Abstract
Purpose
To determine whether dynamic contrast enhancement magnetic resonance imaging (DCE-MRI) characteristics of the breast tumor and background parenchyma can distinguish molecular subtypes (i.e., luminal A/B or basal) of breast cancer.
Materials and Methods
84 patients from one institution and 126 patients from the cancer genome atlas (TCGA) were used for discovery and external validation, respectively. 35 quantitative image features were extracted from DCE-MRI (1.5 or 3T) including morphology, texture, and volumetric features, which capture both tumor and background parenchymal enhancement (BPE) characteristics. Multiple testing was corrected using the Benjamini-Hochberg method to control false discovery rate (FDR). Sparse logistic regression models were built using the discovery cohort to distinguish each of the three studied molecular subtypes versus the rest, and the models were evaluated in the validation cohort.
Results
On univariate analysis in discovery and validation cohorts, two features characterizing tumor and two characterizing BPE were statistically significant in separating luminal A versus non-luminal A cancers; two features characterizing tumor were statistically significant for separating luminal B; one feature characterizing tumor and one characterizing BPE reached statistical significance for distinguishing basal (Wilcoxon P<0.05, FDR<0.25). In discovery and validation cohorts, multivariate logistic regression models achieved an area under the receiver operator characteristic curve (AUC) of 0.71 and 0.73 for luminal A cancer, 0.67 and 0.69 for luminal B cancer, and 0.66 and 0.79 for basal cancer, respectively.
Conclusion
DCE MR imaging characteristics of breast cancer and BPE may potentially be used to distinguish among molecular subtypes of breast cancer.
Keywords: Breast cancer, Molecular subtype, Imaging genomics, Dynamic contrast enhanced MRI, Classification
INTRODUCTION
Breast cancer is a heterogeneous disease. A variety of clinical and pathological factors, such as patient age, tumor size, histological grade, hormone receptor status and human epidermal growth factor receptor 2 (HER2) status, are used to stratify breast cancer patients to assess prognosis and determine therapy. Gene expression profiling of breast cancer has revealed four main intrinsic molecular subtypes, i.e., luminal A, luminal B, HER2-enriched, and basal-like types (1). Each of these subtypes has distinct gene expression patterns, clinical features, response to treatment, and prognosis (2).
Luminal tumors represent a majority (~70%) of invasive breast cancer. They are generally estrogen-receptor (ER) and/or progesterone-receptor (PR) positive, and respond well to endocrine therapy (3). Among the four subtypes, luminal A breast cancer has the best prognosis, whereas luminal B cancer tends to be more proliferative, of higher histologic grade, and is associated with a higher risk of early relapse and poorer disease-free survival (4). On the other hand, basal-like cancer, which is often ER/PR/HER2 negative, is associated with BRCA1 dysfunction and generally has a poor prognosis (3).
Current assessment of molecular subtypes is based on either gene expression profiling (GEP) or immunohistochemistry (IHC) (5). Due to the cost, complexity, and initial requirement of fresh frozen tissue, the use of GEP has been limited in clinical practice. This led to the development of the more practical IHC-based classification of breast cancer, which has high or moderately high concordance with intrinsic molecular subtypes (1,5), and subsequently adopted by the St Gallen International Expert Consensus panel (6) to define breast cancer subtypes.
Molecular testing requires tissue specimens typically obtained by needle biopsy, and thus can provide inconclusive due to inadequate tumor material or sampling of a small portion of the tumor. Under these circumstances, imaging (in particular, dynamic contrast-enhanced magnetic resonance imaging or DCE MRI), which shows the entire tumor, may provide information about the molecular subtype. For instance, if initial biopsy indicates luminal A cancer while imaging suggests a high probability of basal-like cancer, repeat biopsy prior to treatment would be warranted given its more aggressive behavior and distinct response to therapy (7).
Previous studies have investigated the relationship between MR imaging and genomic features (8–13) or molecular subtypes (14–21) of breast cancer. However, most studies use a single measurement platform (GEP or IHC) for molecular subtypes and lack independent validation. Additionally, most only focus on the tumor, ignoring the surrounding breast tissue. A few recent studies have shown that the appearance of background parenchymal enhancement (BPE) on DCE MR imaging is associated with breast cancer risk (22,23), triple-negative breast cancer (15), response to chemotherapy (24,25), local recurrence (26), disease-free survival and overall survival (27), suggesting an important role of BPE in breast cancer.
The purpose of this study was to determine whether DCE MR imaging characteristics of the breast tumor and background parenchyma could distinguish among molecular subtypes of breast cancer, and to validate the findings on an external independent cohort. We aimed to demonstrate the robustness of our models by evaluating on two distinct measurement platforms (GEP and IHC) to determine molecular subtypes.
METHODS
Patient Population
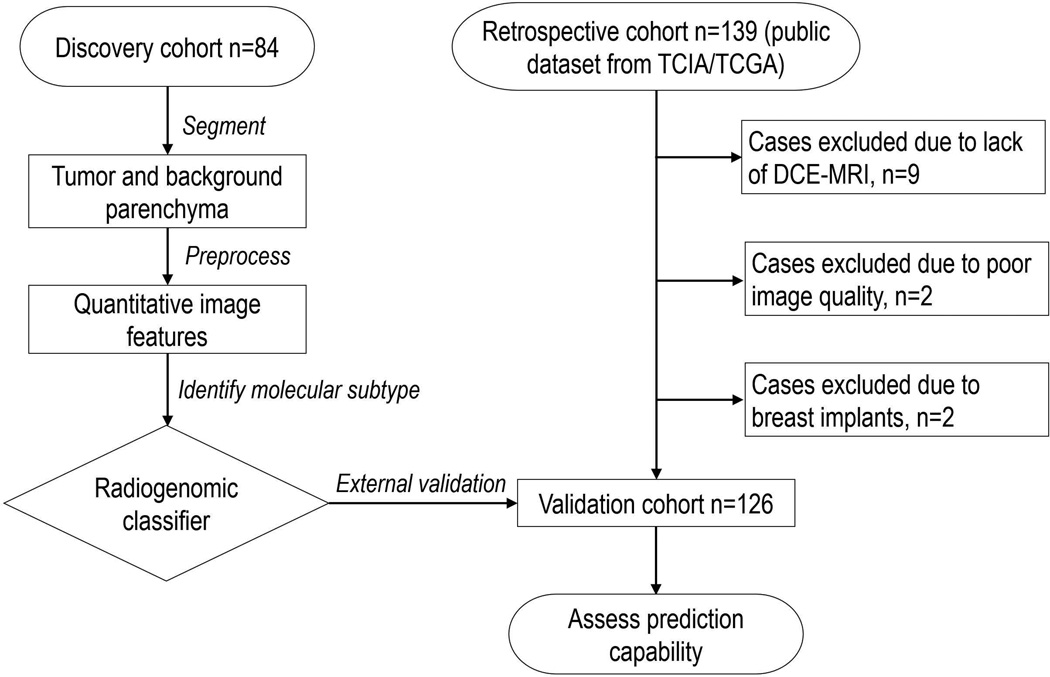
Under approval from the Institutional Review Board of Hokkaido University Hospital, 84 patients with breast cancer were retrospectively collected at Hokkaido University from February 2012 through May 2013. The inclusion criteria were that patients had 1) pathologically proven invasive carcinomas in the unilateral breast, 2) no prior hormonal therapy or neoadjuvant chemotherapy. Informed consent was waived according to Ethical Guidelines for Clinical Studies of the Japanese Ministry of Health, Labour, and Welfare. All patient data were anonymized and de-identified prior to analysis. These 84 patients had pre-operative bilateral DCE-MRI scan and molecular subtype information available, which comprised the discovery cohort. In addition, we downloaded publicly available breast cancer cases (n=139) through the cancer imaging archive (TCIA) for the cancer genome atlas (TCGA). After careful selection (Figure 1), 126 patients whose imaging scans acquired from September 1999 through June 2006 were included in this study as an external validation cohort. Patient characteristics are summarized in Table 1.
Figure 1.
Flowchart of the complete design of the proposed study.
Table 1.
Demographic Data in Discovery Cohort (84 patients) and Validation Cohort (126 patients)
| Discovery Cohort n=84 (internal retrospective data) |
Validation Cohort n=126 (external retrospective data) |
P Value† | ||
|---|---|---|---|---|
| Age (y) | 0.001 | |||
| Median | 60 (32–84) | 53 (29–82) | ||
| Mean ± standard deviation | 59.2 ± 11.0 | 54.0 ± 11.5 | ||
| Menopausal status | 0.004 | |||
| Postmenopausal | 61 (72.6) | 66 (52.4) | ||
| Not postmenopausal | 23 (27.4) | 53 (42.1) | ||
| Not Available | 0 (0) | 7 (5.5) | ||
| Estrogen receptor (ER) | 0.97 | |||
| Positive | 69 (82.1) | 105 (83.3) | ||
| Negative | 15 (17.9) | 21 (16.7) | ||
| Progesterone receptor (PR) | 0.82 | |||
| Positive | 60 (71.4) | 93 (73.8) | ||
| Negative | 24 (28.6) | 33 (26.2) | ||
|
Human epidermal growth factor receptor 2 (HER2) |
0.005 | |||
| Positive | 4 (4.8) | 23 (18.3) | ||
| Negative | 80 (95.2) | 100 (79.4) | ||
| Equivocal | 0 (0) | 3 (2.4) | ||
| Molecular subtype‡ | 0.053 | |||
| Luminal A | 42 (50.0) | 73 (57.9) | ||
| Luminal B | 27 (32.1) | 22 (17.5) | ||
| HER2-enriched | 4 (4.8) | 10 (7.9) | ||
| Basal-like | 11 (13.1) | 16 (12.7) | ||
| Normal-like | 0 (0) | 5 (4.0) | ||
| Histologic type | 0.012 | |||
| Ductal carcinoma | 68 (81.0) | 106 (84.1) | ||
| Lobular carcinoma | 8 (9.5) | 17 (13.5) | ||
| Mixed ductal/lobular carcinoma | 0 (0) | 2 (1.6) | ||
| Other | 8 (9.5) | 1 (0.8) | ||
| MR imaging index lesion type | 0.37 | |||
| Mass | 71 (84.5) | 113 (89.7) | ||
| Nonmass | 13 (15.5) | 13 (10.3) | ||
Note: Unless otherwise indicated, data are number of patients, with percentage in parentheses.
Statistical comparison between two cohorts was computed with χ2 (categorical variables) or Wilcoxon rank sum test (continuous variables).
IHC classification (St Gallen consensus) for discovery cohort, and intrinsic subtypes (gene expression profiling) for validation cohort
Molecular Subtypes
There is high to moderately high agreement between IHC and GEP for classification of luminal (73%–100%) and basal-like 80% breast cancers (5). For the patients collected from Hokkaido University Hospital, the pathology report of each patient was analyzed. Their tumor specimens via core biopsy were assessed with immunohistochemical (IHC) analysis to determine the status of ER, PR, HER2 and Ki67. Each tumor was classified into four molecular subtypes according to the St Gallen Consensus 2011 (6), with 1) luminal A, if ER+ and/or PR+, HER2−, Ki67 < 14%, 2) luminal B, if ER+ and/or PR+, HER2 overexpressed or Ki67 ≥ 14%, 3) Basal-like, if ER−, PR−, HER2−, and 4) HER2-enriched, if ER−, PR− and HER2+.
For patients from TCIA/TCGA, the molecular subtype from IHC test is not available. Instead, we assessed their intrinsic molecular subtype through analyzing the gene expression profiles of their tumor specimens collected during surgery. The gene expression data, i.e., RNA-seqV2 level 3 version 1.1.0 data, were obtained through the UCSC Cancer Genomics Browser (https://genome-cancer.ucsc.edu, accessed April 1, 2016). PAM50 (28) was used to predict the intrinsic molecular subtypes of the breast cancer patients. Specifically, we adopted the 50-gene classifier as defined in PAM50, which consisted of centroids constructed by the PAM algorithm and distances calculated using Spearman’s rank correlation. The final subtype classification was assigned based on the nearest five centroids (28).
Imaging Protocol
For data from Hokkaido University Hospital, MR imaging was performed with Achieva 3.0 T TX system (Philips Healthcare, Best, Netherlands) with a 7-channel breast coil in prone position. The dynamic protocol was in accordance with the American College of Radiology guidelines. In brief, bilateral T1 weighted images were acquired in the axial plane with a fat-suppressed gradient echo sequence: repetition time 4.9 ms, echo time 2.4 ms, flip angle 10°, filed of view: 320 × 320 mm2, voxel size: 0.8 × 0.8 × 1.6 mm3. An intravenous gadolinium-based contrast agent was delivered with the dose of 0.1 mmol/kg and flushed with 20 mL saline. MR images were acquired at four-time points, with the first one taken immediately before contrast injection, and the second, third and fourth time points at 1 minute, 2 minutes and 6 minutes after injection. The quality of the scans was verified for lack of artifacts, patient motion or poor fat suppression that would negatively impact subsequent image analysis by a radiologist (X. S. 11 years of experience in breast imaging).
For dataset from TCIA/TCGA, DCE MR imaging was acquired from six different centers, on a 1.5 T or 3 T GE, Siemens or Philips whole-body MRI system, with a standard double breast coil. The dynamic protocol used was in accordance with the American College of Radiology guidelines (29). The DCE-MR imaging protocols include one pre-contrast image and two to seven post-contrast images (with a gadolinium-based contrast agent), in either the axial or sagittal view. T1 weighted images were acquired with a fat-suppressed gradient echo sequence: repetition time ranged from 3.8 to 8.3 ms, echo time ranged from 1.1 to 4.2 ms, flip angle ranged from 8° to 10°, in-plane resolution ranged from 0.39 to 0.97 mm, and the slice thickness ranged from 0.9 to 3.2 mm.
Tumor and Background Parenchyma Segmentation
Two radiologists (F. K. and X. S. with 14 and 11 years of experience in breast imaging) blinded to tumor subtypes manually delineated the 3D tumor in a slice-by-slice manner, taking into account all available sequences (such as T2-weighted MRI). Both radiologists reached consensus regarding all tumor contours. Then, the ipsilateral parenchyma was segmented in an automated fashion from DCE MRI: first, the breast ROI was segmented, then fuzzy c means clustering separated the entire breast region into fatty tissue and non-fatty tissue (parenchyma and cancer), and lastly the parenchyma tissue was obtained by excluding the predefined tumor region from the non-fatty tissue region. The parenchymal segmentation was manually revised by two radiologists when necessary.
Image Preprocessing
Inconsistency in imaging protocols within TCIA/TCGA and between datasets can cause variability in the quantitative image features. To mitigate this effect and ensure robust feature extraction, we applied several procedures to normalize the imaging data. First, the N4 bias correction (30) was implemented to correct the shading appearing in the MR images. Second, we matched the temporal resolution between the two cohorts. For each patient, we extracted DCE-MRI scans before contrast administration, at an early postcontrast phase (temporal sampling of the center of k-space between two to three minutes) and at a late postcontrast phase (around six minutes) for further analysis, in accordance with ACR BI-RADS and previous multi-institutional study design (31). Third, similar to image normalization that implemented in, for each patient, the DCE-MRI was normalized via the imaging value of precontrast parenchyma, i.e., average of interquartile voxel values from parenchyma before contrast administration, to explicitly account for heterogeneous imaging protocols. Lastly, all the DCE-MR images were resized to have an isotropic voxel resolution of 0.8 mm, which assured meaningful 3D texture feature computation in the following.
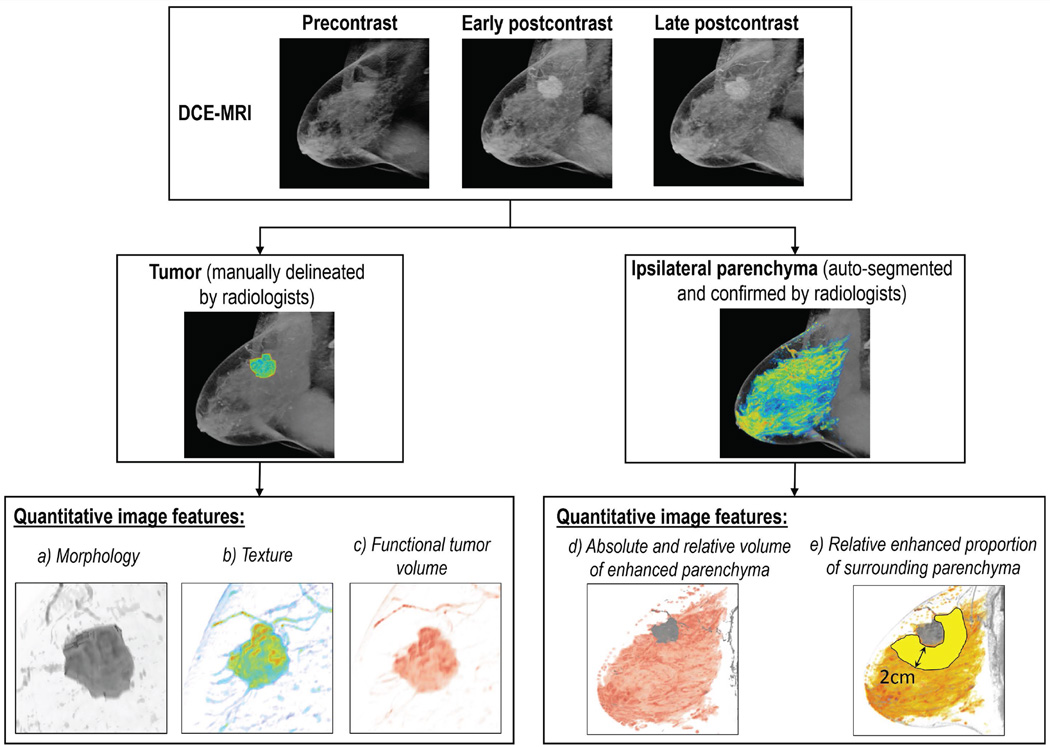
Quantitative Image Feature Extraction
We extracted a total of 35 quantitative image features from each patient’s DCE-MRI to characterize the tumor and parenchymal enhancement as well as intra-tumor heterogeneity, as shown in Figure 2. The proposed feature pool investigates the phenotypical properties of both tumor and parenchyma, including 8 tumor morphological features, 12 tumor texture features, 3 functional tumor volume features, 6 parenchymal enhancement features and 6 tumor-surrounding parenchymal enhancement features. These features and their potential clinical relevance are elaborated in Table 2 and Supplementary Table 1. Here, we used four out of 22 Haralick texture features calculated from the gray-level co-occurrence matrix. These texture features were selected to avoid redundancy and were commonly used in the medical image analysis literature (32,33). The cutoff values for signal enhancement ratio (SER) and percentage enhancement (PE) were chosen a priori, including low, medium and high thresholds matched to values at the bottom 10th percentile, median, and top 10th percentile. The calculation was implemented with MATLAB (MathWorks, Natick, Massachusetts).
Figure 2.
Schematic illustration of quantitative DCE MR imaging features extraction procedure. DCE-MRI: dynamic contrast-enhanced magnetic resonance image.
Table 2.
Names and interpretations of 35 Quantitative Image Features Extracted from DCE-MRI
| Type | No. | Name | Interpretation |
|---|---|---|---|
| Morphology | 8 |
|
Measure of tumor shape, size, and boundary smoothness, i.e., quantitative descriptors according to BIRADS (29) |
| Texture | 12 |
|
Measure the spatial heterogeneity of intensity value of the computed maps within the tumor |
| Functional tumor volume |
3 |
|
A subset of tumor which has quick contrast uptake and washout defined by SER |
| Ipsilateral parenchymal enhancement |
6 |
|
Measure the enhanced subset of ipsilateral breast parenchyma, at the early postcontrast phase, in accordance with the BIRADS (29) |
| Tumor surrounding parenchymal enhancement |
6 |
|
Measure the enhanced parenchyma surrounding the tumor within 2cm distance |
Note: PE: percent enhancement, which is defined as
SER: signal enhancement ratio, which is defined as
mapearly enhance: enhancement map at early postcontrast phase, which is defined as mapearly enhance = Iearly postcontrast − Iprecontrast
maplate enhance: enhancement map at late postcontrast phase, which is defined as maplate enhance = Ilate postcontrast − Iearly postcontrast
Model Discovery for Molecular Subtypes
We constructed separate models to distinguish luminal A versus non-luminal A cancers, luminal B versus non-luminal B cancers, as well as basal-like versus non-basal-like cancers using DCE MR imaging features based on the discovery cohort. We did not analyze the HER2-enriched subtype due to a small number of patients in the discovery cohort (n=4) and relatively poor agreement (41–69%) between gene expression patterns and IHC surrogates for these subtypes (5).
Given the relatively large number of quantitative image features, we combined logistic regression and Least Absolute Shrinkage and Selection Operator (LASSO) (34) to perform feature selection to avoid over-fitting. Briefly, ten-fold internal cross validation was applied and repeated 100 times to minimize the potential selection bias, and the most frequently selected imaging features were used to fit the final model.
For comparison, we assessed the predictive capacity of the clinicopathological information (age, postmenopausal status and histologic type) as the baseline. The baseline model was fitted with logistic regression in the discovery cohort, using R version 3.2.3. In addition, we compared with the predictive capability of the qualitative features by radiologists’ reading according to ACR BI-RADS, including mass shape, margin, and internal enhancement characteristics.
Model Validation and Statistical Analysis
We tested the subtype models on the independent external validation cohort. The receiver operating characteristics (ROC) curve analysis and area under the curve (AUC) was used to assess the prediction capability of the proposed imaging predictors. The threshold to separate different molecular subtypes was defined in the discovery cohort based on the Youden’s J statistics (35), and the same threshold was applied to the validation cohort. The corresponding sensitivity, specificity, and accuracy were reported. Mann-Whitney U statistics tested the statistical significance of the multivariate model (i.e., comparing its AUC to a random guess with AUC=0.5). The method of DeLong test was utilized in the construction of 95% confidence intervals as well as in computing p-values of the pair-wise comparison of ROC curves. To adjust for multiple statistical testing, the Benjamini-Hochberg method was used to control the false discovery rate (FDR) on univariate analysis. P-value less than 0.05 and FDR less than 0.25 was considered to indicate statistically significant differences. Similar to (8), a larger threshold of FDR is adopted to increase the likelihood of positive findings. The statistical analysis was in the R.
RESULTS
Univariate Analysis
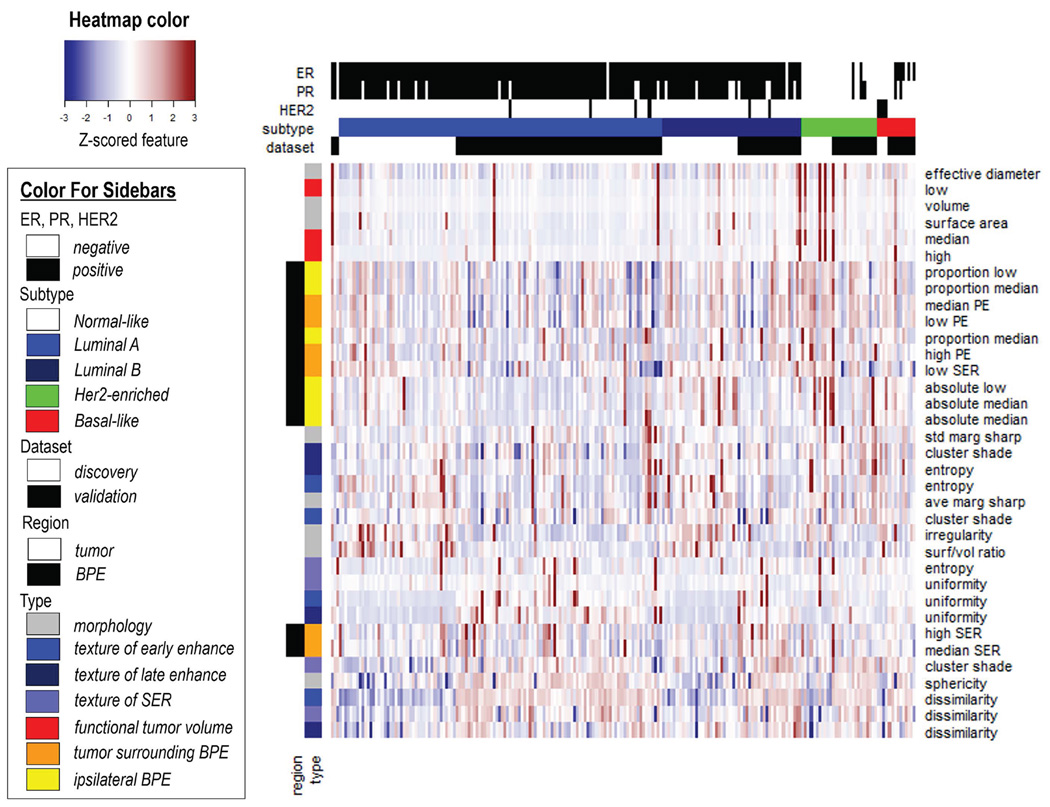
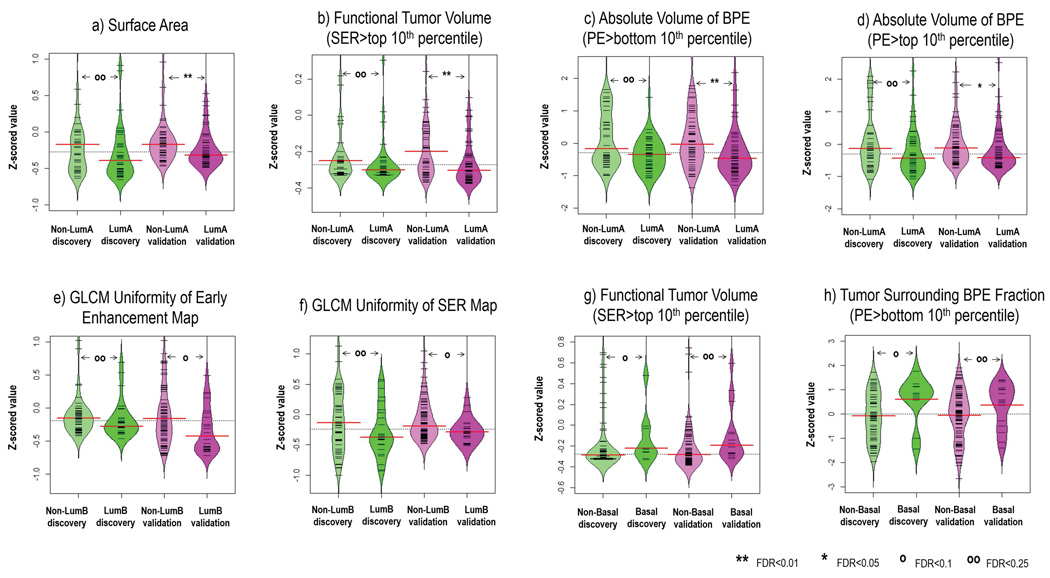
All 35 quantitative imaging features were illustrated in a heat map in Figure 3, and the corresponding Pearson correlation matrix did not indicate that the imaging features were correlated (absolute correlation coefficient<0.8, supplementary Figure 1). On univariate analysis, four imaging features were statistically significant in separating luminal A versus non-luminal A patients in the validation cohort (three features had Wilcoxon P<0.05, false discovery rate [FDR]<0.01, one had Wilcoxon P<0.05, FDR<0.05). Two quantitative imaging features reached significance to differentiate luminal B from non-luminal B cases in the validation cohort (Wilcoxon P<0.05, FDR<0.10). Two quantitative imaging features reached significance to separate basal-like versus non-basal-like cancers (Wilcoxon P<0.05, FDR<0.25). Figure 4 showed the detailed distribution of these 6 features as well as their FDR adjusted for multiple testing.
Figure 3.
Clustering analysis of the quantitative imaging features from both discovery and validation cohorts. In the heat map, all 35 features (presented in different rows and color-coded by the region and type) from all 210 patients (presented in each column) were correlated with their molecular subtype (color-coded on the top bar). All features were standardized to have a zero mean and unit standard deviation. BPE: background parenchymal enhancement.
Figure 4.
Imaging features significantly associated with molecular subtypes (after correction for multiple testing) in both discovery and validation cohorts, a–d) 4 features for distinguishing luminal A versus non-luminal A, e–f) 2 features for distinguishing luminal B versus non-luminal B, and g–h) 2 features for distinguishing basal-like versus non-basal-like. Wilcoxon rank sum test was implemented to investigate pairwise difference. Also, the false discovery rate (FDR) adjusted for multiple testing was reported.
Multivariate Analysis
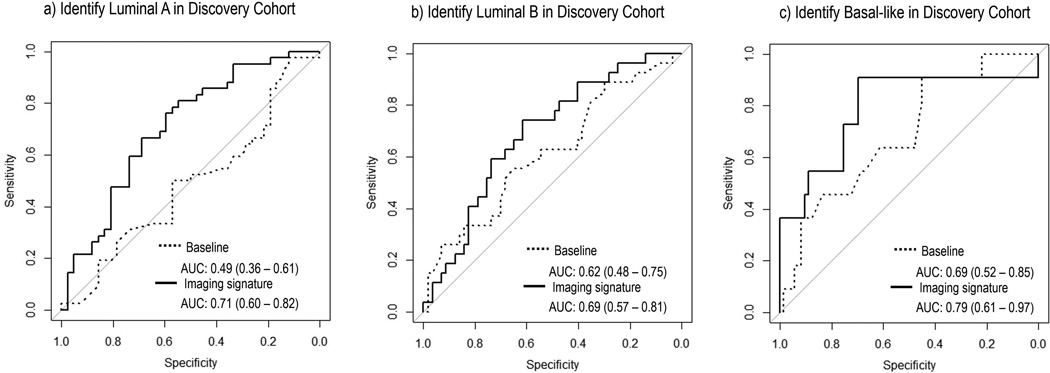
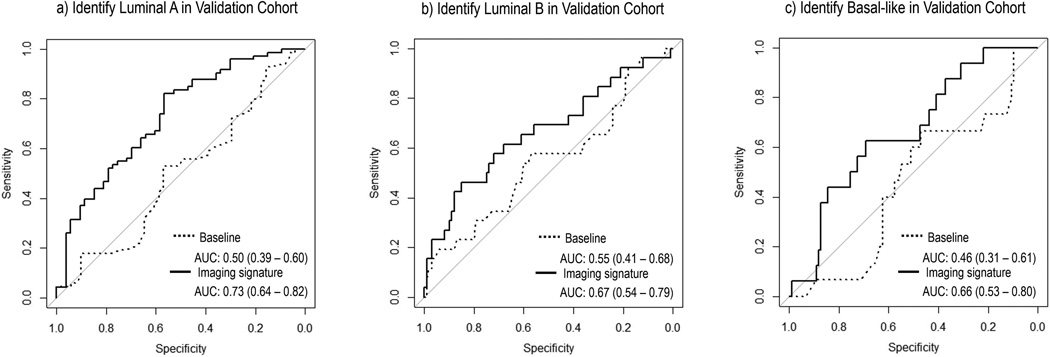
The optimal subtype models respectively for luminal A, luminal B, and basal-like cancer based on the discovery cohort were illustrated in Table 3. Additionally, the corresponding ROC curves for three molecular subtypes were plotted in Figures 5 and 6 separately for discovery and validation cohorts. On independent validation, all three models achieved good accuracy and remained statistically significant (Mann-Whitney P<0.05), as shown in Table 3. The subtype models based on quantitative imaging features were significantly better than the baseline models consisting of three clinical factors, with Delong P of 0.022, 0.048 and 0.039 for luminal A, luminal B, and basal-like breast cancer, respectively.
Table 3.
Subtype Model Construction and Evaluation for Luminal A, Luminal B and Basal-like Breast Cancer.
| Classification Model | P Value† | Threshold‡ | Sensitivity | Specificity | Accuracy | |
|---|---|---|---|---|---|---|
| Lumina A, discovery |
−0.44 × SERClustershade − 0.34 × FTVSER>bottom 10% − 0.30 × BPEsurround,PE>bottom 10% − 0.42 × BPEabs vol PE>bottom 10% + 0.11 |
0.0005 | 0.48 | 0.76 | 0.60 | 0.68 |
| Luminal A, validation |
- | 6.8×10−6 | - | 0.61 | 0.70 | 0.64 |
| Lumina B, discovery |
0.41 × Maplate enhance,dissimilarity + 0.37 × SERdissimilarity − 1.17 × SERuniformity + 0.23 × BPEabs vol,PE>bottom 10% − 1.20 |
0.003 | 0.32 | 0.74 | 0.61 | 0.65 |
| Luminal B, validation |
- | 0.005 | - | 0.62 | 0.68 | 0.67 |
| Basal- like, discovery |
1.36 × Tumorvolume + 0.96 × BPEsurround,PE>median − 2.47 |
0.001 | 0.11 | 0.91 | 0.70 | 0.73 |
| Basal- like, validation |
- | 0.018 | - | 0.63 | 0.69 | 0.68 |
Note:
Note: P value was from Mann-Whitney U statistics;
Threshold was defined solely based on the discovery cohort using the Youden’s J statistics.
SERClusterShade is the cluster shade of SER map; FTVSER > bottom 10% is the functional tumor volume, with SER > bottom 10th percentile; BPEsurround, PE > bottom 10% is the enhanced proportion of tumor surrounding parenchyma, with PE > bottom 10th percentile; BPEabs vol, PE > bottom 10% is the absolute volume of enhanced parenchyma in the ipsilateral breast with tumor, with PE > bottom 10th percentile; Maplate enhance, dissimilarity is the dissimilarity measure of late enhancement map; SERdissimilarity is the dissimilarity measure of SER map; SERuniformity is the uniformity measure of SER map; Tumorvolume is the gross tumor volume; BPEsurround,PE>median is the enhanced proportion of tumor surrounding parenchyma, with PE > median.
Figure 5.
ROC curves of the built imaging signature and the baseline model with three clinical factors (age, menopausal status, and histologic type) trained in the discovery cohort to distinguish a) luminal A, b) luminal B, and c) basal like cancer type. The corresponding AUCs and 95% confidence intervals were reported.
Figure 6.
Validation of previously built models from discovery cohort in the independent cohort to distinguish a) luminal A, b) luminal B, and c) basal like cancer type. The corresponding AUCs and 95% confidence intervals were reported.
BI-RADS Features Were Not Predictive Of Molecular Subtypes
The subtype models based on BI-RADS features were inferior to those based quantitative image features, with AUC ranging from 0.56 to 0.57 (Mann-Whitney P>0.05) for validation (see supplementary Figure 2).
DISCUSSION
In this radiogenomic study, we investigated the relationship between DCE MR imaging characteristics and intrinsic molecular subtypes of breast cancer. Through extensive image analysis, we extracted quantitative image features of the breast tumor as well as surrounding background parenchyma.
The ability to accurately define breast cancer molecular subtypes on an individualized basis has important therapeutic implications. For instance, Luminal B cancer has a distinct profile of therapeutic response in that it is relatively insensitive to endocrine therapy, and relatively resistant to chemotherapy compared with HER2 and basal subtypes (36). While patients with the less aggressive luminal A cancer may be adequately treated with endocrine therapy alone, women with luminal B cancer often require more aggressive therapy, possibly by combining endocrine and molecularly targeted therapy (7). Promising therapeutic targets for luminal B cancers, such as phosphatidylinositol-3-kinase (PI3K) and mammalian target of rapamycin (mTOR) inhibitors, are being actively tested and have shown clinical efficacy in prospective trials (37).
Our data suggest that DCE MR imaging could be used to identify patients who are more likely to harbor the more aggressive luminal B cancer versus luminal A cancer. Our study builds upon previous radiogenomic work (14–21) and adds to the accumulating evidence that imaging may potentially provide useful supplementary information to molecular analysis in breast cancer, especially in situations where the latter is inconclusive because of insufficient tumor sampling in a small needle biopsy.
We showed that luminal A cancer was significantly associated with smaller functional tumor volume, smaller surface area (which is correlated with volume), and smaller volume of enhanced background parenchyma compared to non-luminal A cancer in both cohorts. Previous studies have shown that smaller functional tumor volume and lower BPE around the tumor are associated with superior survival (38) and lower risk of recurrence in breast cancer (26), respectively. As luminal A cancer is known to have the best prognosis among all four subtypes, this lends support for our findings here. Conversely, we showed that basal-like breast cancer was significantly associated with larger functional tumor volume and higher tumor surrounding BPE fraction, which also seems to be consistent with a generally poor prognosis of basal-like cancer.
Our data indicate that the more aggressive luminal B cancer was significantly associated with higher intra-tumor heterogeneity (i.e., the lower uniformity of the gray-level co-occurrence matrix) in both cohorts. This adds to the extensive evidence that imaging heterogeneity is associated with aggressive disease in many cancer types (32). Additionally, higher volume of BPE conferred a higher risk for luminal B cancer on multivariate analysis in our study, which also appears to be consistent with recent studies showing the associations between higher BPE and a higher risk of local recurrence (26) as well as worse recurrence-free survival in breast cancer (39). One recent study specifically looking at the association between DCE MR imaging and luminal B breast cancer also showed the relevance of BPE (17).
To quantify the degree of BPE, we used volume-based measurements in our study. This is different from previous studies (26,27) that calculate the absolute signal enhancement or its ratio of background parenchyma, which may be prone to MR signal artifacts or variations in image acquisition protocols. We believe that volume-based measurements are more robust to such variations (38) and that they are more closely related to how radiologists observe and interpret BPE on MR imaging, as suggested in BIRADS (29).
In our study, the molecular subtypes were defined on two different platforms, i.e., IHC in the discovery cohort and gene expression profiles in the validation cohort. For luminal and basal cancers, there is a high though not perfect concordance between the two assays (73–100% and 80%) (5), which might explain the variations in different subtype models. Despite the potential difference, we were able to validate our results on two independent platforms, attesting the robustness of the radiogenomic relations found here.
There were a small number (4%) of tumors in the validation cohort that fall into the normal breast-like subtype. Unlike the other four intrinsic molecular subtypes, the normal-breast like subtype is less reproducible, and shows low tumor cellularity (<50%) when examined pathologically, explaining why they group with the true normal samples (40). Thus, when a tumor is classified as the normal breast-like subtype, it is most likely caused by a false negative biopsy, where the sample is predominantly composed of normal breast tissue and not tumor tissue. This again points to the potential limitations of molecular analysis based on a needle biopsy, and highlights the value of imaging for depicting the entire tumor volume.
Limitations of our study include the retrospective nature of our external validation cohort. Due to a small sample size, we were not able to study the HER2-enriched subtype. While discovery was benefited by the use of a uniform acquisition protocol for MR imaging, the image data in the validation cohort came from multiple institutions and consisted of several different acquisition protocols and parameters, which might have influenced the image quantification. Nevertheless, we have conducted careful image preprocessing and normalization, as well as automated image analysis to minimize the potential biases (indeed the results were validated on the external cohort). Our findings need to be further validated in larger prospective cohorts.
In conclusion, DCE MR imaging characteristics of the tumor and background parenchyma can be used to distinguish among molecular subtypes of breast cancer. Future studies should be conducted to elucidate the biological mechanism behind these relations.
Supplementary Material
Acknowledgments
The authors thank The Cancer Imaging Archive (TCIA) for providing the breast cancer cases enrolled in The Cancer Genome Atlas (TCGA) study.
GRANT SUPPORT
This research was partially supported by the NIH grant number R01 CA193730.
Footnotes
DISCLOSURE:
None.
REFERENCES
- 1.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Sotiriou C, Neo S-Y, McShane LM, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proceedings of the National Academy of Sciences. 2003;100(18):10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schnitt SJ. Classification and prognosis of invasive breast cancer: from morphology to molecular taxonomy. Modern Pathology. 2010;23:S60–S64. doi: 10.1038/modpathol.2010.33. [DOI] [PubMed] [Google Scholar]
- 4.Loi S, Haibe-Kains B, Desmedt C, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25(10):1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 5.Guiu S, Michiels S, Andre F, et al. Molecular subclasses of breast cancer: how do we define them? The IMPAKT 2012 Working Group Statement. Annals of oncology. 2012;23(12):2997–3006. doi: 10.1093/annonc/mds586. [DOI] [PubMed] [Google Scholar]
- 6.Goldhirsch A, Wood W, Coates A, Gelber R, Thürlimann B, Senn H-J. Strategies for subtypes—dealing with the diversity of breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Annals of oncology. 2011 doi: 10.1093/annonc/mdr304. mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ades F, Zardavas D, Bozovic-Spasojevic I, et al. Luminal B breast cancer: molecular characterization, clinical management, and future perspectives. J Clin Oncol. 2014;32(25):2794–2803. doi: 10.1200/JCO.2013.54.1870. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto S, Maki DD, Korn RL, Kuo MD. Radiogenomic analysis of breast cancer using MRI: a preliminary study to define the landscape. Am J Roentgenol. 2012;199(3):654–663. doi: 10.2214/AJR.11.7824. [DOI] [PubMed] [Google Scholar]
- 9.Ashraf AB, Daye D, Gavenonis S, et al. Identification of intrinsic imaging phenotypes for breast cancer tumors: preliminary associations with gene expression profiles. Radiology. 2014;272(2):374–384. doi: 10.1148/radiol.14131375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamamoto S, Han W, Kim Y, et al. Breast Cancer: Radiogenomic Biomarker Reveals Associations among Dynamic Contrast-enhanced MR Imaging, Long Noncoding RNA, and Metastasis. Radiology. 2015;275(2):384–392. doi: 10.1148/radiol.15142698. [DOI] [PubMed] [Google Scholar]
- 11.Sutton EJ, Oh JH, Dashevsky BZ, et al. Breast cancer subtype intertumor heterogeneity: MRI-based features predict results of a genomic assay. J Magn Reson Imaging. 2015;42(5):1398–1406. doi: 10.1002/jmri.24890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Zhu Y, Burnside ES, et al. MR Imaging Radiomics Signatures for Predicting the Risk of Breast Cancer Recurrence as Given by Research Versions of MammaPrint, Oncotype DX, and PAM50 Gene Assays. Radiology. 2016:152110. doi: 10.1148/radiol.2016152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu J, Cui Y, Sun X, et al. Unsupervised clustering of quantitative image phenotypes reveals breast cancer subtypes with distinct prognoses and molecular pathways. Clin Cancer Res. 2017 doi: 10.1158/1078-0432.CCR-16-2415. clincanres. 2415.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agner SC, Rosen MA, Englander S, et al. Computerized image analysis for identifying triple-negative breast cancers and differentiating them from other molecular subtypes of breast cancer on dynamic contrast-enhanced MR images: a feasibility study. Radiology. 2014;272(1):91–99. doi: 10.1148/radiol.14121031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Kato F, Oyama-Manabe N, et al. Identifying Triple-Negative Breast Cancer Using Background Parenchymal Enhancement Heterogeneity on Dynamic Contrast-Enhanced MRI: A Pilot Radiomics Study. PloS one. 2015;10(11):e0143308. doi: 10.1371/journal.pone.0143308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sutton EJ, Dashevsky BZ, Oh JH, et al. Breast cancer molecular subtype classifier that incorporates MRI features. J Magn Reson Imaging. 2016 doi: 10.1002/jmri.25119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazurowski MA, Zhang J, Grimm LJ, Yoon SC, Silber JI. Radiogenomic analysis of breast cancer: luminal B molecular subtype is associated with enhancement dynamics at MR imaging. Radiology. 2014;273(2):365–372. doi: 10.1148/radiol.14132641. [DOI] [PubMed] [Google Scholar]
- 18.Martincich L, Deantoni V, Bertotto I, et al. Correlations between diffusion-weighted imaging and breast cancer biomarkers. European radiology. 2012;22(7):1519–1528. doi: 10.1007/s00330-012-2403-8. [DOI] [PubMed] [Google Scholar]
- 19.Waugh S, Purdie C, Jordan L, et al. Magnetic resonance imaging texture analysis classification of primary breast cancer. European radiology. 2016;26(2):322–330. doi: 10.1007/s00330-015-3845-6. [DOI] [PubMed] [Google Scholar]
- 20.Kim EJ, Kim SH, Park GE, et al. Histogram analysis of apparent diffusion coefficient at 3.0 t: Correlation with prognostic factors and subtypes of invasive ductal carcinoma. J Magn Reson Imaging. 2015;42(6):1666–1678. doi: 10.1002/jmri.24934. [DOI] [PubMed] [Google Scholar]
- 21.Blaschke E, Abe H. MRI phenotype of breast cancer: kinetic assessment for molecular subtypes. J Magn Reson Imaging. 2015;42(4):920–924. doi: 10.1002/jmri.24884. [DOI] [PubMed] [Google Scholar]
- 22.King V, Brooks JD, Bernstein JL, Reiner AS, Pike MC, Morris EA. Background Parenchymal Enhancement at Breast MR Imaging and Breast Cancer Risk. Radiology. 2011;260(1):50–60. doi: 10.1148/radiol.11102156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dontchos BN, Rahbar H, Partridge SC, et al. Are Qualitative Assessments of Background Parenchymal Enhancement, Amount of Fibroglandular Tissue on MR Images, and Mammographic Density Associated with Breast Cancer Risk? Radiology. 2015;276(2):371–380. doi: 10.1148/radiol.2015142304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hattangadi J, Park C, Rembert J, et al. Breast stromal enhancement on MRI is associated with response to neoadjuvant chemotherapy. Am J Roentgenol. 2008;190(6):1630–1636. doi: 10.2214/AJR.07.2533. [DOI] [PubMed] [Google Scholar]
- 25.Preibsch H, Wanner L, Bahrs S, et al. Background parenchymal enhancement in breast MRI before and after neoadjuvant chemotherapy: correlation with tumour response. European radiology. 2015:1–7. doi: 10.1007/s00330-015-4011-x. [DOI] [PubMed] [Google Scholar]
- 26.Kim SA, Cho N, Ryu EB, et al. Background Parenchymal Signal Enhancement Ratio at Preoperative MR Imaging: Association with Subsequent Local Recurrence in Patients with Ductal Carcinoma in Situ after Breast Conservation Surgery. Radiology. 2014;270(3):699–707. doi: 10.1148/radiol.13130459. [DOI] [PubMed] [Google Scholar]
- 27.van der Velden BHM, Dmitriev I, Loo CE, Pijnappel RM, Gilhuijs KGA. Association between Parenchymal Enhancement of the Contralateral Breast in Dynamic Contrast-enhanced MR Imaging and Outcome of Patients with Unilateral Invasive Breast Cancer. Radiology. 2015;276(3):675–685. doi: 10.1148/radiol.15142192. [DOI] [PubMed] [Google Scholar]
- 28.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards SD, Lipson JA, Ikeda DM, Lee JM. Updates and revisions to the BI-RADS magnetic resonance imaging lexicon. Magnetic resonance imaging clinics of North America. 2013;21(3):483–493. doi: 10.1016/j.mric.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Tustison NJ, Avants BB, Cook PA, et al. N4ITK: improved N3 bias correction. IEEE transactions on medical imaging. 2010;29(6):1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hylton NM, Blume JD, Bernreuter WK, et al. Locally advanced breast cancer: MR imaging for prediction of response to neoadjuvant chemotherapy—results from ACRIN 6657/I-SPY TRIAL. Radiology. 2012;263(3):663–672. doi: 10.1148/radiol.12110748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2015:151169. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu J, Gong G, Cui Y, Li R. Intratumor partitioning and texture analysis of dynamic contrast-enhanced (DCE)-MRI identifies relevant tumor subregions to predict pathological response of breast cancer to neoadjuvant chemotherapy. J Magn Reson Imaging. 2016;44(5):1107–1115. doi: 10.1002/jmri.25279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society Series B (Methodological) 1996:267–288. [Google Scholar]
- 35.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3(1):32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 36.Bhargava R, Beriwal S, Dabbs DJ, et al. Immunohistochemical surrogate markers of breast cancer molecular classes predicts response to neoadjuvant chemotherapy. Cancer. 2010;116(6):1431–1439. doi: 10.1002/cncr.24876. [DOI] [PubMed] [Google Scholar]
- 37.Baselga J, Campone M, Piccart M, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. New Engl J Med. 2012;366(6):520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hylton NM, Gatsonis CA, Rosen MA, et al. Neoadjuvant Chemotherapy for Breast Cancer: Functional Tumor Volume by MR Imaging Predicts Recurrence-free Survival—Results from the ACRIN 6657/CALGB 150007 I-SPY 1 TRIAL. Radiology. 2015:150013. doi: 10.1148/radiol.2015150013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi JS, Ko ES, Ko EY, Han B-K, Nam SJ. Background Parenchymal Enhancement on Preoperative Magnetic Resonance Imaging: Association With Recurrence-Free Survival in Breast Cancer Patients Treated With Neoadjuvant Chemotherapy. Medicine. 2016;95(9) doi: 10.1097/MD.0000000000003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peppercorn J, Perou CM, Carey LA. Molecular subtypes in breast cancer evaluation and management: divide and conquer. Cancer investigation. 2008;26(1):1–10. doi: 10.1080/07357900701784238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.