Summary
Although hydrogen sulfide (H2S) is perhaps best known as a toxic gas, the electron-rich H2S functions as an energy source and electron donor for chemolithotrophic and photosynthetic bacteria, via sulfide oxidation, and is a universal substrate for cysteine biosynthesis. These distinct harmful and beneficial roles of H2S suggest the need to “sense” prevailing concentrations of sulfide and downstream reactive sulfur species (RSS) and regulate the expression of genes mediating sulfide homeostasis. The paper by Li et al. in this issue of Molecular Microbiology adds Cupriavidus FisR to an expanding repertoire of regulatory mechanisms that bacteria have evolved to sense cellular RSS and mitigate their deleterious effects.
It is well established that many organisms are capable of using hydrogen sulfide (H2S, HS−, S2−) as a source of reducing equivalents via sulfide oxidation, which provides sufficient energy to power respiration, energy (ATP) production, and for photosynthetic organisms, photosynthetic electron flow. For example, tetrathionate, a sulfide oxidation product formed by the condensation of two thiosulfate molecules (see Fig. 1A) by thiosulfate dehydrogenases (Brito et al., 2015) can function as a terminal electron acceptor; indeed, in the lumen of the inflamed gut, anaerobic respiratory tetrathionate reduction provides a growth advantage for Salmonella ssp. (Winter et al., 2010). Recent work in eukaryotes describes H2S as a vasorelaxant and signaling molecule (Mustafa et al., 2009, Gadalla & Snyder, 2010) whose biological impact may be intertwined with adaptation to reactive nitrogen species (RNS), including nitric oxide (Kolluru et al., 2015, Cortese-Krott et al., 2015). Although a defined role for microbial H2S signaling has yet to be described, H2S/RNS crosstalk may provide protection against the effects of antibiotic stress (Shatalin et al., 2011).
Figure 1.
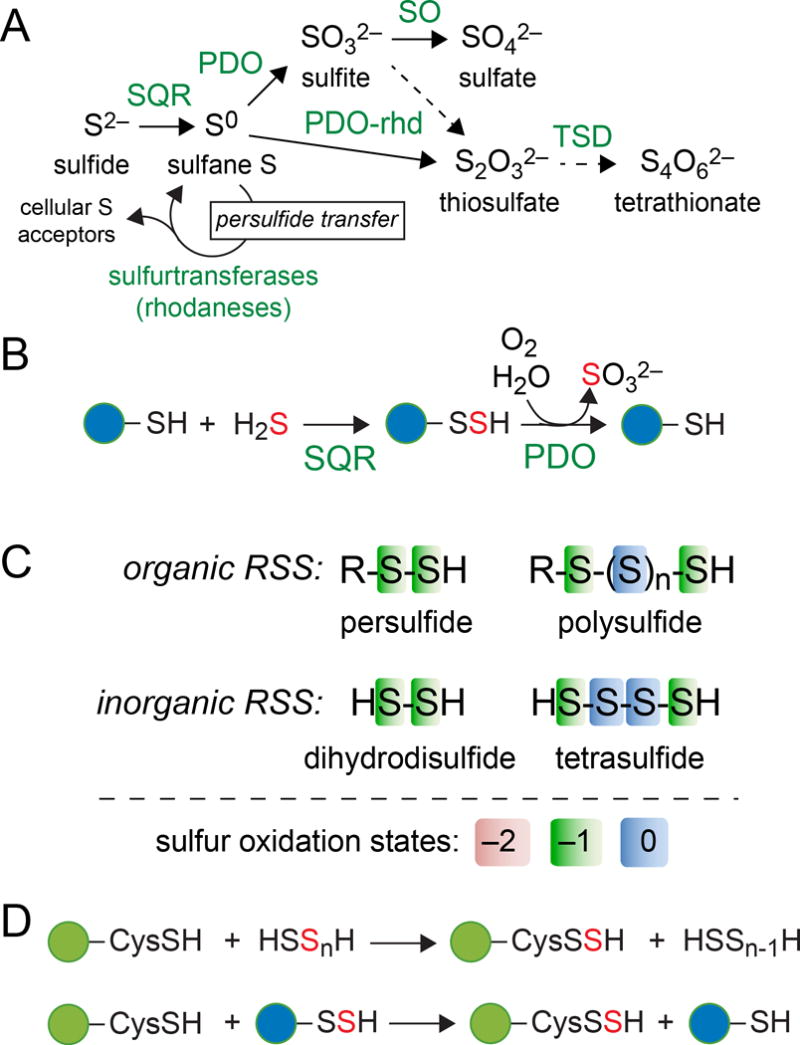
Sulfide oxidation relevant to cellular sulfide homeostasis and sensing of the reactive sulfur species (RSS) in cells. (A) Step-wise oxidation of the most reduced form of S, sulfide (H2S, HS− and S2−) to increasingly more oxidized forms of S, sulfate and tetrathionate, with the enzymes that catalyze these oxidations indicated: SQR, sulfide:quinone oxidoreductase; PDO-persulfide dioxygenase; PDO-rhd, PDO-rhodanese fusion; SO, sulfite oxidase; TSD, thiosulfate dehydrogenase. Sulfurtransferases catalyze persulfide transfer reactions (see panel D) to assimilate bioactive sulfur. (B) Coordinate activity of the SQR and PDO catalyze the oxidation of H2S to sulfite or thiosulfate, with a LMW thiol (blue circle) as an intermediate. (C) Reactive sulfur species organized as organic and inorganic species. The simplest organic RSS is a LMW thiol persulfide, while the simplest inorganic polysulfide is dihydrodisulfide, HSSH (Li et al., 2017). Organic and inorganic polysulfide anions feature S chains (n≥1) with the internal S atoms formally S0 (see panel A). Not shown are organic polydisulfides of the general formula, RS-Sn-SR (n≥1), sulfite and thiosulfate. (D) Examples of potential sulfur transfer reactions between proteome (green circles) or LMW thiols and inorganic (top) and organic (bottom) RSS.
H2S is a potent inhibitor of cytochrome oxidase (Cooper & Brown, 2008) and forms stable coordination complexes with transition metals (Luther et al., 1996) thus potentially limiting their bioavailability. These beneficial yet toxic attributes of H2S therefore suggest the need to regulate intracellular concentrations of this strongly nucleophilic and reducing molecule. This is particularly so since H2S is produced endogenously in many organisms (from bacteria to vertebrates) by two enzymes of the transsulfuration pathway, cystathionine-β-synthase (CBS) and cystathionine-γ-lyase (CSE) (Yadav et al., 2016). Two additional enzymes, 3-mercaptopyruvate sulfurtransferase, which functions in cysteine catabolism, and sulfite reductase, which functions in dissimilatory sulfate reduction, also generate H2S (Yadav et al., 2013, Santos et al., 2015). Sulfide is a substrate for cysteine synthase, which synthesizes cysteine from an acetyl-CoA activated serine, O-acetylserine (OAS); cysteine is the precursor for all S-containing metabolites in the cell, including the major reducing thiol(s), e.g., glutathione or bacillithiol, coenzyme A, thiamine pyrophosphate (TPP), biotin and lipoic acid.
The principal pathway for hydrogen sulfide detoxification and homeostasis is via step-wise oxidation to less toxic products (Libiad et al., 2014). The first step in this process is the two-electron oxidation of sulfide to sulfane or “sulfur-bonded” sulfur (Fig. 1) conjugated to a suitable acceptor nucleophile. When a low molecular weight (LMW) thiolate is the acceptor, sulfide:quinone oxidoreductase (SQR) generates a LMW thiol persulfide (Fig. 1B), with the reducing equivalents shuttled into the quinone pool. Flavocytochrome c:sulfide dehydrogenases (FCSD) catalyze precisely the same reaction as SQRs, but use cytochrome c instead of an oxidized quinone as the electron acceptor (Marcia et al., 2010). Inorganic polysulfides can also be biosynthesized by SQRs when H2S is used as the nucleophilic acceptor (Jackson et al., 2012) (Fig. 1C). LMW thiol persulfides are then subject to further oxidation to sulfite or thiosulfate (see Fig. 1A,B), with the release of the free LMW thiol, by non-heme FeII persulfide dioxygenases (PDO) (Kabil & Banerjee, 2012, Liu et al., 2014, Sattler et al., 2015, Shen et al., 2015, Pettinati et al., 2015), enzymes often mis-annotated as metallo- β-lactamase-like hydrolases (Blh) (Sattler et al., 2015). Further oxidation of sulfite and thiosulfate to sulfate and tetrathionate, respectively, can occur in some organisms (Fig. 1A). Sulfurtransferases or rhodaneses often function as persulfide (activated sulfur) carriers in these processes (Higgins et al., 2015, Mueller, 2006), likely facilitating the production of LMW thiol persulfides (Xin et al., 2016) (Fig. 1A). LMW thiol persulfides and inorganic polysulfides, collectively termed reactive sulfur species (RSS) (Fig. 1C), are detectable in bacterial cells (Shen et al., 2016, Peng et al., 2017) and may well engage in persulfide or RSS transfer reactions (Mishanina et al., 2015) with other small molecule or proteome thiolate acceptors (Wedmann et al., 2016, Gao et al., 2015) (Fig. 1D).
In new work published in this issue of Molecular Microbiology, Liu, Xun and coworkers describe the characterization of FisR from the heterotroph Cupriavidus pinatubonensis JMP134, a Gram-negative β-proteobacterium of industrial and biotechnological importance (Li et al., 2017) (Fig. 2A). FisR is shown to be a transcriptional activator of the divergently transcribed operon, encoding FisR itself and a two–gene cluster of sqr and pdo in C. pinatubonensis. The sqr-pdo cluster, encoding a bona fide SQR and PDO as characterized previously by the same group (Xin et al., 2016), is transcriptionally inducible in FisR-dependent manner by Na2S and inorganic polysulfides (see Fig. 1C; primarily HSSH in this work) and C. pinatubonensis lacking pdo and sqr fails to oxidize Na2S and exhibits a growth defect on sulfide-containing media. Finally, FisR-like regulators are somewhat widespread among proteobacteria and could be further classified into two major clusters on the basis of amino acid similarity, corresponding to Burkholderia spp. and Pseudomonas spp. clades (Li et al., 2017).
Figure 2.
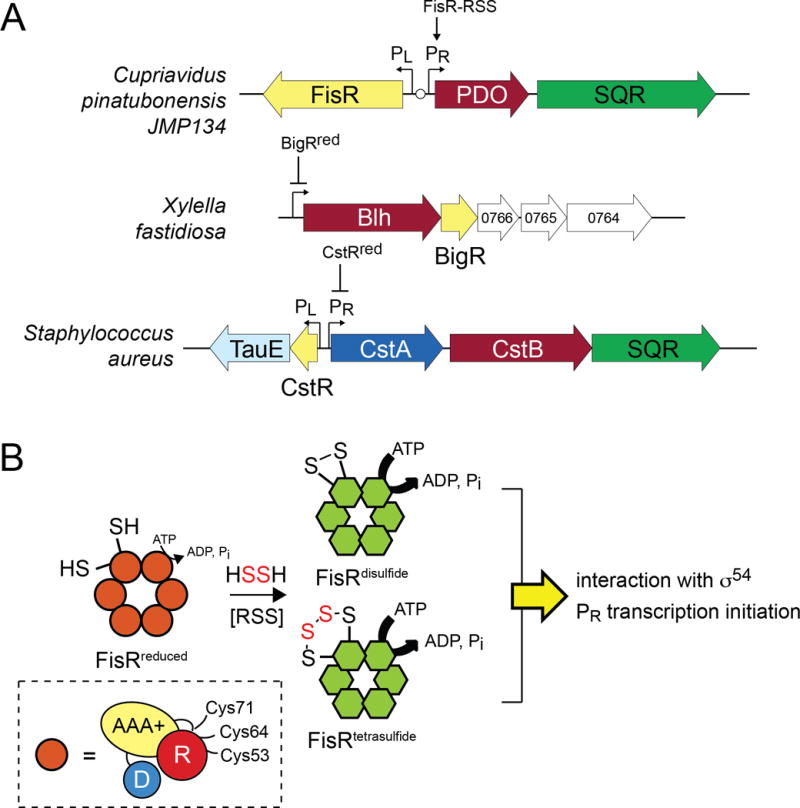
(A) Schematic representations of the operons regulated by FisR (top) (Li et al., 2017), BigR (middle) (Barbosa & Benedetti, 2007) and CstR (bottom) (Grossoehme et al., 2011) (shaded yellow). FisR is an RSS-activated transcriptional activator, while BigR and CstR are repressors in their reduced states; RSS mediates transcriptional depression. All three operons encode a known or presumed persulfide dioxygenase (PDO, shaded maroon), while two encode SQRs (shaded green). Blh, metallo-β-lactamase-like hydrolase. (B) Cartoon representation of hexameric FisR, which in its reduced state has lower ATPase activity, and upon incubation with HSSH (and likely other RSS), forms a mixture of disulfide and tetrasulfide crosslinked species with higher ATPase activity; this in turn activates transcription from the PR promoter (see panel A) via a presumed interaction with RNA polymerase bound σ54. All subunits within the hexamer are anticipated to form crosslinks, but just one is shown for clarity. Inset, domain organization of FisR, with the N-terminal regulatory (R), middle ATPase domain (AAA+) and C-terminal DNA binding domain (D) indicated (based on Bush et al., 2015). The three conserved cysteine residues are indicated located in the R domain or a linker that connects the R and AAA+ domains, with a major site of intrasubunit crosslinking shown to be between Cys53 and Cys64 (Li et al., 2017).
The remarkable findings that connect and contrast the work of Li et al. to previous work are two-fold. First, the striking contrast: unlike RSS-sensing transcriptional repressors already characterized (vide infra), FisR is a member of the bacterial enhancer binding protein (bEBP) family (Fig. 2B). These are σ54-dependent transcription factors that harbor three consensus domains: an N-terminal regulatory or receiver (R) domain that is capable of “sensing” the inducer, a middle AAA+ domain (AAA+) that hydrolyzes ATP, and a C-terminal DNA binding domain (D) which directs a specific interaction with an operator (enhancer) sequence far upstream of the promoter (Bush & Dixon, 2012) (Fig. 2B, inset). This “action at a distance” is mediated by integration host factor (IHF), which introduces a bend in the DNA, positioning bound FisR in close proximity to the promotor-bound RNA polymerase, with ATP hydrolysis stimulating open complex formation and transcription initiation (Hartman et al., 2016). The striking similarity to previous work (Luebke et al., 2014, Shimizu et al., 2017) is that FisR contains three conserved Cys residues (C53, C64 and C71) in the R domain and reduced FisR is activated by exposure to glutathione persulfide and inorganic polysulfide RSS (see Fig. 1C). Major products of the reaction as analyzed by tandem mass spectrometry (Luebke et al., 2014) include a disulfide linkage between C53 and C64, but not C53 and C71, as well as tetrasulfide linkage between the same two cysteine residues (Fig. 2B). These oxidative modifications do not impact the DNA-binding activity or oligomerization state of FisR [FisR, like other bEBPs is an obligatory homohexameric ring structure (Bush et al., 2015), but in this case is in equilibrium with tetramers, dimers and monomers], but do increase the ATPase activity modestly, a stimulation that appears to be abolished in the C53S, C64S and C71S mutants (Li et al., 2017). Left for future work is the mechanism by which RSS-dependent oxidative modification of FisR in the R domain impacts transcriptional activation of the pdo-sqr promoter (Fig. 2B).
This study by Li, Xun and colleagues provides new insights into the evolution of sulfide sensing, homeostasis and detoxification in microbes (Li et al., 2017) (Fig. 2A). Earlier work on the regulation of the sulfur oxidation (sox) genes in the α-proteobacterium Pseudaminobacter salicylatoxidans and related bacteria (Mandal et al., 2007) led to the identification of SoxR, a member of the ubiquitous arsenic repressor (ArsR) family of the homodimeric winged helical repressors that specifically harbors two conserved cysteine residues (Ma et al., 2009). SoxR was next essentially rediscovered and structurally characterized as BigR from the non-sulfur oxidizing plant pathogens Xylella fastidiosa and Agrobacterium tumafaciens where it regulates the expression of what is predicted to be a type II PDO (Blh) (Barbosa & Benedetti, 2007, Sattler et al., 2015) (Fig. 2A). BigR and regulated gene products are reported to impact hypoxic growth in biofilms via unknown chemical and physiological mechanisms (Guimaraes et al., 2011). This was followed by our characterization of Staphylococcus aureus CstR (Luebke et al., 2014, Grossoehme et al., 2011), a dithiol-containing transcriptional repressor like BigR, but derived from an entirely different family of homotetrameric disc-shaped bacterial repressors evolutionarily related to the copper-sensitive operon repressor (CsoR) (Liu et al., 2007) and the E. coli NiII efflux regulator RcnR (Higgins & Giedroc, 2014, Iwig et al., 2006) (Fig. 2A). The CstR-regulated cst operon encodes a sulfurtransferase CstA, a PDO-rhodanese fusion protein CstB, and an SQR (Fig. 2A) that is strongly induced by sulfide or inorganic polysulfides added to cells (Luebke et al., 2014). Reduced CstR reacts directly and rapidly with LMW thiol persulfides and sodium tetrasulfide (S4) to form a mixture of di-, tri- and tetrasulfide crosslinked species involving two conserved cysteines, thus establishing the scope of chemistry that was possible with these RSS (Luebke et al., 2014). Strikingly, a recently described master regulator of sulfide-dependent photosynthesis in Rhodobacter capsulatus, sulfide:quinone oxidoreductase repressor (SqrR), a BigR- and SoxR-like repressor, carries out precisely analogous chemistry when incubated with glutathione persulfide by forming di-, tri- and tetrasulfide crosslinked species between the two conserved Cys residues (Shimizu et al., 2017).
This new work establishes that FisR engages in CstR- and SqrR-like dithiol oxidative chemistry, on yet a third distinct molecular scaffold and therefore raises a number of important questions (Li et al., 2017). For example, to what degree and what conserved aspects of protein microenvironment render the cysteine pair in these three structural scaffolds selective for RSS? Clearly, a minimum of two cysteine residues must be involved, including an attacking thiol, which is selective for RSS over other potential oxidants, and a resolving thiol which would create a disulfide linkage with the release of H2S; further rounds of per- and polysulfide attack of this regulator disulfide or direct initial attack of a polysulfide species would create a range of di-, tri- and tetrasulfide crosslinked products (Mishanina et al., 2015) as experimentally observed (Luebke et al., 2014, Shimizu et al., 2017, Li et al., 2017). The fact that three Cys are functionally required and conserved in most FisRs suggests the possibility of thiol-disulfide exchange after initial formation of a non-native disulfide, as pointed out by the authors (Li et al., 2017). In fact, R. capsulatus SqrR harbors a third cysteine, which although not required in cells for RSS sensing, changes the scope of oxidative reaction products obtained upon incubation with glutathione persulfide (Shimizu et al., 2017). Do organic or inorganic per- and polysulfide species (see Fig. 1) play the major cellular role in RSS sensing? Does this vary in different bacteria? Li et al argue that polysulfides are likely physiological pdo-sqr inducers in C. pinatubonensis given the low Km for sulfide by microbial SQRs (Xin et al., 2016, Shen et al., 2016) and the potent electrophilicity of a tautomer of HSSH, thiosulfoxide HS(=S)H, that is capable of reacting directly with thiolates (Ono et al., 2014).
Finally, both FisR and CstR orchestrate what is essentially an acute phase response to H2S toxicity, with rapid induction following sulfide exposure, with a return to basal transcription 30 minutes post-induction (Luebke et al., 2014, Li et al., 2017). The origin of these induction kinetics is unknown, and may not track with RSS clearance in cells (Peng et al., 2017). Perhaps cells have an overcapacity to “buffer” sulfide as RSS so as to protect critical physiology under conditions of sulfide toxicity while deploying persulfide or RSS transfer chemistries (see Fig. 1D) to rapidly remodel sulfur speciation as needed. This would allow an organism to harness the chemical potential of H2S while mitigating collateral damage associated with sulfide toxicity. Precisely what that damage is, however, is currently unknown in any organism. In any case, the characterization of this new proteobacterial sulfide-responsive regulator suggests that sulfide homeostasis is evolutionarily conserved and provides an organism with a means to utilize both exogenous sulfide sources and endogenous RSS that may be important at the host-bacterial pathogen interface (Luhachack & Nudler, 2014).
Acknowledgments
Work in the author’s laboratory on bacterial sulfide homeostasis is supported by a grant from the NIH (GM118157).
References
- Barbosa RL, Benedetti CE. BigR, a transcriptional repressor from plant-associated bacteria, regulates an operon implicated in biofilm growth. J bacteriol. 2007;189:6185–6194. doi: 10.1128/JB.00331-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito JA, Denkmann K, Pereira IA, Archer M, Dahl C. Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum: Structural and functional insights into thiosulfate oxidation. J Biol Chem. 2015;290:9222–9238. doi: 10.1074/jbc.M114.623397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush M, Dixon R. The role of bacterial enhancer binding proteins as specialized activators of σ54-dependent transcription. Microbiol Mol Biol Rev. 2012;76:497–529. doi: 10.1128/MMBR.00006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush M, Ghosh T, Sawicka M, Moal IH, Bates PA, Dixon R, et al. The structural basis for enhancer-dependent assembly and activation of the AAA transcriptional activator NorR. Mol Microbiol. 2015;95:17–30. doi: 10.1111/mmi.12844. [DOI] [PubMed] [Google Scholar]
- Cooper CE, Brown GC. The inhibition of mitochondrial cytochrome oxidase by the gases carbon monoxide, nitric oxide, hydrogen cyanide and hydrogen sulfide: chemical mechanism and physiological significance. J Bioenerg Biomembr. 2008;40:533–539. doi: 10.1007/s10863-008-9166-6. [DOI] [PubMed] [Google Scholar]
- Cortese-Krott MM, Kuhnle GG, Dyson A, Fernandez BO, Grman M, DuMond JF, et al. Key bioactive reaction products of the NO/H2S interaction are S/N-hybrid species, polysulfides, and nitroxyl. Proc Natl Acad Sci U S A. 2015;112:E4651–4660. doi: 10.1073/pnas.1509277112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadalla MM, Snyder SH. Hydrogen sulfide as a gasotransmitter. J Neurochem. 2010;113:14–26. doi: 10.1111/j.1471-4159.2010.06580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XH, Krokowski D, Guan BJ, Bederman I, Majumder M, Parisien M, et al. Quantitative H2S-mediated protein sulfhydration reveals metabolic reprogramming during the integrated stress response. Elife. 2015;4:e10067. doi: 10.7554/eLife.10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossoehme N, Kehl-Fie TE, Ma Z, Adams KW, Cowart DM, Scott RA, et al. Control of copper resistance and inorganic sulfur metabolism by paralogous regulators in Staphylococcus aureus. J Biol Chem. 2011;286:13522–13531. doi: 10.1074/jbc.M111.220012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes BG, Barbosa RL, Soprano AS, Campos BM, de Souza TA, Tonoli CC, et al. Plant pathogenic bacteria utilize biofilm growth-associated repressor (BigR), a novel winged-helix redox switch, to control hydrogen sulfide detoxification under hypoxia. J Biol Chem. 2011;286:26148–26157. doi: 10.1074/jbc.M111.234039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman CE, Samuels DJ, Karls AC. Modulating Salmonella Typhimurium’s response to a changing environment through bacterial enhancer-binding proteins and the RpoN regulon. Front Mol Biosci. 2016;3:41. doi: 10.3389/fmolb.2016.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins KA, Giedroc D. Insights into protein allostery in the CsoR/RcnR family of transcriptional repressors. Chem Lett. 2014;43:20–25. doi: 10.1246/cl.130965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins KA, Peng H, Luebke JL, Chang FM, Giedroc DP. Conformational analysis and chemical reactivity of the multidomain sulfurtransferase, Staphylococcus aureus CstA. Biochemistry. 2015;54:2385–2398. doi: 10.1021/acs.biochem.5b00056. [DOI] [PubMed] [Google Scholar]
- Iwig JS, Rowe JL, Chivers PT. Nickel homeostasis in Escherichia coli – The rcnR-rcnA efflux pathway and its linkage to NikR function. Mol Microbiol. 2006;62:252–262. doi: 10.1111/j.1365-2958.2006.05369.x. [DOI] [PubMed] [Google Scholar]
- Jackson MR, Melideo SL, Jorns MS. Human sulfide:quinone oxidoreductase catalyzes the first step in hydrogen sulfide metabolism and produces a sulfane sulfur metabolite. Biochemistry. 2012;51:6804–6815. doi: 10.1021/bi300778t. [DOI] [PubMed] [Google Scholar]
- Kabil O, Banerjee R. Characterization of patient mutations in human persulfide dioxygenase (ETHE1) involved in H2S catabolism. J Biol Chem. 2012;287:44561–44567. doi: 10.1074/jbc.M112.407411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolluru GK, Yuan S, Shen X, Kevil CG. H2S regulation of nitric oxide metabolism. Methods Enzymol. 2015;554:271–297. doi: 10.1016/bs.mie.2014.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li J, Lü C, Xia Y, Xin Y, Liu H, et al. FisR activates σ54-dependent transcription of sulfide-oxidizing genes Cupriavidus pinatubonensis JMP134. Mol Microbiol. 2017 doi: 10.1111/mmi.13725. in the press. [DOI] [PubMed] [Google Scholar]
- Libiad M, Yadav PK, Vitvitsky V, Martinov M, Banerjee R. Organization of the human mitochondrial hydrogen sulfide oxidation pathway. J Biol Chem. 2014;289:30901–30910. doi: 10.1074/jbc.M114.602664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Xin Y, Xun L. Distribution, diversity, and activities of sulfur dioxygenases in heterotrophic bacteria. Appl Environ Microbiol. 2014;80:1799–1806. doi: 10.1128/AEM.03281-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Ramesh A, Ma Z, Ward SK, Zhang L, George GN, et al. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat Chem Biol. 2007;3:60–68. doi: 10.1038/nchembio844. [DOI] [PubMed] [Google Scholar]
- Luebke JL, Shen J, Bruce KE, Kehl-Fie TE, Peng H, Skaar EP, et al. The CsoR-like sulfurtransferase repressor (CstR) is a persulfide sensor in Staphylococcus aureus. Mol Microbiol. 2014;94:1343–1360. doi: 10.1111/mmi.12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhachack L, Nudler E. Bacterial gasotransmitters: an innate defense against antibiotics. Curr Opin Microbiol. 2014;21:13–17. doi: 10.1016/j.mib.2014.06.017. [DOI] [PubMed] [Google Scholar]
- Luther GW, Rickard DT, Theberge S, Olroyd A. Determination of metal (Bi)Sulfide stability constants of Mn2+, Fe2+, Co2+, Ni2+, Cu2+, and Zn2+ by voltammetric methods. Environ Sci Technol. 1996;30:671–679. [Google Scholar]
- Ma Z, Jacobsen FE, Giedroc DP. Coordination chemistry of bacterial metal transport and sensing. Chem Rev. 2009;109:4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S, Chatterjee S, Dam B, Roy P, Das Gupta SK. The dimeric repressor SoxR binds cooperatively to the promoter(s) regulating expression of the sulfur oxidation (sox) operon of Pseudaminobacter salicylatoxidans KCT001. Microbiology. 2007;153:80–91. doi: 10.1099/mic.0.29197-0. [DOI] [PubMed] [Google Scholar]
- Marcia M, Ermler U, Peng G, Michel H. A new structure-based classification of sulfide:quinone oxidoreductases. Proteins. 2010;78:1073–1083. doi: 10.1002/prot.22665. [DOI] [PubMed] [Google Scholar]
- Mishanina TV, Libiad M, Banerjee R. Biogenesis of reactive sulfur species for signaling by hydrogen sulfide oxidation pathways. Nat Chem Biol. 2015;11:457–464. doi: 10.1038/nchembio.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller EG. Trafficking in persulfides: delivering sulfur in biosynthetic pathways. Nat Chem Biol. 2006;2:185–194. doi: 10.1038/nchembio779. [DOI] [PubMed] [Google Scholar]
- Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK, et al. H2S signals through protein S-sulfhydration. Sci Signal. 2009;2:ra72. doi: 10.1126/scisignal.2000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Akaike T, Sawa T, Kumagai Y, Wink DA, Tantillo DJ, et al. Redox chemistry and chemical biology of H2S, hydropersulfides, and derived species: implications of their possible biological activity and utility. Free Radic Biol Med. 2014;77:82–94. doi: 10.1016/j.freeradbiomed.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Shen J, Edmonds KA, Luebke JL, Hickey AK, Palmer LD, et al. Sulfide homeostasis and nitroxyl intersect via formation of reactive sulfur species (RSS) in Staphylococcus aureus. mSphere. 2017;3 doi: 10.1128/mSphere.00082-17. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettinati I, Brem J, McDonough MA, Schofield CJ. Crystal structure of human persulfide dioxygenase: structural basis of ethylmalonic encephalopathy. Hum Mol Genet. 2015;24:2458–2469. doi: 10.1093/hmg/ddv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AA, Venceslau SS, Grein F, Leavitt WD, Dahl C, Johnston DT, et al. A protein trisulfide couples dissimilatory sulfate reduction to energy conservation. Science. 2015;350:1541–1545. doi: 10.1126/science.aad3558. [DOI] [PubMed] [Google Scholar]
- Sattler SA, Wang X, Lewis KM, DeHan PJ, Park CM, Xin Y, et al. Characterizations of two bacterial persulfide dioxygenases of the metallo-β-lactamase superfamily. J Biol Chem. 2015 doi: 10.1074/jbc.M115.652537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatalin K, Shatalina E, Mironov A, Nudler E. H2S: a universal defense against antibiotics in bacteria. Science. 2011;334:986–990. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- Shen J, Keithly ME, Armstrong RN, Higgins KA, Edmonds KA, Giedroc DP. Staphylococcus aureus CstB Is a novel multidomain persulfide dioxygenase-sulfurtransferase involved in hydrogen sulfide detoxification. Biochemistry. 2015;54:4542–4554. doi: 10.1021/acs.biochem.5b00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Peng H, Zhang Y, Trinidad JC, Giedroc DP. Staphylococcus aureus sqr encodes a type II sulfide:quinone oxidoreductase and impacts reactive sulfur speciation in cells. Biochemistry. 2016;55:6524–6534. doi: 10.1021/acs.biochem.6b00714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Shen J, Fang M, Zhang Y, Hori K, Trinidad JC, et al. Sulfide-responsive transcriptional repressor SqrR functions as a master regulator of sulfide-dependent photosynthesis. Proc Natl Acad Sci U S A. 2017;114:2355–2360. doi: 10.1073/pnas.1614133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedmann R, Onderka C, Wei S, Szijarto IA, Miljkovic JL, Mitrovic A, et al. Improved tag-switch method reveals that thioredoxin acts as depersulfidase and controls the intracellular levels of protein persulfidation. Chem Sci. 2016;7:3414–3426. doi: 10.1039/c5sc04818d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Y, Liu H, Cui F, Liu H, Xun L. Recombinant Escherichia coli with sulfide:quinone oxidoreductase and persulfide dioxygenase rapidly oxidises sulfide to sulfite and thiosulfate via a new pathway. Environ Microbiol. 2016;18:5123–5136. doi: 10.1111/1462-2920.13511. [DOI] [PubMed] [Google Scholar]
- Yadav PK, Martinov M, Vitvitsky V, Seravalli J, Wedmann R, Filipovic MR, et al. Biosynthesis and reactivity of cysteine persulfides in signaling. J Am Chem Soc. 2016;138:289–299. doi: 10.1021/jacs.5b10494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav PK, Yamada K, Chiku T, Koutmos M, Banerjee R. Structure and kinetic analysis of H2S production by human mercaptopyruvate sulfurtransferase. J Biol Chem. 2013;288:20002–20013. doi: 10.1074/jbc.M113.466177. [DOI] [PMC free article] [PubMed] [Google Scholar]


