Abstract
The MUC1 gene evolved in mammalian species to provide protection of epithelia. The transmembrane MUC1 C-terminal subunit (MUC1-C) signals stress to the interior of the epithelial cell and, when overexpressed as in most carcinomas, functions as an oncoprotein. MUC1-C induces the epithelial-mesenchymal transition (EMT) by activating the inflammatory NF-κB p65 pathway and, in turn, the EMT-transcriptional repressor ZEB1. Emerging evidence has indicated that MUC1-C drives a program integrating the induction of EMT with activation of stem cell traits, epigenetic reprogramming and immune evasion. This mini-review focuses on the potential importance of this MUC1-C program in cancer progression.
Keywords: MUC1-C, EMT, epigenetics, tumor suppressor genes, BMI1, DNMTs, PD-L1, immune evasion
INTRODUCTION
Epithelia are laterally connected layers of cells that evolved in metazoans to provide barriers from the external environment. Stratified layers of epithelia, as found in the epidermis, function as one example of a protective physical barrier. Simple epithelia that line the surface of the gastrointestinal and pulmonary tracts, as well as ducts in specialized organs, also act as barriers. However, in contrast to stratified epithelia, simple epithelia from which most carcinomas arise are largely comprised of single layers of epithelial cells with apical-basal polarity. The apical cell border of simple epithelia faces the luminal space and is thus exposed to diverse forms of stress, such as toxins, microorganisms and inflammation, among others. Accordingly, a robust defense for epithelia with planar cell polarity became necessary during evolution to maintain integrity of the epithelial barrier. The secreted and transmembrane mucins evolved to play such a role [1].
Mucin 1 (MUC1) emerged in mammals for protection of epithelia
MUC1 is a transmembrane mucin that is expressed at the apical surfaces of normal epithelia [1]. MUC1 emerged from the secreted MUC5B mucin late in vertebrate evolution and, interestingly, the expression of MUC1 homologs is restricted to mammalian species [2]. MUC1 consists of two subunits [1]. The MUC1 N-terminal (MUC1-N) subunit is the mucin component of the heterodimer, which contains glycosylated tandem repeats that are characteristic of the mucin family members [1]. MUC1-N is positioned extracellularly at the apical epithelial membrane in a complex with the MUC1 transmembrane C-terminal (MUC1-C) subunit. MUC1-N is also shed from the cell surface. MUC1-N thereby contributes to a mucous barrier that, along with other mucins, such as the secreted forms, physically protects the apical epithelial membrane from damage [1].
MUC1-C functions as a signaling subunit
An important distinction is that, in contrast to the secreted mucins, MUC1 emerged with additional sequences encoding the transmembrane MUC1-C subunit [2], which when activated functions in transducing signals to the interior of the epithelial cell. In this way, MUC1-C has the capacity to promote loss of polarity with induction of the epithelial-mesenchymal transition (EMT) [3, 4] and thereby interact with other cell surface molecules, including receptor tyrosine kinases (RTKs) [5–12]. The available evidence has further supported the premise that (i) MUC1-C functions in activating multiple pathways linked to EMT, self-renewal and survival, and (ii) the overexpression of MUC1-C, as found in diverse human carcinomas, represents a subversion and appropriation of these functions by cancer cells to promote their own growth and immortality [1, 13].
MUC1-C INDUCES MULTIPLE PATHWAYS LINKED TO CANCER
MUC1-C functions as an oncoprotein
Earlier studies of depolarized carcinoma cells showed that MUC1-C interacts with RTKs, such as EGFR and HER2, at the cell membrane and thereby promotes their activation and downstream pathways (Fig. 1A) [9–11, 13]. MUC1-C is targeted to the nucleus [14], where it interacts with diverse activators and repressors of transcription [1, 13] (Fig. 1A). In this way, MUC1-C stabilizes the WNT effector, β-catenin, and induces activation of WNT/β-catenin/TCF4 target genes, including CCND1, MYC and CTGF [15–19]. MUC1-C is also targeted to the mitochondrial outer membrane and suppresses stress- and death receptor-induced apoptosis (Fig. 1A) [20–22]. Moreover, MUC1-C and specifically the cytoplasmic domain (i) is sufficient to confer self-renewal, anchorage-independent growth and tumorigenicity, and (ii) acts as a target for inhibiting MUC1-C-induced transformation [13, 23, 24].
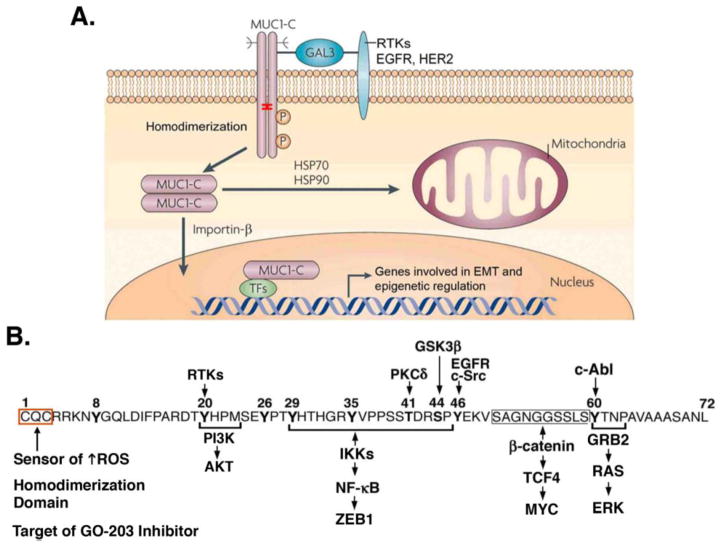
Figure 1. Activation of MUC1-C induces diverse pathways linked to cancer.
A. The MUC1 protein undergoes autocleavage into the shed N-terminal (MUC1-N) and transmembrane C-terminal (MUC1-C) subunits. This nomenclature is used to denote positioning of the subunits and to distinguish them from genetic isoforms, which use Greek symbols; for example, ERα and ERβ, the PKC isoenzymes and PDGF receptors. MUC1-C forms homodimers in the response to stress and transformation that are mediated by a CQC motif in the cytoplasmic domain (highlighted with red bracket). In turn, MUC1-C homodimers are necessary for interactions with RTKs at the cell membrane mediated by galectin-3 bridges, import into the nucleus by an importin-β-mediated mechanism, and transport to the mitochondrial outer membrane by HSP70/HSP90. In the nucleus, MUC1-C forms complexes with transcription factors (TFs), such as NF-κB p65 and ZEB1, which drive EMT and epigenetic regulators. B. The MUC1-C cytoplasmic domain is a 72 aa intrinsically disordered protein that functions as a node for the integration of diverse signaling pathways. The CQC motif is a sensor of oxidative stress that is necessary for MUC1-C homodimerization and function. In addition, the MUC1-C CQC motif is targeted by the GO-203 inhibitor. Highlighted are selected phosphorylation sites, which regulate binding to the indicated effectors of downstream signaling pathways.
MUC1-C cytoplasmic domain is an intrinsically disordered protein
MUC1-C consists of a 58 amino acid (aa) extracellular domain, a 28 aa transmembrane domain and a 72 aa cytoplasmic domain [1]. Given the above evidence in support of diverse MUC1-C functions, one might reasonably ask how this relatively small protein can drive multiple pathways. In response, the MUC1-C cytoplasmic domain has no apparent structure [25], consistent with intrinsically disordered proteins as found in oncogenic molecules, such as p53, that function as nodes to direct the activation of multiple signaling pathways (Fig. 1B). In addition, the MUC1-C cytoplasmic domain contains a CQC motif that functions as sensor of oxidative stress and is necessary for the formation of MUC1-C homodimers (Fig. 1B) [25–27]. The MUC1-C cytoplasmic domain is subject to phosphorylation by certain kinases that, in turn, regulate interactions with effectors, such as p53 [8, 28], PI3K [9] and β-catenin [15], of downstream pathways, which are linked to hallmarks of the cancer cell (Fig. 1B) [1, 13].
MUC1-C ACTIVATES THE EMT PROGRAM
The EMT program is a fundamental process and an important regulator of cancer progression in which polarized epithelial cells acquire mesenchymal and stem cell properties [29–32]. EMT is directed by a group of transcription factors (EMT-TFs), which includes ZEB1, a member of the zinc finger E-box binding homeodomain, basic helix-loop-helix family [31]. EMT is a reversible program during embryonic development and repair of epithelial cell damage [30]. However, aberrant and prolonged activation of EMT in cancer cells contributes to a malignant phenotype with a capacity for plasticity, invasion, metastases, stemness and resistance to apoptosis [29–32]. The available evidence supports a role for MUC1-C in driving EMT by activating pathways linked to epithelial-mesenchymal plasticity and as an oncogenic effector of the EMT program and stem cell traits [3, 33–35].
MUC1-C directly contributes to activation of EMT-associated signaling pathways
Activation of the EMT program can be attributed to a diverse array of autocrine, paracrine and inflammatory signaling cascades [31]. For example, paracrine signaling from the microenvironment resulting in activation of TGF-β, RTKs, WNT or STAT3 signaling, among others, has been associated with induction of EMT-TFs and epithelial-mesenchymal plasticity [31]. In this respect, MUC1-C has been linked to the direct activation of (i) TAK1, an effector of the TGF-β pathway [36], (ii) the LIN28B→let-7→HMGA2 pathway, which is associated with EMT and stemness [35], (iii) RTKs [9–11, 13], (iv) WNT/β-catenin signaling [15, 17, 19], and (v) STAT3 [37]. These findings have supported the notion that MUC1-C has the potential to directly contribute to epithelial-mesenchymal plasticity of normal epithelial cells and their malignant counterparts by circumventing signals from the microenvironment.
MUC1-C directly activates an inflammatory NF-κB p65→ZEB1→EMT pathway
MUC1-C intersects with the inflammatory TAK1→IKK→NF-κB pathway, forms nuclear complexes with NF-κB p65 and promotes activation of NF-κB p65 target genes, including MUC1 itself in an autoinductive circuit (Fig. 2A) [36, 38, 39]. In turn, MUC1-C activates ZEB1 by two mechanisms [3]. MUC1-C induces ZEB1 expression by occupying the ZEB1 promoter with NF-κB p65 and thereby driving ZEB1 transcription (Fig. 2B) [3]. In addition, MUC1-C binds directly to ZEB1 and promotes ZEB1 occupancy on target gene promoters [3]. Of interest in this regard, ZEB1 drives escape from tumor latency in the response to inflammation and is sufficient for carcinoma cells to stably enter a metastatic state [40]. These and the above findings highlighted the notion that MUC1-C functions in directly activating an inflammatory pathway linked to EMT.
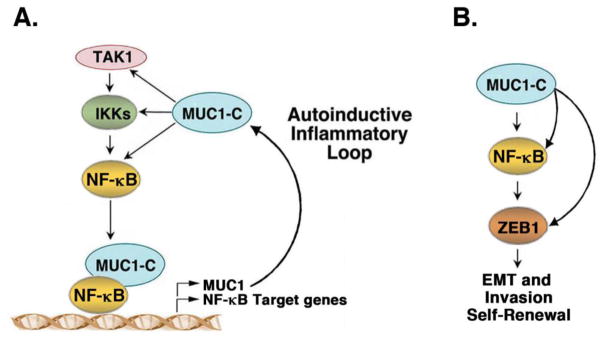
Figure 2. MUC1-C activates an autoinductive inflammatory circuit that drives EMT, invasion and self-renewal capacity.
A. MUC1-C activates the inflammatory NF-κB pathway by interacting directly with TAK1, IKKs and NF-κB p65. MUC1-C promotes occupancy of NF-κB p65 on promoters of its target genes, including MUC1 itself, and drives their transcription. B. The MUC1-C→NF-κB p65 circuit activates the ZEB1 gene. In turn, MUC1-C binds to ZEB1 and promotes ZEB1-mediated repression of target genes, such as CRB3, encoding the CRB3 polarity factor, and miR-200c, which promotes EMT and invasion.
In further support of the importance of the MUC1-C→NF-κB→ZEB1 pathway, the interactions between MUC1-C and ZEB1 (i) downregulate expression of cell polarity factors, such as CRB3, needed for maintenance of apical-basal polarity, (ii) suppress E-cadherin, a key protein for formation of the adherens junction, (iii) repress miR-200c, and (iv) induce EMT and invasion (Fig. 2B) [3, 4, 24]. Moreover and importantly, activation of the MUC1-C→NF-κB p65→ZEB1 pathway is integrated with the induction of self-renewal capacity, a hallmark of cancer stem-like cells [24, 33]. As a result, targeting MUC1-C genetically or pharmacologically is associated with reversal of the EMT phenotype and decreases in self-renewal [3, 4, 24, 33]. These findings thus identified a previously unrecognized role for MUC1-C as an oncogenic effector in induction of the EMT program and stem cell traits.
MUC1-C INTEGRATES EMT WITH EPIGENETIC REGULATORS AND IMMUNE EVASION
Cancer cells undergo reversible transitions between epithelial and mesenchymal phenotypes that are regulated by multiple afferent signals [31]. Activation of the EMT-TFs, such as ZEB1, is in part responsible for the resulting changes in gene expression patterns, such as repression of the CDH1 gene encoding E-cadherin. In this context, EMT-TF function is associated with the recruitment of epigenetic regulators that promote the extent and potential irreversibility of gene repression [31]. EMT-TFs interact with diverse epigenetic regulators [31], among which MUC1-C has been linked to date with regulation of the polycomb repressive complex 1 (PRC1) and to the DNA methyltransferases (DNMTs). PRC1 includes the BMI1 protein, which contributes to the E3 ubiquitin ligase function that catalyzes the mono-ubiquitination of histone H2A and thereby promotes gene silencing [41]. The DNMTs catalyze the methylation of DNA, which has an essential role in the epigenetic regulation of gene expression.
BMI1 and repression of TSG expression
PRC1 includes BMI1, RING1 and the catalytic RING2 subunit [41]. BMI1 is overexpressed in multiple carcinomas and is associated with poor outcomes [41]. BMI1 has also been linked with self-renewal of cancer stem-like cells, and the induction of EMT and invasive tumor phenotypes [41]. Intriguingly, targeting MUC1-C in human carcinoma cells results in the suppression of BMI1, RING1 and RING2, indicating that MUC1-C promotes the expression of multiple PRC1 subunits [42]. With regard to the mechanistic regulation of BMI1, the evidence indicates that MUC1-C activates the BMI1 promoter by a MYC-dependent mechanism [42]. MUC1-C also blocks miR-200c-mediated downregulation of BMI1 expression, linking this mechanism to ZEB1-induced suppression of miR-200c and the EMT program [42]. Analysis of gene expression datasets further demonstrated highly significant correlations between MUC1-C and BMI1, supporting the notion that MUC1-C contributes to the overexpression of BMI1 in breast and other carcinomas [42]. MUC1-C also directs BMI1 function by binding directly to this stem cell factor, promoting occupancy of BMI1 on the CDKN2A promoter and repressing expression of the p16INK4a tumor suppressor [42].
DNMTs and regulation of DNA methylation
DNMT1, DNMT3a and DNMT3b are dysregulated in cancer cells. DNMT1 is primarily responsible for maintaining DNA methylation patterns, whereas DNMT3a and DNMT3b largely establish de novo postreplicative DNA methylation. Interestingly, DNMT1 and DNMT3b are both required for repressing genes in cancer cells [43]. Of further interest in this regard, MUC1-C induces expression of DNMT1 and DNMT3b, but not DNMT3a [44]. Involvement of MUC1-C in activating the TAK1→IKK→NF-κB p65 pathway provided support for MUC1-C-induced integration of the inflammatory response with induction of ZEB1 and EMT [3, 36]. Along these lines, however, there was no evidence to link MUC1-C with changes in DNA methylation patterns. In searching for such an association, subsequent studies found that MUC1-C occupies the DNMT1 and DNMT3b promoters in complexes with NF-κB p65, promotes NF-κB p65 occupancy and thereby drives DNMT1 and DNMT3b transcription [44]. MUC1-C also increases DNA methylation of the CDH1 promoter in association with repression of E-cadherin expression [44], supporting a model in which MUC1-C integrates inflammatory signaling with EMT and epigenetic regulatory mechanisms (Fig. 3A).
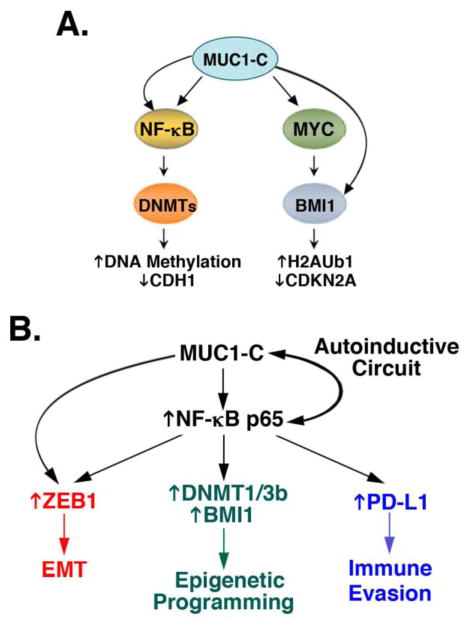
Figure 3. MUC1-C integrates EMT with epigenetic programming and immune evasion.
A. The MUC1-C→NF-B p65 circuit activates transcription of the DNMT1 and DNMT3b, but not DNMT3a, genes and thereby regulates global DNA methylation patterns. The MUC1-C→NF-κB p65→DNMT1/3b pathway also induces DNA methylation of the CDH1 promoter in association with suppression of E-cadherin expression. In addition, MUC1-C induces BMI1 transcription by a MYC-dependent mechanism and increases BMI1 expression by ZEB1-mediated suppression of miR-200c. MUC1-C binds to BMI1 and promotes ubiquitinylation of H2A (H2AUb1) and suppression of the BMI1 target gene, CDKN2A, with downregulation of p16INK4a expression. Activation of BMI1 has been associated with cancer progression as a result of increases in self-renewal capacity, stem-like cells and genomic instability. B. The MUC1-C program activates the NF-κB p65 circuit and in turn integrates induction of (i) ZEB1 and EMT, (ii) the epigenetic regulators DNMT1/3b and BIM1, and (iii) PD-L1, an effector of immune evasion.
Suppression of immune recognition and destruction
The EMT program promotes cancer cell invasion and the acquisition of stem-like properties, which contribute to the metastatic process [31]. The EMT program has also been associated with upregulation of the programmed death ligand 1 (PD-L1) and thereby immune evasion [45, 46], albeit by unclear mechanisms. MUC1 protects cancer cells from killing by (i) tumor necrosis-related apoptosis-inducing ligand (TRAIL), (ii) Fas ligand and (iii) perforin/granzyme-dependent lysis by cytotoxic T cells [22, 47]. Moreover, recent work has shown that MUC1-C is sufficient to induce PD-L1 expression in carcinoma cells by activation of the inflammatory NF-κB p65 pathway [48]. MUC1-C increases NF-κB p65 occupancy on the PD-L1/CD274 promoter and drives CD274 transcription, indicating that MUC1-C contributes to suppression of immune recognition [48] (Fig. 3B). Of additional interest, the MUC1-C→NF-κB p65→ZEB1 pathway represses genes, such as IFNG, which are of importance for activating an anti-tumor immune response [48]. Along these lines, targeting MUC1-C with a pharmacologic inhibitor is associated with downregulation of PD-L1 and induction of IFNγ in vitro and in mouse models. These findings have uncovered a potentially significant role for MUC1-C in integrating EMT with induction of PD-L1 and suppression of immune effectors that confer innate and adaptive responses.
MUC1-C IS AN ATTRACTIVE CANCER TARGET
Why are there no effective agents that target the MUC1-C subunit?
MUC1-C is a highly attractive target based on its importance in driving EMT and other pathways that are associated with cancer and its overexpression in diverse carcinomas. Accordingly, one might reasonably assume that MUC1-C would be a major focus of drug development efforts, which has not been the case. At first glance, this might be surprising; however, early attempts at targeting MUC1 focused on the shed MUC1-N subunit and were unsuccessful for reasons that now seem obvious given an understanding of MUC1-C function in cancer. Nonetheless, the MUC1 heterodimer (MUC1-N/MUC1-C) was largely considered as a “Cancer Marker” that is overexpressed in carcinomas. Such an attribution was in fact correct in that the MUC1-N/MUC1-C heterodimer is overexpressed in cancer cells, but became flawed with the emergence of how MUC1-C overexpression is of importance in driving hallmarks of the cancer cell. Despite these discoveries, which should ignite interest in MUC1-C as a cancer target, there remain challenges in how to develop effective agents against this oncoprotein.
MUC1-C cytoplasmic domain CQC motif is a target for drug development
A paradigm for developing agents against transmembrane receptors, such as EGFR and HER2, has been largely based on generating antibody-based approaches against the extracellular domains and small molecules against the intracellular kinase domains. MUC1-C has a short 58 aa extracellular domain, which to date has been a challenge for the generation of antibodies. The MUC1-C cytoplasmic domain is devoid of a kinase function and is an intrinsically disordered protein, which also poses challenges for the development of small molecules. Nonetheless, the MUC1-C CQC motif in the cytoplasmic domain is necessary for MUC1-C (i) activation by oxidative stress, (ii) homodimerization and (iii) import to the nucleus and mitochondria [14, 25–27]. In addition, findings that mutation of the CQC motif to AQA abrogate MUC1-C function and confer a dominant-negative effect for transformation supported the premise that the CQC motif is indeed an Achilles’ heel of this oncoprotein [49]. Accordingly, cell-penetrating peptides were developed to target the MUC1-C CQC motif and block MUC1-C homodimerization and function [25, 27]. Treatment of cancer cells with these agents is associated with increases in ROS, loss of self-renewal, downregulation of MUC1-C-induced signaling pathways and induction of late apoptotic/necrotic death [13, 24, 27, 50]. Based on these and other findings, the MUC1-C inhibitor GO-203 has been studied in a Phase I trial for patients with advanced carcinomas and, as a result of a favorable safety profile, has been formulated in nanoparticles for the treatment of patients with cancers that overexpress the MUC1-C oncoprotein [51].
HYPOTHETICAL CONSIDERATIONS
Why did MUC1-C emerge with the development of mammals?
The MUC1 gene appeared late in the evolutionary landscape to provide protection of mammals from the external environment. Mammals evolved from cold-blooded reptiles and this progression to warm-bloodedness necessitated substantial changes in metabolism associated with increases in reactive oxygen species (ROS), mutations and cancer. Mammalian epithelia at the intersection with the environment likely became especially vulnerable in maintaining their integrity when balancing increases in metabolic rates and the exposure to stress, such as inflammation and damage, which also promotes increases in ROS. In support of such a contention, Muc1-deficient mice are more susceptible to respiratory and gastrointestinal infections than their normal counterparts [52–54]. As a potential effector for maintaining that balance, the MUC1-C cytoplasmic domain CQC motif functions as a sensor of oxidative stress and, in response to increases in ROS, forms homodimers that are necessary for MUC1-C function [25–27]. MUC1-C expression is also induced in the response to increases in ROS levels [26], which contributes to the formation of autoinductive circuits, such as that involving the MUC1-C→NF-κB pathway. In support of such a model that evolved for protection of mammalian epithelia against oxidative stress, one of the important functions of MUC1-C involves the attenuation of endogenous and stress-induced increases in intracellular ROS levels that promote cell death [26, 27, 55].
Prolonged MUC1-C activation could contribute to a malignant phenotype
Simple epithelia in mammals, such as those lining the respiratory and gastrointestinal tracts, are single cell layers that, if damaged, can pose a threat to survival of the organism. The same threat applies to simple epithelia that line the ducts of specialized organs, such as the breast, prostate, pancreas, liver and other tissues that give rise to carcinomas. Indeed, if epithelial cells are subject to irreparable damage, the induction of apoptosis can serve in eliminating those cells, but in turn creates a void or wound in the epithelial barrier. Such a wound necessitates the activation of an epithelial repair process, which would conceivably require EMT for movement of surrounding cells into the wound and a proliferative response for filling the wound to enable reconstruction of the planar polar cell barrier. MUC1-C appears to have evolved to play such a functional role; however, prolonged or irreversible activation of the MUC1-C program could have deleterious effects that promote EMT, proliferation and cancer progression.
Carcinoma cells have subverted a MUC1-C-induced program that evolved to protect normal epithelia
In conclusion, MUC1-C functions in promoting loss of polarity with induction of EMT and epigenetic reprogramming. MUC1-C also promotes (i) proliferation, such as activation of RTKs, the PI3K/AKT and MEK/ERK pathways, and WNT/β-catenin/TCF4 signaling with induction of cyclin-D1 and MYC, (ii) regulation of the NF-κB p65 inflammatory response, (iii) protection against ROS-induced cell death, and (iv) induction of immune evasion, all of which can be advantageous in repairing damaged epithelial layers. MUC1-C can directly activate these pathways, supporting the notion that the overexpression of MUC1-C in diverse carcinomas represents an exploitation of these growth, repair and survival signals, which evolved to protect their normal epithelial counterparts. The increased expression of MUC1-C in carcinomas also represents a subversion of autoinductive circuits in which MUC1-C activates multiple transcription factors, for example NF-κB p65, which in turn drive MUC1 transcription. Unlike other oncoproteins, such as RAS, which are activated by mutations, MUC1-C overexpression is sufficient to induce hallmarks of the cancer cell. Indeed, mutations in the MUC1-C cytoplasmic domain function as dominant-negatives for transformation [49]. In concert with these findings and remarkably, analysis of over 1000 different carcinomas failed to identify MUC1-C mutations (ICGC CbiPortal, TumorPortal), indicating that fidelity of this subunit is of importance for cancer progression.
Acknowledgments
The authors have attempted to cite work here that first identified a role for MUC1-C in certain aspects of oncogenic signaling, such as direct interactions with effectors and induction of downstream pathways associated with cancer progression. We regret that many of the other MUC1-C publications could not be cited given limitations in the number of permissible references.
This publication was supported by the National Cancer Institute of the National Institutes of Health under award numbers R01CA097098 and R01CA166480.
Abbreviations
- MUC1
mucin 1
- MUC1-C
MUC1 C-terminal subunit
- EMT
epithelial-mesenchymal transition
- MUC1-N
MUC1 N-terminal subunit
- RTK
receptor tyrosine kinase
- EMT-TF
EMT-inducing transcription factor
- PRC1
polycomb repressive complex 1
- DNMT
DNA methyltransferase
- H2AUb1
H2A ubiquitylation
- TSG
tumor suppressor gene
- TRAIL
tumor necrosis-related apoptosis-inducing ligand
- PD-L1
programmed death ligand 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kufe D. Mucins in cancer: function, prognosis and therapy. Nature Reviews Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duraisamy S, Kufe T, Ramasamy S, Kufe D. Evolution of the human MUC1 oncoprotein. Int J Oncology. 2007;31:671–677. [PubMed] [Google Scholar]
- 3.Rajabi H, Alam M, Takahashi H, Kharbanda A, Guha M, Ahmad R, DK MUC1-C oncoprotein activates the ZEB1/miR-200c regulatory loop and epithelial-mesenchymal transition. Oncogene. 2014;33:1680–1689. doi: 10.1038/onc.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alam M, Bouillez A, Tagde A, Ahmad R, Rajabi H, Maeda T, Hiraki M, Suzuki Y, Kufe D. MUC1- C represses the Crumbs complex polarity factor CRB3 and downregulates the hippo pathway. Molecular Cancer Research. 2016:1266–1276. doi: 10.1158/1541-7786.MCR-16-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Ren J, Yu W, Li G, Kuwahara H, Yin L, Carraway KL, Kufe D. The EGF receptor regulates interaction of the human DF3/MUC1 carcinoma antigen with c-Src and β-catenin. J Biol Chem. 2001;276:35239–35242. doi: 10.1074/jbc.C100359200. [DOI] [PubMed] [Google Scholar]
- 6.Ramasamy S, Duraisamy S, Barbashov S, Kawano T, Kharbanda S, Kufe D. The MUC1 and galectin-3 oncoproteins function in a microRNA-dependent regulatory loop. Mol Cell. 2007;27:992–1004. doi: 10.1016/j.molcel.2007.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh PK, Wen Y, Swanson BJ, Shanmugam K, Kazlauskas A, Cerny RL, Gendler SJ, Hollingsworth MA. Platelet-derived growth factor receptor beta-mediated phosphorylation of MUC1 enhances invasiveness in pancreatic adenocarcinoma cells. Cancer Res. 2007;67:5201–5210. doi: 10.1158/0008-5472.CAN-06-4647. [DOI] [PubMed] [Google Scholar]
- 8.Singh PK, Behrens ME, Eggers JP, Cerny RL, Bailey JM, Shanmugam K, Gendler SJ, Bennett EP, Hollingsworth MA. Phosphorylation of MUC1 by Met modulates interaction with p53 and MMP1 expression. J Biol Chem. 2008;283:26985–26995. doi: 10.1074/jbc.M805036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raina D, Kosugi M, Ahmad R, Panchamoorthy G, Rajabi H, Alam M, Shimamura T, Shapiro G, Supko J, Kharbanda S, Kufe D. Dependence on the MUC1-C oncoprotein in non-small cell lung cancer cells. Mol Cancer Ther. 2011;10:806–816. doi: 10.1158/1535-7163.MCT-10-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kharbanda A, Rajabi H, Jin C, Tchaicha J, Kikuchi E, Wong K, Kufe D. Targeting the oncogenic MUC1-C protein inhibits mutant EGFR-mediated signaling and survival in non-small cell lung cancer cells. Clin Cancer Res. 2014;20:5423–5434. doi: 10.1158/1078-0432.CCR-13-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raina D, Uchida Y, Kharbanda A, Rajabi H, Panchamoorthy G, Jin C, Kharbanda S, Scaltriti M, Baselga J, Kufe D. Targeting the MUC1-C oncoprotein downregulates HER2 activation and abrogates trastuzumab resistance in breast cancer cells. Oncogene. 2014;33:3422–3431. doi: 10.1038/onc.2013.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mori Y, Akita K, Yashiro M, Sawada T, Hirakawa K, Murata T, Nakada H. Binding of galectin-3, a β-galactoside-binding lectin, to MUC1 protein enhances phosphorylation of extracellular signal-regulated kinase 1/2 (ERK1/2) and Akt, promoting tumor cell malignancy. J Biol Chem. 2015;290:26125–26140. doi: 10.1074/jbc.M115.651489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kufe D. MUC1-C oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene. 2013;32:1073–1081. doi: 10.1038/onc.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leng Y, Cao C, Ren J, Huang L, Chen D, Ito M, Kufe D. Nuclear import of the MUC1-C oncoprotein is mediated by nucleoporin Nup62. J Biol Chem. 2007;282:19321–19330. doi: 10.1074/jbc.M703222200. [DOI] [PubMed] [Google Scholar]
- 15.Huang L, Chen D, Liu D, Yin L, Kharbanda S, Kufe D. MUC1 oncoprotein blocks GSK3β-mediated phosphorylation and degradation of β-catenin. Cancer Res. 2005;65:10413–10422. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- 16.Behrens ME, Grandgenett P, Bailey J, Singh P, Yi C, Yu F, MAH The reactive tumor microenvironment: MUC1 signaling directly reprograms transcription of CTGF. Oncogene. 2010;29:5667–5677. doi: 10.1038/onc.2010.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajabi H, Ahmad R, Jin C, Kosugi M, Alam M, Joshi M, Kufe D. MUC1-C oncoprotein induces TCF7L2 transcription factor activation and promotes cyclin D1 expression in human breast cancer cells. J Biol Chem. 2012;287:10703–10713. doi: 10.1074/jbc.M111.323311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou CH, Huang MJ, Chen CH, Shyu MK, Huang J, Hung JS, Huang CS, Huang MC. Up-regulation of C1GALT1 promotes breast cancer cell growth through MUC1-C signaling pathway. Oncotarget. 2015;6:6123–6135. doi: 10.18632/oncotarget.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouillez A, Rajabi H, Pitroda S, Jin C, Alam M, Kharbanda A, Tagde A, Wong K, Kufe D. Inhibition of MUC1-C suppresses MYC expression and attenuates malignant growth in KRAS mutant lung adenocarcinomas. Cancer Res. 2016;76:1538–1548. doi: 10.1158/0008-5472.CAN-15-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren J, Agata N, Chen D, Li Y, Yu W-H, Huang L, Raina D, Chen W, Kharbanda S, Kufe D. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anti-cancer agents. Cancer Cell. 2004;5:163–175. doi: 10.1016/s1535-6108(04)00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren J, Bharti A, Raina D, Chen W, Ahmad R, Kufe D. MUC1 oncoprotein is targeted to mitochondria by heregulin-induced activation of c-Src and the molecular chaperone HSP90. Oncogene. 2006;25:20–31. doi: 10.1038/sj.onc.1209012. [DOI] [PubMed] [Google Scholar]
- 22.Agata N, Kawano T, Ahmad R, Raina D, Kharbanda S, Kufe D. MUC1 oncoprotein blocks death receptor-mediated apoptosis by inhibiting recruitment of caspase-8. Cancer Res. 2008;68:6136–6144. doi: 10.1158/0008-5472.CAN-08-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alam M, Ahmad R, Rajabi H, Kharbanda A, Kufe D. MUC1-C oncoprotein activates ERK→C/EBPβ-mediated induction of aldehyde dehydrogenase activity in breast cancer cells. J Biol Chem. 2013;288:30829–30903. doi: 10.1074/jbc.M113.477158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alam M, Rajabi H, Ahmad R, Jin C, Kufe D. Targeting the MUC1-C oncoprotein inhibits self-renewal capacity of breast cancer cells. Oncotarget. 2014;5:2622–2634. doi: 10.18632/oncotarget.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raina D, Agarwal P, Lee J, Bharti A, McKnight C, Sharma P, Kharbanda S, Kufe D. Characterization of the MUC1-C cytoplasmic domain as a cancer target. PLoS One. 2015;10:e0135156. doi: 10.1371/journal.pone.0135156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin L, Kufe D. Human MUC1 carcinoma antigen regulates intracellular oxidant levels and the apoptotic response to oxidative stress. J Biol Chem. 2003;278:35458–35464. doi: 10.1074/jbc.M301987200. [DOI] [PubMed] [Google Scholar]
- 27.Raina D, Ahmad R, Rajabi H, Panchamoorthy G, Kharbanda S, Kufe D. Targeting cysteine-mediated dimerization of the MUC1-C oncoprotein in human cancer cells. Int J Oncol. 2012;40:1643–1649. doi: 10.3892/ijo.2011.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei X, Xu H, Kufe D. Human MUC1 oncoprotein regulates p53-responsive gene transcription in the genotoxic stress response. Cancer Cell. 2005;7:167–178. doi: 10.1016/j.ccr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 29.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 30.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 31.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye X, Weinberg RA. Epithelial-Mesenchymal Plasticity: A Central Regulator of Cancer Progression. Trends Cell Biol. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kharbanda A, Rajabi H, Jin C, Alam M, Wong K, Kufe D. MUC1-C confers EMT and KRAS independence in mutant KRAS lung cancer cells. Oncotarget. 2014;5:8893–8905. doi: 10.18632/oncotarget.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy LD, Sahraei M, Subramani DB, Besmer D, Nath S, Tinder TL, Bajaj E, Shanmugam K, Lee YY, Hwang SI, Gendler SJ, Mukherjee P. MUC1 enhances invasiveness of pancreatic cancer cells by inducing epithelial to mesenchymal transition. Oncogene. 2011;30:1449–1459. doi: 10.1038/onc.2010.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alam M, Ahmad R, Rajabi H, Kufe D. MUC1-C induces the LIN28B→LET-7→HMGA2 axis and self-renewal in NSCLC cells. Mol Cancer Res. 2015;13:449–460. doi: 10.1158/1541-7786.MCR-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi H, Jin C, Rajabi H, Pitroda S, Alam M, Ahmad R, Raina D, Hasegawa M, Suzuki Y, Tagde A, Bronson RT, Weichselbaum R, Kufe D. MUC1-C activates the TAK1 inflammatory pathway in colon cancer. Oncogene. 2015;34:5187–5197. doi: 10.1038/onc.2014.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmad R, Rajabi H, Kosugi M, Joshi M, Alam M, Vasir B, Kawano T, Kharbanda S, Kufe D. MUC1-C oncoprotein promotes STAT3 activation in an auto-inductive regulatory loop. Science Signaling. 2011;4:ra9. doi: 10.1126/scisignal.2001426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahmad R, Raina D, Trivedi V, Ren J, Rajabi H, Kharbanda S, Kufe D. MUC1 oncoprotein activates the IκB kinase β complex and constitutive NF-κB signaling. Nat Cell Biol. 2007;9:1419–1427. doi: 10.1038/ncb1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahmad R, Raina D, Joshi MD, Kawano T, Kharbanda S, Kufe D. MUC1-C oncoprotein functions as a direct activator of the NF-κB p65 transcription factor. Cancer Res. 2009;69:7013–7021. doi: 10.1158/0008-5472.CAN-09-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Cock JM, Shibue T, Dongre A, Keckesova Z, Reinhardt F, Weinberg RA. Inflammation Triggers Zeb1-Dependent Escape from Tumor Latency. Cancer Res. 2016;76:6778–6784. doi: 10.1158/0008-5472.CAN-16-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siddique HR, Saleem M. Role of BMI1, a stem cell factor, in cancer recurrence and chemoresistance: preclinical and clinical evidences. Stem Cells. 2012;30:372–378. doi: 10.1002/stem.1035. [DOI] [PubMed] [Google Scholar]
- 42.Hiraki M, Maeda T, Bouillez A, Alam M, Tagde A, Hinohara K, Suzuki Y, Markert T, Miyo T, Komura K, Ahmad R, Rajabi H, Kufe D. MUC1-C activates BMI1 in human cancer cells. Oncogene. 2016 Nov 28; doi: 10.1038/onc.2016.439. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, Baylin SB, Vogelstein B. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 44.Rajabi H, Tagde A, Alam M, Bouillez A, Pitroda S, Suzuki Y, Kufe D. DNA methylation by DNMT1 and DNMT3b methyltransferases is driven by the MUC1-C oncoprotein in human carcinoma cells. Oncogene. 2016;35:6439–6445. doi: 10.1038/onc.2016.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mak MP, Tong P, Diao L, Cardnell RJ, Gibbons DL, William WN, Skoulidis F, Parra ER, Rodriguez-Canales J, Wistuba, Heymach JV, Weinstein JN, Coombes KR, Wang J, Byers LA. A patient-derived, pan-cancer EMT signature identifies global molecular alterations and immune target enrichment following epithelial-to-mesenchymal transition. Clin Cancer Res. 2016;22:609–620. doi: 10.1158/1078-0432.CCR-15-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lou Y, Diao L, Parra Cuentas ER, Denning WL, Chen L, Fan YH, Byers LA, Wang J, Papadimitrakopoulou VA, Behrens C, Rodriguez J, Hwu P, Wistuba, Heymach JV, Gibbons DL. Epithelial-mesenchymal transition is associated with a distinct tumor microenvironment including elevation of inflammatory signals and multiple immune checkpoints in lung adenocarcinoma. Clin Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-15-1434. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.David JM, Hamilton DH, Palena C. MUC1 upregulation promotes immune resistance in tumor cells undergoing brachyury-mediated epithelial-mesenchymal transition. Oncoimmunology. 2016;5:e1117738. doi: 10.1080/2162402X.2015.1117738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bouillez A, Rajabi H, Jin C, Samur M, Tagde A, Alam M, Hiraki M, Maeda T, Hu X, Adeegbe D, Kharbanda S, Wong K-K, Kufe D. MUC1-C integrates PD-L1 induction with repression of immune effectors in non-small cell lung cancer. Oncogene. 2017 doi: 10.1038/onc.2017.47. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kufe D. Functional targeting of the MUC1 oncogene in human cancers. Cancer Biol Ther. 2009;8:1201–1207. doi: 10.4161/cbt.8.13.8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raina D, Ahmad R, Joshi M, Yin L, Wu Z, Kawano T, Vasir B, Avigan D, Kharbanda S, Kufe D. Direct targeting of the MUC1 oncoprotein blocks survival and tumorigenicity of human breast carcinoma cells. Cancer Res. 2009;69:5133–5141. doi: 10.1158/0008-5472.CAN-09-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hasegawa M, Sinha RK, Kumar M, Alam M, Yin L, Raina D, Kharbanda A, Panchamoorthy G, Gupta D, Singh H, Kharbanda S, Kufe D. Intracellular targeting of the oncogenic MUC1-C protein with a novel GO-203 nanoparticle formulation. Clin Cancer Res. 2015;21:2338–2347. doi: 10.1158/1078-0432.CCR-14-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McAuley JL, Linden SK, Png CW, King RM, Pennington HL, Gendler SJ, Florin TH, Hill GR, Korolik V, McGuckin MA. MUC1 cell surface mucin is a critical element of the mucosal barrier to infection. J Clin Invest. 2007;117:2313–2324. doi: 10.1172/JCI26705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi S, Park YS, Koga T, Treloar A, Kim KC. TNF-alpha is a key regulator of MUC1, an anti-inflammatory molecule, during airway Pseudomonas aeruginosa infection. Am J Respir Cell Mol Biol. 2011;44:255–260. doi: 10.1165/rcmb.2009-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Umehara T, Kato K, Park YS, Lillehoj EP, Kawauchi H, Kim KC. Prevention of lung injury by Muc1 mucin in a mouse model of repetitive Pseudomonas aeruginosa infection. Inflamm Res. 2012;61:1013–1020. doi: 10.1007/s00011-012-0494-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin L, Kharbanda S, Kufe D. MUC1 oncoprotein promotes autophagy in a survival response to glucose deprivation. Int J Oncol. 2009;34:1691–1699. doi: 10.3892/ijo_00000300. [DOI] [PMC free article] [PubMed] [Google Scholar]