Abstract
Diffusion weighted magnetic resonance imaging, or DWI, is one of the most promising tools for the analysis of neural microstructure and the structural connectome of the human brain. The application of DWI to map early development of the human connectome in-utero, however, is challenged by intermittent fetal and maternal motion that disrupts the spatial correspondence of data acquired in the relatively long DWI acquisitions. Fetuses move continuously during DWI scans. Reliable and accurate analysis of the fetal brain structural connectome requires careful compensation of motion effects and robust reconstruction to avoid introducing bias based on the degree of fetal motion. In this paper we introduce a novel robust algorithm to reconstruct in-vivo diffusion-tensor MRI (DTI) of the moving fetal brain and show its effect on structural connectivity analysis. The proposed algorithm involves multiple steps of image registration incorporating a dynamic registration-based motion tracking algorithm to restore the spatial correspondence of DWI data at the slice level and reconstruct DTI of the fetal brain in the standard (atlas) coordinate space. A weighted linear least squares approach is adapted to remove the effect of intra-slice motion and reconstruct DTI from motion-corrected data. The proposed algorithm was tested on data obtained from 21 healthy fetuses scanned in-utero at 22–38 weeks gestation. Significantly higher fractional anisotropy values in fiber-rich regions, and the analysis of whole-brain tractography and group structural connectivity, showed the efficacy of the proposed method compared to the analyses based on original data and previously proposed methods. The results of this study show that slice-level motion correction and robust reconstruction is necessary for reliable in-vivo structural connectivity analysis of the fetal brain. Connectivity analysis based on graph theoretic measures show high degree of modularity and clustering, and short average characteristic path lengths indicative of small-worldness property of the fetal brain network. These findings comply with previous findings in newborns and a recent study on fetuses. The proposed algorithm can provide valuable information from DWI of the fetal brain not available in the assessment of the original 2D slices and may be used to more reliably study the developing fetal brain connectome.
Keywords: Fetal brain, Diffusion-weighted MRI, Registration, Motion Correction, Connectome
1. Introduction
The analysis of white matter structure and neural connectivity is highly desired for the evaluation of normal and abnormal brain growth during the fetal and neonatal periods when the brain undergoes its most rapid development (Dubois et al., 2014; Vasung et al., 2016). Diffusion-weighted magnetic resonance imaging (DW-MRI), or DWI, is considered one of the most promising tools for the analysis of neural structural connectivity and white matter abnormalities. Significant insight has been gained about early brain development through ex-vivo DWI and MRI of postmortem fetal brains as an excellent complement to histology (Huang et al., 2006; Kostovic and Vasung, 2009; Takahashi et al., 2012; Huang et al., 2013; Kolasinski et al., 2013; Xu et al., 2014; Huang and Vasung, 2014). While histology provides much better spatial resolution (~ μm) compared to ex-vivo and in-vivo DWI (~ 100–500 μm, and ~ 1000–2000 μm, respectively), DWI provides non-invasive 3D characterization and analysis of the whole brain anatomy with much less imaging time. Ex-vivo studies have shown proliferation of neuroepithelial stem cells during 9–13 week gestational age (GA) (Kostovic and Vasung, 2009), followed by radial migration and axonal growth which forms a dominant radial organization in the second trimester and gradually turns into emergent fiber bundles by the end of the third trimester (Takahashi et al., 2012). Limbic fibers develop first, association fibers develop last, and commisural and projection fibers develop from anterior to posterior throughout the prenatal developmental period (Huang et al., 2006; Huang and Vasung, 2014).
The advantage of ex-vivo imaging is its relatively high spatial resolution and signal-to-noise ratio that is achieved by using high-field magnets to scan a fixed, non-moving specimen with extended MR imaging time. The reported time to perform such scans ranges from 2 hours per case on 4.7-T MRI scanners (Takahashi et al., 2012; Xu et al., 2014) up to 21 hours per case on 4.7-T or 11.7-T MRI scanners (Huang et al., 2006, 2013). Ex-vivo DWI, however, can only be performed in postmortem specimens. The number of samples in previous ex-vivo studies was hence limited to 3 fetal brains at 19–20 weeks GA by Huang et al. (2006), 11 samples in the 13–21 weeks GA by Huang et al. (2013), and 17 samples in 19–41 weeks GA by Xu et al. (2014). Obviously the ability to perform in-vivo DWI in the fetus will dramatically enhance the use of this powerful imaging technique in analyzing and understanding the mechanisms of brain development and neurodevelopmental abnormalities and their connection to genetic and environmental factors, as well as its clinical use (Takahashi et al., 2012; Huang et al., 2013). In-vivo DWI of the fetal brain, however, is very challenging, mainly because of uncontrollable fetal motion. Practical limits on acquisition time and limited signal-to-noise ratio (SNR) achievable from the small fetal brain through receivers of a body coil, combined with motion artifacts, limit the spatial resolution and accuracy of fetal DWI. Maternal sedation and breath-holds are not desired and not routinely performed in fetal MRI in the United States (U.S.) as most sedatives have been considered toxic or of uncertain harm in animal models by U.S. Food and Drug Administration (FDA).
Fetal DWI was examined in-vivo for the evaluation of white matter development in early 2000s (Righini et al., 2003; Bui et al., 2006). Early studies and current clinical evaluation are based on measurements in 2D slices acquired rapidly with a few diffusion directions. Righini et al. (2003) used short (20-second) breath-hold DWI scans with only 3 non-colinear gradient directions to establish normal values of apparent diffusion coefficient (ADC) from 15 fetuses scanned at 22–35 weeks GA. Bui et al. (2006) used ~ 1-minute DWI scans with 6 non-colinear gradient directions to compute both ADC and fractional anisotropy (FA) values. Despite using oral sedatives and maternal breath-hold or quiet breathing they reported 50% dropout in tests due to excessive motion artifacts. These early studies showed the necessity of technical developments to compensate for motion and achieve better spatial resolution and SNR. Kim et al. (2008) developed a short-repetition time, fast DWI scheme with model-based compensation of longitudinal relaxation effects to achieve 3-directional DWI in 13 and 18 seconds. While this method allows maternal breath-hold and significantly reduces motion effects, it is limited to 3 or very few gradient directions that do not allow more advanced analysis of brain structure with DWI.
Advanced DWI analysis can lead to high-level information about the white matter microstructure and its connections beyond ADC and FA maps. With sufficient number of non-colinear gradient directions (more than 6), DWI can be used for diffusion-tensor imaging (DTI) which can map the white matter tracts in 3D. To achieve white matter tractography, in-utero, Kasprian et al. (2008) were the first who used ~ 2-minute axial, single-shot echo-planar DWI sequences with 32 diffusion gradient directions in 40 unsedated fetuses in the GA range of 18–37 weeks and reported successful visualization of sensorimotor and callosal tracts in 40% of cases. Reduced head mobility due to engagement of the fetal head in the maternal pelvis (90% of studied fetuses were in cephalic presentation) and reduced amniotic fluid due to abnormalities were considered factors that potentially contributed to relatively high success rate despite the relatively long DWI acquisition times in this study. In another study, Zanin et al. (2011) applied 12-directional, ~ 2-minute DWI scans to 61 normal fetuses in the GA range of 23–38 weeks. Despite running the DWI three times to increase the chance of a motion-free scan, they reported a 28% success rate, visualized corticospinal tracts, optic radiations, and callosal tracts in 17 fetuses, and suggested 3 phases for white matter maturation based on DTI measurements including FA and ADC values. In more recent studies, Kasprian et al. (2013) and Jakab et al. (2015) used in-vivo DWI and the commonly-used DTI processing software with volume-to-volume registration to analyze disrupted brain connectivity in fetuses with callosal agenesis. Mitter et al. (2015b) applied similar DTI processing methods to 16-directional DWI and achieved in-vivo tractography of fetal association fibers in 24 living unsedated fetuses in the GA range of 20–34 weeks. They reported 20% success rate (24 of 120 fetal DTI examinations) in this analysis. Mitter et al. (2015a) validated in-vivo tractography of 7 fetal cases with histology. This cohort included 4 cases with callosal agenesis and a case with Joubert syndrome. The analysis showed both the value of in-vivo DWI and its limitations.
Fetal MRI has evolved through the use of more advanced hardware and software (?); for example the use of 3-T for fetal MRI has led to higher SNR and spatial resolution. Routine DTI processing methods and tools, designed for adult brains, including volume-to-volume registration and motion detection, were deemed insufficient and unsuitable for the continuously moving fetal brain, therefore several studies addressed the technical challenges surrounding 3D fetal DWI reconstruction from stacks of multiple 2D slices: Jiang et al. (2009) developed the first motion correction technique for the reconstruction of fetal brain DTI from multiple axial DWI scans. They used slice-to-volume registration for motion correction and scattered data interpolation (SDI) for DTI reconstruction. This technique was inspired by the earlier works on structural fetal brain MRI reconstruction (Rousseau et al., 2006; Jiang et al., 2007), in which slice-to-volume registration was initialized by volume-to-volume registration. Oubel et al. (2012) proposed a groupwise registration approach where diffusion-sensitized and non-diffusion sensitized image slices were registered separately in groups and subsequently to a T2-weighted MR image of the fetal brain, and solved a high-order model of the DWI signal through dual radial basis function interpolation. Fogtmann et al. (2014) proposed adding a robust regularization prior based on Huber norm to a three-stage registration-reconstruction process initialized by the registration approach developed by Oubel et al. (2012). They coupled registration (for motion correction) with a model of the imaging point spread function (PSF) to enable reconstruction from multiple-view scans. In terms of results, Jiang et al. (2009) reported ADC and FA values in 8 fetuses. Fogtmann et al. (2014) showed experimental results for FA and tractography (of the corpus callosum) in a sedated fetal monkey and 4 human fetal cases, and Oubel et al. (2012) presented the results of successful callosal and pyramidal tractography in 4 sedated human fetal cases.
Slice-to-volume registration, which has been the core of the above mentioned techniques for fetal head motion correction, was previously proposed to improve functional MRI of moving subjects (Kim et al., 1999) and has been extensively used in more recent, robust, model-based structural T2-weighted fetal brain MRI reconstruction algorithms (Gholipour et al., 2010; Kuklisova-Murgasova et al., 2012; Kainz et al., 2015). Motion correction in MRI, in general, has also been studied extensively with partial solutions that depend on the type of the sequences and application, and the amount and frequency of motion (Zaitsev et al., 2015). Prospective motion correction techniques (Maclaren et al., 2010; White et al., 2010; Maclaren et al., 2013), in particular those designed for DWI (Aksoy et al., 2011; Kober et al., 2012), cannot be readily used for fetal DWI, and the currently-used retrospective methods, that are based on volume-to-volume registration and volume-level motion detection (Elhabian et al., 2014; Kreilkamp et al., 2015), are not robust and efficient in the case of continuous fetal motion. These technical difficulties motivated the technical works reviewed earlier (Jiang et al., 2009; Oubel et al., 2012; Fogtmann et al., 2014). Nevertheless motion correction using slice-to-volume registration and DWI reconstruction need to be further improved. In our experiments we identified that the ill-posed nature of the slice-to-volume registration process and the effect of motion are the main barriers to achieve robust fetal brain DWI reconstruction.
To address the technical challenges in fetal brain DWI reconstruction, in this paper we introduce an algorithm that relies on two main innovations: 1) robust inter-slice motion estimation through coupling temporal image registration with a discrete-time model of fetal head motion dynamics; and 2) robust DTI model reconstruction through detecting and rejecting motion-corrupted data and weighted linear least squares (WLLS) estimation that mitigates the effect of signal loss caused by intra-slice motion, a critical point that was not addressed in previous works. The motion tracking algorithm we propose here for fetal DWI has been adapted from our recent work on motion-robust DWI reconstruction in young children and adults (Marami et al., 2016a). In contrast to previous works which relied on independent slice-to-volume registration (Oubel et al., 2012) or a regularized version of it (Fogtmann et al., 2014), in our approach we explicitly modeled dynamics of motion with a state space model, and estimated temporal motion trajectories with a robust Kalman filter (Agamennoni et al., 2012). We note that the ultrafast slice acquisition in DWI (slice acquisition time ≈ 100ms), results in a sampling rate close to 10Hz that allows effective use of our approach in DWI of moving subjects. In an extension of our original work we have achieved improved multi-plane coverage of the anatomy using simultaneous multi-slice DWI in adult subjects (Marami et al., 2016b).
To achieve robust performance in fetal brain DWI, which suffers from complex motion effects including intra-slice motion artifacts, limited SNR, limited spatial resolution, and arbitrary orientation, we 1) relax the causality property of our motion tracking approach through a sliding window on the sequence of acquired slices; 2) automatically detect and reject motion-corrupted slices to enhance motion tracking and model reconstruction; and 3) augment motion tracking with our proposed image registration and processing pipeline that allows DTI reconstruction and in turn standard color-coding and analysis of FA, tractography, and groupwise whole-brain connectivity analysis in a standard coordinate space. We examined our proposed algorithm first in adult volunteers who stayed still and moved during DWI scans and then in 21 non-sedated fetuses scanned in the GA range of 22 to 38 weeks. We compared FA values in regions-of-interest and evaluated tractography and fetal brain connectivity analysis. The results indicate robust reconstruction and tractography in the presence of various amounts of motion, and significant improvement in all metrics and analyses, including group connectivity analysis, over methods that do not explicitly take into account the dynamics of motion and the effect of intra-slice motion-induced signal loss. Connectivity analysis based on 21 cases in this study shows that, similar to adult and newborn brains, the fetal brain network has high modularity and clustering, and small world characteristics in the second half of pregnancy. Our results comply with the findings in neural connectivity in preterm infants and newborns (van den Heuvel et al., 2014; De Asis-Cruz et al., 2015), and fetuses (Jakab et al., 2015).
2. Materials and Methods
A fetal DWI acquisition involves a series of 2D spin-echo echo-planar imaging volume acquisitions (each with M slices) that involve N0 b=0 volumes (S0) and N1 diffusion-sensitized scans b≠0 (Si; i = 1…N1). Each Si has a corresponding diffusion gradient direction gi. Typical values of N0, N1, and M for fetal DWI can be 1–3, 12–15, and 15–30, respectively, depending on the age and size of the fetus. A single DWI scan, therefore, includes K = M × (N0 + N1) slices (i.e. 200–500 slices) that are acquired in an interleaved manner in intervals of the repetition time (TR). Often, the fetus moves continuously during slice acquisitions, but the motion is not always so fast that it affects the quality of each DWI slice acquisition. Consequently, intra-slice motion artifacts do not happen frequently as fetal and maternal motion is often much slower than the timing of each DWI slice acquisition (≈ 100ms). Inter-slice motion, on the other hand, almost always exists. The DWI acquisitions may be repeated a few times in the same or different orientations (axial, coronal, or sagittal) with respect to the fetal head providing redundancy in data (in image and q-space) that would help increase the SNR in DTI model estimation if scans are combined.
Image slices are acquired in an interleaved manner, but each slice is associated with a time stamp k at which it is obtained (k = 1, …, K). All geometric properties (including points and gradient directions) are calculated in the world (scanner) coordinate system. Unlike an adult head that is oriented in a specific direction inside a head coil, the fetal head can be in any arbitrary position/orientation and can move continuously during acquisitions. One slice is the smallest packet of acquired k-space (and q-space) data; therefore, we consider each slice as an observation (or measurement) with a transformation Tk that defines the relative position of the fetal head at time k with respect to a target volume S. We further assume transformations that map S to a target S0, , S0 to a reconstructed T2-weighted (T2w) MRI scan R, , and R to an age-matched atlas image A, (TR→A) that is correctly oriented for color coding the FA and tracts. The details of estimating Tks, the target volumes and transformations, and DTI reconstruction technique are discussed in the following subsections.
2.1. Robust inter-slice motion tracking and registration
We propose to track and estimate the motion of the fetal head during DWI slice acquisitions. We consider a linear dynamic model for fetal head motion, which is described as a discrete-time state-space model
| (1) |
This is a model in which the finite sequence of observations yk (DWI slices) are generated as a function of the sequence of hidden states xk that represent the d degrees-of-freedom motion; ωk and νk are the process and measurement noise and represent uncertainty in the modeling of motion dynamics and observations, respectively. IS is a nonlinear function that relates the motion states xk to the observations yk. The goal is to estimate the states xk ∈ ℜd.
Assuming that the analysis is performed in an area around the fetal head, that undergoes 3D rigid-body motion, d = 6 and xk = [θx, θy, θz, tx, ty, tz]T is a vector of 3 translation and 3 rotation parameters. The solution of the motion tracking problem constitutes estimating the a posteriori probability density function p(xk|y0, …, yK). To solve the problem with image registration, we separate the nonlinear part and rewrite the output model of (1) as
| (2) |
where zk ∈ ℜd is the measurement vector; f (IS(p), yk) is a similarity metric between a reference volume transformed with parameters p and the observations; and h is a window size parameter that controls the bias-variance trade-off in the estimation of motion parameters. An appropriate value of h depends on experimental conditions including 1) sampling rate vs. the speed of motion and 2) reliability of image registration, which depends on the degrees-of-freedom of the transformation and the fidelity of the similarity metric based on image features, resolution and SNR. In our experiments we tried different values of h in the range of 0 and 10 and found h = 1 as an appropriate value. Note that h = 0 leads to a causal motion tracking system that can be used for prospective correction, but has a relatively high variance so is not very robust to inaccuracy of slice-to-volume registration. On the other hand, large values of h significantly regularize slice-to-volume registration and make it robust but lead to smooth non-causal motion tracking systems that will be similar to volume-to-volume registration and interpolation.
The process noise in (1) is drawn from a Gaussian distribution, i.e., as we model it with a random walk; however, the measurement noise in (2) may have heavier tails than a Gaussian distribution because of 1) nonlinearity and uncertainties in slice-to-volume registration, and 2) intra-slice motion that may result in signal loss and exclusion of a number of slices. To deal with non-Gaussian measurement noise we used the robust state estimation technique developed by Agamennoni et al. (2012) that allowed accounting for uncertainties in the measurements caused by registration failures. For all practical purposes image registration was performed in the reverse order of source and target images, i.e., the algorithm dynamically tracks motion by registering a target volume to a set of 2h + 1 slices acquired over the k − h to k + h time steps. Note that due to the interleaved nature of DWI acquisition these temporally adjacent slices are not adjacent in image space and provide a distant coverage of the anatomy in 3D. For example in a 3-interleaved acquisition with a slice thickness of 2 mm, the three slices that fall within a temporal window of h = 1 are 6 mm apart so they sample the anatomy in a distance of 12 mm. This provides a good coverage of the small fetal brain in the slice select direction and further enhances the image registration process for motion tracking.
For robust state estimation we used the algorithm developed by Marami et al. (2016a). Motion-free target volumes are not usually available in fetal MRI and should be reconstructed. We reconstructed R (a T2w image of the fetal brain) from multiple sets of T2w MRI slices using the robust algorithm developed by Kainz et al. (2015). S0 was reconstructed through an iterative process by running our inter-slice motion tracking method for all b=0 volumes followed by reconstruction. The reconstructed image at each iteration was used in the next iteration as the target volume for registration. Similar to S0, S was reconstructed using all b≠0 volumes through iterations. Then, for each DWI scan, we estimated Tks (transformation parameters for each slice) using the proposed inter-slice motion tracking method where S was used as the target volume. and TR→A were computed using volume-to-volume registration from S to S0 and from R to an age-matched template from a correctly oriented spatiotemporal fetal brain MRI atlas (Gholipour et al., 2014b, 2017). As a result, we have a reference b=0 image in the atlas space and transformations that map each DWI slice to that image.
The entire image processing and registration pipeline has been shown in Figure 1. This involves the processing of multiple stacks of T2w images of the anatomy (top row) and the processing of the DWI slices (bottom row). The reconstructed T2w image played the role of a reference image to map S0 and consequently the DWI data into the atlas coordinate space through and TR→A. The brain mask and anatomical parcellation achieved on this image were also used to guide similarity metric calculation and improve motion-tracking slice-to-volume registration in DWI, and for tractography and region-based structural connectivity analysis, respectively. DTI reconstruction, tractography, and connectivity analysis, shown in this diagram, are discussed next.
Figure 1.

The processing pipeline for motion-robust fetal brain DTI reconstruction and structural connectivity analysis. The two dashed-line boxes in the figure separate the T2w image processing pipeline (on top) from the DWI processing pipeline (in the bottom). A volumetric T2w image with isotropic resolution of 0.7mm3 is reconstructed from multiple stacks of T2w scans using robust super-resolution volume reconstruction. An approximate ellipsoidal brain mask is used to crop the brain region prior to reconstruction, but a precise manual brain mask is obtained after reconstruction in ITKSNAP (Yushkevich et al., 2006). The brain mask is then used in N4 correction of intensity non-uniformity (Tustison et al., 2010). The corrected masked image is registered to the atlas space using the algorithm discussed by Gholipour et al. (2012), and tissue segmentation and anatomical parcellation is obtained from multi-atlas segmentation using label propagation through ANTS deformable registration (Avants et al., 2008) and probabilistic label fusion (Akhondi-Asl and Warfield, 2013). A spatiotemporal fetal brain MRI atlas (Gholipour et al., 2014b, 2017) is used in this process with anatomical parcellations adopted from the neonatal brain atlases by Blesa et al. (2016). The DWI stacks of slices are processed with the algorithm developed and discussed in section 2.1. Through the combination of the rigid transformations computed throughout this pipeline, the DWI data are mapped into the S0 image for the reconstruction of DTI in standard orientation. This allows standard color-coding of FA and tractography and enables groupwise connectivity analysis.
In addition to motion tracking for inter-slice motion correction, we needed to take into account the effect of intra-slice motion in the slice motion estimation and DTI reconstruction. This is crucial as signal loss in motion-corrupted slices may mislead image registration and adversely affect the performance of the motion tracking algorithm; As shown in Figure 2a fast fetal motion often affects several slices in a row and the temporal registration information may not be reliable in that period. To this end, we implemented a technique to automatically detect and exclude through-slice motion-corrupted DWI slices using statistical learning. In our earlier work (Marami et al., 2016a) we employed a rule-based technique using two image features computed through a morphological closing filter along the slice-select direction to detect inter-slice intensity discontinuity (first and second features in Figure 2b). In addition to using the above mentioned features, to adopt an algorithm for fetal brain DWI which presents more complex intra-slice motion artifacts, we used normalized mean intensity difference (NMID) of each slice with the whole volume (third feature in Figure 2b), and entropy of the individual slices (forth feature in Figure 2b) to model a support vector machine (SVM) classifier. For this purpose we labeled 2676 image slices (60 image volumes obtained from 12 fetuses), among which 735 were visually identified as corrupted by intra-slice motion. Using the labeled images, we trained and tested the SVM classifier achieving 86.6% sensitivity in detecting motion-corrupted slices and 98.2% specificity on the test data (25% of the all labeled image slices). Using the trained classifier motion-corrupted slices were automatically detected and excluded from both motion tracking and DTI reconstruction processes.
Figure 2.

(a) Some of the slices among 25 interleaved axial diffusion-sensitized image slices of a fetal brain DWI scan, shown in this figure, are corrupted by intra-slice fast motion. slices are ordered from top-left to bottom-right. (b) Input features and the output of the SVM classifier for the example test volume shown in (a). Seven slices were classified as motion-corrupted for this volume.
2.2. Robust diffusion tensor image reconstruction
To reconstruct DWI models from registered slices, we construct a data structure of physical voxels from the mapped DWI data. This data structure stores the intensity value, point-spread-function (PSF), b value, and the corrected diffusion gradient direction in the local neighborhood of regular grid points on the reference b=0 image. For a DTI model based on the Stejskal-Tanner equation: , where B0 and Bi are the intensity values of the b=0 image and the diffusion-sensitized images (b≠0), respectively, we compute the DTI model using a WLLS estimation method (Koay et al., 2006; Marami et al., 2016a) by minimizing:
| (3) |
where γ = [Dxx, Dxy, Dxz, Dyy, Dyz, Dzz] is a vector of 6 DTI model parameters which is estimated in a way that guarantees a positive definite diffusion tensor D, and Mi,j is the n × 6 DTI design matrix that is built from the (x, y, z) components of the diffusion gradient directions (gis) and the b value; Bi is the observed diffusion-sensitized signal at any registered slice voxel, and is the corresponding b=0 signal, which is interpolated from the b=0 image at the slice voxel location, and n is the total number of voxels in the neighborhood of the regular grid point.
The weights wis in (3) balance the contribution of the observed voxels based on their distance to the tensor location, which are calculated based on the PSF kernel function. The shape of the PSF is acquisition dependent and its exact calculation yields improved image contrast in image reconstruction (Jiang et al., 2009; Gholipour et al., 2010; Kuklisova-Murgasova et al., 2012; Kainz et al., 2015). For fetal DWI we used a Gaussian slice profile
| (4) |
where dxi, dyi and dzi are the distance between the center of the ith voxel and the center of the grid point (reconstructed voxel) transformed to the slice image domain using the transformation estimated for the slice image; hence, dxi and dyi are the offsets from the center of the voxel on the slice image plane, and dzi is the offset in slice acquisition direction. σx, σy and σz determine the width of the bell-shaped Gaussian function in the x, y (in-plane) and z (out-of-plane) direction. As suggested by Jiang et al. (2009) and Kainz et al. (2015) these values are obtained based on the image spacing as
| (5) |
where sx, sy and sz are image spacing in the x, y (in-plane) and z (out-of-plane) direction. The weights αi are set to Bi, not only to contribute to nonlinear DTI model estimation as suggested by prior work, e.g., Salvador et al. (2005), but also to make the estimation robust to complete or partial signal loss due to intra-slice motion. The gis in Mi,j are the corrected diffusion gradient directions based on the Ttk of each slice; i.e., gi = Rtkg0i, where g0i is the ith predefined gradient direction in the scanner coordinates and Rtk is the rotation matrix corresponding to Ttk. The WLLS solution in (3) provides robust DTI reconstruction from motion-corrected data.
2.3. Tractography and connectivity analysis
Unlike most of the previous works that generated ROI-based tracts without standard color-coding, standard color-coding of the FA maps and tracts was achieved in this study thanks to the mapping to the correctly-oriented atlas coordinates. While deterministic region-based tractography was used in previous works on fetal DWI, whole-brain stochastically-initialized locally deterministic step tractography was performed in this study. This was performed by the method described by Peters et al. (2012) and Lewis et al. (2013), with 8 tract seeds and 5 steps per voxel, FA and direction momentum both equal to 0.5, minimum FA threshold of 0.1 (Mukherjee et al., 2002), and maximum fiber angle of 40 degrees. The range of potential streamlines examined in this approach was broad compared to conventional deterministic tractography. Stochastic sampling was continued until a predetermined number of streamlines (equal to 8 here) were created for each seed voxel. Constrained quaternion interpolation was used and the tracts were stopped at the boundary of the brain mask.
Structural connectivity analysis was performed through atlas-based parcellation of the reconstructed T2w images (through the process described in the caption of Figure 1) and mapping the regions into the space of S0 using the inverse . Atlas-based parcellation was performed based on 90 anatomical regions mapped to the spatiotemporal fetal brain MRI atlas (Gholipour et al., 2017) from the neonatal brain MRI atlas developed by Blesa et al. (2016). We studied structural connectivity based on streamlines connecting all pairs of anatomical regions. A symmetric 90×90 connectivity matrix was obtained for each brain. Streamlines connecting each region to itself were ignored so diagonal elements of the matrix were zero. Using the structural connectivity matrices, we defined structural connectomes of the fetal brains as networks, where regions and connections were considered nodes and edges, respectively. Connectivity strength was calculated between regions ROIi and ROIj by
| (6) |
where Cij is the streamline count between the two regions and Vi and Vj are the volumes of the regions.
To study the properties of the fetal brain structural connectome, we used graph theory and calculated graph theoretical measures. In particular, we calculated measures of connectivity, integration, segregation, and small worldness of the brain network (Rubinov and Sporns, 2010). We measured network connectivity by mean connectivity strength over all nodes. We calculated the node degree as the number of connections between a node and all other nodes. We also computed the degree distribution which is the probability distribution of node degrees over the entire network. We measured network integration using characteristic path length and global efficiency. Characteristic path length quantified the length of the shortest paths through the network and was calculated as the average of the inverse sum of the edge weights for edges along a path (shortest path length) for all pairs of nodes. Global efficiency was calculated as the average inverse of the shortest path lengths. With integration we aimed at quantifying the density of the network. A fully connected network has high integration and a sparsely connected network has low integration. For segregation, which quantifies how much a network is organized into sub-networks, we used two measures: the modularity index and the clustering coefficient. Modularity was calculated as the maximum of the ratio of the number of connections within a sub-network to the number of connections exiting that network over all possible sub-networks. Clustering coefficient was calculated as the average of the fractions of a node’s neighbors that were also connected to each other, weighted by the product of the relevant edge strengths.
One of the most interesting characteristics of efficient complex networks, which has shown to exist in animal and human brain networks, is the small worldness property. A small-world network has both high connectivity and high clustering coefficient or modularity. Such a network has high connectivity while keeping low number of connections. It is not completely understood when the small world sub-networks of the human brain form during brain development but small world characteristics have been observed in neonatal brains (Brown et al., 2014). We measured the small worldness of the fetal brain as the ratio of the clustering coefficient to the characteristic path length. In order to test the degree of significance of these measures, we compared measures of fetal brain connectivity with average measures obtained from 100 random networks generated with similar number of nodes and edges.
Finally, to study the similarity and variability of the connectome between subjects, we calculated the edge space similarity (ESS) of the connectivity networks. Given the connectivity graph networks G and H for two subjects, the ESS is computed as
| (7) |
where is the Dice coefficient of binary overlap between graphs G and H at threshold value d; and Gd and Hd are unweighted binary graphs obtained from thresholding G and H at the graph density value of d (Jakab et al., 2015). ESS is a measure of network similarity between two subjects and takes values between 0 and 1.
All steps of the proposed technique were implemented in C++ using the Insight Toolkit (ITK) (Johnson et al., 2013). For the registration of diffusion-weighted images, we used Mattes mutual information intensity-based image similarity metric (Mattes et al., 2003), a versor type rigid 3D transformation and a gradient-descent approach for optimization. We also used the Computational Radiology Laboratory Toolkit (CRKit) for diffusion MRI analysis, tractography, and the evaluation and visualization of results. Manual work for DWI reconstruction was minimal, limited to creating an approximate ellipsoidal brain mask in the brain region on one of the original images and regular quality control for generating this mask, choosing an initial reference image, and ensuring validity of data and results from initial DICOM conversion to connectivity analysis. Computation time depends on the implementation, computational resources, the number of scans acquired and analyzed for each case, size of the brain (gestational age) and the desired spacing for DTI reconstruction. We ran all steps of the DWI reconstruction and tractography on a Linux system with Intel(R) Xeon(R) CPU X5690 @ 3.47GHz and 64 GB RAM. It required approximately 110 minutes for an example case with three DWI scans, 12 gradient directions (S images) and one S0 image for each scan.
3. Results
We compared DTI reconstruction using our algorithm (motion-tracking slice-to-volume registration; called MT-SVR) with 1) original analysis without motion correction or robust reconstruction (Orig); 2) volume-to-volume registration (VVR) that is the state-of-the-art in DTI analysis (Elhabian et al., 2014), and has been used previously along with motion detection and volume rejection for fetal DTI reconstruction (Kasprian et al., 2008; Zanin et al., 2011; Kasprian et al., 2013; Jakab et al., 2015); and 3) hierarchical slice-to-volume registration (SVR) initialized by volume-to-volume registration followed by temporal registration between sets of S0 and S images, that follows, in steps, the techniques developed by Jiang et al. (2009) and Oubel et al. (2012). We used WLLS estimation in all four DTI reconstruction methods. For evaluation and comparison of techniques, we first conducted controlled motion experiments with healthy adult volunteers, and then applied the techniques to in-vivo DWI of 21 fetuses, in which we examined FA values in different brain regions, performed tractography, and analyzed group connectivity and edge space similarity. The results of these experiments are presented and discussed in the following subsections.
3.1. Quantitative evaluation by controlled motion volunteer experiments
For quantitative evaluation in controlled experiments with gold standard (GS), i.e., motion-free scans, we performed volunteer experiments, where healthy adult volunteers were asked to stay still during one DWI scan and move during several other scans. The total number of 8 DWI scans with intentional motion of different types and magnitude up to 30° (measured with an electromagnetic sensor) were acquired from two volunteers. Imaging was performed on a 3T Skyra Siemens scanner (Siemens Healthineers, Erlangen, Germany) using a 32-channel head coil, and involved DWI with 6 b=0 images and 30 diffusion directions, with b = 1000s/mm2, TR/TE = 9000–13200/88 ms, flip angle =90°, slice thickness =2mm, matrix size =128 × 128, and field-of-view of 256 mm. The study was approved by the Boston Children’s Hospital Institutional Review Board Committee and written informed consent was obtained from all participants.
Figure 3 compares the color-coded FA of different methods for a volunteer experiment. The results indicate that even in the presence of extensive continuous head motion (up to 15 mm in translation and 30° of rotation), our proposed method (MT-SVR) generated superior results that are comparable to the gold standard (GS). Statistics of the L2 norm of FA value differences (ΔFA) (Marami et al., 2016a) and the angle between the principal eigenvectors of the tensors of each method and the GS (ΔDir) (Marami et al., 2016a), shown in Table 1 for three fiber-rich brain regions, showed significantly lower errors by MT-SVR compared to the other methods.
Figure 3.
Axial view of the color-coded FA in an adult volunteer experiment. Our method (MT-SVR) generated FA that is very similar to the GS (based on motion-free scans) and outperformed SVR (note the red artifact on PLIC in (c)), VVR, and Orig.
Table 1.
FA value differences (ΔFA) and the angular difference of the principal eigenvectors of the tensors (ΔDir) in corpus callosum (CC), cingulum (Cin), and the limbs of the internal capsule (LIC) regions for the volunteer subject studies, compared in four methods. Lower values of these dissimilarity metrics indicate better performance. The best values in each comparison have been highlighted with bold text.
| ΔFA
× 102 (mean ± standard deviation)
|
ΔDir
× 102 (mean ± standard deviation)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Orig | VVR | SVR | MT-SVR | Orig | VVR | SVR | MT-SVR | |
| CC | 37±15 | 11±6 | 13±4 | 10±2 | 71±20 | 48±16 | 42±13 | 37±11 |
| Cin | 39±12 | 23±8 | 23±8 | 18±5 | 85±14 | 55±12 | 42±23 | 26±9 |
| LIC | 31±6 | 15±3 | 12±2 | 10±2 | 80±8 | 24±10 | 21±6 | 18±4 |
3.2. Fetal DTI reconstruction
We analyzed MRI scans of 21 fetuses scanned at GA range of 22–38 weeks, 30.6 ± 5.3 weeks. Fetal MRI for these cases was performed on 3T Siemens Skyra scanners using 16 channel body matrix and spine coils with a protocol that involved a 3-plane localizer, multiple repeated T2-weighted (T2w) half-Fourier acquisition single shot turbo spin echo scans of the fetal brain, and 2–8 DWI scans in the axial and/or coronal planes. Each DWI scan was performed with 1–3 b=0 volumes and 12 diffusion-sensitized volumes with b=500 s/mm2, TR/TE=3000–4000/60 ms, variable slice thickness between 2–4 mm, and in-plane resolution of 2 mm. The total acquisition time for each DWI scan was between 50 to 90 seconds, depending on the size of the fetal brain, number of slices, and TR. No maternal sedation or breath-hold was used. The study was approved by the Boston Children’s Hospital Institutional Review Board Committee and written informed consent was obtained from all participants.
Figure 4 shows color-coded FA of a 32 week GA fetus obtained from different techniques as well as the corresponding region of the reconstructed T2w image. This comparison, in specific, shows the effectiveness of motion correction and robust reconstruction technique (MT-SVR). Note that in these experiments we rejected motion corrupted data and used WLLS reconstruction in Orig, VVR, and SVR, but the advantage of MT-SVR in robust dynamic motion tracking and robust reconstruction led to its superior performance compared to other configurations. For quantitative evaluation we calculated FA values obtained from different reconstructions in small regions-of-interest in the corpus callosum (CC) and the anterior/posterior limbs of the internal capsule (LIC), and plotted them for all fetal cases as a function of GA (in days) in Figure 5. This figure shows a steady increase in FA with GA in these fiber-rich regions where high FA values are expected. Uncorrected motion, however, results in blur artifacts and a reduction in FA values; therefore, robust motion correction and reconstruction should result in increased FA values. This analysis indicates improved results by MT-SVR compared to other reconstructions including SVR, which also performed well compared to Orig and VVR. The improvement using MT-SVR over SVR was more prominent in the lower GA range where the fetus usually moves more frequently and faster.
Figure 4.

(a–d) From top to bottom: axial, coronal, and sagittal views of the color-coded FA in a 32-week GA fetus using different DTI reconstructions. (e) The corresponding region of reconstructed T2w image. By correcting motion and robust reconstruction, MT-SVR generates meaningful FA maps of structures like CC, internal capsule, cingulum, and corticospinal tracts that comply much better with our knowledge of the anatomy. The effects of uncompensated motion artifacts are observed in FA maps obtained from Orig, VVR and SVR.
Figure 5.
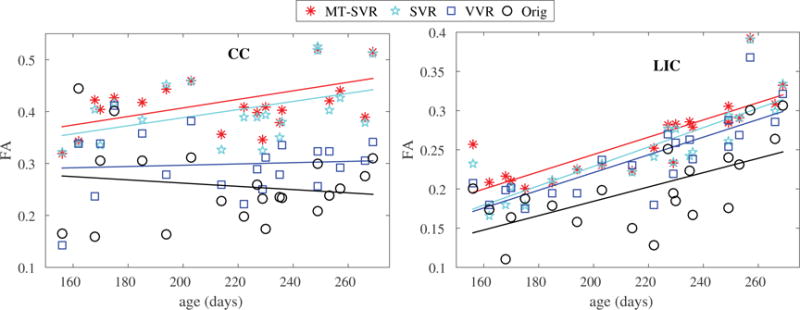
Comparison of FA values in the CC and LIC regions as a function of GA using different DTI reconstructions. Uncompensated motion resulted in reduced FA in fiber-rich regions such as CC and LIC. This figure shows 1) superior results using MT-SVR compared to other methods, and 2) increased FA with GA which can be attributed to increased anisotropic diffusion due to brain maturation. Linear regression equations for FA vs. GA (in days), obtained from MT-SVR results, were FACC = 0.2424 + 0.0008GA and FALIC = 0.0234 + 0.0011GA.
We performed multiple regression analysis to investigate the relationship between FA (in CC and LIC) with GA, number of scans and gradient directions (N1), and the magnitude and speed of motion based on estimated slice motion parameters. Although FA showed negative correlation with the magnitude and speed of motion for all techniques, the effect was non-significant (p > 0.1) for MT-SVR and SVR. The reduction in FA with higher magnitude and speed of motion was, however, significant for Orig and VVR. The results are shown in Figure 6. The dependence to both GA and N1 was significant and positive at p < 0.05, with R2 = 0.45 and 0.82 for the multiple regression models for CC and LIC, respectively. This analysis showed that MT-SVR was robust to motion, while Orig and VVR were not; and the results were improved with higher number of scans, which is expected as more scans improve the SNR and may also improve registration. Linear regression equations for FA vs. GA (in days), obtained from MT-SVR results, were FACC = 0.2424 + 0.0008GA and FALIC = 0.0234 + 0.0011GA.
Figure 6.

Comparison of FA values obtained from different DTI reconstructions in the CC and LIC regions as a function of the average absolute value and speed (differential) of motion. Uncompensated motion resulted in reduced FA in fiber-rich regions such as CC and LIC. This is evident from the reduction in FA in these regions with higher average absolute value and speed of motion. The rate of reduction in FA with increased magnitude and speed of motion was significant for Orig and VVR, but was not significant for MT-SVR. This shows the robustness of MT-SVR to motion in these regions.
As previously mentioned, we used the WLLS estimation technique in Equation (3) for DTI reconstruction. The weights, set as the diffusion-sensitized signal at each voxel, made the estimation robust to complete or partial loss of signal due to fast motion. The effect of WLLS is shown in Figure 7, in which color-coded FA of a 28-week GA fetus obtained from WLLS estimation is compared against FA obtained from linear least square (LLS) estimation (where the weights αi in Equation (3) were set to 1 for LLS). While all image slices were used for reconstruction in Orig, VVR and SVR methods, intra-slice motion-corrupted slices were detected and excluded in the MT-SVR reconstruction. As can be seen from (a) and (e), the fetus moved significantly during the scan (both intra- and inter-slice motion). Intra-slice motion artifacts can be easily seen in (b) and (c) where LLS estimation was used, where these artifacts are much reduced in (f) and (g) with WLLS estimation. Comparing (c) with (d), and (g) with (h) shows the effectiveness of our motion-tracking slice-to-volume registration technique against the SVR method.
Figure 7.

Sagittal views of the color-coded FA in the CC region in a 28-week GA fetus using different motion correction and DTI reconstructions, LLS (a–d) and WLLS (e–h). This figure illustrates the effectiveness of WLLS estimation in mitigating intra-slice motion artifacts in comparison to LLS. It should be noted that although artifacts caused by intra-slice fast motion can be seen in Orig, VVR, and SVR using LLS estimation, since motion-corrupted slices were excluded from the estimation in MT-SVR (both LLS and WLLS), less artifacts are seen in MT-SVR images.
3.3. Fetal tractography and connectivity analysis
Figures 8 and 9 show whole-brain tractography results for two fetuses scanned at 24 and 36 weeks GA, respectively. While tractography based on Orig was significantly degraded by uncompensated motion (Figures 8a and 9a), by using MT-SVR (Figures 8d and 9d), we were able to reconstruct major fiber pathways despite large and fast fetal motion. With MT-SVR we were consistently able to visualize the corticospinal and callosal tracts, as well as corona radiata and cingulum tracts that can easily be obscured or distorted if Orig or VVR reconstructions are used. Visual assessment of tractography results complied with quantitative analyses based on FA values reported earlier, and was also confirmed through groupwise connectivity analysis presented and discussed next. Connectivity analysis shows the impact of MT-SVR and the accuracy that is practically gained by using our proposed method as opposed to using VVR or SVR.
Figure 8.

Tractography of the brain of a 24-week fetus using different motion correction methods. While tractography based on Orig failed, MT-SVR revealed the highest number of major fiber pathways that are formed as early as 24 weeks GA.
Figure 9.

Tractography of the brain of a 36-week fetus using different motion correction methods. Our method (MT-SVR) generated the most detailed tractography of major fiber pathways. Tracts obtained from the other methods were degraded because of residual motion artifacts.
Figure 10 shows color-coded edge space similarity (ESS) matrices of the study subjects obtained from the four reconstruction methods. Grayscale-coded average magnitude of motion has also been shown for each case in the bottom x-axis of the matrices. It is observed that cases with higher amounts of motion typically showed lower ESS values (corresponding to blueish lines) in the matrices. While the effect of uncompensated motion was remarkable in Orig matrices, it was much less in VVR and SVR, and was further decreased in MT-SVR matrices. In particular, the block diagonal values in the bottom right corner of MT-SVR matrix show high similarity of structural connectivity between fetuses at GAs higher than 34 weeks.
Figure 10.

Edge space similarity (ESS) of fetal structural connectivity in the study subjects (ordered by age) for four methods: Orig, VVR, SVR, and MT-SVR. Each color-coded element of the square matrix shows the edge space similarity between two subjects. The subjects (1–21 in the y axis of matrices) have been ordered by age. The grayscale color bar in the bottom of the matrix shows the average absolute magnitude of subject motion calculated from estimated motion parameters (brighter corresponds to larger motion). Rows 2, 9, and 14 correspond to subjects 2, 9, and 14 which had largest magnitudes of motion during image acquisition. These subjects showed low ESS corresponding to bluish lines in the matrices for Orig and VVR methods. Similarity matrices with DTI images, corrected and reconstructed using SVR and MT-SVR, clearly show improved similarity between subjects at similar GA ranges (i.e. block diagonal elements). In particular subjects 9 and 14 show higher similarity to other subjects within the same GA range after motion correction. The block diagonal values in the MT-SVR matrix show high structural similarity of fetuses at GAs larger than 34 weeks. This complies with previous work (van den Heuvel et al., 2014) that showed higher ESS in infants at higher GA-equivalent ages. MT-SVR showed better results than all other methods. The similarity was much diminished in matrices corresponding to Orig and VVR due to uncompensated motion effects.
Finally, using connectivity analysis results obtained from MT-SVR DTI reconstruction, we analyzed the properties of the fetal connectome based on graph-theoretic network measures. Figure 11 shows small-world properties of the fetal connectome based on a high level of binary clustering, short average characteristic path lengths, and a high degree of modularity. These have been compared to baseline measures calculated for 100 random networks for each subject. Figure 12 shows that there is a small number of densely connected hub nodes in the fetal connectome, which results in a heavy-tailed (truncated power law) distribution of hub degrees, which is another evidence for the small world properties of the fetal brain network.
Figure 11.

Small-world properties of the fetal connectome: The results show high level of binary clustering (0.49 ± 0.046) in the fetal brain network compared to the corresponding random graph (0.38±0.024) with same number of nodes and degrees for each node (100 random networks for each subject). Furthermore, topological analysis indicates short average path length between graph nodes (2.60 ± 0.17). Taken together the normalized clustering efficiency and normalized path length along with the small world index (2.43 ± 0.27) indicate that the fetal brain structural network at second and third trimesters shows small-world network characteristics. The modularity value for fetal connectome (0.46 ± 0.04) is significantly higher than the corresponding random graph (100 random networks) (p < 0.001) which indicates that the fetal connectome is modularly developed.
Figure 12.

Heavy-tailed degree distribution of fetal connectome: The graph on the left shows the degree distribution, p(k), of the group-averaged fetal connectome thresholded and binarized at ratio 0.15. It can be seen that the group averaged degree distribution is heavy-tailed. There is a small number of densely connected hub nodes located on the right tail of the distribution. This complies with the previous studies on functional connectivity in neonatal brain networks (De Asis-Cruz et al., 2015) and neonatal connectome analysis (van den Heuvel et al., 2014). Right: degree distribution follows an exponential truncated power law, i.e., , using Akaike information criterion (AIC) for finding the best model, contrary to a power law, p(k) k−α, or exponential distribution, p(k) = eαk. We used the R package Brainwaver (Achard et al., 2006) (https://cran.r-project.org/web/packages/brainwaver/index.html) to find the best fit for the degree distribution.
4. Discussion and Conclusion
We introduced a novel technique for robust reconstruction and processing of fetal brain DTI, evaluated it with adult volunteer experiments, and examined on DWI of 21 non-sedated human fetuses scanned in-utero. Two main innovations distinguish our technique from previous works: 1) a robust formulation of slice-to-volume registration using an explicit dynamic model of fetal motion and registration to a common coordinate system, and 2) robust reconstruction based on detecting and rejecting motion-corrupted slices and a weighted linear least squares (WLLS) solution of the DTI model that mitigates the effect of intra-slice motion artifacts. We note that temporal adjacency of slice information was implicitly used in previous SVR methods, but we have developed an explicit dynamic motion tracking approach that uses slice timing and robust state estimation to estimate motion at the slice level. Moreover, we detected and filtered the effect of intra-slice motion which was not considered in the previous works. In all experiments our method (MT-SVR) performed much better than the reconstructions based on original data (Orig) and volume-to-volume registration (VVR). The improvement using MT-SVR over SVR with similar reconstruction technique was more prominent in the lower GA ranges. This is because 1) the magnitude and speed of motion is lower at higher GAs as older fetuses typically have less space to move, and 2) DW images of older fetal brains have more features that naturally make the slice-to-volume registration more robust, thus improve the performance of SVR.
We regressed FA values with the average magnitude and speed of motion calculated from estimated motion parameters, the number of scans, and GA. Similar to previous studies we observed an increase in FA values in fiber-rich regions with increased GA. Our results complied with the FA values and their increase by age reported previously by Jiang et al. (2009) based on 3 normal fetuses compared against curves obtained from serial DTI analysis of 14 preterm neonates by Partridge et al. (2004). Our motion analysis results showed significant reduction in FA with increased magnitude and speed of motion for the Orig and VVR methods, but not for SVR and MT-SVR. These results, presented in Figure 6, showed that MT-SVR was the most robust technique to motion. With MT-SVR we were able to perform whole-brain tractography at unprecedented details (sample results shown in Figures 8 and 9). Groupwise connectivity analysis, also, independently showed improved edge space similarity between subjects through the use of MT-SVR (compared to other methods), which implied that more meaningful connectivity was achieved through effective motion compensation and robust reconstruction versus not compensating for inter-slice and intra-slice motion. This analysis, shown in 10, supports the hypothesis that more meaningful and stronger structural connectivity is shaping throughout the course of in-utero brain maturation; and may be compared to the results obtained from similar groupwise structural connectivity analysis in preterm infants (van den Heuvel et al., 2014).
Using the results obtained from MT-SVR we analyzed the characteristics of the fetal brain connectome using graph theory and observed small world properties. These results for the prenatal period complies with the findings based on neonatal brain connectivity analysis by De Asis-Cruz et al. (2015) and van den Heuvel et al. (2014). The modules and the degree and betweenness hubs of the fetal connectome based on the group averaged structural connectome in our study are shown in Figure 13. These results also comply with results previously reported by De Asis-Cruz et al. (2015) based on functional connectome in neonates and by Jakab et al. (2015) in fetuses. All in all, our results and analyses indicate that robust motion correction and reconstruction can significantly improve the analysis of in-vivo fetal DWI and structural fetal brain connectome. The approach we have developed can be extended to other models of the diffusion signal, but to strengthen the results and evaluation in the absence of sufficient DWI data (i.e. high number of gradient directions and b values typically needed for higher-order models), we focused on a DTI model. We obtained brain tractography with unprecedented details in fetuses in a wide age range. Our results showed that uncompensated motion led to reduced FA values in regions of dense unidirectional fiber tracts such as corpus callosum and the limbs of the internal capsule. Our proposed algorithm based on MT-SVR consistently resulted in highest FA values in those fiber-rich regions.
Figure 13.

Group averaged fetal structural connectome can be separated in 6 modules (shown with different colors). Bigger nodes have higher degrees. These modules comply with the modules detected previously in functional connectome in neonates (De Asis-Cruz et al., 2015). The analysis of the degree and betweenness hub of the group averaged connectome show that the top 14 (15% of total) of highest connected nodes are: left and right precuni, left and right thalami, left and right insula, left and right lenticular nuclei, putamen, left and right anterior cingulate and paracingulate gyri, left and right median cingulate and paracingulate gyri, and left pallidum. These result confirm previous findings reported by Jakab et al. (2015). We used BrainNet Viewer (Xia et al., 2013) to visualize the brain network.
Invaluable work has been done in the past decade on the analysis of prenatal development of human brain connectome using both ex-vivo (Huang et al., 2006; Kostovic and Vasung, 2009; Takahashi et al., 2012; Kolasinski et al., 2013; Xu et al., 2014; Huang and Vasung, 2014; Vasung et al., 2016) and in-vivo (Kasprian et al., 2008, 2010; Zanin et al., 2011; Jakab et al., 2015; Mitter et al., 2015b) fetal MRI, and their correlation and validation with histology (Huang et al., 2013; Mitter et al., 2015a). While routine DTI analysis pipelines were adopted for in-vivo fetal DWI analysis in these studies, our results in this paper and the previous works by Jiang et al. (2009), Oubel et al. (2012), and Fogtmann et al. (2014) show that slice-level motion correction and reconstruction lead to more accurate results, especially in the presence of continuous motion which frequently happens in fetal DWI. Our algorithm, in particular, showed robust performance due to 1) the use of a robust state estimation technique (Agamennoni et al., 2012) for inter-slice motion correction through dynamic motion tracking, and 2) detection and rejection of intra-slice motion and robust reconstruction through weighted linear least squares estimation. The processing pipeline developed in this study for fetal brain DWI analysis, technical advances in fetal brain MRI reconstruction, new advances in fetal brain functional MRI analysis (Seshamani et al., 2016), along with spatiotemporal atlases of the fetal brain (Gholipour et al., 2014a, 2017; Serag et al., 2012) and neonates (Blesa et al., 2016; Makropoulos et al., 2016) can significantly improve the use of in-vivo MRI and DWI to study the development of human brain connectome in-utero. This in-turn facilitates the use of MRI as a powerful imaging modality for the analysis of neurodevelopmental disorders caused by preterm birth, growth restriction, or congenital anomalies.
Acknowledgments
This work was supported in part by the National Institutes of Health (NIH) grants R01 EB018988, R01 EB019483, and R01 NS079788, Intel(C) IPCC, and a faculty career development fellowship award from the Office of Faculty Development at Boston Childrens Hospital to A. Gholipour. B. Scherrer was also supported by Boston Childrens Hospital TRP Pilot Grant and Boston Childrens Hospital K-To-R Award. C.K. Rollins was supported by a scholar award from the Pediatric Heart Network (PHN) under award U10 HL068270 from the National Heart, Lung, and Blood Institute of NIH, and a Neurological Sciences Academic Development Award from the National Institutes of Neurological Disorders and Stroke. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
The authors also acknowledge the use of the CRKit software tools1 and the CRL spatiotemporal fetal brain MRI atlas2 developed in the Computational Radiology Laboratory (CRL) at Boston Children’s Hospital; and the ANTS software toolkit, N4ITK, and ITKSNAP developed in Penn Image Computing and Science Laboratory (PICSL)3 at the University of Pennsylvania.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Preprint submitted to Neuroimage
References
- Achard S, Salvador R, Whitcher B, Suckling J, Bullmore E. A resilient, low-frequency, small-world human brain functional network with highly connected association cortical hubs. The Journal of neuroscience. 2006;26(1):63–72. doi: 10.1523/JNEUROSCI.3874-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agamennoni G, Nieto J, Nebot EM, et al. Approximate inference in state-space models with heavy-tailed noise. Signal Proces, IEEE T. 2012;60(10):5024–5037. [Google Scholar]
- Akhondi-Asl A, Warfield S. Simultaneous truth and performance level estimation through fusion of probabilistic segmentations. Medical Imaging, IEEE Transactions on. 2013 Oct;32(10):1840–1852. doi: 10.1109/TMI.2013.2266258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksoy M, Forman C, Straka M, Skare S, Holdsworth S, Hornegger J, Bammer R. Real-time optical motion correction for diffusion tensor imaging. Magnet Reson Med. 2011;66(2):366–378. doi: 10.1002/mrm.22787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants B, Epstein C, Grossman M, Gee J. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis. 2008;12(1):26. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blesa M, Serag A, Wilkinson AG, Anblagan D, Telford EJ, Pataky R, Sparrow SA, Macnaught G, Semple SI, Bastin ME, Boardman JP. Parcellation of the healthy neonatal brain into 107 regions using atlas propagation through intermediate time points in childhood. Frontiers in neuroscience. 2016;10 doi: 10.3389/fnins.2016.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Miller S, Booth B, Andrews S, Chau V, Poskitt K, Hamarneh G. Structural network analysis of brain development in young preterm neonates. Neuroimage. 2014;101:667–680. doi: 10.1016/j.neuroimage.2014.07.030. [DOI] [PubMed] [Google Scholar]
- Bui T, Daire JL, Chalard F, Zaccaria I, Alberti C, Elmaleh M, Garel C, Luton D, Blanc N, Sebag G. Microstructural development of human brain assessed in utero by diffusion tensor imaging. Pediatric radiology. 2006;36(11):1133–1140. doi: 10.1007/s00247-006-0266-3. [DOI] [PubMed] [Google Scholar]
- De Asis-Cruz J, Bouyssi-Kobar M, Evangelou I, Vezina G, Limperopoulos C. Functional properties of resting state networks in healthy full-term newborns. Scientific reports. 2015;5 doi: 10.1038/srep17755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois J, Dehaene-Lambertz G, Kulikova S, Poupon C, Hüppi PS, Hertz-Pannier L. The early development of brain white matter: a review of imaging studies in fetuses, newborns and infants. Neuroscience. 2014;276:48–71. doi: 10.1016/j.neuroscience.2013.12.044. [DOI] [PubMed] [Google Scholar]
- Elhabian S, Gur Y, Vachet C, Piven J, Styner M, Leppert IR, Pike GB, Gerig G. Subject–motion correction in HARDI acquisitions: choices and consequences. Front Neurol. 2014;5 doi: 10.3389/fneur.2014.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogtmann M, Seshamani S, Kroenke C, Cheng X, Chapman T, Wilm J, Rousseau F, Studholme C. A unified approach to diffusion direction sensitive slice registration and 3-D DTI reconstruction from moving fetal brain anatomy. Med Imaging, IEEE T. 2014;33(2):272–289. doi: 10.1109/TMI.2013.2284014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholipour A, Akhondi-Asl A, Estroff JA, Warfield SK. Multi-atlas multi-shape segmentation of fetal brain MRI for volumetric and morphometric analysis of ventriculomegaly. NeuroImage. 2012;60(3):1819–1831. doi: 10.1016/j.neuroimage.2012.01.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholipour A, Estroff J, Warfield SK. Robust super-resolution volume reconstruction from slice acquisitions: application to fetal brain MRI. Med Imaging, IEEE T. 2010;29(10):1739–1758. doi: 10.1109/TMI.2010.2051680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholipour A, Estroff JA, Barnewolt CE, Robertson RL, Grant PE, Gagoski B, Warfield SK, Afacan O, Connolly S, Neil JJ, Wolfberg A, Mulkern RV. Fetal MRI: A technical update with educational aspirations. Concepts in Magnetic Resonance Part A. 2014a;43(6):237–266. doi: 10.1002/cmr.a.21321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholipour A, Limperopoulos C, Clancy S, Clouchoux C, Akhondi-Asl A, Estroff JA, Warfield SK. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2014b. Construction of a deformable spatiotemporal MRI atlas of the fetal brain: evaluation of similarity metrics and deformation models; pp. 292–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholipour A, Rollins CK, Velasco-Annis C, Ouaalam A, Akhondi-Asl A, Afacan O, Ortinau CM, Clancy S, Limperopoulos C, Yang E, et al. A normative spatiotemporal MRI atlas of the fetal brain for automatic segmentation and analysis of early brain growth. Scientific Reports. 2017;7(1):476. doi: 10.1038/s41598-017-00525-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Jeon T, Sedmak G, Pletikos M, Vasung L, Xu X, Yarowsky P, Richards LJ, Kostović I, Šestan N, et al. Coupling diffusion imaging with histological and gene expression analysis to examine the dynamics of cortical areas across the fetal period of human brain development. Cerebral cortex. 2013;23(11):2620–2631. doi: 10.1093/cercor/bhs241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Vasung L. Gaining insight of fetal brain development with diffusion MRI and histology. International Journal of Developmental Neuroscience. 2014;32:11–22. doi: 10.1016/j.ijdevneu.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Zhang J, Wakana S, Zhang W, Ren T, Richards LJ, Yarowsky P, Donohue P, Graham E, van Zijl PC, et al. White and gray matter development in human fetal, newborn and pediatric brains. Neuroimage. 2006;33(1):27–38. doi: 10.1016/j.neuroimage.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Jakab A, Kasprian G, Schwartz E, Gruber GM, Mitter C, Prayer D, Schöpf V, Langs G. Disrupted developmental organization of the structural connectome in fetuses with corpus callosum agenesis. Neuroimage. 2015;111:277–288. doi: 10.1016/j.neuroimage.2015.02.038. [DOI] [PubMed] [Google Scholar]
- Jiang S, Xue H, Counsell S, Anjari M, Allsop J, Rutherford M, Rueckert D, Hajnal JV. Diffusion tensor imaging of the brain in moving subjects: Application to in-utero fetal and ex-utero studies. Magnet Reson Med. 2009;62(3):645–655. doi: 10.1002/mrm.22032. [DOI] [PubMed] [Google Scholar]
- Jiang S, Xue H, Glover A, Rutherford M, Rueckert D, Hajnal JV. MRI of moving subjects using multislice snapshot images with volume reconstruction (SVR): application to fetal, neonatal, and adult brain studies. Med Imaging, IEEE T. 2007;26(7):967–980. doi: 10.1109/TMI.2007.895456. [DOI] [PubMed] [Google Scholar]
- Johnson HJ, McCormick M, Ibáñez L, Consortium T. I. S. The ITK Software Guide. 3rd. Kitware, Inc.; 2013. In press. [Google Scholar]
- Kainz B, Steinberger M, Wein W, Kuklisova-Murgasova M, Malamateniou C, Keraudren K, Torsney-Weir T, Rutherford M, Aljabar P, Hajnal J, Rueckert D. Fast Volume Reconstruction From Motion Corrupted Stacks of 2D Slices. Med Imaging, IEEE T. 2015;34:1901–1913. doi: 10.1109/TMI.2015.2415453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprian G, Brugger PC, Schöpf V, Mitter C, Weber M, Hainfellner JA, Prayer D. Assessing prenatal white matter connectivity in commissural agenesis. Brain. 2013;136(1):168–179. doi: 10.1093/brain/aws332. [DOI] [PubMed] [Google Scholar]
- Kasprian G, Brugger PC, Weber M, Krssák M, Krampl E, Herold C, Prayer D. In utero tractography of fetal white matter development. Neuroimage. 2008;43(2):213–224. doi: 10.1016/j.neuroimage.2008.07.026. [DOI] [PubMed] [Google Scholar]
- Kasprian G, Del Río M, Prayer D. Fetal diffusion imaging: pearls and solutions. Topics in Magnetic Resonance Imaging. 2010;21(6):387–394. doi: 10.1097/RMR.0b013e31823e6f80. [DOI] [PubMed] [Google Scholar]
- Kim B, Boes J, Bland P, Chenevert T, Meyer CR. Motion correction in fMRI via registration of individual slices into an anatomical volume. Magnet Reson Med. 1999;41(5):964–972. doi: 10.1002/(sici)1522-2594(199905)41:5<964::aid-mrm16>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Kim DH, Chung S, Vigneron DB, Barkovich AJ, Glenn OA. Diffusion-weighted imaging of the fetal brain in vivo. Magnetic resonance in medicine. 2008;59(1):216–220. doi: 10.1002/mrm.21459. [DOI] [PubMed] [Google Scholar]
- Koay CG, Chang LC, Carew JD, Pierpaoli C, Basser PJ. A unifying theoretical and algorithmic framework for least squares methods of estimation in diffusion tensor imaging. J Magn Reson. 2006;182(1):115–125. doi: 10.1016/j.jmr.2006.06.020. [DOI] [PubMed] [Google Scholar]
- Kober T, Gruetter R, Krueger G. Prospective and retrospective motion correction in diffusion magnetic resonance imaging of the human brain. Neuroimage. 2012;59(1):389–398. doi: 10.1016/j.neuroimage.2011.07.004. [DOI] [PubMed] [Google Scholar]
- Kolasinski J, Takahashi E, Stevens AA, Benner T, Fischl B, Zöllei L, Grant PE. Radial and tangential neuronal migration pathways in the human fetal brain: anatomically distinct patterns of diffusion MRI coherence. Neuroimage. 2013;79:412–422. doi: 10.1016/j.neuroimage.2013.04.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostovic I, Vasung L. Seminars in perinatology. Vol. 33. Elsevier; 2009. Insights from in vitro fetal magnetic resonance imaging of cerebral development; pp. 220–233. [DOI] [PubMed] [Google Scholar]
- Kreilkamp BA, Zacà D, Papinutto N, Jovicich J. Retrospective head motion correction approaches for diffusion tensor imaging: Effects of preprocessing choices on biases and reproducibility of scalar diffusion metrics. J Magn Reson Im. 2015 doi: 10.1002/jmri.24965. [DOI] [PubMed] [Google Scholar]
- Kuklisova-Murgasova M, Quaghebeur G, Rutherford MA, Hajnal JV, Schnabel JA. Reconstruction of fetal brain MRI with intensity matching and complete outlier removal. Med Im Anal. 2012;16(8):1550–1564. doi: 10.1016/j.media.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis WW, Sahin M, Scherrer B, Peters JM, Suarez RO, Vogel-Farley VK, Jeste SS, Gregas MC, Prabhu SP, Nelson CA, et al. Impaired language pathways in tuberous sclerosis complex patients with autism spectrum disorders. Cerebral cortex. 2013;23(7):1526–1532. doi: 10.1093/cercor/bhs135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclaren J, Herbst M, Speck O, Zaitsev M. Prospective motion correction in brain imaging: a review. Magnet Reson Med. 2013;69(3):621–636. doi: 10.1002/mrm.24314. [DOI] [PubMed] [Google Scholar]
- Maclaren J, Speck O, Stucht D, Schulze P, Hennig J, Zaitsev M. Navigator accuracy requirements for prospective motion correction. Magnet Reson Med. 2010;63(1):162–170. doi: 10.1002/mrm.22191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makropoulos A, Aljabar P, Wright R, Hüning B, Merchant N, Arichi T, Tusor N, Hajnal JV, Edwards AD, Counsell SJ, et al. Regional growth and atlasing of the developing human brain. Neuroimage. 2016;125:456–478. doi: 10.1016/j.neuroimage.2015.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marami B, Scherrer B, Afacan O, Erem B, Warfield S, Gholipour A. Motion-robust diffusion-weighted brain MRI reconstruction through slice-level registration-based motion tracking. Med Imaging, IEEE T. 2016a;35(10):2258–2269. doi: 10.1109/TMI.2016.2555244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marami B, Scherrer B, Afacan O, Warfield SK, Gholipour A. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2016b. Motion-robust reconstruction based on simultaneous multi-slice registration for diffusion-weighted MRI of moving subjects; pp. 544–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattes D, Haynor DR, Vesselle H, Lewellen TK, Eubank W. PET-CT image registration in the chest using free-form deformations. Med Imaging, IEEE T. 2003;22(1):120–128. doi: 10.1109/TMI.2003.809072. [DOI] [PubMed] [Google Scholar]
- Mitter C, Jakab A, Brugger P, Ricken G, Gruber G, Bettelheim D, Scharrer A, Langs G, Hainfellner J, Prayer D, Kasprian G. Validation of in utero tractography of human fetal commissural and internal capsule fibers with histological structure tensor analysis. Frontiers in neuroanatomy. 2015a;9 doi: 10.3389/fnana.2015.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitter C, Prayer D, Brugger PC, Weber M, Kasprian G. In vivo tractography of fetal association fibers. PloS one. 2015b;10(3):e0119536. doi: 10.1371/journal.pone.0119536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee P, Miller JH, Shimony JS, Philip JV, Nehra D, Snyder AZ, Conturo TE, Neil JJ, McKinstry RC. Diffusion-tensor MR imaging of gray and white matter development during normal human brain maturation. American Journal of Neuroradiology. 2002;23(9):1445–1456. [PMC free article] [PubMed] [Google Scholar]
- Oubel E, Koob M, Studholme C, Dietemann JL, Rousseau F. Reconstruction of scattered data in fetal diffusion mri. Med Im Anal. 2012;16(1):28–37. doi: 10.1016/j.media.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge SC, Mukherjee P, Henry RG, Miller SP, Berman JI, Jin H, Lu Y, Glenn OA, Ferriero DM, Barkovich AJ, et al. Diffusion tensor imaging: serial quantitation of white matter tract maturity in premature newborns. Neuroimage. 2004;22(3):1302–1314. doi: 10.1016/j.neuroimage.2004.02.038. [DOI] [PubMed] [Google Scholar]
- Peters JM, Sahin M, Vogel-Farley VK, Jeste SS, Nelson CA, Gregas MC, Prabhu SP, Scherrer B, Warfield SK. Loss of white matter microstructural integrity is associated with adverse neurological outcome in tuberous sclerosis complex. Academic radiology. 2012;19(1):17–25. doi: 10.1016/j.acra.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Righini A, Bianchini E, Parazzini C, Gementi P, Ramenghi L, Baldoli C, Nicolini U, Mosca F, Triulzi F. Apparent diffusion coefficient determination in normal fetal brain: a prenatal MR imaging study. American journal of neuroradiology. 2003;24(5):799–804. [PMC free article] [PubMed] [Google Scholar]
- Rousseau F, Glenn OA, Iordanova B, Rodriguez-Carranza C, Vigneron DB, Barkovich JA, Studholme C. Registration-based approach for reconstruction of high-resolution in utero fetal MR brain images. Acad Radiol. 2006;13(9):1072–1081. doi: 10.1016/j.acra.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52(3):1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Salvador R, Peña A, Menon D, Carpenter A, Pickard J, Bullmore E. Formal characterization and extension of the linearized diffusion tensor model. Hum brain mapp. 2005;24(2):144–155. doi: 10.1002/hbm.20076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serag A, Aljabar P, Ball G, Counsell SJ, Boardman JP, Rutherford MA, Edwards AD, Hajnal JV, Rueckert D. Construction of a consistent high-definition spatio-temporal atlas of the developing brain using adaptive kernel regression. NeuroImage. 2012;59(3):2255–2265. doi: 10.1016/j.neuroimage.2011.09.062. [DOI] [PubMed] [Google Scholar]
- Seshamani S, Blazejewska AI, Mckown S, Caucutt J, Dighe M, Gatenby C, Studholme C. Detecting default mode networks in utero by integrated 4D fMRI reconstruction and analysis. Human Brain Mapping. 2016 doi: 10.1002/hbm.23303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi E, Folkerth RD, Galaburda AM, Grant PE. Emerging cerebral connectivity in the human fetal brain: an MR tractography study. Cerebral Cortex. 2012;22(2):455–464. doi: 10.1093/cercor/bhr126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tustison NJ, Avants BB, Cook P, Zheng Y, Egan A, Yushkevich P, Gee JC, et al. N4ITK: improved N3 bias correction. Medical Imaging, IEEE Transactions on. 2010;29(6):1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Kersbergen KJ, de Reus MA, Keunen K, Kahn RS, Groenendaal F, de Vries LS, Benders MJ. The neonatal connectome during preterm brain development. Cerebral cortex. 2014:bhu095. doi: 10.1093/cercor/bhu095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasung L, Lepage C, Radoš M, Pletikos M, Goldman JS, Richiardi J, Raguž M, Fischi-Gómez E, Karama S, Huppi PS, et al. Quantitative and qualitative analysis of transient fetal compartments during prenatal human brain development. Frontiers in neuroanatomy. 2016;10 doi: 10.3389/fnana.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White N, Roddey C, Shankaranarayanan A, Han E, Rettmann D, Santos J, Kuperman J, Dale A. PROMO: Real-time prospective motion correction in MRI using image-based tracking. Magnet Reson Med. 2010;63(1):91–105. doi: 10.1002/mrm.22176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, Wang J, He Y. Brainnet viewer: a network visualization tool for human brain connectomics. PloS one. 2013;8(7):e68910. doi: 10.1371/journal.pone.0068910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Takahashi E, Folkerth RD, Haynes RL, Volpe JJ, Grant PE, Kinney HC. Radial coherence of diffusion tractography in the cerebral white matter of the human fetus: neuroanatomic insights. Cerebral Cortex. 2014;24(3):579–592. doi: 10.1093/cercor/bhs330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Zaitsev M, Maclaren J, Herbst M. Motion artifacts in MRI: A complex problem with many partial solutions. J Magn Reson Im. 2015 doi: 10.1002/jmri.24850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanin E, Ranjeva JP, Confort-Gouny S, Guye M, Denis D, Cozzone PJ, Girard N. White matter maturation of normal human fetal brain. an in vivo diffusion tensor tractography study. Brain and behavior. 2011;1(2):95–108. doi: 10.1002/brb3.17. [DOI] [PMC free article] [PubMed] [Google Scholar]



