Abstract
TBC1D4 (also known as AS160) is a Rab·GTPase-activating protein (RabGAP) which functions in insulin signaling. TBC1D4 is critical for translocation of glucose transporter 4 (GLUT4), from an inactive, intracellular, vesicle-bound site to the plasma membrane, where it promotes glucose entry into cells. The TBC1D4 protein is structurally subdivided into two N-terminal phosphotyrosine-binding (PTB) domains, a C-terminal catalytic RabGAP domain, and a disordered segment in between containing potential Akt phosphorylation sites. Structural predictions further suggest that a region C-terminal to the RabGAP domain adopts a coiled-coil motif. We show that C-terminal region (CTR) region is largely α-helical and mediates TBC1D4 RabGAP dimerization. RabGAP catalytic activity and thermal stability appear to be independent of CTR-mediated dimerization.
Keywords: TBC1D4, AS160, TBC1D1, Dimerization, Coiled-coil, RabGAP
1. Introduction
Insulin signaling is critical to total body nutrient homeostasis. Increases in circulating glucose levels that follow meals induce insulin release from pancreatic β-cells. Circulating insulin acts at insulin receptors present on insulin responsive tissues, including liver, fat and muscle, which respond by increasing the uptake of glucose. Insulin-stimulated glucose uptake is the prototypical action of insulin, although insulin also has many other actions in cells. Years of intense investigation have mapped out the intracellular signaling pathways leading to enhanced glucose transport as well as many other insulin actions. The upstream events always involve insulin binding to insulin receptors, which activates its intrinsic tyrosine kinase activity leading to phosphorylation of both the receptor itself and its primary substrates, the insulin receptor substrate (IRS-1, IRS-2, etc.) proteins. By contrast, the downstream cellular functions of insulin diverge from the scaffolding effects of the IRS proteins. Insulin-stimulated glucose uptake requires vesicular trafficking. The glucose transporter 4 (GLUT4) proteins reside in GLUT4 vesicles, a specialized organelle that responds to insulin. In its absence most of the GLUT4 is inactive as it resides inside the cell in the GLUT4 vesicles. Upon insulin stimulation, the vesicles translocate to the cell surface where they mediate glucose entry [1].
Vesicle trafficking generally involves budding, un-coating, docking and fusion steps that are regulated by the Rab·GTPases (Rabs), which can be shuttled between GTP-bound (active) or GDP-bound (inactive) states. The Rab·GTPase-activating proteins (RabGAPs) enhance the intrinsically low rates of GTP-to-GDP hydrolysis to inactivate the Rabs [2,3].
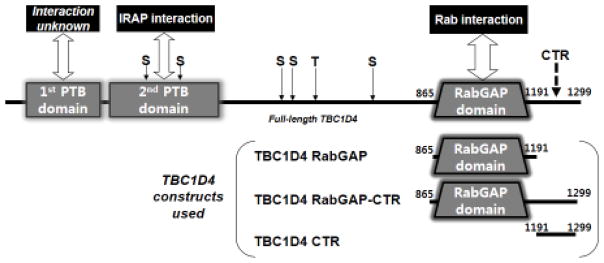
TBC1D4, which has also been called AS160, and TBC1D1 are the RabGAPs with roles in GLUT4 vesicle translocation in adipocytes and the skeletal myocytes, respectively [4–6]. TBC1D4 and TBC1D1 are homologous multi-domain proteins of ~1300 residues. Each contains two phosphotyrosine binding (PTB) domains predicted from the protein sequence (Fig. 1). While the physiological and biochemical roles of the first PTB domain are unknown, the second PTB domain binds insulin-regulated aminopeptidase (IRAP), a single-membrane spanning protein marker for GLUT4 vesicles [7,8]. The second PTB domain also appears to bind phospholipids, which targets it to cellular membranes [9]. The second PTB domain may therefore serve not only in localizing TBC1D4 to GLUT4 vesicles but also as a phosphorylation dependent regulatory switch during the docking of GLUT4 vesicles at the plasma membrane [9]. Many of the serine and threonine residues phosphorylated by Akt are located C-terminal to the PTB domain. The catalytic RabGAP domain is at the C-terminus of the protein (Fig. 1). In the basal state, TBC1D4 catalyzes the hydrolysis of Rab-bound GTP, generating Rab·GDP, which is inactive and leaves GLUT4 vesicles inside the cell. Under insulin stimulation, activated Akt phosphorylates TBC1D4, which down-regulates its RabGAP activity leaving Rab·GTP intact. This promotes GLUT4 vesicle translocation to the plasma membrane and enhances the glucose uptake.
Fig. 1. Domains of TBC1D4 protein and the boundaries of recombinant proteins used in this study.
TBC1D4 (also known as AS160) is a multi-domain protein with two N-terminal PTB domains and a catalytic RabGAP domain. Serine (S) and threonine (T) residues undergoing Akt-mediated phosphorylation are indicted at the approximate regions by arrows. A region predicted to form a coiled-coil is located at the C-terminal region (CTR). Three different TBC1D4 recombinant proteins were expressed in E. coli to analyze the oligomeric behavior of TBC1D4. TBC1D4 RabGAP (TBC1D4 865-1191, monomeric molecular mass ~41 kDa) includes only the catalytic RabGAP domain, TBC1D4 RabGAP-CTR (TBC1D4 865-1299, monomeric molecular mass ~53 kDa) includes the catalytic RabGAP and CTR, and TBC1D4 CTR (TBC1D4 1192-1299, monomeric molecular mass ~15 kDa) includes only the CTR.
TBC1D4 also contains a C-terminal region of ~100 amino acids at the protein end (Fig. 1) whose function has not been ascribed. Secondary structural predictions [10] indicate that the C-terminal region (CTR) of TBC1D4 adopts a coiled-coil configuration. In this study, we use recombinant TBC1D4 proteins to show that TBC1D4 dimerizes via the CTR. Moreover, we show that the in vitro catalytic activity of the RabGAP domain is independent of CTR-mediated dimerization, and that CTR has no effect in thermal stability of the protein.
2. Results
2.1. Oligomer formation of TBC1D4 via the C-terminal region (CTR)
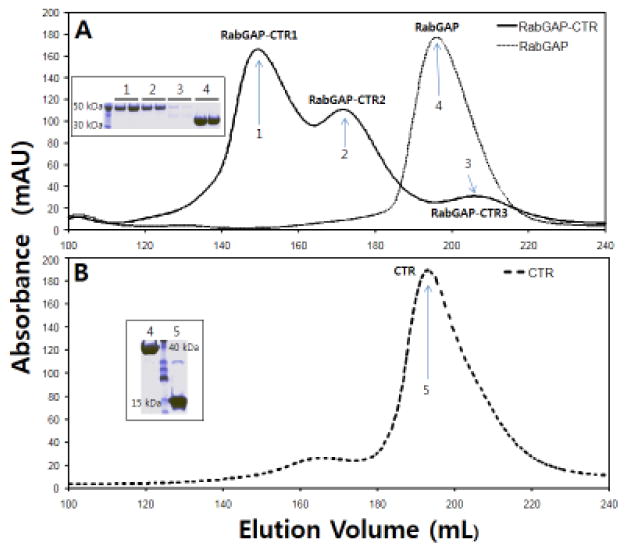
The C-terminal region (CTR, residues 1192-1299) of TBC1D4, which is C-terminal to its catalytic RabGAP domain (RabGAP, 865-1191), is predicted to adopt a coiled-coil structure (http://www.ch.embnet.org/software/COILS_form.html) [10]. We initially used size-exclusion chromatography to assess the oligomeric states of three TBC1D4 recombinant proteins, RabGAP and CTR alone, and the collinear RabGAP-CTR segment encompassing both regions (Fig. 1). The proteins were expressed in E. coli and purified prior to analysis. The RabGAP itself eluted as a monomer (Fig. 2A), consistent with previous findings using analytical ultra-centrifugation and X-ray crystallographic analyses [11,12]. By contrast, RabGAP-CTR eluted as a mixture of mainly two molecular mass forms (Fig. 2A, RabGAP-CTR1 and RabGAP-CTR2). Absorbance intensity and SDS-PAGE analyses suggest a third potential aggregation state (RabGAP-CTR3) whose amount is much less than the other two species (Fig. 2A and inset). No further studies were performed on this minor RabGAP-CTR3.
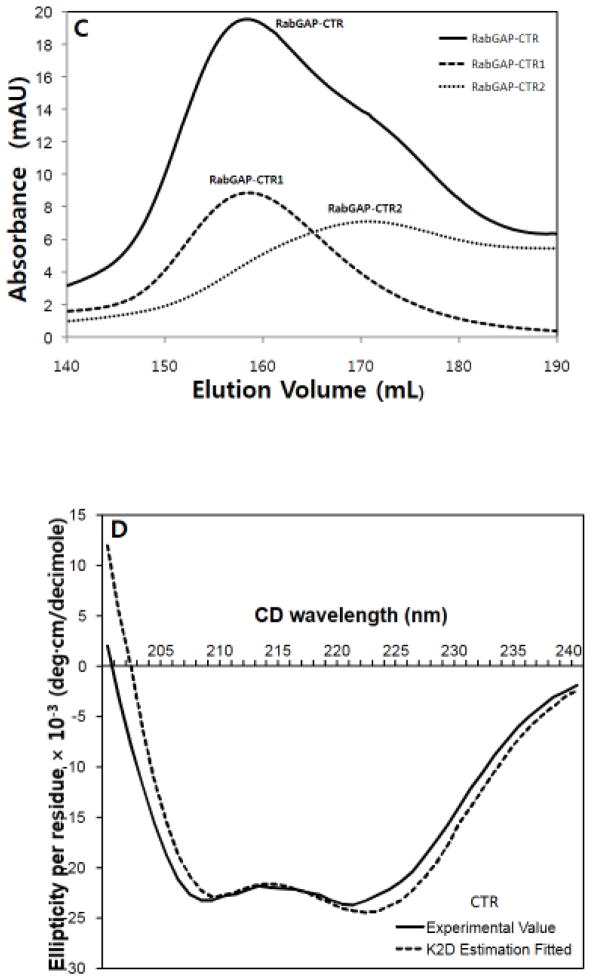
Fig. 2. TBC1D4 oligomerization is mediated by the α-helical CTR.
TBC1D4 RabGAP, RabGAP-CTR and CTR were eluted on a size-exclusion column, and their elution fractions were analyzed by SDS-PAGE (inset). (A) RabGAP eluted as a single component while RabGAP-CTR eluted as a mixture of mainly two components (RabGAP-CTR1 and RabGAP-CTR2). A minor elution peak (RabGAP-CTR3) was negligible as indicated by SDS-PAGE analysis (inset). (B) Despite having the smallest molecular mass, CTR eluted as a single component at a volume comparable to RabGAP. (C) Elution fractions of the two TBC1D4 RabGAP-CTR (RabGAP-CTR1 & RabGAP-CTR2) peaks were separately concentrated and re-eluted. Segregation of RabGAP-CTR1 and RabGAP-CTR2 peaks (dotted line) indicates that the two oligomeric RabGAP-CTR components did not interchange. (D) Circular dichroism analysis suggests that the TBC1D4 CTR is largely α-helical. The experimental ellipticity-wavelength values were analyzed by K2D secondary structure estimation program, which suggests CTR with 78% α-helical content.
CTR alone elutes primarily as a single peak with an elution volume comparable to that of RabGAP. Thus, at physiological pH and ionic strength, and at the protein concentrations studied, the ~15 kDa CTR protein adopts an apparent hydrodynamic radius similar to that of the ~41 kDa RabGAP protein, suggesting CTR is non-globular (Fig. 2B).
2.2. Irreversible oligomerization between TBC1D4 RabGAP-CTR1 and RabGAP-CTR2
We next explored the oligomeric reversibility of RabGAP-CTR1 and RabGAP-CTR2. When collected separately and re-analyzed using size-exclusion chromatograph, the two forms retained their original elution characteristics suggesting that interchange between oligomeric forms occurs slowly (Fig. 2C). The decreased resolution of RabGAP-CTR1 and RabGAP-CTR2 on the sizing column compared to Fig. 2A is due to larger sample volumes for protein loading.
2.3. Dynamic light scattering of TBC1D4 RabGAP and CTR
The comparable elution volumes for CTR (~15 kDa monomeric molecular mass) and RabGAP (~41 kDa monomeric molecular mass) during size-exclusion chromatography suggested comparable hydrodynamic radii for these proteins. Dynamic light scattering (DLS) was used for further assessment. Using this independent method for estimating hydrodynamic radius, the molecular sizes of RabGAP and CTR were again found to be highly similar to one another, whereas RabGAP-CTR1 and RabGAP-CTR2 were significantly larger than RabGAP and CTR (Table 1).
Table 1.
Hydrodynamic radii estimated by dynamic light scattering (DLS).
| TBC1D4 | Hydrodynamic radius (Å) |
|---|---|
| RabGAP (865-1191) | 43 |
| CTR (1192-1299) | 40 |
| RabGAP-CTR1 (865-1299) | 85 |
| RabGAP-CTR2 (865-1299) | 67 |
2.4. The α-helicity of TBC1D4 CTR
Secondary structure prediction algorithms predicted the CTR of TBC1D4 is largely α-helical. Circular dichroism (CD) analysis of CTR confirms this prediction (Fig. 2D). The experimental ellipticity values determined at different wavelengths (200–240 nm) were used for secondary structure estimations. The results indicate that >60% of His6-CTR is α-helical, with the remainder adopting random coil (helix estimations: K2D, 78%; K2D2, 84–87%; SELCON3, 62%). Because ~16% of the His6-CTR protein corresponds to its His6-tag, which is a random coil, we estimate that CTR itself is 76–100% α-helical (Fig. 2D). The N-terminal His6-tag of the recombinant CTR protein was left intact to assist in protein concentration (see Methods).
2.5. TBC1D4 dimerization via CTR
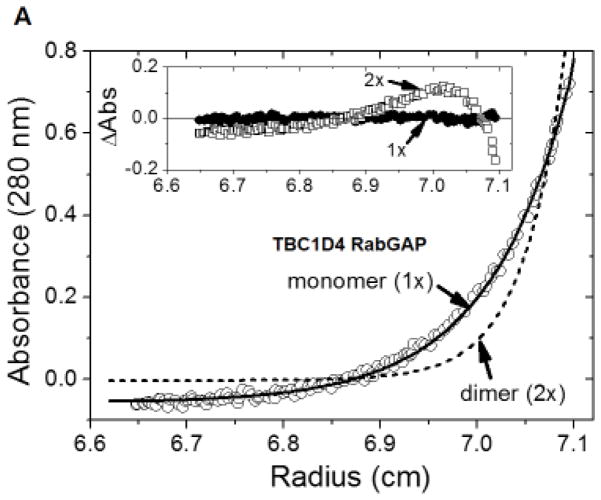
The oligomeric states of TBC1D4-derived RabGAP, CTR, and RabGAP-CTR proteins with His6-tags were further investigated using equilibrium sedimentation. For RabGAP-CTR, only RabGAP-CTR1 which eluted with a larger molecular mass than RabGAP-CTR2 on the size-exclusion column was studied. Equilibrium distributions and model fitting of RabGAP indicated that RabGAP behaves as a homogeneous monomer in solution as previously shown (Fig. 3A) [12]. The weighted root-mean-square (RMS) error for the dimeric fit (5.85 × 10−2) with systematic deviations is shown in a residual distribution plot (Fig. 3A inset). When the data were fit to a monomeric model, RMS improved (8.66 × 10−3) with substantially better residual distributions, further illustrating that RabGAP is likely monomeric in solution. Also, the result showed that the presence of His6-tag on RabGAP does not mediate dimerization.
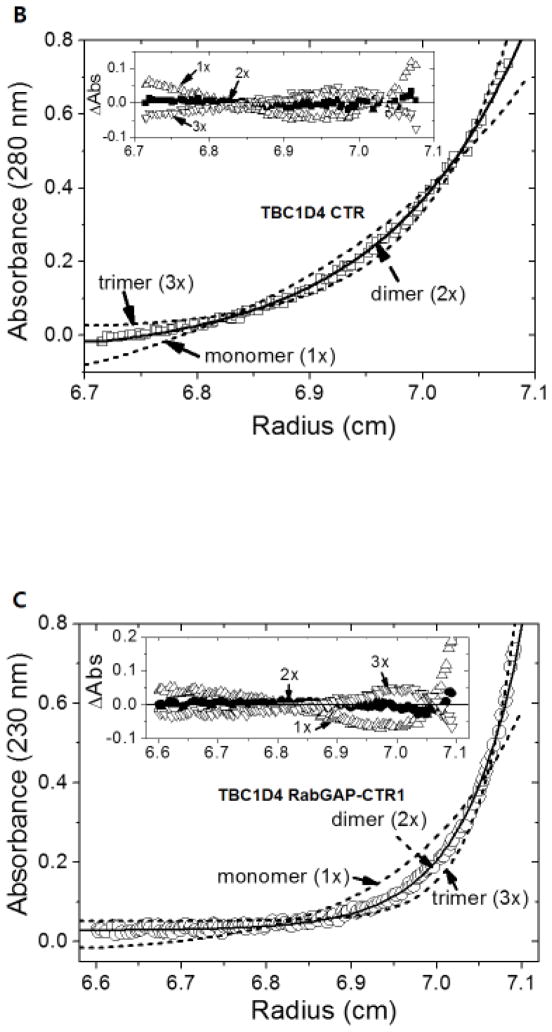
Fig. 3. Dimerization of TBC1D4 is mediated by CTR.
The oligomeric states of TBC1D4 RabGAP (A), CTR (B), and RabGAP-CTR1 (C) were analyzed by sedimentation equilibrium. (A) Sedimentation equilibrium distributions of RabGAP (TBC1D4 865-1191) measured at 18,000 rpm and 280 nm (□) are fitted for both an ideal monomer (solid line, 1×) and a dimer (dotted line, 2×). The random residual distributions shown for a monomer (□, 1×) in comparison to a dimer (○, 2×) indicate that RabGAP exists as homogeneous monomers in solution (inset). (B) Sedimentation equilibrium distributions of CTR (TBC1D4 1192-1299) measured at 18,000 rpm and 280 nm (□) are fitted for an ideal dimer (solid line, 2×), monomer, and a trimer (dotted line, 1×, 3×). Random residual distributions shown for a dimer (●, 2×) in comparison to either a monomer (△, 1×), or a trimer (▽, 3×) indicate that CTR exists as homogeneous dimers in solution (inset). (C) Sedimentation equilibrium distributions of RabGAP-CTR1 (TBC1D4 865-1299) measured at 13,000 rpm and 230 nm (○) are fitted for an ideal dimer (solid line, 2×), monomer, and a trimer (dotted line, 1×, 3×). Random residual distributions shown for a dimer (●, 2×) in comparison to either a monomer (△, 1×), or a trimer (▽, 3×) indicate that RabGAP-CTR1 exists as homogeneous dimers in solution (inset).
Equilibrium sedimentation distribution data for CTR and RabGAP-CTR1 indicate that in solution these proteins form homogeneous dimers (Fig. 3B & 3C). The sedimentation data for CTR fit to a dimer model, with a better RMS value (9.61 × 10−3) and improved residual distributions, compared to monomer (3.97 × 10−2) or trimer (2.50 × 10−2) models (Fig. 3B). Similarly, the equilibrium data for RabGAP-CTR1 fit a dimer model with a better RMS (9.62 × 10−3) and improved random residual distributions, than monomer (4.87 × 10−2) and trimer (2.56 × 10−2) models (Fig. 3C). These data indicate that the CTR region of TBC1D4 mediates dimerization of the RabGAP-CTR protein. Because the dimeric size of RabGAP-CTR1 further implies that the smaller sized RabGAP-CTR2 is in a state of a monomer, no further AUC study on RabGAP-CTR2 was performed. Also, AUC investigations at different protein concentrations and centrifugal speeds do not support rapidly reversible monomer-dimer transitions for CTR or RabGAP-CTR1.
2.6. Catalytic activities of TBC1D4 RabGAP with and without CTR
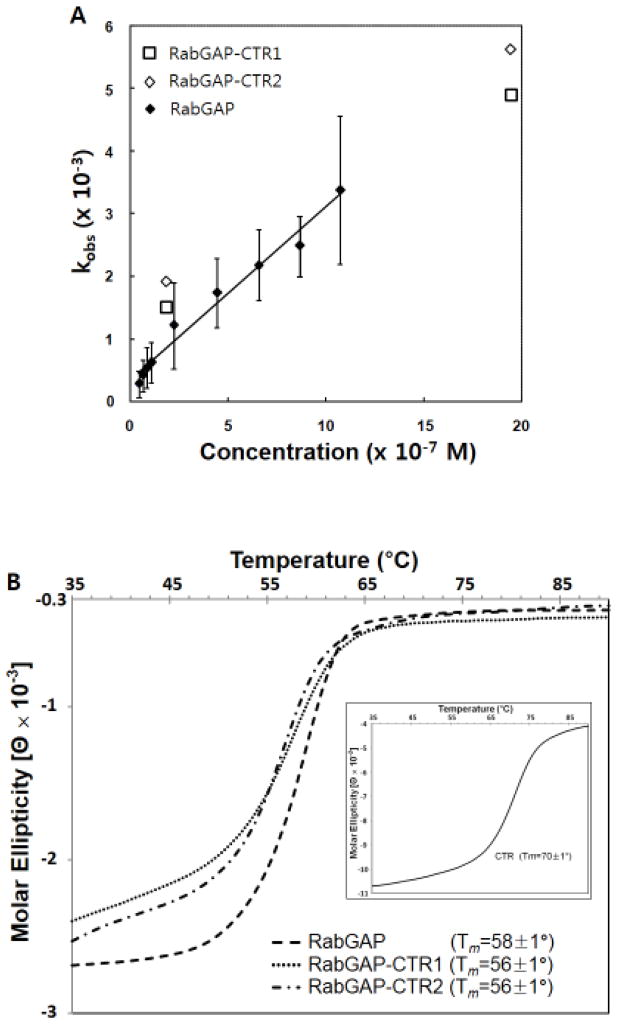
Rates of GTP hydrolysis were assessed for the TBC1D4 RabGAP-CTR proteins using Rab14·GTP as the substrate, as previously described for the RabGAP domain by itself [11]. Values for kobs were similar for dimeric RabGAP-CTR1 and monomeric RabGAP and RabGAP-CTR2 proteins (Fig. 4A), suggesting that CTR-mediated dimerization does not influence in vitro RabGAP catalytic activity.
Fig. 4. In vitro RabGAP activities and thermal denaturation curves of TBC1D4 RabGAP domain with and without CTR.
(A) RabGAP activities were measured for both the dimeric and monomeric TBC1D4 RabGAP-CTR components (□: RabGAP-CTR1; ◇: RabGAP-CTR2) using Rab14·GTP as the substrate. Values of kobs measured at two different concentrations are comparable to the previously reported kobs of only the RabGAP domain (◆, kobs data are taken directly from ref.11). (B) Thermal denaturation studies performed using CD indicate that CTR does not thermally stabilize the RabGAP domain. Fitted melting temperatures (Tm) for each protein are shown. CTR by itself without RabGAP domain has higher Tm than the RabGAP domain containing proteins (inset).
2.7. Thermal stabilities of TBC1D4 RabGAP with and without CTR
The thermal stabilities of the TBC1D4 RabGAP, RabGAP-CTR1, RabGAP-CTR2, and CTR were measured by carrying out temperature induced unfolding experiments using CD (Fig. 4B). The denaturation results with similar melting temperature for RabGAP by itself and RabGAP with CTR indicate that CTR does not thermally stabilize the RabGAP domain. Interestingly, CTR alone show a higher Tm compared to the RabGAP domain containing proteins.
3. Discussion
TBC1D4 and TBC1D1 are closely related RabGAPs which regulate GLUT4 vesicle trafficking in adipocytes and skeletal myocytes, respectively. These proteins contain regions C-terminal to their catalytic RabGAP domains predicted to adopt coiled-coil configurations. These ~100 amino acid “CTRs” of TBC1D4 and TBC1D1 (residues 1195–1272 and 1076–1152, respectively) share 41% sequence identity and 68% sequence similarity. Circular dichroism spectral properties of recombinant TBC1D4 CTR protein suggest it has high α-helix content. Size-exclusion chromatography of expressed TBC1D4 proteins containing both RabGAP and CTR regions indicate that CTR mediates oligomerization. The large hydrodynamic radius of CTR, given its molecular mass, suggests it adopts an extended or elongated configuration consistent with an α-helix. Analytical ultracentrifugation experiments further demonstrate that CTR mediates dimer formation.
It is interesting that recombinant RabGAP-CTR is a monomer/dimer mixture and that interconversions between them were unapparent on an hour to several day timescale. Nevertheless, there was no evidence for covalent bond formation between monomers in either RabGAP-CTR1 or CTR dimers. Given the various RabGAP and RabGAP-CTR proteins studied have comparable catalytic activities, we concluded that dimerization is neither necessary for nor influences catalytic efficiency. Also, CTR has no effect in thermal stability of the RabGAP domain. We have not identified alternative functions for CTR other than dimerization. Rab14 was chosen to be the substrate for these studies because it is a potential in vivo substrate and has previously been used as an in vitro substrate of TBC1D4 [11,13]. While Rab10 has been suggested to be a cellular substrate [14,15], it is not expressed in sufficient quantities for the assay.
The cellular functions of dimerization were doubtless not reflected by our in vitro enzymatic assays using protein fragments. The recombinant proteins containing the RabGAP domain alone (327 residues) or in tandem with CTR (435 residues) are much shorter than 1299-residue, full-length TBC1D4. TBC1D4 is also predicted to contain two PTB domains, whose functions may either require or be altered by protein dimerization. While beyond the scope of this study, the functions and consequences of dimerization may need to be studied in cells by assessing the effects of active and dominant-inhibitory forms of TBC1D4 in GLUT4 translocation assays.
There is precedence in the literature for the dimerization of RapGAP and RasGAP proteins. For example, CAPRI, a protein that contains both RapGAP and RasGAP activities, has been also shown to form homodimers via a C-terminal helical motif [16]. Interestingly, both dimeric and monomeric CAPRI proteins co-exist in cells, and Ca2+-dependent dimerization reportedly acts as a switch to enhance the RapGAP activity over the RasGAP activity. Another structural study revealed that Rap1GAP, a distant relative of the RabGAP protein family, also has a dimerization domain which is not required for in vitro catalytic activity [17,18]. And the crystal structure of the DHR-2 domain of DOCK2, a guanine nucleotide exchange factor (GEF), also suggests that dimerization occurs [19]. Although deletion of the dimerization region did not affect in vitro RacGEF activity, it influenced the Rac-mediated cell-migration [20]. These additional findings support potential roles for dimerization in the activities and cellular functions of GAP and GEF proteins, consistent with potential cellular roles for dimerization in TBC1D4.
4. Material and methods
4.1. Protein expression and purification
Fragments of cDNA encoding human TBC1D4 (RabGAP-CTR, 865-1299; RabGAP, 865-1191; CTR, 1192-1299; DNA from Takahiro Nagase, Kazusa DNA Research Institute) and mouse Rab14 (1–175; DNA from Gus Leinhard, Dartmouth University) were PCR-cloned into pET28a (Novagen) vector. The plasmids were transformed into BL21 (DE3) (Stratagene) strain and were grown in LB medium under kanamycin selection (25 μg/mL). Recombinant protein expressions were induced with 0.5 mM isopropyl β-D-thiogalactopyranoside (IPTG) at OD600 = ~0.8, and the cells were further grown at 25 °C for 16 hours. The proteins were expressed with His6-tags and were purified on Ni2+- nitrilotriacetic beads with imidazole elution. The His6-tags were either removed or left intact depending on different studies as described below. When removed, the proteins were treated with thrombin (Roche) as previously reported [10].
Rab14 was additionally purified on Superdex® 200 HR26/60 sizing column (GE Healthcare) pre-equilibrated in GF buffer (50 mM Tris pH 7.5 and 150 mM NaCl) using the ÄKTA FPLC system (GE Healthcare). Rab14 was concentrated by Amicon (Millipore) concentrator, and further loaded with GTP using methods previously published [11]. Briefly, ~1–10 mg of purified Rab14 was incubated with 10-fold molar excess of GTP at 4 °C for 2–3 hours in GF buffer with 5 mM EDTA and 5 mM dithiothreitol (DTT). Unbound free GTP was removed using a desalting column (BioRad) pre-equilibrated in GF buffer. Rab14 were aliquoted and stored at −80 °C for subsequent RabGAP assays. The Rab14 concentration was quantified by absorption (λ = 280 nm) by employing the calculated molar extinction coefficient (24,200 M−1cm−1) [21].
4.2. Detection of protein oligomerization using size-exclusion chromatography
His6-cleaved recombinant TBC1D4 RabGAP-CTR, RabGAP and CTR were eluted from a Superdex® 200 HR 26/60 sizing column pre-equilibrated with GF buffer and 5 mM DTT. The elution profile was monitored using absorbance (λ = 260 nm) using the ÄKTA FPLC system. Elution fractions at the peaks were analyzed by SDS-PAGE for further protein identification. Irreversible monomer-to-dimer interchange between the two species of TBC1D4 RabGAP-CTR was assessed by collecting and concentrating elution fractions corresponding to each species, and eluting them on the same column.
4.3. Dynamic light scattering for estimation of hydrodynamic radii
Hydrodynamic radii of four proteins (RabGAP, RabGAP-CTR1, RabGAP-CTR2 and CTR) were estimated by using dynamic light scattering (DLS). The purified His6-cleaved recombinant proteins in GF buffer were analyzed using DynaPro Titan (Wyatt Technology Corporation) instrument.
4.4. Circular dichroism for secondary structure estimation and thermal stability comparison
The contents of secondary structural elements in CTR were determined by scanning ellipticity over wavelength using JASCO spectropolarimeter (Model J-810). Since determination of the protein concentration is critical for the secondary structure estimation, CTR with a small molecular mass had to be concentrated for quantification. Hence, the His6-tag of the CTR protein was left intact. The concentration of His6-CTR was quantified by employing the calculated molar extinction coefficient at λ = 280 nm (2,560 M−1cm−1). CTR protein with the final concentration of 6 μg/mL was analyzed using a 0.1 cm path-length cuvette to obtain the spectrum. Various secondary structure estimation programs such as K2D [22] K2D2 [23] and SELCON [24] were applied on the CD data for approximation of the helical content.
The thermal stabilities of the TBC1D4 proteins were measured (λ = 222 nm) by carrying out temperature induced unfolding experiments with ~0.5 mg/mL of TBC1D4 RabGAP, RabGAP-CTR1, RabGAP-CTR2, and CTR proteins in GF buffer. Molar ellipticity values were calculated and fitted to a sigmoidal curve for determination of the melting temperature.
4.5. Equilibrium sedimentation studies using analytical ultracentrifugation (AUC)
Recombinant TBC1D4 RabGAP, CTR and RabGAP-CTR1 with His6-tags were used for the sedimentation equilibrium experiments. Since the thrombin treatment to remove His6-tag may induce non-specific cleavages of the proteins during the course of near 3-day, 20 °C equilibration necessary for the AUC experiments, the His6-tags in the proteins were all left intact. The proteins were purified similarly on the ÄKTA FPLC system in GF buffer with 5 mM DTT, and were concentrated by centrifugation. During concentration the proteins were washed several times with GF buffer containing 1.5 mM Tris(2-carboxyethyl)phosphine (TCEP) to minimize the formation of non-specific cysteine-mediated disulfide bonds. Non-specific cysteine-mediated disulfide bonds and proteolytic degradation of the proteins were monitored over 3 days at 4 °C on a non-reducing SDS-PAGE, where there appeared to be none. The final protein concentrations for the analytical ultracentrifugation experiments were adjusted to an optimal absorbance signal of 0.1–0.25 units.
Equilibrium sedimentation studies were performed using a Beckman ProteomeLab XL-A analytical ultracentrifuge with proteins in GF buffer with 1.5 mM TCEP at 20 °C as previously explained [12]. Briefly, equilibrium sedimentation of RabGAP (6.69 mM from calculated extinction coefficient of 37,360 cm−1M−1), CTR (for this fragment high concentrations of 33.6–83.9 mM were used due to low extinction coefficient of 2,980 cm−1M−1), and RabGAP-CTR1 (1.24 mM from calculated extinction coefficient of 40,340 cm−1M−1) were measured at 230 nm and 280 nm using a two-sector cell (loading volume of 180 μL) at two speeds (13,000 and 18,000 rpms). All measured data fit well to given models and representative results measured at 13,000 rpm (RabGAP-CTR1) and 18,000 rpm (RabGAP and CTR) are presented. Since the solubility of RabGAP-CTR1 was rather low when compared to other proteins, data collected at 230 nm are presented. The time required for attainment of equilibrium was established by running the rotor until the scans were invariant for 4 hours. This time was achieved at most by 70 hours at the given rotor speed. Partial specific volumes of the proteins and the buffer were calculated using Sednterp [25]. Calculated monomeric molecular masses (RabGAP, 40,503 Da; CTR, 14,748 Da; RabGAP-CTR1, 52,963 Da) were used for the model fittings.
Mathematical data analyses were performed using a non-linear least-squares fitting for a homogeneous model on equations below [26]. The selection of the model was made by examining the number of weighted sum or the combination of square values and weighted root mean square error values. Data analysis and mathematical modeling were performed using MLAB [27].
(Cr, total concentration at the radial position r; Cb, concentration of protein at the cell bottom; Mp, molecular mass of protein; v, partial specific volume of protein; ρ, solution density; ω, rotor angular velocity; ε, baseline error)
4.6. GTPase assay
Although the presence of His6-tag does not alter the TBC1D4 RabGAP activity, the His6-cleaved RabGAP-CTR1 and RabGAP-CTR2 were used for the GTPase assay as in the previous study [11]. The two states of the protein were purified from the expressed RabGAP-CTR using Superdex® 200 sizing column, separately concentrated, and the concentrations quantified by employing the calculated molar extinction coefficient (37,600 M−1 cm−1). The Rab14·GTP hydrolysis kinetics were measured using EnzChek phosphate assay kit (Invitrogen) [11]. Briefly, solutions containing 50 mM Tris pH 7.5, 11 mM MgCl2, 0.2 mM 2-amino-6-mercapto-7-methylpurine riboside (MESG), 1 U/mL purine nucleoside phosphorylase (PNP), and 30 μM GTP-loaded Rab14 were mixed with TBC1D4 RabGAP-CTR in a 96-well microplate (Corning). Absorbance changes at 360 nm were monitored using a microplate reader. The time-course data were fitted to a pseudo 1st order Michaelis-Menten model to obtain kobs. kobs measured at two different TBC1D4 concentrations (2.0 μM and 19 μM) were compared with the previously reported data of TBC1D4 RabGAP activity [11].
Acknowledgments
This work was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2016R1D1A1A09918187 to SY.P.), Korea Research Foundation Grant 2011-0010437 (to SJ.K.), National Institutes of Health Grants R01 DK51729 (to S.E.S.), and P30 DK36836 (to Joslin Diabetes Center).
Abbreviations
- AS160
Akt substrate of 160 kDa
- GLUT
glucose transporter
- PTB
phosphotyrosine-binding
- RabGAP
Rab GTPase-activating protein
- TBC
Tre-2, Bub2 and Cdc16
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Watson RT, Pessin JE. Bridging the GAP between insulin signaling and GLUT4 translocation. Trends Biochem Sci. 2006;31:215–222. doi: 10.1016/j.tibs.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–119. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 3.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;8:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 4.Kane S, Sano H, Liu SCH, Asara JM, Lane WS, Garner CW, Lienhard GE. A method to identify serine kinase substrates. Akt phosphorylates a novel adipocyte protein with a Rab GTPase-activating protein (GAP) domain. J Biol Chem. 2002;277:22115–22118. doi: 10.1074/jbc.C200198200. [DOI] [PubMed] [Google Scholar]
- 5.Sano H, Kane S, Sano E, Mîinea CP, Asara JM, Lane WS, Garner CW, Lienhard GW. Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation. J Biol Chem. 2003;278:14599–14602. doi: 10.1074/jbc.C300063200. [DOI] [PubMed] [Google Scholar]
- 6.Dugani CB, Klip A. Glucose transporter 4: cycling, compartments and controversies. EMBO Rep. 2005;6:1137–1142. doi: 10.1038/sj.embor.7400584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peck GR, Ye S, Pham V, Fernando RN, Macaulay SL, Chai SY, Albiston AL. Interaction of the Akt substrate, AS160 with the glucose transporter 4 vesicle marker protein insulin-regulated aminopeptidase. Mol Endocrinol. 2006;20:2576–2583. doi: 10.1210/me.2005-0476. [DOI] [PubMed] [Google Scholar]
- 8.Park S, Kim KY, Kim S, Yu YS. Affinity between TBC1D4 (AS160) phosphotyrosine-binding domain and insulin-regulated aminopeptidase cytoplasmic domain measured by isothermal titration calorimetry. BMB Rep. 2012;45:360–364. doi: 10.5483/bmbrep.2012.45.6.030. [DOI] [PubMed] [Google Scholar]
- 9.Tan SX, Ng Y, Burchfield JG, Ramm G, Lambright DG, Stöckli J, James DE. The Rab GTPase-activating protein TBC1D4/AS160 contains an atypical phosphotyrosine-binding domain that interacts with plasma membrane phospholipids to facilitate GLUT4 trafficking in adipocytes. Mol Cell Biol. 2012;32:4946–4959. doi: 10.1128/MCB.00761-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lupas A, Van Dyke M, Stock J. Predicting Coiled Coils from Protein Sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 11.Park SY, Jin W, Woo JR, Shoelson SE. Crystal structures of Human TBC1D1 and TBC1D4 (AS160) RabGAP domains reveal critical elements for GLUT4 translocation. J Biol Chem. 2011;286:18130–18138. doi: 10.1074/jbc.M110.217323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park SY, Kim S-J. TBC1D1 and TBC1D4 (AS160) RabGAP domains are characterized as monomers in solution by analytical ultracentrifugation. Bull Korean Chem Soc. 2011;32:2125–2128. [Google Scholar]
- 13.Mîinea CP, Sano H, Kane S, Sano E, Fukuda M, Peränen J, Lane WS, Lienhard GE. AS160, the Akt substrate regulating GLUT4 translocation, has a functional Rab GTPase-activating protein domain. Biochem J. 2005;391:87–93. doi: 10.1042/BJ20050887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sano H, Roach WG, Peck GR, Fukuda M, Lienhard GE. Rab10 in insulin-stimulated GLUT4 translocation. Biochem J. 2008;411:89–95. doi: 10.1042/BJ20071318. [DOI] [PubMed] [Google Scholar]
- 15.Sano H, Eguez L, Teruel MN, Fukuda M, Chuang TD, Chavez JA, Lienhard GE, McGraw TE. Rab10, a target of the AS160 Rab GAP, is required for insulin-stimulated translocation of GLUT4 to the adipocyte plasma membrane. Cell Metab. 2007;5:293–303. doi: 10.1016/j.cmet.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Dai Y, Walker SA, de Vet E, Cook S, Welch HC, Lockyer PJ. Ca2+-dependent monomer and dimer formation switches CAPRI Protein between Ras GTPase-activating protein (GAP) and RapGAP activities. J Biol Chem. 2011;286:19905–19916. doi: 10.1074/jbc.M110.201301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daumke O, Weyand M, Chakrabarti PP, Vetter IR, Wittinghofer A. The GTPase-activating protein Rap1GAP uses a catalytic asparagine. Nature. 2004;429:197–201. doi: 10.1038/nature02505. [DOI] [PubMed] [Google Scholar]
- 18.Scrima A, Thomas C, Deaconescu D, Wittinghofer A. The Rap-RapGAP complex: GTP hydrolysis without catalytic glutamine and arginine residues. EMBO J. 2008;27:1145–1153. doi: 10.1038/emboj.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulkarni K, Yang J, Zhang Z, Barford D. Multiple factors confer specific Cdc42 and Rac protein activation by dedicator of cytokinesis (DOCK) nucleotide exchange factors. J Biol Chem. 2011;286:25341–25351. doi: 10.1074/jbc.M111.236455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terasawa M, Uruno T, Mori S, Kukimoto-Niino M, Nishikimi A, Sanematsu F, Tanaka Y, Yokoyama S, Fukui Y. Dimerization of DOCK2 is essential for DOCK2-mediated Rac activation and lymphocyte migration. PLoS One. 2012;7:e46277. doi: 10.1371/journal.pone.0046277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 22.Andrade MA, Chacón P, Merelo JJ, Morán F. Evaluation of secondary structure of proteins from UV circular dichroism using an unsupervised learning neural network. Prot Eng. 1993;6:383–390. doi: 10.1093/protein/6.4.383. [DOI] [PubMed] [Google Scholar]
- 23.Perez-Iratxeta C, Andrade-Navarro MA. K2D2: estimate of protein secondary structure from circular dichroism spectra. BMC Struct Biol. 2007;8:25. doi: 10.1186/1472-6807-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sreerama N, Venyaminov SYU, Woody RW. Estimation of the number of α-helical and βstrand segments in proteins using CD spectroscopy. Protein Sci. 1999;8:370–380. doi: 10.1110/ps.8.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, et al., editors. Analytical ultracentrifugation in biochemistry and polymer science. The Royal Society of Chemistry; Cambridge, UK: 1992. pp. 90–125. [Google Scholar]
- 26.Jung WS, Hong CK, Lee S, Kim CS, Kim SJ, Kim SI, Rhee S. Structural and functional insights into intramolecular fructosyl transfer by inulin fructotransferase. J Biol Chem. 2007;282:8414–8423. doi: 10.1074/jbc.M607143200. [DOI] [PubMed] [Google Scholar]
- 27.Knott GD. MLAB - a mathematical modeling tool. Comput Programs Biomed. 1979;10:271–280. doi: 10.1016/0010-468x(79)90075-8. [DOI] [PubMed] [Google Scholar]