Trend test | 0.62 (0.44-0.87) | 0.006 | 0.58 (0.37-0.92) | 0.021 | ||||
| GA+AA vs GG | 0.57 (0.38-0.87) | 0.009 | 0.45 (0.25-0.81) | 0.007 | ||||
Abbreviations: SNP, single-nucleotide polymorphism; CMSS, cutaneous melanoma-specific survival; CM, cutaneous melanoma; MDACC, MD Anderson Cancer Center; HR: hazards ratio; CI: confidence interval.
Adjusted for age, sex, Breslow thickness, regional/distant metastasis, ulceration of tumor and tumor cell mitotic rate.
Adjusted for age and sex.
Figure 2. Selected SNPs and survival prediction.
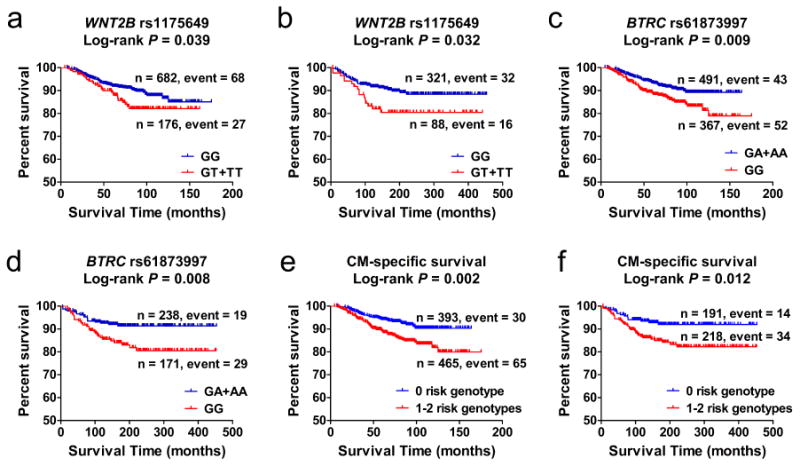
(a-b) Kaplan–Meier curves of cutaneous melanoma-specific survival (CMSS) stratified by WNT2B rs1175649, assuming a dominant model in the MDACC study (a) and in the Harvard study (b). (c-d) Kaplan–Meier curves of CMSS stratified by BTRC rs61873997, assuming a dominant model in the MDACC study (c) and in the Harvard study (d). (e-f) Kaplan–Meier survival curves of the combined risk genotypes: dichotomized groups of the NUGs in the MDACC study (e) and in the Harvard study (f).
Survival of CM patients with unfavorable genotypes
To better estimate the joint effect of the two tag SNPs on risk of death, we combined the unfavorable genotypes (those all associated with an increased death risk) of WNT2B rs1175649 GT/TT and BTRC rs61873997 GG into one variable (or a genetic score), which we termed the ‘number of unfavorable genotypes’ (NUG) score. The P value for trend test for a per-unit increase in the NUG score was < 0.0001 for CMSS in the MDACC study. In the Harvard study, a similar trend was observed in the association of NUGs and CMSS (P = 0.0009, Table 3). Prognosis was the worst in patients with a NUG score of 2 for CMSS in the MDACC study (adjHR = 3.01; 95% CI = 1.56–5.78, P = 0.001) and the Harvard study (adjHR = 3.84; 95% CI = 1.74–8.47, P = 0.0008), compared with a NUG score of 0. We next dichotomized all patients into a low-risk group (NUG score 0) and a high-risk group (NUG score 1–2). We found that, compared with the low-risk group, the high-risk group had a more than 2-fold higher CM-death risk in the MDACC study (adjHR = 2.27, 95% CI = 1.44–3.57, P = 0.0004) and the Harvard study (adjHR = 2.30, 95% CI = 1.23–4.29, P = 0.009, Table 3). For the illustrative purpose, Kaplan–Meier curves of these associations of the NUG score with CMSS are shown in Figure 2e-2f.
Table 3. HRs for associations between NUGs and CMSS in CM patients of the MDACC study and the Harvard study.
| Genotype | MDACC study | Harvard study | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| All | Death (%) | HR (95% CI)2 | P2 | All | Death (%) | HR (95% CI)3 | P3 | |
| NUG1 | ||||||||
| 0 | 393 | 30 (7.6) | 1.00 | 191 | 14 (7.3) | 1.00 | ||
| 1 | 387 | 51 (13.2) | 2.12 (1.33-3.40) | 0.002 | 177 | 23 (13.0) | 1.92 (0.99-3.74) | 0.055 |
| 2 | 78 | 14 (17.9) | 3.01 (1.56-5.78) | 0.001 | 41 | 11 (26.8) | 3.84 (1.74-8.47) | 0.0008 |
| Trend test | <0.0001 | 0.0009 | ||||||
|
| ||||||||
| Dichotomized | ||||||||
| 0 | 393 | 30 (7.6) | 1.00 | 191 | 14 (7.3) | 1.00 | ||
| 1-2 | 465 | 65 (14.0) | 2.27 (1.44-3.57) | 0.0004 | 218 | 34 (15.6) | 2.30 (1.23-4.29) | 0.009 |
Abbreviations: HR, hazards ratio; NUGs, number of unfavorable genotypes; CMSS, cutaneous melanoma-specific survival; CM, cutaneous melanoma; MDACC, MD Anderson Cancer Center; CI: confidence interval.
Risk genotypes were rs1175649 TT+GT, and rs61873997 GG;
Adjusted for age, sex, Breslow thickness, regional/distant metastasis, ulceration of tumor and tumor cell mitotic rate.
Adjusted for age and sex.
Stratified analyses of the NUG score as a predictor of survival in CM patients
We further performed stratified analyses to investigate whether the combined effect of unfavorable genotypes on CMSS was modified by the clinical variables factors in MDACC study. We found that patients with the high-risk NUG score (1–2), but not the low-risk NUG score (0), showed a substantially increased risk of CM death in the presence or absence of concomitant clinical variables (e.g., age, sex, regional/distant metastasis, Breslow thickness, ulceration and tumor cell mitotic rate), except for three subgroups (male; patients without ulceration; patients with tumor cell mitotic rate ≤ 1/mm2), However, no heterogeneity was observed among all the subgroups (Supplementary Table S5 online).
Genotype-phenotype correlation analyses
To further understand the molecular mechanisms underlying the observed risk associations with the genotypes, we evaluated the correlations between SNPs and their corresponding mRNA expression levels in normal cells using the publically available RNA-seq data of lymphoblastoid cell lines from 373 European descendants included in the 1000 Genomes Project (Genomes Project et al., 2012; Lappalainen et al., 2013). We found that rs1175649 genotype demonstrated a significant association with an increased mRNA expression of WNT2B in both additive and dominant models (P = 0.006 and P = 0.042, respectively, Figure 3a-b). However, no significant correlation was found between rs61873997 genotypes and BTRC mRNA expression (for additive model, P = 0.900; for recessive model, P = 0.152; Supplementary Figure S2 online). We further performed expression quantitative trait loci (eQTLs) analysis using data from the Genotype-Tissue Expression (GTEx) Project (http://www.gtexportal.org/home) (Consortium, 2013), which includes WNT2B rs1175649 in subcutaneous adipose tissues from 298 donors. It was shown that rs1175649 T was associated with a significant increased WNT2B mRNA expression level (P = 4.4E-8) in an additive genetic model (Figure 3c), which confirms our initial findings.
Figure 3. Associations between rs1175649 genotypes and WNT2B mRNA expression levels.
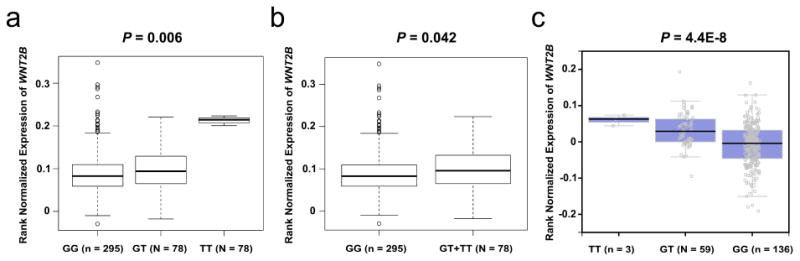
The expression quantitative trait loci analysis (eQTLs) for WNT2B rs1175649 in 373 Europeans from the 1000 Genomes Project in the additive model (a) and in the dominant model (b). (c) The expression quantitative trait loci analysis (eQTLs) from the Genotype-Tissue Expression (GTEx) project for WNT2B rs1175649 in an additive genetic model.
Discussion
In the present study, we found that genetic variants WNT2B rs1175649 and BTRC rs61873997, were likely to independently or jointly modulate the survival of CM patients. Moreover, rs1175649 was found to influence WNT2B mRNA expression levels. These findings suggest that Wnt pathway gene variants may have biological roles in CM progression, possibly through a mechanism of modulating expression of these genes.
Although the prevalence of the Wnt signaling pathway deregulation in melanoma is known, the effect of this deregulation is complex. Several studies have shown that nuclear β-catenin levels decrease as melanoma progresses and that this decrease is associated with a poor prognosis (Bachmann et al., 2005; Meyer et al., 2012). Conversely, other studies reported that the Wnt pathway activation stimulates cell growth (Widlund et al., 2002), bypasses senescence (Delmas et al., 2007), promotes tumor cell survival and chemoresistance (Sinnberg et al., 2011), resulting in a worsened prognosis (Kielhorn et al., 2003; Murakami et al., 2001). Likewise, β-catenin activation in melanocytes might restrict cellular motility and migration and promote lung metastasis in the NRAS-driven melanoma murine model (Gallagher et al., 2013). Besides, β-catenin activation substantially increases metastasis to the lungs in GEMMs with mutant Braf and inactivated Pten, indicating that β-catenin may have conflicting roles in the metastasis of melanoma, repressing migration while promoting metastasis (Damsky et al., 2011). Therefore, clinical decisions should be made by taking into the consideration the context-dependent role of the Wnt signaling in melanoma development.
In the present study, we found some significant associations of CMSS with genetic variants in WNT2B and BTRC. Patients with increasing the NUG score, which represents the combined effect of unfavorable genotypes of these genetic variants, had poorer survival. Notably, the effect was consistent across different analyses and most subgroup comparisons, supporting a robust association of the NUG score with CM survival. Furthermore, we believe that our results are likely biologically plausible. The genotype-phenotype correlation indicates that WNT2B expression levels may be modulated by rs1175649, which provides a possible explanation for the observed association with CM survival.
WNT2B is located on chromosome 1p13, participates in the canonical Wnt pathway and transduces signals through Frizzled and LRP5/LRP6 receptors, promoting the release of β-catenin from the AXIN-APC degradation complex (Korinek et al., 1997). WNT2B has been involved in several aspects of tumor biology, including metastatic ability and drug resistance. For example, silencing of WNT2B inhibits metastasis and enhances cisplatin sensitivity in ovarian cancer cells (Wang et al., 2012); WNT2B has also been shown to be overexpressed in two cisplatin-resistant oral cancer cell lines, and the silencing of WNT2B suppresses the proliferation and colony forming ability of cancer cells (Li et al., 2015).
BTRC is located on chromosome 10q24.32. As a cytoplasmic negative regulator of the Wnt pathway, BTRC is important for the ubiquitin-mediated degradation of β-catenin by the proteasome. Although we could find little prior evidence linking BTRC to CM prognosis, it has been shown to influence prognosis in other cancers. For instance, it is reported that down-regulation of BTRC were associated with poor prognosis in nasopharyngeal carcinoma patients (Yan et al., 2015). Besides, BTRC gene expression levels predicted postoperative recurrence of lung cancer in patients (Tseng et al., 2010).
The current study has several important strengths. A major strength of this study is the comprehensive analyses of associations between SNPs in the Wnt pathway genes and CM survival using two published GWASs datasets that had strict quality control procedures. In our analyses, we adjusted for variables that could confound our observations of a genetic effect on CMSS. Although compared with the MDACC survival analysis (with adjustment for age, sex, Breslow thickness, regional/distant metastasis, ulceration and mitotic rate), the adjusted information was only available for age and sex in the Harvard study, and no heterogeneity was observed in the results from the two studies, which indicates that the observed effect of each SNP on CMSS from the two studies is consistent. We also performed the FPRP correction to assess the possibility of false-positive associations as a result of multiple testing. Furthermore, the pathway-based analysis allowed us to explore the polygenic effects and properly identifying the truly functional variants from the available high-dimensional data and also can improve detection of the combined effects of these SNPs on survival. Our findings demonstrated the potential importance of assessing CM prognosis by combining clinicopathological characteristics with the added genetic information.
However, the present study has some limitations. First, among our participants, treatment plans likely varied, and we could not control for the differences in treatment. Nevertheless, systemic therapies have had only a modest impact on patient survival, and it is highly unlikely that treatment plans varied systematically by germline genotype. We also performed stratified analyses by regional/distant metastasis, Breslow thickness, ulceration and mitosis to minimize the effect of different treatments. The second caveat is that the prognosis-predicting model was built in a US non-Hispanic white population, and therefore its application to different ethnic groups requires further investigation and validation. Another limitation is that our analysis did not take into account gene-gene interactions that widely exist in cancer. Our ability to perform such an evaluation was limited by the sample size and study power, given the relatively limited number of outcome events observed in the study population.
In summary, we performed a comprehensive assessment of genetic variants in genes involved in the Wnt pathway in two independent GWAS datasets, identified two SNPs (WNT2B rs1175649 and BTRC rs61873997) that may independently or jointly affect the survival of CM patients. However, our findings need to be validated in an independent, larger patient population, preferably with different ethnic groups.
Materials and Methods
Study population
This study contained a discovery dataset (the GWAS dataset from MDACC) and a replication dataset [the Nurses'Health Study (NHS) and the Health Professionals Follow-up Study (HPFS) from the Harvard University]. The study protocols were approved by Institutional Review Boards at both MD Anderson and Brigham and Women's Hospital with a written informed consent obtained from each of the subjects.
Discovery Dataset
Participant recruitment and patients' characteristics in the MDACC GWAS study have been described elsewhere (Amos et al., 2011; Li et al., 2013). In brief, patients were accrued for a hospital-based case-control study of CM at MDACC between March 1998 and August 2008. All cases were diagnosed with histologically confirmed cutaneous melanoma. The stage of the disease and length of the follow-up were determined from the date of diagnosis. The final analysis included 858 patients for whom information for clinicopathological variables was available. All individuals provided a written informed consent under an Institutional Review Board-approved protocol. The genotype data used in this study can be accessed using the National Center for Biotechnology Information (NCBI) Database of Genotypes and Phenotypes (dbGaP; http://www.ncbi.nlm.nih.gov/gap), with the study accession number phs000187.v1.p1. The detailed genotyping information and data quality control have been reported (Simard et al., 2012). Genome-wide imputation was performed using the MACH software program based on 1000 Genomes Project, phase I V2 CEU data (Li et al., 2010).
Replication Dataset
The Harvard dataset consisted of two studies: NHS and HPFS. Sampling, genotyping and quality control procedures have been described previously (Song et al., 2012). Information on melanoma development was first collected in the 1984 questionnaire and 1986 questionnaire of NHS and HPFS, respectively. Eligible cases in both the NHS and HPFS cohorts were participants with histopathologically confirmed invasive melanoma, diagnosed at any time after baseline up to the 2008 follow-up cycle for both cohorts. All subjects were US non-Hispanic white. In the final analysis, 409 patients were kept in the data after quality control. Genotyping in the Harvard dataset was performed using the Illumina HumanHap610 array. Imputation was performed based on genotyped SNPs and haplotype information from phase II HapMap CEU data using the program MACH (Biernacka et al., 2009). Only SNPs with imputation quality r2 > 0.8 were included, and a total of 1,579,307 SNPs passed through the filter.
SNP selection for the Wnt pathway analysis
On the basis of the databases of KEGG (http://www.genome.jp/kegg/) and Biocarta (http://www.biocarta.com/), we selected the following 151 Wnt pathway genes that are located on autosomes. Common SNPs (minor allele frequency ≥ 0.05, genotyping rate ≥ 95% and Hardy– Weinberg equilibrium P value ≥ 0.00001) within these genes or their ± 2 kb flanking regions were selected for association analysis. As a result, 19,830 common SNPs in the Wnt pathway were extracted from our CM GWAS dataset. For illustrative purposes, a flow chart of detailed SNP selection among Wnt pathway genes is shown in Figure 1a.
Statistical methods
Further details are available in the Supplementary Materials (online).
Supplementary Material
Acknowledgments
We thank the individuals who participated in this project. We thank the Johns Hopkins University Center for Inherited Disease Research for conducting high-throughput genotyping for this study. We would like to thank the participants and staff of the Nurses' Health Study and Health Professionals Follow-up Study for their valuable contributions as well as the following state cancer registries for their assistance: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND,OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data. This work was support by NIH/NCI R01 CA100264, 2P50CA093459, P30CA014236, R01 CA49449, P01 CA87969, UM1 CA186107, UM1 CA167552, The University of Texas MD Anderson Cancer Center Various Donors Melanoma and Skin Cancers Priority Program Fund, the Miriam and Jim Mulva Research Fund, the McCarthy Skin Cancer Research Fund, the Marit Peterson Fund for Melanoma Research and National Natural Science Foundation of China (no.81502863).
Abbreviation
- CM
cutaneous melanoma
- SNPs
single-nucleotide polymorphisms
- GWAS
genome-wide association study
- WNT2B
Wnt family member 2B
- BTRC
beta-transducin repeat containing E3 ubiquitin protein ligase
- CMSS
cutaneous melanoma-specific survival
- HR
hazards ratio
- adjHR
adjusted hazards ratio
- CI
confidence interval
- FPRP
false positive report probability
- NUG
number of unfavorable genotypes
Footnotes
Conflict of Interest: The authors state no conflict of interest.
Supplementary Material: Supplementary material is linked to the online version of the paper at http://www.nature.com/jid
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amos CI, Wang LE, Lee JE, Gershenwald JE, Chen WV, Fang S, et al. Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum Mol Genet. 2011;20:5012–23. doi: 10.1093/hmg/ddr415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann IM, Straume O, Puntervoll HE, Kalvenes MB, Akslen LA. Importance of P-cadherin, beta-catenin, and Wnt5a/frizzled for progression of melanocytic tumors and prognosis in cutaneous melanoma. Clin Cancer Res. 2005;11:8606–14. doi: 10.1158/1078-0432.CCR-05-0011. [DOI] [PubMed] [Google Scholar]
- Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27:6199–206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berwick DC, Harvey K. The importance of Wnt signalling for neurodegeneration in Parkinson's disease. Biochem Soc Trans. 2012;40:1123–8. doi: 10.1042/BST20120122. [DOI] [PubMed] [Google Scholar]
- Biernacka JM, Tang R, Li J, McDonnell SK, Rabe KG, Sinnwell JP, et al. Assessment of genotype imputation methods. BMC Proc. 2009;3(Suppl 7):S5. doi: 10.1186/1753-6561-3-s7-s5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien AJ, Moore EC, Lonsdorf AS, Kulikauskas RM, Rothberg BG, Berger AJ, et al. Activated Wnt/beta-catenin signaling in melanoma is associated with decreased proliferation in patient tumors and a murine melanoma model. Proc Natl Acad Sci U S A. 2009;106:1193–8. doi: 10.1073/pnas.0811902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilosi M, Poletti V, Zamo A, Lestani M, Montagna L, Piccoli P, et al. Aberrant Wnt/beta-catenin pathway activation in idiopathic pulmonary fibrosis. Am J Pathol. 2003;162:1495–502. doi: 10.1016/s0002-9440(10)64282-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damsky WE, Curley DP, Santhanakrishnan M, Rosenbaum LE, Platt JT, Gould Rothberg BE, et al. beta-catenin signaling controls metastasis in Braf-activated Pten-deficient melanomas. Cancer Cell. 2011;20:741–54. doi: 10.1016/j.ccr.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de La Coste A, Romagnolo B, Billuart P, Renard CA, Buendia MA, Soubrane O, et al. Somatic mutations of the beta-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc Natl Acad Sci U S A. 1998;95:8847–51. doi: 10.1073/pnas.95.15.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas V, Beermann F, Martinozzi S, Carreira S, Ackermann J, Kumasaka M, et al. Beta-catenin induces immortalization of melanocytes by suppressing p16INK4a expression and cooperates with N-Ras in melanoma development. Genes Dev. 2007;21:2923–35. doi: 10.1101/gad.450107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong QZ, Zhang XF, Zhao Y, Jia HL, Zhou HJ, Dai C, et al. Osteopontin promoter polymorphisms at locus -443 significantly affect the metastasis and prognosis of human hepatocellular carcinoma. Hepatology. 2013;57:1024–34. doi: 10.1002/hep.26103. [DOI] [PubMed] [Google Scholar]
- Gallagher SJ, Rambow F, Kumasaka M, Champeval D, Bellacosa A, Delmas V, et al. Beta-catenin inhibits melanocyte migration but induces melanoma metastasis. Oncogene. 2013;32:2230–8. doi: 10.1038/onc.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Orte E, Saenz-Narciso B, Moreno S, Cabello J. Multiple functions of the noncanonical Wnt pathway. Trends Genet. 2013;29:545–53. doi: 10.1016/j.tig.2013.06.003. [DOI] [PubMed] [Google Scholar]
- Hari L, Brault V, Kleber M, Lee HY, Ille F, Leimeroth R, et al. Lineage-specific requirements of beta-catenin in neural crest development. J Cell Biol. 2002;159:867–80. doi: 10.1083/jcb.200209039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarrola-Villava M, Kumar R, Nagore E, Benfodda M, Guedj M, Gazal S, et al. Genes involved in the WNT and vesicular trafficking pathways are associated with melanoma predisposition. Int J Cancer. 2015;136:2109–19. doi: 10.1002/ijc.29257. [DOI] [PubMed] [Google Scholar]
- Inestrosa NC, Montecinos-Oliva C, Fuenzalida M. Wnt signaling: role in Alzheimer disease and schizophrenia. J Neuroimmune Pharmacol. 2012;7:788–807. doi: 10.1007/s11481-012-9417-5. [DOI] [PubMed] [Google Scholar]
- Jemal A, Saraiya M, Patel P, Cherala SS, Barnholtz-Sloan J, Kim J, et al. Recent trends in cutaneous melanoma incidence and death rates in the United States, 1992-2006. J Am Acad Dermatol. 2011;65:S17–25 e1-3. doi: 10.1016/j.jaad.2011.04.032. [DOI] [PubMed] [Google Scholar]
- Kielhorn E, Provost E, Olsen D, D'Aquila TG, Smith BL, Camp RL, et al. Tissue microarray-based analysis shows phospho-beta-catenin expression in malignant melanoma is associated with poor outcome. Int J Cancer. 2003;103:652–6. doi: 10.1002/ijc.10893. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–7. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–11. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Yin M, Wang LE, Amos CI, Zhu D, Lee JE, et al. Polymorphisms of nucleotide excision repair genes predict melanoma survival. J Invest Dermatol. 2013;133:1813–21. doi: 10.1038/jid.2012.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SJ, Yang XN, Qian HY. Antitumor effects of WNT2B silencing in GLUT1 overexpressing cisplatin resistant head and neck squamous cell carcinoma. Am J Cancer Res. 2015;5:300–8. [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Fuchs TJ, Bosserhoff AK, Hofstadter F, Pauer A, Roth V, et al. A seven-marker signature and clinical outcome in malignant melanoma: a large-scale tissue-microarray study with two independent patient cohorts. PLoS One. 2012;7:e38222. doi: 10.1371/journal.pone.0038222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Toda S, Fujimoto M, Ohtsuki M, Byers HR, Etoh T, et al. Constitutive activation of Wnt/beta-catenin signaling pathway in migration-active melanoma cells: role of LEF-1 in melanoma with increased metastatic potential. Biochem Biophys Res Commun. 2001;288:8–15. doi: 10.1006/bbrc.2001.5719. [DOI] [PubMed] [Google Scholar]
- Palacios J, Gamallo C. Mutations in the beta-catenin gene (CTNNB1) in endometrioid ovarian carcinomas. Cancer Res. 1998;58:1344–7. [PubMed] [Google Scholar]
- Purdue MP, Freeman LE, Anderson WF, Tucker MA. Recent trends in incidence of cutaneous melanoma among US Caucasian young adults. J Invest Dermatol. 2008;128:2905–8. doi: 10.1038/jid.2008.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani P, Takeo M, Chou W, Myung P, Bosenberg M, Chin L, et al. Coordinated activation of Wnt in epithelial and melanocyte stem cells initiates pigmented hair regeneration. Cell. 2011;145:941–55. doi: 10.1016/j.cell.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimm DL, Caca K, Hu G, Harrison FB, Fearon ER. Frequent nuclear/cytoplasmic localization of beta-catenin without exon 3 mutations in malignant melanoma. Am J Pathol. 1999;154:325–9. doi: 10.1016/s0002-9440(10)65278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinner S. Wnt-signalling and the metabolic syndrome. Horm Metab Res. 2009;41:159–63. doi: 10.1055/s-0028-1119408. [DOI] [PubMed] [Google Scholar]
- Schramm SJ, Mann GJ. Melanoma prognosis: a REMARK-based systematic review and bioinformatic analysis of immunohistochemical and gene microarray studies. Mol Cancer Ther. 2011;10:1520–8. doi: 10.1158/1535-7163.MCT-10-0901. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- Simard EP, Ward EM, Siegel R, Jemal A. Cancers with increasing incidence trends in the United States: 1999 through 2008. CA Cancer J Clin. 2012;62:118–28. doi: 10.3322/caac.20141. [DOI] [PubMed] [Google Scholar]
- Sinnberg T, Menzel M, Ewerth D, Sauer B, Schwarz M, Schaller M, et al. beta-Catenin signaling increases during melanoma progression and promotes tumor cell survival and chemoresistance. PLoS One. 2011;6:e23429. doi: 10.1371/journal.pone.0023429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F, Qureshi AA, Zhang J, Amos CI, Lee JE, Wei Q, et al. Exonuclease 1 (EXO1) gene variation and melanoma risk. DNA Repair (Amst) 2012;11:304–9. doi: 10.1016/j.dnarep.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng RC, Lee SH, Hsu HS, Chen BH, Tsai WC, Tzao C, et al. SLIT2 attenuation during lung cancer progression deregulates beta-catenin and E-cadherin and associates with poor prognosis. Cancer Res. 2010;70:543–51. doi: 10.1158/0008-5472.CAN-09-2084. [DOI] [PubMed] [Google Scholar]
- Wang H, Fan L, Xia X, Rao Y, Ma Q, Yang J, et al. Silencing Wnt2B by siRNA interference inhibits metastasis and enhances chemotherapy sensitivity in ovarian cancer. Int J Gynecol Cancer. 2012;22:755–61. doi: 10.1097/IGC.0b013e3182540284. [DOI] [PubMed] [Google Scholar]
- Widlund HR, Horstmann MA, Price ER, Cui J, Lessnick SL, Wu M, et al. Beta-catenin-induced melanoma growth requires the downstream target Microphthalmia-associated transcription factor. J Cell Biol. 2002;158:1079–87. doi: 10.1083/jcb.200202049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Kraft P, Stolzenberg-Solomon R, Steplowski E, Brotzman M, Xu M, et al. Genome-wide association study of survival in patients with pancreatic adenocarcinoma. Gut. 2014;63:152–60. doi: 10.1136/gutjnl-2012-303477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Zeng Z, Gong Z, Zhang W, Li X, He B, et al. EBV-miR-BART10-3p facilitates epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma by targeting BTRC. Oncotarget. 2015;6:41766–82. doi: 10.18632/oncotarget.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Xie W, Mostaghel E, Nakabayashi M, Werner L, Sun T, et al. SLCO2B1 and SLCO1B3 may determine time to progression for patients receiving androgen deprivation therapy for prostate cancer. J Clin Oncol. 2011;29:2565–73. doi: 10.1200/JCO.2010.31.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawel RH, Chiappa SA, Allen C, Raffel C. Sporadic medulloblastomas contain oncogenic beta-catenin mutations. Cancer Res. 1998;58:896–9. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


