Abstract
The PI3-kinase/AKT pathway integrates signals from external cellular stimuli to regulate essential cellular functions, and is frequently aberrantly activated in human cancers. Recent research demonstrates that tight regulation of the epigenome is critical in preserving and restricting transcriptional activation, which can impact cellular growth and proliferation. In this review we examine mechanisms by which the PI3K/AKT pathway regulates the epigenome to promote oncogenesis, and highlight how connections between PI3K/AKT and the epigenome may impact the future therapeutic treatment of cancers featuring a hyperactivated PI3K/AKT pathway.
Introduction
Known mutational activation or inactivation of chromatin modifiers has demonstrated that the epigenome can promote oncogenic growth and cancer. Recent research suggests that the activity of signaling pathways that drive oncogenesis indirectly regulate the epigenome via the modulation of proteins and enzymes required for chromatin reading, writing, and erasure. In fact, the term epigenetic modulator was recently coined to describe oncogenic driver genes that modulate signal transduction and are also implicated in epigenomic modification [1]. Deregulation of the PI3K signaling pathway is a key event in cancers due to the prevalence of oncogenic activating mutations and genetic inactivation of tumor suppressors regulating the pathway. Epigenetic modulators participate in the PI3K/AKT pathway and have been shown to regulate the epigenome and contribute to the oncogenicity of PI3K in cancer. In this review we outline the progress in elucidating PI3K pathway-dependent epigenetic modulators and the prospect of therapeutic targeting of the epigenome in the treatment of PI3K pathway-activated cancers.
The PI3K/AKT signaling pathway in oncogenesis
The class IA phosphatidylinositol 3-kinase (PI3K)/AKT pathway integrates signals from growth factors and cytokines, relaying these signals through multiple downstream cellular effectors. In turn, these effectors regulate essential cellular functions including growth, metabolism, survival, and proliferation [2]. Class IA PI3Ks function as heterodimers that consist of a p110 catalytic subunit and a regulatory p85 subunit. Three highly homologous genes (PIK3CA, PIK3CB, and PIK3CD) encode the catalytic isoforms [3]. In response to exogenous growth factor or cytokine stimulation, receptor tyrosine kinases (RTKs) or G protein-coupled receptors (GPCRs) are activated, and class IA PI3Ks are recruited to the cellular membrane. At the membrane PI3K catalyzes the phosphorylation of PtdIns 4,5-bisphoshate (PIP2) to generate the lipid second messenger PtdIns 3,4,5-triphosphosate (PIP3) [4]. As such, the PIP3 second messenger activates downstream signaling pathways, many of which diverge downstream of AKT. The activity of PI3K is reversed by the lipid phosphatase and tumor suppressor, PTEN, which catalyzes the removal of a phosphate group from PIP3 to restore PIP2, thereby inactivating PI3K signaling.
The serine/threonine protein kinase AKT, encoded by three separate genes for the isoforms AKT1, AKT2, and AKT3, is a central PI3K signaling conduit [5]. PI3K modulates essential cellular functions utilizing AKT-dependent and AKT-independent mechanisms; significantly more is understood about AKT-dependent PI3K signaling, which is the focus of this review. AKT modulates diverse cellular functions including cell survival, growth, proliferation, metabolism, migration and proliferation through the phosphorylation of more than 200 identified substrates. In most cases, AKT-mediated effector phosphorylation at the consensus R-x-R-x-x-S/T motif negatively regulates effector function [6]. For example, AKT-mediated phosphorylation of FOXO transcription factors enhances FOXO nuclear export, and phosphorylation of pro-apoptotic protein BAD results in its functional inactivation, highlighting a redundant approach by which PI3K/AKT promotes cellular survival [7, 8]. AKT-mediated substrate phosphorylation activates mTORC1 and increases protein synthesis, positively regulating cell growth. However, AKT increases cellular growth through the negative regulation of TSC2: AKT-mediated TSC2 phosphorylation prevents TSC2 from functioning as GTPase-activating protein (GAP) for Rheb, causing Rheb-GTP to accumulate and activate mTORC1 [9]. Phosphorylation of PRAS40 by AKT also activates mTORC1 by relieving direct inhibition on the mTORC1 complex [10].
The PI3K/AKT pathway promotes transcriptional competence via epigenomic regulation
The examples above highlight a small fraction of PI3K/AKT effectors that control cellular processes that are frequently deregulated in human cancers. Importantly, changes to the chromatin landscape are also associated with the development of cancer, some of which are mediated by the PI3K/AKT signaling pathway. Extensive research has unveiled novel AKT substrates that when phosphorylated by AKT, promote or poise cells for transcriptional activation, thus regulating oncogenesis via chromatin modifications. Several key examples are highlighted below (Figure 1).
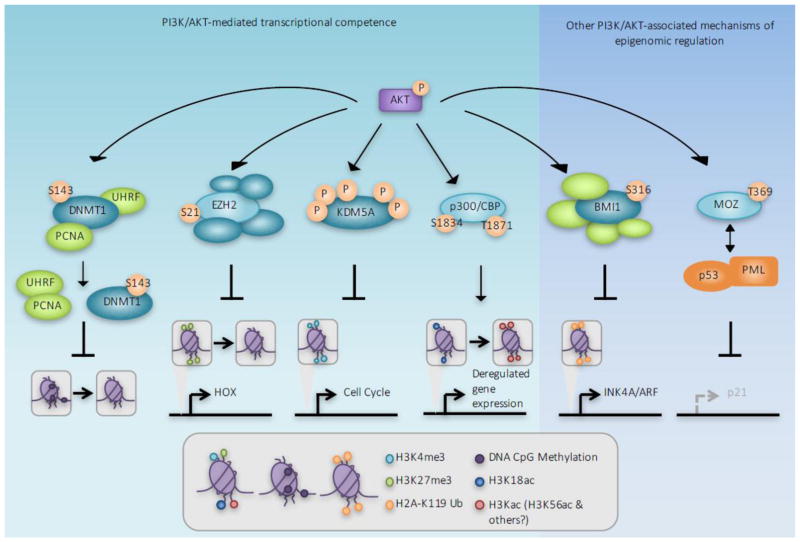
Figure 1. AKT-mediated substrate phosphorylation promotes transcriptional competence.
Using multiple mechanisms, PI3K/AKT pathway activation primes chromatin for subsequent transcriptional activation. From Left to Right: The DNA methyltransferase DNMT1/3a dissociates with chromatin following phosphorylation by AKT, causing DNA hypomethylation at promoter CpG islands, which has been shown to increase gene expression. AKT-mediated phosphorylation of the histone methyltransferase EZH2 decreases EZH2 affinity for chromatin, reducing the repressive promoter-associated H3K27me3 modification. Phosphorylation of the histone demethylase KDM5A increases its cytoplasmic localization, thereby increasing promoter H3K4me3 and transcriptional competence. Histone acetyltransferase activity and substrate affinity of p300/CBP is enhanced with AKT phosphorylation, increasing H3K56 and other lysine acetylation and transcriptional activation. CBP phosphorylation at T1871 has also been shown to specifically reduce H3K18ac, thereby contributing to oncogenic growth. PI3K/AKT has also been reported to have other effects on chromatin; Bmi1 phosphorylation by AKT promotes H2A ubiquitination and AKT-mediated phosphorylation of the histone acetyltransferase MOZ affects alters p53 acetylation and function, both of which independently modulate tumor suppressor expression.
AKT influences DNA replication-associated DNA methylation
The preservation and, thus, heritability of DNA CpG dinucleotide methylation patterns throughout the genome is essential during DNA replication. A high frequency of CpG dinucleotides, termed CpG islands, is present at or near promoters, whereas the remainder of the genome is typically characterized by minimal CpG dinucleotides. Aberrant CpG methylation patterns are common in human cancer; promoter-associated CpG islands are prone to aberrant hypermethylation; and hypomethylation of CpG poor genomic regions frequently occurs. CpG dinucleotides within developmental genes, non-relevant lineage-specific genes, some proto-oncogenes and oncogenes, and endogenous retroviral sequences are hypermethylated, as a mechanism to suppress DNA transcription [11]. The PI3K/AKT pathway promotes transcriptional activation by reducing global genome DNA methylation. S-phase-associated DNA methylation is maintained by DNA methyltransferase 1 (DNMT1) and deregulation of DNMT1 and other DNMTs has been reported in multiple cancer types [12]. The PI3K/AKT pathway regulates DNMT1 through AKT-mediated phosphorylation at S143. While DNMT1 S143 phosphorylation by AKT has not been shown to directly affect DNMT1 methyltransferase activity, it somewhat paradoxically increases DNMT1 stability [13]. Thus DNMT1 S143 phosphorylation impedes subsequent SETD7-mediated DNMT1 monomethylation at K142, an event that normally triggers DNMT1 proteasome-mediated degradation. However, DNMT1 S143 phosphorylation also reduces its association with DNA-replication associated proteins PCNA and UHRF1 at the replication fork [14]. Together these facts suggest that while DNMT1 S143 phosphorylation by AKT increases DNMT1 half-life, phosphorylated DNMT1 is unable to associate with and subsequently methylate DNA.
The PI3K/AKT pathway has also been reported to regulate locus-specific DNA hypomethylation such that transcriptional activation occurs at specific genes. The AKT-target genes GSK3α/β regulate DNA methylation of the regulatory regions of the imprinted genes IGF2 and IGF2R by reducing expression of the DNA methyltransferase DNMT3a2 [15]. AKT activation, loss of GSK3α/β expression, or GSK3 inhibition promotes the hypomethylation of the IGF2/IGF2R genomic loci, which aberrantly activates IGF2/IGF2R transcription [15]. Since IGF2/IGF2R expression can in turn upregulate PI3K/AKT signaling, this system may constitute a feed-forward loop. PI3K/AKT may also regulate DNMT3 activity independently of GSK3α/β through direct phosphorylation of at least one putative AKT phosphorylation consensus motif on DNMT3 [16]. It has therefore been proposed that the PI3K/AKT pathway may support the hypermethylated DNA state frequently observed in cancer.
AKT regulates histone methylation by targeting the H3K27 methyltransferase EZH2
An appropriate balance of activating and inhibitory histone methylation events is critical for maintaining context dependent and appropriate gene transcription. This balance is frequently disrupted in cancer, in a manner favoring transcriptional activation that is associated with PI3K/AKT pathway activation. EZH2 is the histone methyltransferase in the polycomb repressive complex 2 (PRC2) and is responsible for silencing transcription by trimethylating promoter-associated Histone H3 Lys27 (H3K27me3). H3K27me3 is antagonized by promoter-localized trimethylation of Histone H3 Lys4 (H3K4me3), which is indicative of transcriptional competence. Simultaneous H3K4me3 and H3K27me3 occurs at some genomic loci; these bivalent domains are transcriptionally repressed but poised for rapid transcriptional activation following the loss of H3K27me3. AKT-mediated phosphorylation of EZH2 at S21 inhibits EZH2 methyltransferase activity and reduces H3K27me3 [17]. The lack of H3K27me3 at bivalent loci then promotes transcriptional activation and erroneous gene expression. As a result, EZH2 provides a central node by which the PI3K pathway promotes transcriptional competence of bivalent, and therefore poised, genomic loci. Loss of EZH2 methyltransferase activity towards non-bivalent loci may also reduce transcriptional silencing due to the lack of H3K27me3. AKT-mediated EZH2 S21 phosphorylation can also augment EZH2-mediated gene transcription independently of its effects on the methyltransferase activity of EZH2. Phosphorylation of EZH2 enhances its function as an oncogenic transcriptional co-activator by facilitating its association with androgen receptor and other transcription factors [18]. Together these studies suggest that transcriptional competence is in part established by AKT-mediated EZH2 phosphorylation thereby reducing repressive H3K27 methylation at both bivalent and monovalent genomic loci.
H3K4me3 as a PI3K/AKT target
The PI3K/AKT pathway favors transcriptional activation through other means in addition to the reduction of H3K27me3. Promoter associated H3K4me3 is characteristic of transcriptionally active euchromatin and has been reported to be elevated in breast and colorectal cancers [19, 20], which are commonly associated with PI3K-pathway activation. The PI3K pathway was recently shown to be essential in regulating H3K4me3 in in vivo models of PI3K-activated breast cancer [21]. H3K4 methylation is regulated by the enzymatic activity of the MLL/KMT2 histone methyltransferases (HMTs) [22] and the KDM5 family of histone demethylases (KDMs)[23, 24]. Promoter associated H3K4me3 has emerged as a key posttranslational histone modification indicative of transcriptional activation. AKT-mediated KDM5A phosphorylation on up to five amino acids promotes KDM5A nuclear exit, reminiscent of AKT-mediated FOXO3 phosphorylation [7]. A shift in KDM5A subcellular localization is dependent on PI3K activation in models of breast cancer in vivo. Loss of nuclear KDM5A limits exposure of KDM5A to its substrates, H3K4me2/3, which may explain the increase in H3K4me3 observed in PIK3CAH1047R-activated breast cancer models. Moreover, AKT and KDM5A collaborate to regulate cell cycle-promoting genes, which may have implications in the oncogenic growth of PI3K-activated cancers.
Crosstalk between p300/CBP and PI3K signaling
Histone acetylation of lysine residues, which is critical to promote transcriptional activation at euchromatic sites within the genome, is also regulated by PI3K/AKT. Acetylation neutralizes the charge of the N terminal histone tail, which increases the accessibility of DNA to transcription factors. The lysine histone acetyltransferase (HAT) p300, and its paralog CBP, commonly referred to as p300/CBP, acetylate over 100 histone and non-histone substrates [25]. P300/CBP regulate the expression of genes controlling cellular processes that are frequently deregulated in cancers by independently functioning as a protein scaffold, transcriptional co-activator, and HAT. In response to upstream signaling events, AKT stimulates p300 and CBP HAT activity following AKT-mediated phosphorylation of p300 and CBP at S1834 and T1871, respectively [26]. Phosphorylation of p300/CBP also increases transcription factor assembly on euchromatin and subsequently helps to recruit basal transcriptional machinery. Specific acetylation of H3 lysine 56 (H3K56ac) is critical for packaging of DNA into chromatin during DNA replication and DNA damage repair, which is mediated by p300/CBP [27]. While it has not been shown that PI3K/AKT activation is required for p300/CBP-mediated H3K56ac, high H3K56ac is detected in breast, thyroid, and skin cancers [27]. P300/CBP also acetylates H3K18, and AKT-mediated phosphorylation of CBP, but not p300, reduces H3K18ac, which is reduced or lost during oncogenesis and may correlate with a poor clinical outcome [28]. Indeed, loss of H3K18ac correlates with an increase in oncogenic growth measured in in vitro transformation assays in liver cell lines [29]. These data suggest at least one AKT-dependent CBP function in promoting oncogenic growth that is not shared with its paralog p300. It is therefore possible that PI3K/AKT-dependent p300 and/or CBP regulation promotes transcription and oncogenic growth by modulating substrate acetylation, including, but not necessarily limited to, H3K18ac and H3K56ac.
H2A ubiquitination in response to PI3K signaling
EZH2 and PRC2-mediated H3K27me3 contribute to transcriptional silencing and this process is negatively regulated by PI3K/AKT. However, AKT also promotes the subsequent polycomb/PRC1-mediated H2A ubiquitination, which contributes to features of transcriptionally inactive and repressed chromatin at specific genomic loci. After PRC2-mediated H3K27me3, the PRC1 containing form of the polycomb complex recognizes H3K27me3, which triggers H2A K119 ubiquitination by PRC1 [30]. Specifically the PRC1 core complex member Bmi1 stimulates the E3 ubiquitin ligase activity of its heterodimeric partner Ring1B, directing its ubiquitination to H2A K119 [31]. H2A K119 ubiquitination contributes to polycomb-mediated gene repression. Bmi1 has been linked to the specific silencing of the INK4A/ARF locus that encodes the p16INK4A and p19ARF tumor suppressors. AKT-mediated Bmi1 S316 phosphorylation dissociates Bmi1 from the INK4A/ARF locus, thereby reducing H2A ubiquitination and inhibiting transcriptional silencing of the INK4A/ARF locus [18, 31]. Bmi1 S316 phosphorylation has been associated with reduced cell proliferation and increased senescence [18]. While reduced proliferation is typically incompatible with the oncogenic growth observed from aberrant PI3K-pathway activation, oncogene induced senescence (OIS) is a phenomena observed in cancer, and AKT-mediated Bmi1 phosphorylation is one possible mechanism that may explain this process.
Regulation of non-histone substrates by the histone acetyltransferase MOZ
The histone acetyltransferase monocytic leukemic zinc-finger (MOZ) functions as part of a quaternary histone acetyltransferase (HAT) complex with BRPF1, ING5, and hEAF6 that uses multiple chromatin reader domains to recognize posttranslational modifications on the N terminal unstructured regions of histone tails. Using its tandem PHD domains, MOZ recognizes H3 when acetylated at H3K14, enabling subsequent promoter acetylation of target genes [32]. Other members of the quaternary HAT complex use their reader domains to direct MOZ acetyltransferase activity towards H3K14, as well as H4K5, H4K8, and H4K12 [33]. However MOZ also possesses at least one described acetyltransferase activity that is independent of histones, and this activity is negatively regulated by AKT. MOZ associates with p53 in promyelocytic leukemia (PML) nuclear bodies [34]. Forming a ternary complex with p53 and PML, MOZ acetylates p53 at K120 and K382, which subsequently stimulates p53-mediated transcription of p21 to induce cell-cycle arrest and senescence [34]. AKT-mediated phosphorylation of MOZ at T369 prevents MOZ-PML association, thereby inhibiting ternary complex formation with p53 and hence p53 acetylation [35]. Disruption of the MOZ-PML association renders MEFs resistant to PML-induced senescence, suggesting a new mechanism by which the PI3K/AKT pathway may contribute to oncogenic growth. However, it is unclear whether MOZ T369 phosphorylation alters MOZ affinity for histone substrates as well as for p53/PML.
Clinically targeting the PI3K pathway in PI3K-activated solid cancers
Constitutive activation of the PI3K/AKT pathway commonly occurs in human cancers through mutational activation or amplification of receptor tyrosine kinases, mutational activation of the RAS or PIK3CA genes, mutational activation or amplification of AKT1/AKT2/AKT3, as well as via inactivation of PTEN or the regulatory PI3K p85 subunits (PIK3R1 and PIK3R2). Deregulation of this pathway occurs across cancer types, by some estimates occurring in 70% or more of diagnosed cases of uterine and lung cancers, and in more than 50% of breast, prostate, cervical, ovarian, and glioblastoma multiforme cancers. The high frequency of genomic alteration of this pathway across cancer types, coupled with its central role in regulating pathways critical for cellular growth and survival has prompted the development of PI3K-pathway specific inhibitors as anti-cancer therapeutics.
Previous generation therapeutic modalities for the treatment of PI3K-activated solid malignancies
Preclinical development and clinical testing of PI3K pathway inhibitors is ongoing, with an emphasis on pan-PI3K inhibitors that target all class I PI3K isoforms, isoform selective PI3K inhibitors that target individual class I PI3K isoforms, AKT inhibitors, and mTOR-targeted inhibitors. Many of the early PI3K inhibitors were reversible ATP-competitive pan-PI3K inhibitors that target all class I PI3K isoforms with similar efficacy such as GDC0941 [36], the dual pan-PI3K/catalytic mTOR inhibitor BEZ235 [37], or dual mTORC1/2 inhibitors such as MLN-0128 [38] (Figure 2). Administered as single agents, these inhibitors have generally failed to produce robust results in the clinic despite promising anti-tumorigenic results in preclinical studies. Allosteric and catalytic AKT inhibitors, such as MK2206 and GSK690693, respectively, have also been tested in phase I/II clinical studies of varying solid tumor types. Generally AKT monotherapy is also associated with low objective response rates [39, 40], although next-generation AKT inhibitors are in clinical development.
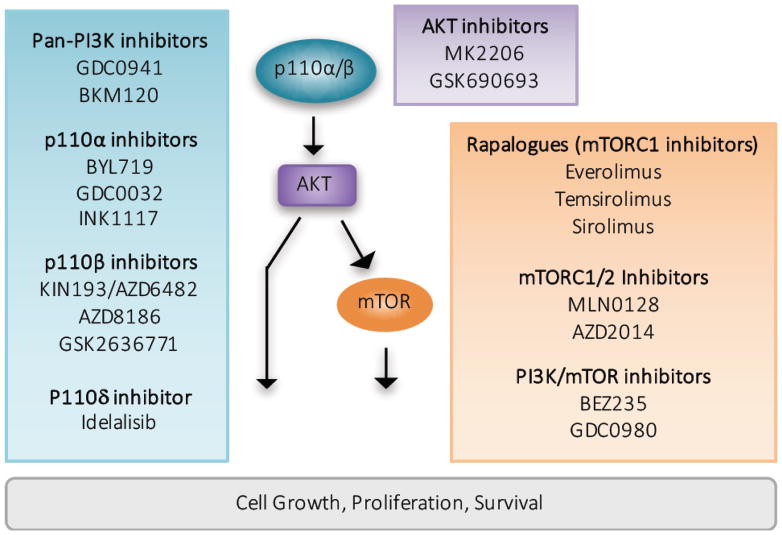
Figure 2. Therapeutic inhibition of the PI3K/AKT pathway in cancer.
PI3K/AKT pathway hyperactivation represents a viable therapeutic modality in cancer. The blue box indicates PI3K inhibitors with preclinical and/or clinical activity. Non-selective, “pan” PI3K inhibitors target all class I PI3K isoforms and p110α-selective inhibitors are biologically active against tumors with p110α hyperactivation through upstream RTK activation or direct p110α mutational activation. P110β-selective inhibitors are efficacious in the treatment of tumors characterized by PTEN genomic alteration, and the p110δ-selective inhibitor Idelalisib is effective in blocking PI3K-mediated B cell receptor signaling and is FDA approved for the treatment of relapsed chronic lymphoid leukemia (CLL). The purple box highlights AKT inhibitors, which are most effective in the treatment of tumors characterized by AKT genomic alteration but may also possess therapeutic benefit in PI3K hyperactivated tumors. mTORC1, dual mTORC1/2, and dual-specificity mTOR/PI3K inhibitors are represented in the orange box. Currently several rapalogues are FDA-approved for multiple cancer indications.
Because PIK3CA is frequently mutated in solid tumors (most commonly via E542K/E545K helical domain mutants or H1047R catalytic domain mutants), isoform-selective PI3K inhibitors (BYL719, GDC0032) that preferentially target the PIK3CA protein product p110α are under preclinical evaluation and clinical testing. These p110α-selective inhibitors have shown promising clinical responses in tumors featuring PIK3CA mutations while preclinical studies suggest that tumors characterized by RTK activation, for instance via HER2 amplification, may also respond to p110α-selective inhibition [41, 42]. In contrast, PTEN-deficient solid tumors are primarily dependent on the PIK3CB protein product p110β for PI3K signaling [43]; and p110β-selective PI3K inhibitors are in preclinical development [44] and in phase I/II clinical testing for the treatment of solid tumors with genetic loss of PTEN or loss of PTEN expression by epigenetic or post-transcriptional means. Lastly, B cell signaling is upregulated in chronic lymphocytic leukemia (CLL), frequently occurring through PI3K pathway hyperactivation. The PIK3CD protein product p110δ is highly expressed in lymphoid tissue and expression is restricted to hematopoietic cells, prompting the development of the p110δ-selective inhibitor Cal-101 (Idelalisib). Recently Idelalisib became the first PI3K inhibitor approved for the treatment of relapsed CLL in combination with the CD-20 antibody rituximab [45].
The future of clinical PI3K/AKT pathway inhibition in the treatment of solid tumors
To date, the limited efficacy and durability of monotherapeutic PI3K/AKT/mTOR inhibition has driven research into the development of suitable combination therapies to pair with existing or next-generation PI3K/AKT/mTOR-selective inhibitors. A large number of labs have reported success combining PI3K/AKT inhibitors with inhibitors of multiple targets, including RTKs.
The collaboration between the PI3K/AKT pathway and chromatin modification through the activity of the epigenetic modulator AKT suggests that PI3K/AKT inhibition in combination with epigenetic modifier inhibition may be an attractive and synergistic strategy for the treatment of PI3K-pathway activated solid tumors. Inhibitors targeting epigenetic modifiers are under preclinical development and clinical testing and in some cases, have showed modest response rates in some cancer types. Combinations of PI3K/AKT/mTOR and chromatin modifier inhibition are currently under exploration in preclinical studies and, in a select few cases, clinical trials. Below we discuss several examples of advances made towards therapeutic inhibition of epigenetic modifiers. Where relevant, we discuss the development and testing of combination therapies targeting the PI3K pathway and epigenetic modifiers, such that PI3K/AKT activation and aberrant transcriptional activation is abrogated.
DNA methyltransferase (DNMT) inhibition
Aberrant CpG methylation of tumor suppressor genes transcriptionally silences their expression and is commonly observed in cancer. Elevated PTEN promoter methylation, which silences PTEN expression and promotes AKT hyperactivation, has been observed in diverse cancer cell lines and patient samples. Preclinical studies have found that increased PTEN expression and, hence, reduced AKT signaling is observed following treatment with the DNMT inhibitor decitabine in xenograft models of tamoxifen-resistant ER+ breast cancer. DNMT inhibitors, including decitabine and azacitidine, are cytidine antimetabolite analogues that function by inhibiting DNMT-mediated DNA CpG methylation. While these and other DMNT inhibitors show some anti-tumorigenic activity as single agents in solid and hematological malignancies, preclinical studies suggest synergistic anti-tumorigenic activity when combined with the EGFR inhibitor gefitinib or the mTOR inhibitor rapamycin in colorectal cancer cell lines [46, 47]. As a result, decitabine and azacitidine are both under clinical investigation for the treatment of solid tumors in combination with the mTOR inhibitors sirolimus or everolimus.
Preclinical studies have found that nasopharyngeal cell lines resistant to the PI3K/mTOR inhibitor BEZ235 are characterized by low expression of PTEN and the PP2A subunit PPP2R2B as a result of promoter hypermethylation [48]. BEZ235 refractory cell lines were modestly re-sensitized with the addition of decitabine [48]. Further research suggests the importance of DNMT3b function in promoting mTORC2 activation in melanoma; genetic loss of DNMT3 expression results in promoter hypomethylation and increased expression of mir-196b, which targets the mTORC2 component RICTOR. Loss of RICTOR expression in turn prevents mTORC2 activation and abrogates cellular growth [49]. These data suggest DNMT3b promotes mTORC2 activation during transformation. It is possible that combined pharmaceutical inhibition of DNMT3b and mTORC2, either directly via mTORC1/2 catalytic inhibitors or indirectly via PI3K/AKT inhibition, may provide durable and efficacious inhibition of DNA methylation and PI3K/mTOR signaling in human cancers.
HDAC inhibition
Next generation sequencing has identified genome-wide changes to histone acetylation in cancer. General histone acetylation is associated with enhanced gene transcription, and genetic ablation of the chromatin erasers responsible for histone deacetylation (histone deacetylases, HDACs) is anti-tumorigenic in models of many solid cancers [50]. Several studies have demonstrated the epigenetic repression of key tumor suppressors (CDKN1A) and DNA damage repair enzymes (BRCA, ATR) in tumors characterized by high HDAC expression. Indeed, elevated HDAC expression has been observed in breast, lung, liver, and other cancer types, and knockdown of several HDACs in these and other cancers induces apoptosis and cell cycle arrest [50], suggesting aberrant HDAC activity may be associated with cancer. As a result, HDAC inhibitors have entered clinical trials for the treatment of various cancers; Panobinostat is under phase III clinical testing for the treatment of cutaneous T cell lymphoma (CTCL), and numerous other HDAC inhibitors are currently in phase I/II clinical trials for the treatment of both solid and hematological malignancies. While most preclinical and clinically tested HDAC inhibitors function as nonselective pan HDAC inhibitors, they largely function by inhibiting HDAC function in multimeric protein complexes. Importantly, HDAC inhibitors such as Panobinostat and Vorinostat inhibit the HDAC1/2 containing complex co-REST, which also regulates H3K4 methylation through the LSD1 histone demethylase. Indeed, HDAC1/2 inhibition reduces H3K4me3 and transcriptional competence by indirectly inhibiting co-REST activity and LSD1 [51]. Since PI3K/AKT activation promotes H3K4me3, it is tempting to speculate that dual PI3K and HDAC1/2 inhibition may offer durable inhibition of both PI3K and H3K4me3 in cancers characterized by hyperactivated PI3K signaling and increased histone acetylation.
Preclinical evidence demonstrates anti-tumorigenic effects of combined HDAC and PI3K inhibition in the treatment of MYC-amplified cancers [52]. Dual pan-HDAC and pan-PI3K inhibition with Panobinostat/BKM120 increased HDAC-mediated FOXO1 expression and activation. These events were associated with increased cell death in patient derived xenograft models of MYC-amplified medulloblastoma [52]. These results demonstrate that targeting non-redundant pathways such that FOXO1 is reactivated may be a promising approach for the treatment of some types of cancer. Recently a single-agent dual HDAC/PI3K inhibitor (CUDC-907) showed promising antitumor activity in a phase I clinical trial for patients with diffuse large-B cell lymphoma [53]. It is tempting to speculate that subsequent generations of single-agent HDAC/PI3K inhibitors may increase in efficacy if isoform-selective HDAC or PI3K inhibition is achieved, or if combination therapy with other therapeutic targets is attained.
Inhibition of EZH2 enzymatic activity
EZH2 genomic amplification and overexpression occurs across many types of cancers including breast and prostate and results in excessive EZH2 histone methyltransferase (HMT) activity [54, 55], which increases H3K27me3 and gene silencing. EZH2 overexpression and increased H3K27me3 is associated with the transcriptional silencing of genes required for differentiation and lineage specificity as well as tumor suppressors INK4A/ARF. To date, cancer-associated gain-of-function mutations have been characterized. Mutation of EZH2 tyrosine 641 or alanines 677 and 687, located in the catalytic SET domain increases H3K27me3, and has been identified in non-Hodgkin’s lymphoma [56, 57]. EZH2 also possesses oncogenic activity independent of its HMT activity and PRC2 function. EZH2 promotes transcriptional activation through diverse mechanisms via its association with transcription factors ERα, WNT, AR, amongst others. EZH2 activity is antagonized by other histone modifying enzymes, including the SWI/SNF complex. Genetic loss of SWI/SNF or other EZH2 antagonists poises EZH2 for activation. As a result, the development and clinical testing of EZH2 small molecule inhibitors is underway for use in solid and hematological malignancies characterized by EZH2 amplification, overexpression, or gain-of-function mutations.
First generation EZH2 inhibitors were non-selective compounds that inhibited the processing of S-adenosyl-L-homocysteine hydrolase (SAH). The resulting increase in SAH levels represses the activity of S-adenosyl-L-methionine (SAM)-dependent histone lysine methyltransferases, the class of enzyme to which EZH2 belongs. One such inhibitor, 3-deazaneplanocin A (DZNep), effectively inhibits EZH2 and H3K27me3 and induces apoptosis in cell culture models, but its nonspecific function and toxicity in animal models have prevented its further development [58]. To increase efficacy and reduce toxicity, researchers have emphasized identifying potent and durable SAM-competitive inhibitors that selectively inhibit either wild type EZH2 or gain-of-function EZH2 mutations, including the orally bioavailable compounds UNC1999 and EPZ-6438 (Tazemetostat). Treatment of preclinical models of EZH2-activated tumors (SMARCB1/INI1 loss, EZH2 gain-of-function mutation) with Tazemetostat demonstrate a reduction in H3K27me3 and tumor regression [59, 60]. Objective response rates in a phase I Tazemetostat trial in non-Hodgkin’s lymphoma (NHL) were observed in 9 of 15 (60%) of evaluable patients, with phase II trials in NHL currently enrolling [61]. Moreover, phase II clinical trials testing Tazemetostat in solid tumors characterized by SMARCA4 or SMARCB1/INI1 loss are currently underway.
MLL1 inhibition via disruption of the MLL1-MEN1 or MLL1-WDR5 interface
Methylation of H3K4 is primarily mediated by the mixed-lineage leukemia (MLL) enzyme family. Genomic translocations characterized by MLL1 truncation coupled with in-frame fusion to more than 70 identified proteins produce oncogenic MLL1 fusion proteins that frequently occur in acute myeloid and lymphoid leukemias in adult and pediatric patients. However, MLL1 basal histone methyltransferase activity is low and is only enhanced following MLL1 association with the core complex members WDR5, ASH2L, and RbBP5 [62]. To this purpose, MLL1-selective inhibitors under preclinical development (MM-102, MM-401) target the MLL1-WDR5 interface for the treatment of hematological malignancies characterized by MLL-rearrangement and show promising antitumor activity in by reducing tumor growth and increasing apoptosis in MLL1-rearraned leukemias [63]. Separately, the association of the N-terminal region of MLL1 with Menin (MEN1) is required for the regulation of MLL1 target genes and mediates the oncogenicity of MLL1 translocations [64]. While MEN1 itself lacks a DNA binding domain, association with DNA binding proteins including MLL1 enhances MEN1 oncogenicity. Thus, small molecules targeting the MLL1-MEN1 interface are under preclinical development (MI-463, MI-503) and have been shown to prevent the MLL1-MEN1 association and in so doing, block the progression of MLL1-rearranged leukemias [65]. As a result, at least one MLL1-MEN1 inhibitor is poised to enter early stage clinical trials as early as 2017. The most frequent MLL1 translocations lose the SET domain that is responsible for H3K4 methyltransferase activity as a result of the in-frame fusion to sequences from ENL, AF4, AF9, or AF10. These specific MLL-fusion proteins directly or indirectly recruit the H3K79 histone methyltransferase Dot1L and direct Dot1L histone methyltransferase activity to MLL1 targets. As a result, many MLL1-rearranged leukemias are sensitive to Dot1L inhibition [66]. The Dot1l inhibitor EPZ-5676 (Pinometostat) has been shown to reduce H3K79 methylation and MLL1 target gene expression, which inhibits cell proliferation in in vitro and in vivo models of MLL1-rearranged leukemias and has since moved into Phase I and II clinical trials [66]. Recently, MLL1 rearrangement in primary human patient samples and human cancer derived cell lines originating from acute myeloid leukemia (AML) was shown to enhance sensitivity to PI3K-pathway inhibition [67]. Dual PI3K/mTOR inhibition reduced tumor burden and prolonged survival in a preclinical AML model characterized by MLL1-AF9 translocation. It is tempting to speculate that combined PI3K/mTOR and MLL1 or Dot1L inhibition may be an efficacious and durable therapeutic strategy for leukemias characterized by MLL1 translocations, irrespective of PI3K pathway activation.
While the majority of preclinical studies to date utilize MLL1-selective inhibition in MLL1-rearranged leukemias, some solid tumors may also be suitable targets for MLL1 inhibition. For example, loss of MLL1 expression in breast cancer cell lines characterized by gain of function p53 mutations and wild type MLL1 reduces tumor formation [68]. Recent reports suggest MEN1 itself may be an oncogene, as high expression in Estrogen Receptor (ER)-positive breast cancer correlates with poor clinical outcome [69]. Moreover, MEN1 has been shown to associate with ER and facilitate the recruitment of MLL1, which increases promoter-associated H3K4me3 and the transcription of ER-responsive genes [69].
Similarly, MEN1 associates with Androgen Receptor (AR) and the MLL1 complex in castration resistant prostate cancer (CRPC). This MEN1-MLL1 complex binds AR on the promoters of AR target genes, and inhibition of the MLL1-MEN1 interaction reduces prostate cancer growth in preclinical models of CRPC [70]. These data suggest that solid tumors characterized by MLL1 amplification, MEN1 overexpression, or elevated H3K4me3 may be candidates for MLL1 inhibition, either through small molecule inhibitors targeting the MLL1-MEN1 or the MLL1-WDR5 interface.
Conclusion
The contribution of PI3K/AKT on epigenomic modulation, transcriptional activation, and cancer is considerable. Future progress in the treatment of PI3K pathway activated solid tumors should explore additional mechanism-based therapy combinations that are driven by restoring the normal balance of repressive and activating chromatin modifications. Progress in which PI3K pathway-targeted agents are combined with inhibitors targeting chromatin modification may produce invaluable preclinical mechanistic insight and antitumor activity in the clinic.
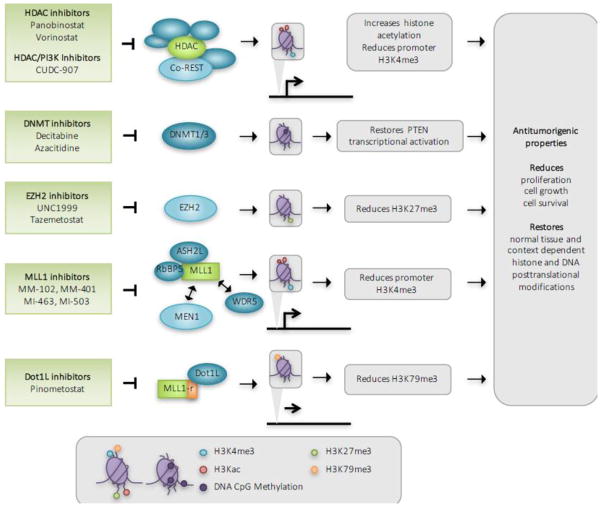
Figure 3. Therapeutic modulation of chromatin in cancer.
Inhibitors targeting chromatin modification also show anti-tumorigenic activity in preclinical and clinical studies and have limited FDA-approval to date. Highlighted here, non-selective HDAC inhibitors maintain histone acetylation, which in some cases also inhibit H3K4me3 to reduce transcriptional activation. Preclinical synergistic activity observed with combined HDAC and PI3K inhibitors launched the development of the dual specificity HDAC/PI3K inhibitor CUDC-907. DNMT inhibitors reduce CpG island methylation and in so doing, restore gene expression. This is especially critical at genomic loci encoding tumor suppressors such as PTEN, where transcriptional silencing is associated with promoter DNA hypermethylation and is a common event in cancer. The Dot1L inhibitor Pinometostat has shown in vivo efficacy for the treatment of MLL1-rearranged (MLL1-r) leukemias and is currently in Phase I/II clinical trials.
Acknowledgments
The authors apologize to the many colleagues whose work they were unable to cite due to space limitations. This work was supported by the American Cancer Society (125303-PF-13-097-01-CCE) to J.M.S, the National Institutes of Health (NIH) P50 CA168504 (to T.M.R. and J.J.Z), P50 CA165962 (to T.M.R. and J.J.Z), CA187918-02 (to T.M.R. and J.J.Z), CA210057-01 (to J.J.Z), CA172461-04 (to J.J.Z), and Breast Cancer Research Foundation (to J.J.Z).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Feinberg AP, Koldobskiy MA, Gondor A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet. 2016;17:284–299. doi: 10.1038/nrg.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 3.Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15:7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alessi DR, Caudwell FB, Andjelkovic M, Hemmings BA, Cohen P. Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett. 1996;399:333–338. doi: 10.1016/s0014-5793(96)01370-1. [DOI] [PubMed] [Google Scholar]
- 7.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 8.Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 9.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 10.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 11.Luczak MW, Jagodzinski PP. The role of DNA methylation in cancer development. Folia Histochem Cytobiol. 2006;44:143–154. [PubMed] [Google Scholar]
- 12.Chik F, Szyf M, Rabbani SA. Role of epigenetics in cancer initiation and progression. Adv Exp Med Biol. 2011;720:91–104. doi: 10.1007/978-1-4614-0254-1_8. [DOI] [PubMed] [Google Scholar]
- 13.Esteve PO, Chang Y, Samaranayake M, Upadhyay AK, Horton JR, Feehery GR, Cheng X, Pradhan S. A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability. Nat Struct Mol Biol. 2011;18:42–48. doi: 10.1038/nsmb.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hervouet E, Lalier L, Debien E, Cheray M, Geairon A, Rogniaux H, Loussouarn D, Martin SA, Vallette FM, Cartron PF. Disruption of Dnmt1/PCNA/UHRF1 interactions promotes tumorigenesis from human and mice glial cells. PLoS One. 2010;5:e11333. doi: 10.1371/journal.pone.0011333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popkie AP, Zeidner LC, Albrecht AM, D’Ippolito A, Eckardt S, Newsom DE, Groden J, Doble BW, Aronow B, McLaughlin KJ, White P, Phiel CJ. Phosphatidylinositol 3-kinase (PI3K) signaling via glycogen synthase kinase-3 (Gsk-3) regulates DNA methylation of imprinted loci. J Biol Chem. 2010;285:41337–41347. doi: 10.1074/jbc.M110.170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Badeaux AI, Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nat Rev Mol Cell Biol. 2013;14:211–224. doi: 10.1038/nrm3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, Ping B, Otte AP, Hung MC. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310:306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- 18.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, Wu X, Stack EC, Loda M, Liu T, Xu H, Cato L, Thornton JE, Gregory RI, Morrissey C, Vessella RL, Montironi R, Magi-Galluzzi C, Kantoff PW, Balk SP, Liu XS, Brown M. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benard A, Goossens-Beumer IJ, van Hoesel AQ, de Graaf W, Horati H, Putter H, Zeestraten EC, van de Velde CJ, Kuppen PJ. Histone trimethylation at H3K4, H3K9 and H4K20 correlates with patient survival and tumor recurrence in early-stage colon cancer. BMC Cancer. 2014;14:531. doi: 10.1186/1471-2407-14-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mungamuri SK, Murk W, Grumolato L, Bernstein E, Aaronson SA. Chromatin modifications sequentially enhance ErbB2 expression in ErbB2-positive breast cancers. Cell Rep. 2013;5:302–313. doi: 10.1016/j.celrep.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spangle JM, Dreijerink KM, Groner AC, Cheng H, Ohlson CE, Reyes J, Lin CY, Bradner J, Zhao JJ, Roberts TM, Brown M. PI3K/AKT Signaling Regulates H3K4 Methylation in Breast Cancer. Cell Rep. 2016;15:2692–2704. doi: 10.1016/j.celrep.2016.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tenney K, Shilatifard A. A COMPASS in the voyage of defining the role of trithorax/MLL-containing complexes: linking leukemogensis to covalent modifications of chromatin. J Cell Biochem. 2005;95:429–436. doi: 10.1002/jcb.20421. [DOI] [PubMed] [Google Scholar]
- 23.Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin WG., Jr The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Cohen I, Poreba E, Kamieniarz K, Schneider R. Histone modifiers in cancer: friends or foes? Genes Cancer. 2011;2:631–647. doi: 10.1177/1947601911417176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang WC, Chen CC. Akt phosphorylation of p300 at Ser-1834 is essential for its histone acetyltransferase and transcriptional activity. Mol Cell Biol. 2005;25:6592–6602. doi: 10.1128/MCB.25.15.6592-6602.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Cerbo V, Schneider R. Cancers with wrong HATs: the impact of acetylation. Brief Funct Genomics. 2013;12:231–243. doi: 10.1093/bfgp/els065. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Xing ZB, Zhang JH, Fang Y. Akt kinase targets the association of CBP with histone H3 to regulate the acetylation of lysine K18. FEBS Lett. 2013;587:847–853. doi: 10.1016/j.febslet.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 31.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Qiu Y, Liu L, Zhao C, Han C, Li F, Zhang J, Wang Y, Li G, Mei Y, Wu M, Wu J, Shi Y. Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription. Genes Dev. 2012;26:1376–1391. doi: 10.1101/gad.188359.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poplawski A, Hu K, Lee W, Natesan S, Peng D, Carlson S, Shi X, Balaz S, Markley JL, Glass KC. Molecular insights into the recognition of N-terminal histone modifications by the BRPF1 bromodomain. J Mol Biol. 2014;426:1661–1676. doi: 10.1016/j.jmb.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rokudai S, Aikawa Y, Tagata Y, Tsuchida N, Taya Y, Kitabayashi I. Monocytic leukemia zinc finger (MOZ) interacts with p53 to induce p21 expression and cell-cycle arrest. J Biol Chem. 2009;284:237–244. doi: 10.1074/jbc.M805101200. [DOI] [PubMed] [Google Scholar]
- 35.Rokudai S, Laptenko O, Arnal SM, Taya Y, Kitabayashi I, Prives C. MOZ increases p53 acetylation and premature senescence through its complex formation with PML. Proc Natl Acad Sci U S A. 2013;110:3895–3900. doi: 10.1073/pnas.1300490110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raynaud FI, Eccles SA, Patel S, Alix S, Box G, Chuckowree I, Folkes A, Gowan S, De Haven Brandon A, Di Stefano F, Hayes A, Henley AT, Lensun L, Pergl-Wilson G, Robson A, Saghir N, Zhyvoloup A, McDonald E, Sheldrake P, Shuttleworth S, Valenti M, Wan NC, Clarke PA, Workman P. Biological properties of potent inhibitors of class I phosphatidylinositide 3-kinases: from PI-103 through PI-540, PI-620 to the oral agent GDC-0941. Mol Cancer Ther. 2009;8:1725–1738. doi: 10.1158/1535-7163.MCT-08-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maira SM, Stauffer F, Brueggen J, Furet P, Schnell C, Fritsch C, Brachmann S, Chene P, De Pover A, Schoemaker K, Fabbro D, Gabriel D, Simonen M, Murphy L, Finan P, Sellers W, Garcia-Echeverria C. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol Cancer Ther. 2008;7:1851–1863. doi: 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- 38.Gokmen-Polar Y, Liu Y, Toroni RA, Sanders KL, Mehta R, Badve S, Rommel C, Sledge GW., Jr Investigational drug MLN0128, a novel TORC1/2 inhibitor, demonstrates potent oral antitumor activity in human breast cancer xenograft models. Breast Cancer Res Treat. 2012;136:673–682. doi: 10.1007/s10549-012-2298-8. [DOI] [PubMed] [Google Scholar]
- 39.Gonzalez-Angulo AM, Krop I, Akcakanat A, Chen H, Liu S, Li Y, Culotta KS, Tarco E, Piha-Paul S, Moulder-Thompson S, Velez-Bravo V, Sahin AA, Doyle LA, Do KA, Winer EP, Mills GB, Kurzrock R, Meric-Bernstam F. SU2C phase Ib study of paclitaxel and MK-2206 in advanced solid tumors and metastatic breast cancer. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/dju493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oki Y, Fanale M, Romaguera J, Fayad L, Fowler N, Copeland A, Samaniego F, Kwak LW, Neelapu S, Wang M, Feng L, Younes A. Phase II study of an AKT inhibitor MK2206 in patients with relapsed or refractory lymphoma. Br J Haematol. 2015;171:463–470. doi: 10.1111/bjh.13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castellano E, Sheridan C, Thin MZ, Nye E, Spencer-Dene B, Diefenbacher ME, Moore C, Kumar MS, Murillo MM, Gronroos E, Lassailly F, Stamp G, Downward J. Requirement for interaction of PI3-kinase p110alpha with RAS in lung tumor maintenance. Cancer Cell. 2013;24:617–630. doi: 10.1016/j.ccr.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Utermark T, Rao T, Cheng H, Wang Q, Lee SH, Wang ZC, Iglehart JD, Roberts TM, Muller WJ, Zhao JJ. The p110alpha and p110beta isoforms of PI3K play divergent roles in mammary gland development and tumorigenesis. Genes Dev. 2012;26:1573–1586. doi: 10.1101/gad.191973.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jia S, Liu Z, Zhang S, Liu P, Zhang L, Lee SH, Zhang J, Signoretti S, Loda M, Roberts TM, Zhao JJ. Essential roles of PI(3)K-p110beta in cell growth, metabolism and tumorigenesis. Nature. 2008;454:776–779. doi: 10.1038/nature07091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ni J, Liu Q, Xie S, Carlson C, Von T, Vogel K, Riddle S, Benes C, Eck M, Roberts T, Gray N, Zhao J. Functional characterization of an isoform-selective inhibitor of PI3K-p110beta as a potential anticancer agent. Cancer Discov. 2012;2:425–433. doi: 10.1158/2159-8290.CD-12-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Furman RR, Sharman JP, Coutre SE, Cheson BD, Pagel JM, Hillmen P, Barrientos JC, Zelenetz AD, Kipps TJ, Flinn I, Ghia P, Eradat H, Ervin T, Lamanna N, Coiffier B, Pettitt AR, Ma S, Stilgenbauer S, Cramer P, Aiello M, Johnson DM, Miller LL, Li D, Jahn TM, Dansey RD, Hallek M, O’Brien SM. Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N Engl J Med. 2014;370:997–1007. doi: 10.1056/NEJMoa1315226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lou YF, Zou ZZ, Chen PJ, Huang GB, Li B, Zheng DQ, Yu XR, Luo XY. Combination of gefitinib and DNA methylation inhibitor decitabine exerts synergistic anti-cancer activity in colon cancer cells. PLoS One. 2014;9:e97719. doi: 10.1371/journal.pone.0097719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang YJ, Zhao SL, Tian XQ, Sun DF, Xiong H, Dai Q, Li XQ, Fang JY. Combined inhibition of Dnmt and mTOR signaling inhibits formation and growth of colorectal cancer. Int J Colorectal Dis. 2009;24:629–639. doi: 10.1007/s00384-009-0664-8. [DOI] [PubMed] [Google Scholar]
- 48.Qian XJ, Li YT, Yu Y, Yang F, Deng R, Ji J, Jiao L, Li X, Wu RY, Chen WD, Feng GK, Zhu XF. Inhibition of DNA methyltransferase as a novel therapeutic strategy to overcome acquired resistance to dual PI3K/mTOR inhibitors. Oncotarget. 2015;6:5134–5146. doi: 10.18632/oncotarget.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Micevic G, Muthusamy V, Damsky W, Theodosakis N, Liu X, Meeth K, Wingrove E, Santhanakrishnan M, Bosenberg M. DNMT3b Modulates Melanoma Growth by Controlling Levels of mTORC2 Component RICTOR. Cell Rep. 2016;14:2180–2192. doi: 10.1016/j.celrep.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014;124:30–39. doi: 10.1172/JCI69738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 52.Pei Y, Liu KW, Wang J, Garancher A, Tao R, Esparza LA, Maier DL, Udaka YT, Murad N, Morrissy S, Seker-Cin H, Brabetz S, Qi L, Kogiso M, Schubert S, Olson JM, Cho YJ, Li XN, Crawford JR, Levy ML, Kool M, Pfister SM, Taylor MD, Wechsler-Reya RJ. HDAC and PI3K Antagonists Cooperate to Inhibit Growth of MYC-Driven Medulloblastoma. Cancer Cell. 2016;29:311–323. doi: 10.1016/j.ccell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Younes A, Berdeja JG, Patel MR, Flinn I, Gerecitano JF, Neelapu SS, Kelly KR, Copeland AR, Akins A, Clancy MS, Gong L, Wang J, Ma A, Viner JL, Oki Y. Safety, tolerability, and preliminary activity of CUDC-907, a first-in-class, oral, dual inhibitor of HDAC and PI3K, in patients with relapsed or refractory lymphoma or multiple myeloma: an open-label, dose-escalation, phase 1 trial. The Lancet. Oncology. 2016 doi: 10.1016/S1470-2045(15)00584-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 55.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, Sabel MS, Livant D, Weiss SJ, Rubin MA, Chinnaiyan AM. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, Yap D, Humphries RK, Griffith OL, Shah S, Zhu H, Kimbara M, Shashkin P, Charlot JF, Tcherpakov M, Corbett R, Tam A, Varhol R, Smailus D, Moksa M, Zhao Y, Delaney A, Qian H, Birol I, Schein J, Moore R, Holt R, Horsman DE, Connors JM, Jones S, Aparicio S, Hirst M, Gascoyne RD, Marra MA. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, Copeland RA. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci U S A. 2010;107:20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi RK, Tan PB, Liu ET, Yu Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knutson SK, Kawano S, Minoshima Y, Warholic NM, Huang KC, Xiao Y, Kadowaki T, Uesugi M, Kuznetsov G, Kumar N, Wigle TJ, Klaus CR, Allain CJ, Raimondi A, Waters NJ, Smith JJ, Porter-Scott M, Chesworth R, Moyer MP, Copeland RA, Richon VM, Uenaka T, Pollock RM, Kuntz KW, Yokoi A, Keilhack H. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol Cancer Ther. 2014;13:842–854. doi: 10.1158/1535-7163.MCT-13-0773. [DOI] [PubMed] [Google Scholar]
- 60.Xu B, On DM, Ma A, Parton T, Konze KD, Pattenden SG, Allison DF, Cai L, Rockowitz S, Liu S, Liu Y, Li F, Vedadi M, Frye SV, Garcia BA, Zheng D, Jin J, Wang GG. Selective inhibition of EZH2 and EZH1 enzymatic activity by a small molecule suppresses MLL-rearranged leukemia. Blood. 2015;125:346–357. doi: 10.1182/blood-2014-06-581082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vincent Ribrag J-CS, Michot Jean-Marie, Schmitt Anna, Postel-Vinay Sophie, Bijou Fontanet, Thomson Blythe, Keilhack Heike, Blakemore Stephen J, Reyderman Larisa, Kumar Pavan, Fine Greg, McDonald Alice, Ho Peter T, Italiano Antoine. Phase 1 Study of Tazemetostat (EPZ-6438), an Inhibitor of Enhancer of Zeste-Homolog 2 (EZH2): Preliminary Safety and Activity in Relapsed or Refractory Non-Hodgkin Lymphoma. Blood. 2015;126:473. [Google Scholar]
- 62.Li Y, Han J, Zhang Y, Cao F, Liu Z, Li S, Wu J, Hu C, Wang Y, Shuai J, Chen J, Cao L, Li D, Shi P, Tian C, Zhang J, Dou Y, Li G, Chen Y, Lei M. Structural basis for activity regulation of MLL family methyltransferases. Nature. 2016;530:447–452. doi: 10.1038/nature16952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Senisterra G, Wu H, Allali-Hassani A, Wasney GA, Barsyte-Lovejoy D, Dombrovski L, Dong A, Nguyen KT, Smil D, Bolshan Y, Hajian T, He H, Seitova A, Chau I, Li F, Poda G, Couture JF, Brown PJ, Al-Awar R, Schapira M, Arrowsmith CH, Vedadi M. Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5. Biochem J. 2013;449:151–159. doi: 10.1042/BJ20121280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 65.Borkin D, He S, Miao H, Kempinska K, Pollock J, Chase J, Purohit T, Malik B, Zhao T, Wang J, Wen B, Zong H, Jones M, Danet-Desnoyers G, Guzman ML, Talpaz M, Bixby DL, Sun D, Hess JL, Muntean AG, Maillard I, Cierpicki T, Grembecka J. Pharmacologic inhibition of the Menin-MLL interaction blocks progression of MLL leukemia in vivo. Cancer Cell. 2015;27:589–602. doi: 10.1016/j.ccell.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Daigle SR, Olhava EJ, Therkelsen CA, Basavapathruni A, Jin L, Boriack-Sjodin PA, Allain CJ, Klaus CR, Raimondi A, Scott MP, Waters NJ, Chesworth R, Moyer MP, Copeland RA, Richon VM, Pollock RM. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood. 2013;122:1017–1025. doi: 10.1182/blood-2013-04-497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sandhofer N, Metzeler KH, Rothenberg M, Herold T, Tiedt S, Groiss V, Carlet M, Walter G, Hinrichsen T, Wachter O, Grunert M, Schneider S, Subklewe M, Dufour A, Frohling S, Klein HG, Hiddemann W, Jeremias I, Spiekermann K. Dual PI3K/mTOR inhibition shows antileukemic activity in MLL-rearranged acute myeloid leukemia. Leukemia. 2015;29:828–838. doi: 10.1038/leu.2014.305. [DOI] [PubMed] [Google Scholar]
- 68.Zhu J, Sammons MA, Donahue G, Dou Z, Vedadi M, Getlik M, Barsyte-Lovejoy D, Al-awar R, Katona BW, Shilatifard A, Huang J, Hua X, Arrowsmith CH, Berger SL. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature. 2015;525:206–211. doi: 10.1038/nature15251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dreijerink KM, Mulder KW, Winkler GS, Hoppener JW, Lips CJ, Timmers HT. Menin links estrogen receptor activation to histone H3K4 trimethylation. Cancer Res. 2006;66:4929–4935. doi: 10.1158/0008-5472.CAN-05-4461. [DOI] [PubMed] [Google Scholar]
- 70.Malik R, Khan AP, Asangani IA, Cieslik M, Prensner JR, Wang X, Iyer MK, Jiang X, Borkin D, Escara-Wilke J, Stender R, Wu YM, Niknafs YS, Jing X, Qiao Y, Palanisamy N, Kunju LP, Krishnamurthy PM, Yocum AK, Mellacheruvu D, Nesvizhskii AI, Cao X, Dhanasekaran SM, Feng FY, Grembecka J, Cierpicki T, Chinnaiyan AM. Targeting the MLL complex in castration-resistant prostate cancer. Nat Med. 2015;21:344–352. doi: 10.1038/nm.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]