Abstract
Aligning brain structures across individuals is a central prerequisite for comparative neuroimaging studies. Typically, registration approaches assume a strong association between the features used for alignment, such as macro-anatomy, and the variable observed, such as functional activation or connectivity. Here, we propose to use the structure of intrinsic resting state fMRI signal correlation patterns as a basis for alignment of the cortex in functional studies. Rather than assuming the spatial correspondence of functional structures between subjects, we have identified locations with similar connectivity profiles across subjects. We mapped functional connectivity relationships within the brain into an embedding space, and aligned the resulting maps of multiple subjects. We then performed a diffeomorphic alignment of the cortical surfaces, driven by the corresponding features in the joint embedding space. Results show that functional alignment based on resting state fMRI identifies functionally homologous regions across individuals with higher accuracy than alignment based on the spatial correspondence of anatomy. Further, functional alignment enables measurement of the strength of the anatomo-functional link across the cortex, and reveals the uneven distribution of this link. Stronger anatomo-functional dissociation was found in higher association areas compared to primary sensory- and motor areas. Functional alignment based on resting state features improves group analysis of task based functional MRI data, increasing statistical power and improving the delineation of task-specific core regions. Finally, a comparison of the anatomo-functional dissociation between cohorts is demonstrated with a group of left and right handed subjects.
Introduction
Accurate alignment of brain structures is essential for the quantitative comparison of local characteristics across individuals. Anatomical alignment is a standard procedure for fMRI analysis, and has had a substantial impact by enabling location specific comparison of individuals or cohorts (Fischl, 2012). However, relying on macro-anatomy suffers from limitations if the association between anatomy and function is weak. Here, we propose to align imaging data across individuals based on the global functional connectivity structure observed during resting state functional magnetic resonance imaging data (rs-fMRI), while at the same time constraining the transformations to diffeomorphisms on the cortical surface.
The potential dissociation between structure and function has been addressed in only a small number of studies which used functional features for inter-individual alignment (Sabuncu et al., 2010; Conroy et al., 2013; Robinson et al., 2014; Langs et al., 2015). Anatomo-functional dissociation occurs if the variability of functional architecture across subjects cannot be entirely explained by anatomical differences (Mueller et al., 2013; Tootell et al., 1995; Smith et al., 2005; Brett et al., 2002). Group studies that rely on a static anatomical relationship can be biased by reduced spatial overlap of functionally similar regions across individuals and corresponding reduced group-level activation. Individual variability in functional architecture could become more complex if diseases affect both function and anatomy, thus making anatomical alignment more prone to these confounding factors (Brett et al., 2002).
We propose a functional alignment approach based on a diffeomorphic registration of functional resting state features along a spherical projection of the cortical surfaces. The proposed alignment is independent of specific tasks, and uses only rs-fMRI signals as a basis for alignment. We quantify global functional network characteristics by spectral embedding, which provides a functional signature of the entire cortex based on resting state fMRI. In this embedding, each dimension represents different aspects of the rs-fMRI architecture.
Functional alignment enables the separate analysis of functional variability and differences in anatomical location. Alignment based on resting state functional connectivity provides a complementary source for correspondence, and allows for the analysis of task-fMRI experiments based on unified correspondences across individuals. While constraining the alignment to a diffeomorphic surface transform limits the variability the method can capture, it disambiguates possible similarities of connection characteristics in widely distributed regions forming functional networks with an anatomical constraint.
In this work, we test if (1) cortical alignment guided by rs-fMRI information is repeatable across scan sessions, (2) if it captures differences across individuals, (3) if the variability is constant or heterogeneous across the cortex, (4) if this alignment improves group-level task fMRI evaluation, and (5) if it can quantify differences across two cohorts of left- and right-handers.
State of the art
Existing standard registration approaches establish correspondence across multiple individuals by assuming anatomical comparability. Typically, in fMRI studies, spatial normalization is employed to map brain regions to a common template space, e.g., the MNI atlas (Evans et al., 1993). This ensures comparability across different studies and atlas labeling. Volume-based normalization approaches are standard and provided by widely used tools such as FSL (Jenkinson et al., 2012) or SPM (Friston et al., 2007). Non-rigid, volume-based transformation models, such as diffeomorphic demons (Vercauteren et al., 2009) or angular interpolation (Duarte et al., 2013), can account for inter-subject anatomical variability and achieve more accurate anatomical registration. Surface-based registration techniques, such as in FreeSurfer (Fischl, 2012) and Spherical Demons (Yeo et al., 2010), use spherical representations of surfaces and align them based on cortical features such as curvature or sulcal depth. Lombaert et al. (2013a) and Lombaert et al. (2013b) introduced diffeomorphic alignment based on spectral features of cortical anatomy providing a speed advantage over traditional surface based registration techniques. Lombaert et al. (2015) extended the spectral alignment approach and incorporated features based on retinotopy on the visual cortex. While we follow a similar paradigm - using a representation of features in a spectral embedding space - the proposed approach uses the global resting-state connectivity structure instead of anatomical features as a basis for alignment. Current volumetric and surface registration approaches provide highly accurate alignment of the macroanatomy; however, they neglect the variability in function across and within subjects (Tootell et al., 1995; Smith et al., 2005), hampering group-level functional analyses especially when the anatomo-functional association is weak.
Alignment approaches that take functional information into account can improve the correspondence across subjects in group analyses. Sabuncu et al. (2010) proposed fine-tuning of anatomical alignment via non-rigid registration that maximizes inter-subject correlation of fMRI signals recorded during a synchronized movie-viewing stimulus. Similarly, Conroy et al. (2013) used spatial patterns of functional responses to movie stimuli to guide anatomical registration with a focus on areas involved in visual processing. Jiang et al. (2013) introduced a progressive matching of multi-range functional connectivity patterns for spatial normalization, and showed that functional alignment improves the overlap of resting-state networks, such as the default mode network. Extending this approach, Langs et al. (2014) proposed decoupling anatomy from function, and aligned fMRI signals recorded during language tasks in an embedding space, improving the match of corresponding task-activation across individuals. Langs et al. (2015) also identified shared resting-state functional networks despite varying spatial footprints across individuals, by parcellation in a population-level embedding space. Nenning et al. (2015) addressed noise during group-wise alignment in the embedding space by a joint-diagonalization approach. By assuming a single activation peak whose position varies across individuals, Gramfort et al. (2015) aligned cortical maps via optimal transport. More recently, Wang et al. (2015) established an individualized functional network parcellation strategy that may provide landmarks of functional networks for cross-subject alignment. Robinson et al. (2014) proposed a surface based multi-modal alignment technique, which allows for anatomical and local functional features to drive cortical alignment. Here we de-couple anatomy and function, and use the distributed connectivity structure represented in a spectral embedding space as a basis for alignment, and investigate how anatomical- and functional alignment differ.
Contribution
Here, we describe diffeomorphic alignment of the cortical surface based on rs-fMRI connectivity characteristics. We tested whether this would improve the accuracy of alignment of functionally similar regions identified on t-fMRI, by evaluating the overlap of homologous regions mapped across subjects and evaluating group-level task activation analysis. The proposed alignment improved overlap and group-level analysis. Aligning repeat rs-fMRI scans of the same individuals showed that the method is stable, while still detecting displacements across subjects. Further investigation of this anatomo-functional dissociation revealed that it was unevenly distributed across the cortex. To test whether this might be a feasible technique with which to study differences between cohorts, we evaluated the difference between left- and right-handed subjects. Results from the experiments support a strong link between functional resting state and task responses, and that functional alignment with resting state networks improve the group analysis of t-fMRI.
Diffeomorphic functional surface alignment
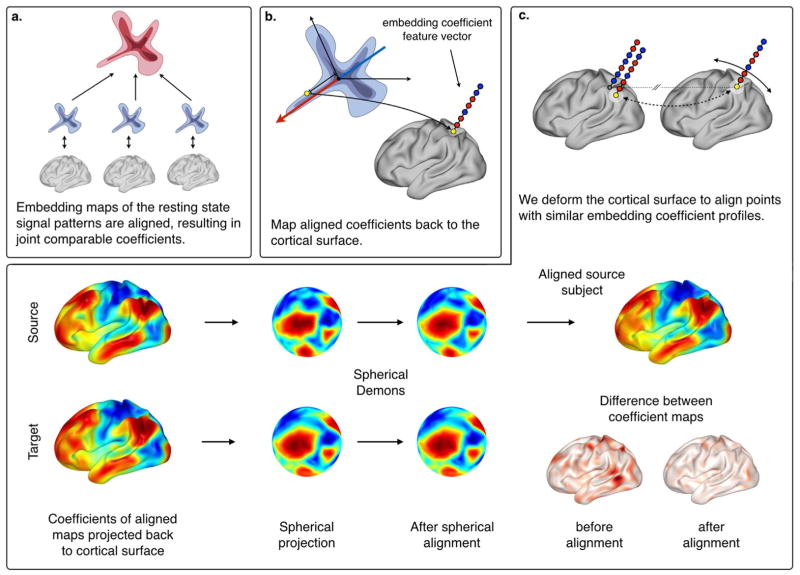
We briefly review diffusion map embedding for the representation of the rs-fMRI data, then explain the alignment of individual subject maps in the embedding space, and, finally, detail how this joint embedding map can drive a diffeomorphic alignment of the cortical surface. An illustrative overview of the method is displayed in Fig. 1.
Fig. 1.
Overview of the functional alignment approach. (a) We mapped individual rs-fMRI functional connectivity architectures into an embedding space, and performed an alignment of these subject-specific functional maps. (b) We projected the resulting aligned embedding coefficients back to the cortical surface obtaining a feature vector for each vertex, and used them to (c) perform diffeomorphic functional alignment along the surface, based on spherical demons.
Embedding map representation of functional connectivity
We review embedding of signal correlation patterns by diffusion maps (Coifman and Lafon, 2006) subsequent to the embedding map alignment described in (Langs et al., 2010, 2011).
For each of S subjects, rs-fMRI data IS ∈ ℝT×N consists of N surface nodes V with a BOLD signal observed at T time points. The connectivity structure of the resting state data can be viewed as a graph, where the vertices represent the surface nodes, and a similarity measure between the corresponding signal time courses defines the edges. We define the edges of this graph as an affinity matrix, WS, where is the Pearson correlation coefficient between the time-series of nodes i and j in subject s, and we keep only the connections with a correlation of r > 0.25 to discard weak and negative connections. Subsequently, the graph is checked to ensure that it is a connected graph.
The spectral representation of the functional connectivity structure in each subject is based on a transition probability graph PS, with P = D−αWD−α. D is the diagonal matrix of node degrees, i.e., di = Σj Wij, and α is a normalization parameter tuning the influence of the data point density, typically set to 0.5 (Coifman and Lafon, 2006). Eigenvalue decomposition of the normalized Laplacian of this graph results in eigenvalues Λ = λ1,…,λn, with λ1 = 1 > λ2 > … > λn, and corresponding eigenvectors Φ. The embedding map representation, or functional geometry of all surface nodes in one subject is defined by new embedding coordinates Ψt of the diffusion map representation. Each vertex, v, in the original graph is represented by new coordinates in the d dimensional embedding space:
| (1) |
where t is the diffusion time, which can be viewed as a scaling parameter. A high diffusion time leads to an embedding that primarily captures coarse network structure, while a low diffusion time captures connectivity with finer granularity. At the same time the diffusion time limits the influence of the neighboring network parts influencing the distance between two nodes. Limiting this influence to a local neighborhood leads to stable results for very large graphs. We fixate the diffusion time with t=1, which considers strong connectivity between nodes, and follows previous literature in this choice (Langs et al., 2014, 2015). For further processing, we only keep the first 50 embedding coefficients. This embedding space reflects the functional organization of the fMRI data decoupled from spatial positions, since the graph is defined only by correlation between the time-series. The functional connectivity structure is encoded as an embedding map, Ψ, where the functional relationships of the fMRI signals are captured by the diffusion distance, Dt. The Euclidean distance in this new embedding space approximates the diffusion distance
| (2) |
and represents the probability of traveling from node i to node j in t steps, by considering all possible paths between these two nodes. The embedding reflects the functional organization of the brain and allows the uncoupling of function from spatial position, and modeling the connectivity structure as a graph forces brain regions with high correlation to be close in this map.
Functional alignment of resting-state networks
Subject-specific spectral embedding maps are not directly comparable, since the eigenvalue decomposition can result in flips of coefficient signs, rotations, and differences in the ordering of the embedding dimensions. Thus an initial orthonormal alignment is necessary. For each individual, a functional geometry representation is computed and functional registration is performed with an orthonormal manifold alignment approach proposed by Langs et al. (2014). A source subject-specific functional geometry, Ψs, is aligned to a template embedding map, Ψt, using an orthonormal transformation matrix, Qst. This transformation matrix accounts for rotations, sign flips, and reordering of the spectral components. The optimal transformation can be calculated in closed form with
| (3) |
where wc is the Euclidean distance between the spatial coordinates of a corresponding vertex pair c. Since the core regions of resting-state networks show spatial consistency across healthy subjects (Damoiseaux et al., 2006), correspondences based on spatial position can be used as the initialization for the alignment. An example of the functional alignment is illustrated in Figure S3 in the Supplementary material.
Aligning the individual embedding maps of a target subject t, Ψt, and a source subject s, Ψs, results in a joint embedding space of comparable spectral representations with new coordinates Θt and Θs, denoting the aligned embedding maps. The coordinates of each surface node can be viewed as a spectral signature, which is comparable across subjects, and can be used as a functional feature, characterizing the connectivity structure. In our experiments, we aligned the embedding maps of all subjects to the functional geometry of one subject, resulting in aligned embedding coordinates, 〈Θ1, Θ2,…,ΘS〉, representing all subjects.
Functional surface alignment
We adapted the Spherical Demons (SD) algorithm (Yeo et al., 2010) for diffeomorphic alignment of functional connectivity structures.
Spherical Demons (SD) registers brain surfaces by extending Diffeomorphic Demons (Vercauteren et al., 2009) from Euclidean space to the unit sphere. SD uses a spherical representation of a brain hemisphere for diffeomorphic registration and aligns regions with similar anatomical features describing cortical topology such as curvature or sulcal depth. We used SD for surface alignment, but the cost function is driven by the similarity of functional features rather than of anatomical characteristics. Specifically, we used the spectral signature of the embedding maps as features, and aligned the surface so that the difference between the embeddings, Θi and Θj, of corresponding data points i and j was minimized. The transformation of the Demons algorithm is defined as
| (4) |
where Θt and Θs represent the aligned embedding maps of a target subject and a source subject. The desired transformation, Γ, between the source and the target should approximate the hidden transformation Y, where dist (ϒ, Γ) = ||ϒ − Γ||2 ensures that they are close to each other. We used Θt − Θs○Γ for the difference between the embedding coefficients at the target subject and the embedding coefficients at corresponding positions in the source target, where Γ is the spherical coordinate transformation that establishes correspondence. The regularization term, Reg (ϒ), penalizes the gradient magnitude of the displacement field, defined as Reg (ϒ) = ||δ(ϒ − Id)||2 To extend the Demons algorithm to the unit sphere S2, Yeo et al. (2010) defined the distance term between Γ and Y as the distance between sets of N tangent vectors, and , with . The regularization term is defined as Reg(ϒ) ≜ ||ϒ⃗||V, where ϒ⃗ is restricted to belong to the Hilbert space, V, of vector fields obtained by the closure of the space of smooth vector fields on S2.
We used the diffeomorphic properties of the Spherical Demons algorithm to register brain surfaces based on functional signatures, specified by the spectral representations of the functional resting state architecture. In a multi-resolution fashion, each dimension of the embedding space is iteratively used as a feature for the registration. This ensures that principal dimensions that describe global features, e.g., task-positive and task-negative networks, are used first and fine-tuning occurs with local features, encoded in subsequent dimensions.
Vertex to vertex correspondence
The surface alignment results in new spherical coordinates, Ω, for each vertex and subject. We establish correspondences across subjects in the functional domain of aligned spheres, Ω. For each vertex, I, on a target sphere, Ωt, a corresponding vertex, î, on a source sphere, Ωs, is defined as
| (5) |
where dist is a distance function between the spherical coordinates of Ωt (i) and Ωs (j). During group-level analysis, for each vertex we collected corresponding signals or labels on a template by assigning the signal or label of the nearest neighbor on the aligned spheres of multiple subjects.
Data and preprocessing
Two different datasets are used for our experiments
The HCP dataset comprised 100 randomly chosen subjects of the Human Connectome Project (HCP) (Essen et al., 2012) 500-Subject data release, and contained 54 females and 46 males. The data consists of 3 Tesla MRI acquisitions including two sessions with repeated resting-state measurements (two runs per session), with multiband, accelerated, echo-planar imaging sequencing with a repetition time of 720 ms, providing a total of 1200 frames for each run of resting state acquisition, corresponding to 14 min and 33 s. We used the time-series of two repeated rs-fMRI runs for alignment and repeatability evaluation. In addition, we used t-fMRI data for seven different paradigms (working memory, gambling, motor, language, social cognition, relational processing, emotion processing), each with multiple conditions. We used the t-fMRI data to evaluate the alignment of functionally homologous regions, and the effect of alignment on group-wise t-fMRI analysis. The HCP data release provides already minimally preprocessed data (Glasser et al., 2013), including anatomical surface data normalized to a common average template space, and results from GLM activation analysis. For this work, we used the time-series from the resting state acquisitions mapped onto the surface, consisting of 32492 time-series per hemisphere. We performed nuisance regression of the motion artifacts and bandpass filtering in the 0.01–0.1 Hz frequency range. For the task-based data, we used the z-scores of the GLM activation analysis to evaluate the overlap of active regions on the group level.
The handedness dataset included rs-fMRI data from 52 left-handed and 52 matched (age, gender, ethnicity, education, fMRI data acquisition, head motion, data quality, and other parameters), right-handed subjects (Wang et al., 2015). Each subject performed two resting-state (eyes open) runs in the MRI scanner (6 m 12 s per run). All data were collected on matched 3 T Tim Trio scanners (Siemens, Erlangen, Germany), using a 12-channel phased-array head coil. Images were acquired using the gradient-echo echo-planar pulse sequence (TR=3000 ms, TE=30 ms, flip angle=85°, 3×3×3 mm voxels, FOV=216 and 47 slices collected with interleaved acquisition with no gap between slices). Structural data included a high-resolution multiecho T1-weighted magnetization-prepared gradient-echo image (TR=2200 ms, TI=1100 ms, TE=1.54 ms for image 1 to 7.01 ms for image 4, FA=7°, 1.2×1.2×1.2 mm voxels and FOV=230). Subjects were instructed to stay awake, keep their eyes open, and minimize head movement; no other task instruction was provided. The handedness of each subject was assessed via the Edinburgh handedness inventory (Oldfield, 1971). We used the time-series from the resting state acquisitions mapped onto the FreeSurfer average surface, consisting of 2562 time-series per hemisphere.
Evaluation
We evaluated four aspects of diffeomorphic functional alignment based on resting-state networks: (a) Is the alignment valid and stable? (b) How is the dissociation between anatomy and function distributed across the cortex? (c) Does the alignment of resting-state networks improve group-level analysis of task fMRI data? (d) Can we compare the dissociation between function and anatomy between two cohorts of left- and right-handed subjects?
Stability
To validate the stability of functional surface alignment, we aligned data from two repeated resting-state acquisitions of the same subject, and calculated the spatial displacement between the aligned rs-fMRI networks. We detected corresponding vertices on the aligned spheres, and calculated the spatial distance between their coordinates on the cortical surface template for both rs-fMRI scans of each subject. If the functional alignment reflects a stable architecture for a subject, this distance should be 0 for the entire cortex. We compared this within-subject variability with the displacement detected across subjects, the functional alignment of each subject, and a random different subject.
Distribution of the displacement across the cortex
We evaluated the distribution of average cross-subject displacement, to test whether the link between anatomical location and rs-fMRI structure was weak, and if it varied across the cortex. For within-subject and cross-subject stability analysis, we reported the average displacement and the displacement across the entire cortex, in order to detect the most variable brain regions. In addition, we quantified the spatial displacement for seven basic resting-state networks defined by Yeo et al. (2011) to achieve results comparable to the existing literature.
Improvement of t-fMRI group studies
A dissociation between anatomical location and function affects t-fMRI group studies, since spatial normalization can compensate only for those differences linked to anatomy across subjects. We evaluated whether functional alignment of rs-fMRI networks could improve group-level analysis of t-fMRI data. Improvement indicates whether there is a sufficiently strong link between rs-fMRI and t-fMRI structures.
First, we performed group-level activation analysis (one-sample t-test, fdr 0.05) of the General Linear Model (GLM) contrasts available in the Human Connectome Project (HCP) data set on the entire population. We compared the results obtained after functional alignment with those obtained after standard anatomical alignment in the FreeSurfer average surface space. Functional alignment is based on rs-fMRI, and no information about the t-fMRI is used. We showed qualitative results, and quantified the change in group-activation in different sections across the cortex. We expected the specificity of the group-level activation to increase, i.e., we expected better defined activated areas, whose activation was not reduced by the spatial variability across individuals. We evaluated in which of the seven basic resting-state networks (Yeo et al., 2011) different experimental conditions benefited the most from resting-state alignment.
Second, we measured whether group activation generalizes better to a new individual subject after functional alignment. We quantified how well we could predict an individual activation pattern in a held-out subject based on the group activation of the other subjects. Again, we compared functional alignment to standard anatomical normalization. In a leave-one-out cross validation, we compared the activation in the brain regions of a held-out subject with the group average activation, where brain regions with a GLM z-score >3 were labeled as active. We calculated the Dice coefficient between the prediction labeling and the region measured in the held-out subject. We reported the percentage of overlap improvement between the prediction obtained after functional alignment compared to anatomical alignment in the FreeSurfer surface space.
Comparing the anatomy-function link between different cohorts
Functional alignment reveals the extent to which the brain regions of one subject must be displaced in order to functionally match those of another subject. Since we used spatially normalized data for initialization, we could quantify the distance between initially corresponding brain regions after functional alignment on the spheres. This distance reflects the anatomical discrepancy of the functional anchor locations across subjects. Using this, we could compare the location of corresponding functional areas in different cohorts. We evaluated the difference in functional anchoring in left- and right-handed individuals. First, two repeated resting-state fMRI runs were spatially normalized to a FreeSurfer surface template. For each resting-state run, we performed a pairwise functional alignment between random pairs of left-handed subjects, and right-handed subjects respectively. We then performed a paired t-test between the within cohort displacement measure for each vertex on the cortical surface. The results indicated differences in the spatial distribution of corresponding functional areas. We regressed out anatomical features such as cortical thickness, curvature, and sulcal depth, and used two repeated resting-state acquisitions to evaluate the repeatability of the results, and reported the location and magnitude of the dissociation between the cohorts.
Results
Functional surface alignment is stable over repeated measurements
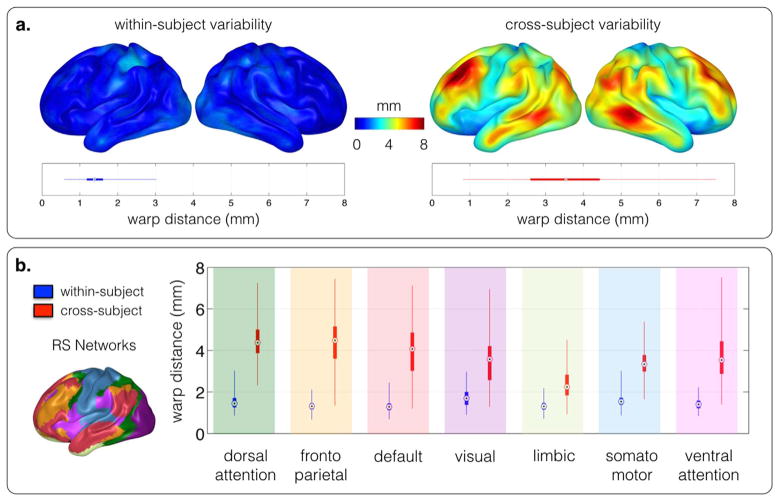
A cortical map of the within-subject average displacement is shown in Fig. 2. The average displacement between two runs of the same subject was 1.4 mm with a standard deviation of 0.3 mm, whereas the average displacement between two runs of random different subjects was 3.5 mm, with a standard deviation of 1.3 mm. The functional displacement was far from evenly distributed across the cortex, and large, average across-subject displacement regions (>8 mm) were not present for within-subject displacement. The average within-subject displacement was significantly smaller than the average cross-subject displacement (two-sample t-test, p < 0.001).
Fig. 2. (a) Stability.
when applied to two repeated rs-fMRI acquisitions of the same subject (within-subject), the group-average spatial displacement estimated by functional demons was low (a, left). Variability across the cortex: the group-average spatial cross-subject displacement captured the location variability of functional networks across subjects. It was distributed un-evenly across the cortex (a, right). The difference between within- and cross-subject displacement was significant (two-sample t-test, p < 0.001). (b) Compared to the spatial within-subject displacement, the spatial cross-subject displacement is un-evenly distributed across the seven basic resting-state networks defined by Yeo et al. (2011).
Functional surface alignment reveals a nonuniform link between anatomy and function across the cortex
While the within-subject analysis showed that the approach is stable, the cross-subject results reflected the variability in functional architecture between the subjects. The dissociation between anatomy and function was distributed unevenly across the cortex (Fig. 2). Fig. 2(b) shows the cross-subject variability in the seven basic resting-state networks (Yeo et al., 2011). This variability was particularly high in the dorsal attention-, fronto parietal-, and default networks, while relatively low in the limbic-, and somato-motor areas.
To confirm that the cross-subject displacement is not due to difficulties in aligning inconsistent cortical folding patterns, we calculated the residual variance of cortical curvature and sulcal depth after alignment by FreeSurfer and after functional alignment (see Figure S1 in the Supplementary material). If functional alignment would primarily help to correct for anatomical alignment errors, or deal with local minima, we expect the residual cortical curvature and sulcal depth variance to decrease. I.e., the functional alignment fixes errors of the anatomical alignment. The residual variance in cortical curvature and sulcal depth is not reduced from anatomical to functional alignment. This supports our interpretation of deformation maps relating to the dissociation between function and anatomy.
Functional alignment based on rs-fMRI improves t-fMRI group analysis
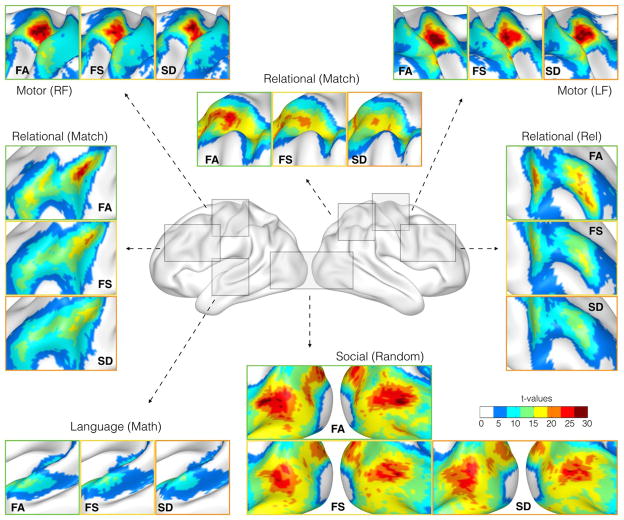
Diffeomorphic functional alignment improves the correspondence of task-specific group activation patterns. Region-specific results for different task conditions are illustrated in Fig. 3. After functional alignment, the overlap of homologous regions across the group increases. This provides higher statistical power for task-specific core regions, and enables a more fine-grained analysis of group-level activation compared to spatial normalization. Fig. 3 shows improved task-specific delineation of the functional core regions and an increase in t-values. For the math condition of the language task, temporal language-relevant regions were more strongly defined. For the left foot condition of the motor task, we observed an increase in the t-statistic in the motor cortex on the right hemisphere, similar to that in the left hemisphere for the right foot condition. Multiple conditions of the relational task showed that the functional alignment resulted in distinct peaks, with increased t-values on the left and the right hemisphere in the frontal regions, whereas spatial normalization was not able to detect those regions optimally.
Fig. 3. Group analysis of t-fMRI data.
Functional alignment (FA) of resting-state networks supports group analysis of t-fMRI data, compared to common anatomical surface alignment with FreeSurfer (FS) and Spherical Demons (SD). It improved the overlap of t-fMRI specific activation patterns, increasing the statistical power of functional core regions and resulting in a sharpened delineation of smaller activated areas. A quantitative assessment of the improvement is provided in Fig. 4.
Compared to the standard spatial normalization, the central regions of the group activations exhibited higher t-values after functional alignment. Subject specific activation foci become closer to each other with functional alignment, compared to anatomical alignment (see Figure S4 in the Supplementary material).
Functional alignment increases the overlap of functional regions across individuals
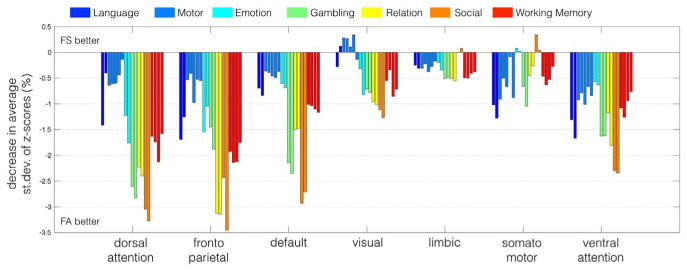
For each resting-state network, Yeo et al. (2011), we calculated the average standard deviation of the task-specific z-scores between subjects, and quantified the percent change between spatial normalization and functional alignment. Results are shown in Fig. 4. Functional alignment decreased the standard deviation across subjects for almost all task conditions and resting-state networks. The largest decrease occurred in the dorsal attention, fronto parietal and default networks. Not surprisingly, these are also the networks with the highest spatial displacement across subjects, as displayed in Fig. 2.
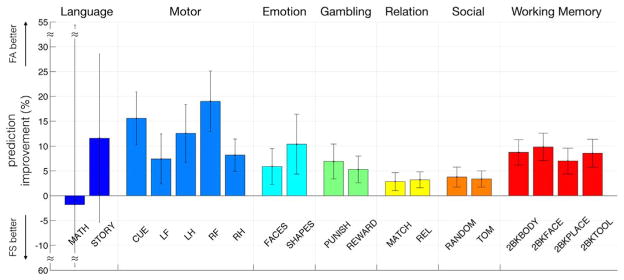
Fig. 4.
Functional alignment decreased the standard deviation of the z-scores for most task conditions, suggesting an improved match of similarly activated areas across the population. The network-specific decrease in standard deviation was the largest in regions associated with the dorsal attention, fronto-parietal, and default network, and the improvement for different conditions within the same experimental tasks was consistent. The overall decrease in standard deviation was the highest in regions that exhibited the largest displacement during the functional alignment procedure.
Functional alignment based on rs-fMRI improves the prediction of a single subject activation pattern
Compared to spatial normalization, functional alignment of resting state features improves the prediction of a subject-specific activation pattern. Functional alignment supports the generalization of a group activation to a new individual subject. Fig. 5 shows the percent change for prediction results for all task conditions of the HCP dataset. Except for the language task conditions, we observed a stable prediction improvement over all experimental conditions. The motor task benefited the most, where we achieved up to a 19% improvement in predicting the activation for the right foot condition.
Fig. 5.
The generalization of the group activation to subject-specific activation patterns is improved with functional alignment. Compared to spatial normalization, the prediction of subject-specific activation patterns, based on group activation analysis of a population showed improvement in all except the language task conditions. This is further evidence that functional alignment improves the overlap of functionally equivalent regions across subjects.
Differences in spatial distribution of function between left- and right-handed subjects
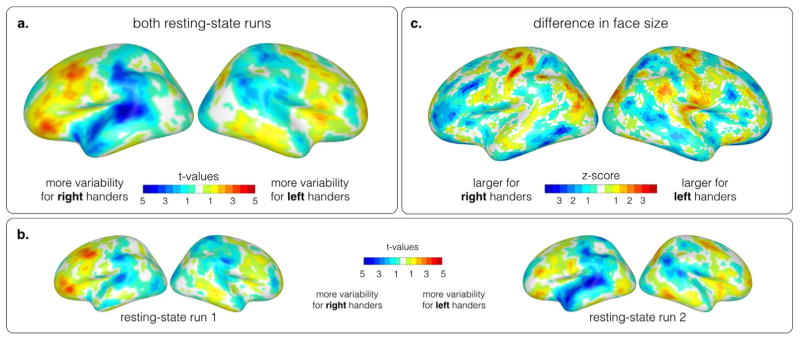
When comparing the displacement between left- and right-handed cohorts, a distinct pattern occurs. Most noticeable is the difference in the frontal cortex, the inferior parietal cortex around the supra-marginal gyrus and the temporal cortex. Overall, the left hemisphere shows a higher displacement compared to the right hemisphere. For left-handed subjects, a higher displacement is found in the frontal regions of the brain, whereas right-handers exhibit a higher displacement in the inferior parietal cortex around the supra-marginal gyrus and the temporal cortex. The higher displacement in the left hemisphere, and in the frontal and tempo-parietal regions indicate a relationship to the language related functional variability found between left and right handers. Fig. 6(a) shows the results for both resting-state runs, and Fig. 6(b) illustrates the results for each resting-state run separately to demonstrate the stability of the located core regions despite intra subject variability. Fig. 6(c) illustrates differences in face size of the surface mesh between the two cohorts after alignment. To confirm that the differences between the two cohorts are not an artifact of anatomical differences affecting anatomical registration, we compared the residual variance of sulcal depth and cortical curvature after alignment across the two cohorts with a paired t-test. Neither of these measures exhibits a significant difference between left-and right handed subjects (see FigureS2 in the Supplementary material).
Fig. 6.
(a) Differences in the functional variability between left-handers and right-handers over two repeated resting-state runs. The colormap illustrates the t-values of a paired t-test between the within cohort variability. Blue colors represent a higher variability for right-handers, and red colors a higher variability for left-handers. Overall, the left hemisphere exhibits higher variability than the right hemisphere. Frontal regions are more variable for left-handers, while tempo-parietal regions show a higher variability between right-handed subjects. (b) Individual analysis of two repeated resting-state runs reveals similar core regions, despite existing functional variability. (c) Differences in face size of the surface mesh between left- and right handers after functional alignment.
Discussion
Understanding the variability - differences in spatial distribution and function - of the functional brain architecture across individuals is important for fMRI studies. In this work, we have shown that diffeomorphic alignment of cortical surfaces based on rs-fMRI signal correlation patterns is feasible. This type of alignment reflects the stable characteristics of individuals, and captures differences across subjects. Using rs-fMRI information can align t-fMRI activation patterns better than sole anatomical normalization, resulting in increased overlap between group- and individual level activation, and higher specificity of the delineation of activated areas than a group-level analysis. Further, the method enables the comparison of cohorts, and reveals the region specificity of the anatomo-functional dissociation across the cortex.
Diffeomorphic functional alignment captures the dissociation between anatomy and function
Spectral embedding maps the global connectivity structure observed in rs-fMRI data to a space that represents signal relationships within subjects. The resulting maps are specific enough to enable matching them across subjects based on their shape in the embedding space. This results in a set of maps where representatives of cortical points that share a similar connectivity profile are spatially close. The resolution of the correspondence in the embedding space is determined by the specificity of the functional connectivity profile of each cortex point (Langs et al., 2014, 2015; Nenning et al., 2015). While this enables the alignment of points based on their role in functional networks, decoupled from anatomy, it is limited because points with a very similar connectivity profile cannot be separated. We constrained the mapping between individuals to diffeomorphic deformations of the cortical surface. If, after anatomical registration, regions have the same anatomical location but are functionally different, then they have different embedding features, and the alignment results in a deformation approximating the difference in spatial locations of functionally similar regions. While the diffeomorphic constraint reduces the ability to map substantial differences across subjects, e.g. due to a pathology such as a brain tumor, it can disambiguate the registration of regions that are part of the same functional network, but are spatially separated, such as different areas of the default network. Similar approaches have been used in spatial normalization with spherical demons (Yeo et al., 2010), and in alignment based on across-individual signal correlations (Conroy et al., 2013).
The small displacement of functional alignment between repeated scans of the same individual show that the method is stable, and can be due to either noise or variability in the resting-state activity at different scan times. Notably, the results indicate a link between the resting-state functional connectivity architecture and the location of task specific activation areas. In line with this, the increase in task-specific area overlap across the cortex, largely coincides with the amount of average displacement across individuals.
Overall, the residual variance of cortical folding patterns after spatial alignment with FreeSurfer is not decreased after functional alignment (see Figure S1 in the Supplementary material). This indicates that the cross-subject displacement and corresponding improvement after functional alignment is not primarily due to difficulties in aligning inconsistent cortical folding patterns, but due to a dissociation between function and anatomy captured by the proposed alignment. However, note that diffeomorphic deformation maps cannot capture functional variability involving inconsistent topology, which is present even across healthy subjects (Glasser et al., 2016).
Anatomo-functional dissociation is distributed unevenly across the cortex
Functional alignment of random pairs of subjects reveals substantial differences in the cortical location of functional areas. The average displacement is not evenly distributed across the cortex. Areas with particularly high discrepancy include the fronto-parietal network, the dorsal attention network, and the default network (Margulies et al., 2016), whereas regions associated with the somato-motor or visual networks have to be warped less to be functionally aligned. These results are related to previous studies, which investigated inter-subject variability, evolutionary timing, and development. In Mueller et al. (2013), the parieto-frontal-, attention, and default networks showed high inter-subject variability of functional connectivity, but the study did not investigate to what extent this was caused by location differences of functional areas, or by the functional connectivity itself. In Langs et al. (2015), shared brain areas were studied by clustering in an embedding space that was constructed similarly to this work. Clusters in the parieto-frontal areas exhibited the highest differences across subjects. In Jakab et al. (2014), the authors studied the timing of emerging functional networks during fetal development, with motor and vision areas emerging first, and temporal- and parieto-frontal networks later. In the present paper, we could specifically assign differences to a displacement of cortical areas involved in a specific network.
Functional alignment improves group-level t-fMRI studies
The proposed diffeomorphic alignment is purely driven by the functional rs-fMRI connectivity structure, without information about task conditions or corresponding fMRI data. Nevertheless, there is a sufficiently strong relationship between t-fMRI and rs-fMRI to yield consistent improvements in the t-fMRI group activation patterns, and the overlap of homologous regions identified by t-fMRI based solely on rs-fMRI. We do not fully understand the relationship between function during rest and tasks (Buckner et al., 2013), although these results suggest a connection between functional resting-state architecture and the neuronal responses to specific tasks. Previous studies have shown that spontaneous brain activity during a resting state contributes to the variability of stimulus-evoked brain responses (Fox et al., 2007; Northoff et al., 2010; He, 2013), revealing a functional link between those conditions. However, the relationship between resting-state and task-based network architecture is largely unexplored. Our results show that the variability in task fMRI activation patterns and resting-state connectivity structure is related.
In addition to helping to understand the relationship of rs-fMRI networks and anatomy, this has direct utility for t-fMRI group studies. Functional alignment normalizes with regard to displacement of functional areas on the cortex, and improves the corresponding specificity of individual regions on the group level.
Comparison of anatomo-functional dissociation between cohorts
The most noticeable difference of anatomo-functional dissociation between left- and right-handed subjects is found in the left hemisphere, and in regions that can be associated with the arcuate fasciculus (Catani et al., 2005; Catani and Mesulam, 2008) and the uncinate fasciculus. We observed distinct differences in the inferior parietal cortex around the supra-marginal gyrus, the lateral temporal cortex, and in the lateral frontal cortex. Takaya et al. (2015) showed in a study group of right-handers, that functional connectivity along the arcuate fasciculus is asymmetric. Similar to Takaya et al. (2015), our results also show the highest displacement for right-handers in the inferior parietal cortex and the lateral temporal cortex. This indicates that our functional alignment approach reveals the functional variability in language related regions. A higher displacement in the frontal regions of the left hemisphere for left-handed subjects could be related to heterogeneous language lateralization. It has been shown that handedness influences the lateralization of language-related brain activity (Knecht et al., 2000) and language-related resting-state networks (Wang et al., 2015). Similar, but less variable, core regions associated with the arcuate fasciculus could be observed on the right hemispheres. Since the arcuate fasciculus connects brain regions that are part of the language network (Catani et al., 2005; Catani and Mesulam, 2008), it can be assumed that the largest difference between the cohorts is related to the connectivity structure of the language connectome.
Comparison to similar approaches
Functional alignment as proposed by Langs et al. (2014) allows us to fully decouple function from spatial position, since the embedding is purely based on functional connectivity. Comparable manifold alignment techniques, such as Lombaert et al. (2013b) or Lombaert et al. (2015) rely on the incorporation of cortical features and geometrical properties. And while these methods show remarkable results in terms of alignment and computational speed, they are in particular interesting for anatomical surface alignment. Robinson et al. (2014) introduced a flexible framework for multi-modal surface matching, which also offers the possibility to incorporate functional characteristics. While we focus on vertex-wise connectivity, Robinson et al. (2014) derives functional features from a group ICA on rs-fMRI data, followed by dual regression to achieve comparability across subjects. Conroy et al. (2013) uses inter-subject correlation to drive cortical alignment. While this is feasible for stimulus induced task fMRI data, it may fall short for resting-state data, since fluctuations in the BOLD signal are not comparable across subjects.
Limitations
The proposed method has several limitations. The functional alignment is restricted to diffeomorphic deformations of cortical surfaces between subjects, and for the diffeomorphic constraint, the topology of the functional networks matters. This can capture only spatially regularized displacements along the cortex, e.g. alignment of areas that are part of the same network, and not substantial changes, such as the unknown migration of a functional area to another hemisphere. The diffeomorphic constraint is a regularization that limits the variability we are able to capture, at the same time it provides a transparent constraint on the inter-individual differences the method can capture. Although core regions of resting-state networks show topological similarity at coarse scales across healthy subjects (Wang et al., 2015), topological variability in function and structure is evident (Glasser et al., 2016; van Essen, 2005). Therefore, the diffeomorphic constraint may limit the ability of the alignment to capture topological inconsistencies. However, it is a means to regularize and disambiguate mappings. Further research is necessary to determine if this constraint should be relaxed, and if diffeomorphic alignment is suited for task related fMRI data. It may fail for a heterogeneous study group with substantial functional variability, such as in patients affected by brain tumors, or epilepsy (Desmurget et al., 2007; Elbert and Rockstroh, 2004).
Another limitation of the present work is the restriction to an analysis of the cortical surface, which does not capture differences in the entire volume. While the surface representation corresponds to many t-fMRI studies, and allows reporting of relevant results, an extension to the entire volume is a clear next step.
Conclusion
In this paper we have proposed a diffeomorphic functional alignment approach based on the spectral signatures of functional connectivity structures. We applied this approach on rs-fMRI data and showed that our approach is stable, and enables a quantification of the dissociation between anatomy and function. It also addresses the shortcomings of spatial normalization, and enables an evaluation of the differences between cohorts of interest. In particular, we have demonstrated that functional alignment based on rs-fMRI characteristics improves group analysis of task fMRI data, indicating a link between functional resting-state and task architecture.
Supplementary Material
Acknowledgments
We are grateful to Prof. Polina Golland for fruitful in-depth discussions during the development of the ideas for this work. This work was supported by NIH grants R01NS091604, R01EB020740, NIBIB NAC P41EB015902, NICHD U01HD087211, 1K25EB013649-01, R41AG052246-01, 1R21AG050122-01A1, and P50MH106435, by FWF KLI 544-B27, and I2714-B31, by OeNB 14812 and 15929, and EU FP7 2012-PIEF-GA-33003. This research was partly supported by Beijing Municipal Science & Technology Commission No. Z161100002616009, and by the Medical Imaging Cluster at Medical University of Vienna.
Appendix A. Supplementary data
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.neuroimage.2017.04.028.
References
- Brett M, Johnsrude IS, Owen AM. The problem of functional localization in the human brain. Nat Rev Neurosci. 2002;3(3):243–249. doi: 10.1038/nrn756. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Krienen FM, Yeo BTT. Opportunities and limitations of intrinsic functional connectivity mri. Nat Neurosci. 2013;16(7):832–837. doi: 10.1038/nn.3423. [DOI] [PubMed] [Google Scholar]
- Catani M, Jones DK, ffytche DH. Perisylvian language networks of the human brain. Ann Neurol. 2005;57(1):8–16. doi: 10.1002/ana.20319. [DOI] [PubMed] [Google Scholar]
- Catani M, Mesulam M. The arcuate fasciculus and the disconnection theme in language and aphasia: History and current state. Cortex. 2008;44(8):953–961. doi: 10.1016/j.cortex.2008.04.002. special Issue on Brain Hodology - Revisiting Disconnection Approaches to Disorders of Cognitive Function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coifman RR, Lafon S. Diffusion maps. Appl Comput Harmon Anal. 2006;21(1):5–30. (special Issue: Diffusion Maps and Wavelets) [Google Scholar]
- Conroy BR, Singer BD, Guntupalli JS, Ramadge PJ, Haxby JV. Inter-subject alignment of human cortical anatomy using functional connectivity. NeuroImage. 2013;81(0):400–411. doi: 10.1016/j.neuroimage.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SARB, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci. 2006;103(37):13848–13853. doi: 10.1073/pnas.0601417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmurget M, Bonnetblanc F, Duffau H. Contrasting acute and slow-growing lesions: a new door to brain plasticity. Brain. 2007;130(Pt 4):898–914. doi: 10.1093/brain/awl300. [DOI] [PubMed] [Google Scholar]
- Duarte J, Sapiro G, Harel N, Lenglet C. A framework for linear and non-linear registration of diffusion-weighted mris using angular interpolation. Front Neurosci. 2013;7:41. doi: 10.3389/fnins.2013.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbert T, Rockstroh B. Reorganization of human cerebral cortex: the range of changes following use and injury. Neuroscientist. 2004;10(2):129–141. doi: 10.1177/1073858403262111. [DOI] [PubMed] [Google Scholar]
- Essen DV, Ugurbil K, Auerbach E, Barch D, Behrens T, Bucholz R, Chang A, Chen L, Corbetta M, Curtiss S, Penna SD, Feinberg D, Glasser M, Harel N, Heath A, Larson-Prior L, Marcus D, Michalareas G, Moeller S, Oostenveld R, Petersen S, Prior F, Schlaggar B, Smith S, Snyder A, Xu J, Yacoub E. The human connectome project: a data acquisition perspective. NeuroImage. 2012;62(4):2222–2231. doi: 10.1016/j.neuroimage.2012.02.018. (connectivityConnectivity) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans A, Collins D, Mills S, Brown E, Kelly R, Peters T. 3d statistical neuroanatomical models from 305 mri volumes. Nuclear Science Symposium and Medical Imaging Conference 199, 3, 1993, IEEE Conference Record; Oct, 1993. pp. 1813–1817. [Google Scholar]
- Fischl B. Freesurfer. NeuroImage. 2012;62(2):774–781. doi: 10.1016/j.neuroimage.2012.01.021. 20 YEARS OF fMRI 20 YEARS OF fMRI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Raichle ME. Intrinsic fluctuations within cortical systems account for intertrial variability in human behavior. Neuron. 2007;56(1):171–184. doi: 10.1016/j.neuron.2007.08.023. [DOI] [PubMed] [Google Scholar]
- Friston K, Ashburner J, Kiebel S, Nichols T, Penny W, editors. Statistical Parametric Mapping: The Analysis of Functional Brain Images. Academic Press; 2007. [Google Scholar]
- Glasser M, Coalson T, Robinson E, Hacker C, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann C, Jenkinson M, Smith S, Van Essen D. A multimodal parcellation of human cerebral cortex. Nature. 2016;536(7615):171–178. doi: 10.1038/nature18933. (8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, Essen DCV, Jenkinson M. The minimal preprocessing pipelines for the human connectome project. NeuroImage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. (mapping the Connectome) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramfort A, Peyré G, Cuturi M. Fast optimal transport averaging of neuroimaging data. Inf Process Med Imaging. 2015:261–272. doi: 10.1007/978-3-319-19992-4_20. [DOI] [PubMed] [Google Scholar]
- He BJ. Spontaneous and task-evoked brain activity negatively interact. J Neurosci: Off J Soc Neurosci. 2013;33(11):4672–4682. doi: 10.1523/JNEUROSCI.2922-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakab A, Schwartz E, Kasprian G, Gruber GM, Prayer D, Schöpf V, Langs G. Fetal functional imaging portrays heterogeneous development of emerging human brain networks. Front Human Neurosci. 2014;8:852. doi: 10.3389/fnhum.2014.00852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl. NeuroImage. 2012;62(2):782–790. doi: 10.1016/j.neuroimage.2011.09.015. 20 YEARS OF fMRI 20 YEARS OF fMRI. [DOI] [PubMed] [Google Scholar]
- Jiang D, Du Y, Cheng H, Jiang T, Fan Y. Groupwise spatial normalization of fmri data based on multi-range functional connectivity patterns. NeuroImage. 2013;82(0):355–372. doi: 10.1016/j.neuroimage.2013.05.093. [DOI] [PubMed] [Google Scholar]
- Knecht S, Dräger B, Deppe M, Bobe L, Lohmann H, Flöel A, Ringelstein EB, Henningsen H. Handedness and hemispheric language dominance in healthy humans. Brain. 2000;123(12):2512–2518. doi: 10.1093/brain/123.12.2512. [DOI] [PubMed] [Google Scholar]
- Langs G, Lashkari D, Sweet A, Tie Y, Rigolo L, Golby A, Golland P. Learning an atlas of a cognitive process in its functional geometry. In: Székely G, Hahn H, editors. Information Processing in Medical Imaging. Vol. 6801 of Lecture Notes in Computer Science. Springer; Berlin Heidelberg: 2011. pp. 135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langs G, Sweet A, Lashkari D, Tie Y, Rigolo L, Golby AJ, Golland P. Decoupling function and anatomy in atlases of functional connectivity patterns: language mapping in tumor patients. NeuroImage. 2014;103(0):462–475. doi: 10.1016/j.neuroimage.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langs G, Tie Y, Rigolo L, Golby A, Golland P. Functional geometry alignment and localization of brain areas. In: Lafferty J, Williams C, Shawe-Taylor J, Zemel R, Culotta A, editors. Advances in Neural Information Processing Systems. Vol. 23. Curran Associates, Inc; 2010. pp. 1225–1233. [PMC free article] [PubMed] [Google Scholar]
- Langs G, Wang D, Golland P, Mueller S, Pan R, Sabuncu MR, Sun W, Li K, Liu H. Identifying shared brain networks in individuals by decoupling functional and anatomical variability. Cereb Cortex Bhv. 2015:189. doi: 10.1093/cercor/bhv189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombaert H, Arcaro M, Ayache N. Brain transfer: Spectral analysis of cortical surfaces and functional maps. In: Ourselin S, Alexander DC, Westin C-F, Cardoso MJ, editors. Information Processing in Medical Imaging: 24rd International Conference, IPMI 2015, vol. 9123 of Lecture Notes in Computer Science. Springer; 2015. pp. 474–487. [DOI] [PubMed] [Google Scholar]
- Lombaert H, Grady L, Polimeni J, Cheriet F. Focusr: feature oriented correspondence using spectral regularization-a method for precise surface matching. IEEE Trans Pattern Anal Mach Intell. 2013a;35(9):2143–2160. doi: 10.1109/TPAMI.2012.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombaert H, Sporring J, Siddiqi K. Information Processing in Medical Imaging: Proceedings of the 23rd International Conference, IPMI 2013. Springer Berlin Heidelberg; Berlin, Heidelberg: 2013b. Diffeomorphic spectral matching of cortical surfaces; pp. 376–389. [DOI] [PubMed] [Google Scholar]
- Margulies D, Ghosh S, Goulas A, Falkiewicz M, Huntenburg J, Langs G, Bezgin G, Eickhoff S, Castellanos F, Petrides M, Jefferies E, Smallwood J. Situating the default-mode network along a principal gradient of macroscale cortical organization. Proc Natl Acad Sci USA. 2016;113(44):12574–12579. doi: 10.1073/pnas.1608282113. (11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Wang D, Fox MD, Yeo BTT, Sepulcre J, Sabuncu MR, Shafee R, Lu J, Liu H. Individual variability in functional connectivity architecture of the human brain. Neuron. 2013;77(3):586–595. doi: 10.1016/j.neuron.2012.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nenning K-H, Kollndorfer K, Schöpf V, Prayer D, Langs G. Information Processing in Medical Imaging. Springer; 2015. Multi-subject manifold alignment of functional network structures via joint diagonalization; pp. 462–473. [DOI] [PubMed] [Google Scholar]
- Northoff G, Qin P, Nakao T. Rest-stimulus interaction in the brain: a review. Trends Neurosci. 2010;33(6):277–284. doi: 10.1016/j.tins.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Oldfield R. The assessment and analysis of handedness: the edinburgh inventory. Neuropsychologia. 1971;9(1):97–113. doi: 10.1016/0028-3932(71)90067-4. [DOI] [PubMed] [Google Scholar]
- Robinson EC, Jbabdi S, Glasser MF, Andersson J, Burgess GC, Harms MP, Smith SM, Essen DCV, Jenkinson M. Msm: a new flexible framework for multimodal surface matching. NeuroImage. 2014;100:414–426. doi: 10.1016/j.neuroimage.2014.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabuncu MR, Singer BD, Conroy B, Bryan RE, Ramadge PJ, Haxby JV. Function-based intersubject alignment of human cortical anatomy. Cereb Cortex. 2010;20(1):130–140. doi: 10.1093/cercor/bhp085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Beckmann CF, Ramnani N, Woolrich MW, Bannister PR, Jenkinson M, Matthews PM, McGonigle DJ. Variability in fmri: a re-examination of inter-session differences. Human Brain Mapp. 2005;24(3):248–257. doi: 10.1002/hbm.20080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaya S, Kuperberg G, Liu H, Greve D, Makris N, Stufflebeam SM. Asymmetric projections of the arcuate fasciculus to the temporal cortex underlie lateralized language function in the human brain. Front Neuroanat. 2015;9:119. doi: 10.3389/fnana.2015.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tootell RB, Reppas JB, Kwong KK, Malach R, Born RT, Brady TJ, Rosen BR, Belliveau JW. Functional analysis of human MT and related visual cortical areas using magnetic resonance imaging. J Neurosci: Off J Soc Neurosci. 1995;15(4):3215–3230. doi: 10.1523/JNEUROSCI.15-04-03215.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Essen DC. A population-average, landmark- and surface-based (pals) atlas of human cerebral cortex. NeuroImage. 2005;28(3):635–662. doi: 10.1016/j.neuroimage.2005.06.058. 〈 http://www.sciencedirect.com/science/article/pii/S1053811905004945〉) [DOI] [PubMed] [Google Scholar]
- Vercauteren T, Pennec X, Perchant A, Ayache N. Diffeomorphic demons: Efficient non-parametric image registration. NeuroImage. 2009;45(1, Supplement 1):S61–S72. doi: 10.1016/j.neuroimage.2008.10.040. mathematics in Brain Imaging. [DOI] [PubMed] [Google Scholar]
- Wang D, Buckner RL, Fox MD, Holt DJ, Holmes AJ, Stoecklein S, Langs G, Pan R, Qian T, Li K, Baker JT, Stufflebeam SM, Wang K, Wang X, Hong B, Liu H. Parcellating cortical functional networks in individuals. Nature Neuroscience Advance Online Publication. 2015 doi: 10.1038/nn.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo B, Sabuncu M, Vercauteren T, Ayache N, Fischl B, Golland P. Spherical demons: fast diffeomorphic landmark-free surface registration. IEEE Trans Med Imag. 2010;29(3):650–668. doi: 10.1109/TMI.2009.2030797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zöllei L, Polimeni JR, Fischl B, Liu H, Buckner RL. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106(3):1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.