Abstract
Transcription factors c-Jun and Fra-1 have been reported to play a role during the initiation and progression in oral squamous cell carcinoma (OSCC). However, cohort studies are rarely reported. Here is an integrative analysis of their prognostic value in OSCC through a multicenter cohort study.313 OSCC patients were included in this study and received regular follow-up. The survival rate and hazard ratios(HR) were generated by survival analysis. The concordance probability and receiver operating characteristic curve area were chosen to measure the model discrimination. High expressions of c-Jun or Fra-1 were associated with poor prognosis, meanwhile the high expression of Fra-1 meant worse prognosis of patients than the high expression of c-Jun. Besides, the interaction effect of c-Jun and Fra-1 was antagonism, when the expression of c-Jun and Fra-1 was both high, the HR was lower than the hazard ratio when only the Fra-1 was at high expression. c-Jun and Fra-1 were both proved to be high risky predictors of death in OSCC, the antagonistic effect suggested that these biomarkers’ activities could be influenced by each other. It may provide a new sight for the studies of OSCC prognosis and treatment.
Introduction
Oral squamous cell carcinoma (OSCC), a major devastating head and neck cancer subtype, is one of the most common cancers worldwide1–3. OSCC can migrate into the maxillary and mandibular bones and has a potent capacity to invade locally and metastasize distantly4. The five-year survival rate of patients with OSCC is less than 50%5, 6. In the clinical practice, the clinical tumor-node-metastasis (TNM) stage which includes tumor stage, lymph nodal stage and metastasis is usually used to predict the progression of OSCC7, 8. But the TNM staging system seems like the outcome of tumor prognosis instead of prediction, and patients with same TNM stages of OSCC may result in dramatically different survival time7, 9, 10. Hence, identification of novel and effective biomarkers is in need, which may serve as prognostic predictors, and also be used to guide treatment of OSCC patients1. To date, the application of gene-based biomarkers in diagnosis and prognosis of OSCC seem to be very promising11–14.
Activator protein 1 (AP-1) is one of the first identified transcription factors that regulates gene expression Signals like cytokines, hormone, infection and reactive oxygen species (ROS) can activate AP-115, mainly through mitogen-activated protein kinase (MAPK)16. The activated AP-1 could increase the transcription of target genes and play roles in intercellular events including cell division, proliferation, differentiation, apoptosis and so on17–19. It is reported that the AP-1 was related to the tumorigenesis20–22, and the overexpression of c-Jun and Fra-1 promotes the invasive growth and metastasis of various tumors23, 24, such as breast cancer24, 25, liver cancer26, skin cancer23 and squamous-cell carcinoma27, 28, and indicated that they may be the potential therapeutic targets for SCC28.
AP1 is characterized as basic leucine-zipper domain, including Jun proteins (c-Jun, JunB, JunD), Fos proteins (c-Fos, FosB, Fra-1, Fra-2), activating transcription factor (ATF) proteins and musculoaponeurotic fibrosarcoma (MAF) proteins16. Most proteins which constitute the AP-1 belong to the JUN proteins, and among these, c-Jun is unique in regulating the cell proliferation29, 30. c-Jun exhibits the highest activation potential in the DNA binding affinities and transactivation capacities of the JUN proteins15, 31. FOS proteins are another main group of AP-1, which are just behind JUN proteins, it is best characterized as immediate early genes16, 32. Among the FOS proteins, apart from c-Fos, Fra-1 is best studied subunit of the Fos proteins, the transcriptional activity of Fra-1 is regulated both transcriptionally and post translationally33, 34. However, current studies explained the influences of c-Jun and Fra-1 on OSCC mainly with experimental models35–38. The hazard risks of c-Jun and Fra-1 overexpression in the OSCC prognosis and the correlations between them and cellular biological processes in OSCC with tissue microarray (TMA) which derived from multicenter cohort study are rarely reported.
Thus, in this study, we focused on the two subunits of AP-1, c-Jun and Fra-1. We performed an integrative analysis of the association between the two subunits of AP-1 and the prognosis of OSCC with multicenter cohort study. And as the intricate relationship of the AP-1 subunits on the tumorigenesis and tumor prognosis15, 39, the interaction effect of c-Jun and Fra-1 on the prognosis was also investigated.
Materials and Methods
Patients’ cohorts
The West China Hospital of Stomatology (Chengdu, China), Guangdong Provincial Stomatological Hospital (Guangzhou, China) and the General Hospital of the People’s Liberation Army (Beijing, China) participated this study. The study was approved by the ethics committees of all the three hospitals, and was conducted in agreement with the Helsinki Declaration. Written informed consent was provided by all participants at baseline and during follow-up.
A total of 313 postoperative patients from three hospitals with primary OSCC tumors constituted this multicenter cohort study and received regular follow-up. Beside the regular visits, all patients could initiate follow-up visits if they were concerned that they had recurrence or a new primary tumor. Information was collected during the follow-up visits, which included the medical history and clinical examination, such as age, gender, smoking status, drinking status, tumor differentiation, clinical TNM stage and primary site of tumor. The survival time of each patient was recorded from the day of surgery until the time of cancer-related death or the end of the follow-up period (5 years), death for other reasons led to censoring of data.
Immunohistochemistry
For immunohistochemical analysis of c-JUN and Fra-1, tissue microarray (TMA) slides of the patients from the three hospitals were used, containing 313 evaluable samples from formalin fixed, paraffin-embedded OSCC cases. EnVision system was used on the staining, described as previous studies40. The results were reviewed by two pathologists independently, and the discrepancies in immunostaining reviewing were solved by consensus. The stained slides were scanned using an Aperio Scanscope (Aperio, USA) and quantified with the available Aperio algorithms41, the original immunohistochemical staining of patients’ tissues the West China Hospital of Stomatology were showed in Supplementary Fig. S1. The immunostaining results of c-Jun and Fra-1 could be divided into high expression and low expression according to percent of cells stained and staining intensity8. The intensity of staining was scored as follows: 0, no color; 1, light yellow; 2, light brown; 3, brown. The number of positive cells was scored as follows: 0, <5%; 1, 5–25%; 2, 25–50%; 3, >50%. The two grades were multiplied together, producing scores from 0 to 9 that were classified as follows: weak staining (0–4 scores); strong staining (6–9 scores).
Statistical analysis
The differences in expression levels among different baseline characteristics of the patients were detected by the t test for continuous variables, the χ 2 test or Fisher’s exact test for categorical variables, the Kruskal-Wallis H test for ordinal variables. The correlation between the expression of c-Jun and Fra-1 was explored by Kendall’s tau. Overall survival (OS) at 5 years was evaluated by the Kaplan–Meier method, with the log-rank test in the univariate analysis between high and low expression of c-Jun and Fra-1. Multivariate survival analysis was performed with the Cox proportional hazards model, and the interaction effect of Fra-1 and c-Jun was tested, the Hazard Ratio (HR) of Cox model was used for evaluate the survival risk, which is the ratio of the hazard rates corresponding to the conditions described by two levels of an explanatory variable42. The concordance probability and receiver operating characteristic (ROC) curve area were chosen to validate the Cox model discrimination among different models with different independent variables43–45. Statistical analyses were performed in R packages (version 3.1.2), mostly with the “survival” package46. Unless stated otherwise, two-sided significance level was 0.05.
Results
Demographic characteristics
The median follow-up time of these OSCC patients was 21 months. 69% of these patients were male, which are twice as much as female. About 50 percent of them were over 60 years old. The immunostaining showed that most tumor cells had a very bright nuclear positive c-Jun expression, a nuclear and cytoplasm positive Fra-1 expression (Fig. 1). High level of c-Jun was detected in 72.5% of the patients, and high level of Fra-1 was detected in 57.2% of the patients from multi centers.
Figure 1.
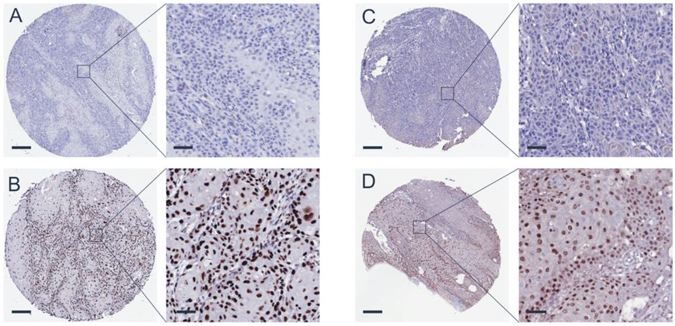
The immunohistochemical staining of c-Jun and Fra-1. Performed by EnVision system, protein immunoreactive substances were mainly displayed in the nucleus. (A) Low expression of c-Jun. Black scale bars at the bottom represent 200 µm (left side) or 50 µm (right side); (B) high expression of c-Jun; (C) low expression of Fra-1; (D) high expression of Fra-1.
Associations between the immunostaining results and the patients’ characteristics
Next, we evaluated the association of expression levels of two AP-1 subunits and other characteristics in OSCC. The results of χ 2 test, Fisher’s exact test and Kruskal-Wallis H test, showed that the expression status of c-Jun and Fra-1 varies among different tumor stages, nodal stages or clinical TNM stage (Table 1), and the differences were statistically significant. We also noticed that there was a trend that the association between and the tumor clinical TNM stage and the expressions of c-Jun and Fra-1 was positive, which showed that the higher TNM stages’ patients had higher level of the two protein expressions. In the contrary, gender, age, smoking status, drinking status, primary sites of OSCC and cell differentiation are not associated with expression of c-Jun and Fra-1 at the 5% significance level.
Table 1.
The association between the expression of c-Jun and Fra-1 with the baseline characteristics of the patients with OSCC.
| Total (100%) | c-Jun | P value | Fra1 | P value* | ||||
|---|---|---|---|---|---|---|---|---|
| low expression | high expression | low expression | high expression | |||||
| NO. patients (all) | 313 (100) | 86 (27.5) | 227 (72.5) | 134 (42.8) | 179 (57.2) | |||
| gender | male | 213 (68.05) | 57 (26.76) | 156 (73.24) | 0.598 | 89 (41.78) | 124 (58.22) | 0.635 |
| female | 100 (31.95) | 29 (29) | 71 (71.00) | 45 (45.00) | 55 (55.00) | |||
| age | ≤60 yr | 159 (50.80) | 48 (30.19) | 111 (69.81) | 0.213 | 70 (44.03) | 89 (55.97) | 0.596 |
| >60 yr | 154 (49.20) | 38 (24.68) | 116 (75.32) | 64 (41.56) | 90 (58.44) | |||
| smoking | never | 167 (53.35) | 50 (29.94) | 117 (70.06) | 0.303 | 72 (43.11) | 95 (56.89) | 0.976 |
| ever | 146 (46.65) | 36 (24.66) | 110 (75.34) | 62 (42.47) | 84 (57.53) | |||
| drinking | never | 182 (58.15) | 53 (29.12) | 129 (70.88) | 0.521 | 78 (42.86) | 104 (57.14) | 0.917 |
| ever | 131 (41.85) | 33 (25.19) | 98 (74.81) | 56 (42.75) | 75 (57.25) | |||
| primary site | cheek | 52 (16.61) | 13 (25.00) | 39 (75.00) | 0.54 | 20 (38.46) | 32 (61.54) | 0.145 |
| tongue | 114 (36.42) | 35 (30.70) | 79 (69.30) | 60 (52.63) | 54 (47.37) | |||
| gum | 66 (21.09) | 15 (22.73) | 51 (77.27) | 26 (39.39) | 40 (60.61) | |||
| others | 81 (25.88) | 23 (28.4) | 58 (71.60) | 28 (34.57) | 53 (65.43) | |||
| cell differentiation | high | 193 (61.34) | 56 (29.17) | 137 (70.83) | 0.696 | 83 (42.71) | 110 (57.29) | 0.921 |
| moderate | 92 (29.39) | 22 (23.91) | 70 (76.09) | 39 (42.39) | 53 (57.61) | |||
| low | 28 (8.95) | 8 (28.57) | 20 (71.43) | 12 (42.86) | 16 (57.14) | |||
| tumor stage | T1 | 45 (14.38) | 17 (37.78) | 28 (62.22) | 0.002 | 22 (48.89) | 23 (51.11) | 0.029 |
| T2 | 139 (44.41) | 48 (34.53) | 91 (65.47) | 68 (48.92) | 71 (51.08) | |||
| T3 | 61 (19.49) | 9 (14.75) | 52 (85.25) | 24 (39.34) | 37 (60.66) | |||
| T4 | 68 (21.73) | 12 (17.65) | 56 (82.35) | 20 (29.41) | 48 (70.59) | |||
| nodal stage | N0 | 168 (53.67) | 57 (33.93) | 111 (66.07) | 0.011 | 81 (48.21) | 87 (51.79) | 0.023 |
| N1-3 | 145 (46.33) | 29 (20) | 116 (80) | 53 (36.55) | 92 (63.45) | |||
| clinical TMN stage | I | 37 (11.82) | 18 (48.65) | 19 (51.35) | 0.003 | 24 (64.86) | 13 (35.14) | 0.001 |
| II | 94 (30.03) | 30 (31.91) | 64 (68.09) | 46 (48.94) | 48 (51.06) | |||
| III | 127 (40.58) | 30 (23.62) | 97 (76.38) | 52 (40.94) | 75 (59.06) | |||
| IV | 55 (17.57) | 8 (14.55) | 47 (85.45) | 12 (21.82) | 43 (78.18) | |||
*The p value was generated using χ2 test, Fisher’s exact test and Kruskal-Wallis H test.
As appeared in Table 1, the distribution trend of c-Jun expression among different population characteristics was similar with Fra-1. The Kendall’s tau was calculated for evaluating the correlation between the expression of c-Jun and Fra-1. And the tau was 0.313 (P < 0.001), indicated that there was a positive correlation between the expression of c-Jun and Fra-1 in the OSCC.
Univariate analysis of the patients’ overall survival
The overall survival at 5 years was 0.345 (95%CI: 0.286, 0.416) in the multicenter cohort, and different groups of cohorts owned different 5 years OS, but some of them had no statistical significances (P > 0.05 under the log-rank test), as shown in Table 2. The 5 years OS of the group without lymphatic metastasis (0.451, 95%CI: 0.364, 0.560) was higher than in the group with lymphatic metastasis (0.238, 95%CI: 0.171, 0.332, log-rank test P < 0.001). And from the Table 2 we could also see that the groups with lower tumor clinical TMN stage had the higher 5 years OS than the groups with higher tumor clinical TMN stage (P = 0.001). The 5 years OS values in different groups of genders, age groups, smoking status, drinking status, primary sites of tumor, cell differentiation or tumor stages had no statistical differences.
Table 2.
The Overall Survival (OS) at 5 years in each subgroup population with OSCC.
| OS at 5 years (95%CI) | p value* | ||
|---|---|---|---|
| gender | male | 0.326 (0.257, 0.413) | 0.547 |
| female | 0.384 (0.283, 0.52) | ||
| age | ≤60 yr | 0.298 (0.221, 0.402) | 0.851 |
| >60 yr | 0.402 (0.319, 0.505) | ||
| smoking | never | 0.339 (0.266, 0.431) | 0.624 |
| ever | 0.349 (0.258, 0.473) | ||
| drinking | never | 0.336 (0.261, 0.433) | 0.742 |
| ever | 0.355 (0.267, 0.471) | ||
| primary site | cheek | 0.408 (0.262, 0.637) | 0.176 |
| tongue | 0.33 (0.232, 0.471) | ||
| gum | 0.331 (0.223, 0.492) | ||
| others | 0.318 (0.224, 0.451) | ||
| cell differentiation | high | 0.336 (0.264, 0.429) | 0.33 |
| moderate | 0.408 (0.298, 0.558) | ||
| low | 0.252 (0.13, 0.49) | ||
| tumor stage | T1 | 0.397 (0.26, 0.606) | 0.068 |
| T2 | 0.399 (0.311, 0.512) | ||
| T3 | 0.295 (0.179, 0.486) | ||
| T4 | 0.243 (0.145, 0.409) | ||
| nodal stage | N0 | 0.451 (0.364, 0.56) | <0.001 |
| N1-3 | 0.238 (0.171, 0.332) | ||
| clinical TMN stage | I | 0.58 (0.425, 0.791) | 0.001 |
| II | 0.413 (0.301, 0.566) | ||
| III | 0.26 (0.181, 0.375) | ||
| IV | 0.259 (0.155, 0.434) | ||
| c-Jun | Low expression | 0.555 (0.445, 0.693) | <0.001 |
| High expression | 0.258 (0.193, 0.345) | ||
| Fra-1 | Low expression | 0.543 (0.45, 0.655) | <0.001 |
| High expression | 0.191 (0.13, 0.282) |
* p value was generated using log-rank test.
The 5 years OS value (0.258, 95%CI: 0.193, 0.345) in the group with high level expression of c-Jun was lower than the value (0.555, 95%CI: 0.445, 0.693) in the group with low level expression (P < 0.001). Contrasted with the group with low expression of c-Jun, the survival risk of the high level expression group is higher, which the HR of high level expression is 2.295(95%CI: 1.552, 3.394). Analogously, the group with high level expression of Fra-1 owned a lower 5 years OS value than the group with the low-level expression, the HR of high level expression was 2.789 (95% CI: 1.994, 3.899) contrasted with the low expression of Fra-1, which mean higher survival risk. The survival curves of OSCC patients with different expression of c-Jun and Fra-1 were showed in Fig. 2 (the survival curves of OSCC patients separated by tumor clinical TNM stages could be found in Supplementary Fig. S2).
Figure 2.
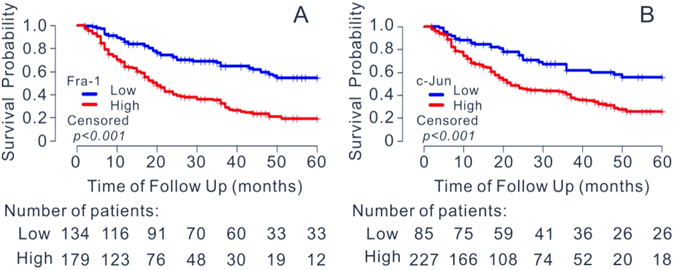
Survival curves of OSCC patients with different expression of c-Jun and Fra-1. (A) Correlation between 5-yr survival rate with the expression of Fra-1. (B) Correlation between 5-yr survival rate with the expression of c-Jun. The survival curves were defined by the Kaplan–Meier method, the tests of survival rates were performed by log-rank test.
Multivariate analysis of patients’ overall survival
To evaluate the influence of c-Jun and Fra-1 on the 5 years OS, multivariate Cox models were performed which were adjusted for demographic and clinical characteristics. One of them was considered the interaction effect of c-Jun and Fra-1, results were provided in Table 3. In the model without interaction factors, the group with high expression of Fra-1 had a poor prognosis compared to the group with low level expression of Fra-1, and the HR was 2.424 (95%CI: 1.651, 3.559). However, there was no statistical significance in the effect of c-Jun (P = 0.092). Since both c-Jun and Fra-1 are the subunits of the AP-1, we next evaluated the interaction effect of c-Jun and Fra-1. The model with interaction factor showed that the high expression of c-Jun and Fra-1 resulted in higher risk of death, and the HRs were 2.511(95%CI: 1.345, 4.688) and 5.625(95%CI: 2.685, 11.788) respectively, suggesting that both factors could promote progression OSCC. And the interaction effect between c-Jun and Fra-1 was 0.333 (95%CI: 0.144, 0.769), suggesting that the effect of c-Jun on the prognosis of OSCC could be obstructed by Fra-1, and vice versa (the interaction prognostic effects between c-Jun and Fra-1 during each clinical stage were showed in Supplementary Fig. S3). When the expression of c-Jun and Fra-1 was both high, the hazard risk of them was 4.703(2.511* 5.625 * 0.333), the interaction effect of c-Jun and Fra-1 was antagonism. The differences in the effects of c-Jun and Fra-1 between the Cox model without interaction factor and the Cox model with the interaction factor may be caused by the interaction effect between the c-Jun and Fra-1. The results in the model with the interaction factor would be chosen as it was comprehensive.
Table 3.
Multivariate analysis of OSCC patients’ overall survival at 5 years.
| Multivariate Cox | Multivariate Cox with interaction | ||||
|---|---|---|---|---|---|
| HR (95%CI) | P value* | HR (95%CI) | P value* | ||
| c-Jun | Low expression | 1 ref | 0.092 | 1 ref | 0.004 |
| High expression | 1.463 (0.940, 2.278) | 2.511 (1.345, 4.688) | |||
| Fra-1 | Low expression | 1 ref | <0.001 | 1 ref | <0.001 |
| High expression | 2.424 (1.651, 3.559) | 5.625 (2.685, 11.788) | |||
| c-Jun*Fra-1 | at least one is low | — | 1 ref | 0.01 | |
| both are high | — | 0.333 (0.144, 0.769) | |||
| gender | male | 1 ref | 0.652 | 1 ref | 0.629 |
| female | 0.905 (0.585, 1.398) | 0.898 (0.58, 1.39) | |||
| age | ≤60 yr | 1 ref | 0.881 | 1 ref | 0.816 |
| >60 yr | 0.976 (0.708, 1.345) | 0.963 (0.699, 1.325) | |||
| smoking | never | 1 ref | 0.634 | 1 ref | 0.434 |
| ever | 0.906 (0.603, 1.360) | 0.85 (0.566, 1.277) | |||
| drinking | never | 1 ref | 0.39 | 1 ref | 0.462 |
| ever | 0.828 (0.539, 1.273) | 0.849 (0.55, 1.312) | |||
| primary site | cheek | 1 ref | 1 ref | ||
| tongue | 1.906 (1.144, 3.176) | 0.013 | 2.018 (1.211, 3.364) | 0.007 | |
| gum | 1.517 (0.865, 2.659) | 0.146 | 1.662 (0.945, 2.924) | 0.078 | |
| others | 1.740 (1.042, 2.905) | 0.034 | 1.847 (1.104, 3.091) | 0.019 | |
| cell differentiation | high | 1 ref | 1 ref | ||
| moderate | 0.832 (0.565, 1.224) | 0.35 | 0.843 (0.573, 1.242) | 0.388 | |
| low | 1.338 (0.791, 2.266) | 0.278 | 1.437 (0.844, 2.445) | 0.182 | |
| tumor stage | T1 | 1 ref | 1 ref | ||
| T2 | 0.742 (0.404, 1.363) | 0.336 | 0.8 (0.431, 1.483) | 0.478 | |
| T3 | 0.661 (0.333, 1.312) | 0.236 | 0.725 (0.36, 1.458) | 0.367 | |
| T4 | 0.998 (0.495, 2.011) | 0.995 | 1.021 (0.503, 2.073) | 0.955 | |
| nodal stage | N0 | 1 ref | 0.018 | 1 ref | 0.018 |
| N1-3 | 2.012 (1.128, 3.590) | 2.015 (1.128, 3.599) | |||
| clinical TMN stage | I | 1 ref | 1 ref | ||
| II | 1.815 (0.814, 4.045) | 0.145 | 1.677 (0.743, 3.782) | 0.213 | |
| III | 1.382 (0.551, 3.462) | 0.49 | 1.268 (0.502, 3.207) | 0.616 | |
| IV | 1.084 (0.382, 3.078) | 0.879 | 1.029 (0.357, 2.965) | 0.958 | |
However, the other factors in the models were not influenced by the interaction effect, the effects of the other factors which consisted by the demographic and clinical characteristics were all similarity between the two models. The primary sites of tumor and the lymphatic metastasis were another two factors with statistical significance on the prognosis of OSCC among the demographic and clinical characteristics. The patients were more likely to die if their primary sites of tumor were tongue and others, contrasted with the group with the primary site of tumor in cheek, the HRs were 2.018(95%CI: 1.211, 3.364) and 1.847(95%CI: 1.104, 3.091) in the model with interaction factor, respectively. The lymphatic metastasis could also lead to the worse prognosis, and the HR was 2.012(95%CI: 1.128, 3.590).
Evaluation of the prognostic effects of c-Jun and Fra-1
Two multivariate Cox models were performed to estimate the effects of c-Jun and Fra-1 on the 5 years OS with adjusting for the confounding bias. To evaluate the predictive value of the two proteins, concordance probability and ROC were used to assess the discriminatory power and the predictive value of the two Cox models, especially the model with interaction factor. The higher concordance probability and the area under the ROC (AUC) of the model, the higher discriminatory power and predictive value of the model would be, and the higher predictive value on the prognosis of c-Jun and Fra-1 would be. The concordance probability of the Cox model without or with interaction factor were 0.689(95%CI: 0.641, 0.737) and 0.699(95%CI: 0.651, 0.747), and the ROC plots were showed in Fig. 3, the AUC of the model without interaction factor was 0.752(95%CI: 0.699, 0.805), and the other was 0.766(95%CI: 0.742, 0.818), they were all higher than 0.5. So the Cox models with c-Jun and Fra-1 owned high predictive value of the prognosis of OSCC. Besides, the concordance probability and AUC of the model with interaction factor were both higher than the values of the model without interaction factor, even though there was no statistical significance(P > 0.05). It suggested that the Cox model considered the interaction effect of c-Jun and Fra-1 was more valued than the model without interaction factor. the interaction factor which consisted by c-Jun and Fra-1, played an important role in the prognosis of OSCC, the same as the c-Jun and Fra-1.
Figure 3.
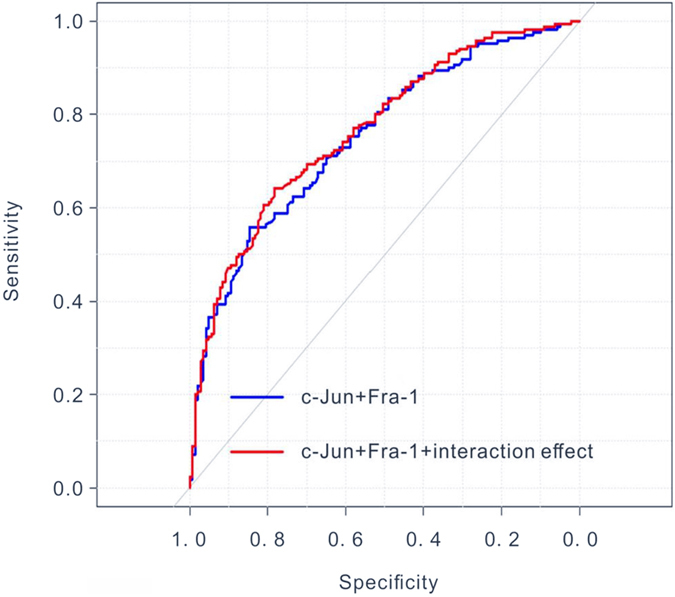
ROC curves for overall survival of OSCC patients with or without interaction effect of c-Jun and Fra-1.
Discussion
AP-1 is involved in a wide range of cellular events, such as cell growth, proliferation, differentiation and apoptosis16, 17. It consists of various dimers of either homodimers or heterodimers. c-Jun and Fra-1 are the two most important subunits of AP-1. and c-Jun can regulate the cell processes as homodimers or heterodimers, but Fra1 needs to combine with JUN proteins to be functional29, 33. They both are reported to be cancer promoters20. However, few studies were focused on the associations between the two proteins and survival prognosis of OSCC with multi cohort study. This study was designed to explore the prognostic value of the two proteins on the survival prognosis of OSCC patients, and detect the interaction effect between c-Jun and Fra-1, by following up three cohorts in China. Three cohorts included 313 postoperative patients, were respectively located in the north, west and south of China, which could minimize the selection bias of patients.
According to the log-rank test, the population with high expression level of c-Jun or Fra-1 own lower survival rates. The demographic characters and clinical characters of patients were adjusted in the Cox model to reduce the confounding bias, such as gender, age, and smoking status, drinking status, tumor differentiation, clinical TNM stage and the primary site of tumor. Several Cox models have been made with different explanatory variables, after that AUC and concordance probability of the Cox model were used to establish the best model, which also help us to find the significance of the interaction effect between c-Jun and Fra-145, 47. From the statistical analysis, the population of OSCC with high expression levels of the two proteins have worse survival prognosis, the hazard of survival in the population with high expression of c-Jun and Fra-1 were all higher than the low expression. It indicated that the c-Jun and Fra-1 are both valuable prognostic biomarkers in OSCC. However, the Cox model showed that the HR of high expression of c-Jun in the model is lower than the Fra-1, which suggested the high expression of Fra-1 would result in worse prognosis than the high expression of c-Jun in OSCC. It differs from Robert Eferl (2003)20, who reported c-Jun may have stronger transforming activity, our study indicates that Fra-1 may play a more important role in OSCC.
Besides, the AUC and concordance probability confirmed the model with interaction factor is the best predictive and discrimination model in this study, which means the interaction effect of c-Jun and Fra-1 exist truly in OSCC. And the interaction effect is antagonism, the hazard in the situation which c-Jun and Fra-1 are both at the high expression is lower than situation which only Fra-1 is at high expression for the OSCC patients. To date, the antagonistic prognostic effect of c-Jun and Fra-1 on OSCC patients has not been reported in other studies, as tumorigenesis of c-Jun and Fra-1 is not fully understood, the mechanism of this antagonism is still unclear. Maybe they are competing for binding to the AP-1 sites or by forming “inactive” heterodimers when they are all at high expression15, 39 and the cellular biological process of c-Jun and Fra-1 are complex, they may be also influenced by tumor or tissue types20, 48, 49.
The classification of the TMA staining was based on the percent of positive cells and staining intensity in our study, while c-Jun and Fra-1 also present in cytoplasm, for example, in certain types of cancers including breast, lung and thyroid cancer cytoplasmic Fra-1 over-expression has been reported, and there is also evidence showing that Fra-1 and c-Fos support growth of human malignant breast tumors by activating membrane biogenesis at the cytoplasm50. Specifically, in HNSCC, Serewko et al. describes Fra-1 expressed predominantly in nuclear51, similar in our study, c-Jun and Fra-1 were mainly appeared in the nucleus. As dispute exists37, it would be necessary to present the cytoplasm and nuclear expressions of these two proteins in HNSCC and their correlations with OSCC patients’ survival in the future study.
To further validated the results displayed by this study, studies with larger sample size of OSCC patients in different counties are needed. And to fully understand the mechanisms of possible antagonistic effect between c-JUN and Fra-1, functional evidences are also needed, more in vivo and in vitro experimental studies should be conducted. And it will be valuable to perform the longitudinal study to explore whether the expression of these proteins would be changed during the tumor progression.
In summary, c-Jun and Fra-1 could be another two valuable prognostic biomarkers in OSCC, and they may help to know more about the prognosis of OSCC. Meanwhile, we could try to find some new diagnostic methods and treatments through these two biomarkers. Besides, this study also indicated that the transforming activity of the AP-1 subunits could be influenced by each other, the interaction of tumor biomarkers may provide a new sight for the studies of tumor prognosis and tumor treatment in OSCC22, 28.
Electronic supplementary material
Acknowledgements
We thank the National Natural Science Foundations of China (document no.: 81621062, 81472533, 81500855, 81602376, 81520108009), and 111 Project of MOE China (B14038) and the Open Foundation (SKLOD2016OF01) from the State Key Laboratory of Oral Diseases Sichuan University for the financial support. We also thank the West China Hospital of Stomatology (Chengdu, China), Guangdong Provincial Stomatological Hospital (Guangzhou, China) and the General Hospital of the People’s Liberation Army (Beijing, China) for the supports. And we also appreciate the participating of all the patients.
Author Contributions
H.X. and Q.C. designed the study, X.J., L.J. and X.Z. collected the samples and recorded the follow-up information. X.J., Y.Y. and P.D. performed the experimental work. H.X. and X.L. did the statistical analysis. H.X., X.J., Z.W. and Q.C. wrote the manuscript. All of the authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Hao Xu and Xin Jin contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-05106-5
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhi-Yong Wang, Email: zhiyong.wang@scu.edu.cn.
Qian-Ming Chen, Email: qmchen@scu.edu.cn.
References
- 1.Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Podlodowska J, Szumiło J, Podlodowski W, Starosławska E, Burdan F. [Epidemiology and risk factors of the oral carcinoma] Polski merkuriusz lekarski: organ Polskiego Towarzystwa Lekarskiego. 2012;32:135–137. [PubMed] [Google Scholar]
- 4.Noguti J, et al. Metastasis from oral cancer: an overview. Cancer Genomics-Proteomics. 2012;9:329–335. [PubMed] [Google Scholar]
- 5.Panzarella V, et al. Diagnostic delay in oral squamous cell carcinoma: the role of cognitive and psychological variables. International journal of oral science. 2014;6:39–45. doi: 10.1038/ijos.2013.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avirovic M, et al. Osteopontin expression is an independent factor for poor survival in oral squamous cell carcinoma: a computer-assisted analysis on TMA sections. J Oral Pathol Med. 2013;42:620–626. doi: 10.1111/jop.12055. [DOI] [PubMed] [Google Scholar]
- 7.Bitu CC, et al. HOXA1 is overexpressed in oral squamous cell carcinomas and its expression is correlated with poor prognosis. BMC cancer. 2012;12:1. doi: 10.1186/1471-2407-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, et al. Associations between proteasomal activator PA28γ and outcome of oral squamous cell carcinoma: Evidence from cohort studies and functional analyses. EBioMedicine. 2015;2:849–856. doi: 10.1016/j.ebiom.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, et al. Tissue microarray analysis reveals the expression and prognostic significance of phosphorylated Akt Thr308 in oral squamous cell carcinoma. Oral surgery, oral medicine, oral pathology and oral radiology. 2013;116:591–597. doi: 10.1016/j.oooo.2013.06.031. [DOI] [PubMed] [Google Scholar]
- 10.Armitage JN, van der Meulen JH. Identifying co-morbidity in surgical patients using administrative data with the Royal College of Surgeons Charlson Score. British Journal of Surgery. 2010;97:772–781. doi: 10.1002/bjs.6930. [DOI] [PubMed] [Google Scholar]
- 11.Chuang J-Y, et al. Syk/JNK/AP-1 Signaling Pathway Mediates Interleukin-6-Promoted Cell Migration in Oral Squamous Cell Carcinoma. International Journal of Molecular Sciences. 2014;15:545–559. doi: 10.3390/ijms15010545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leelahavanichkul K, et al. A role for p38 MAPK in head and neck cancer cell growth and tumor-induced angiogenesis and lymphangiogenesis. Mol Oncol. 2014;8:105–118. doi: 10.1016/j.molonc.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mishra, A. Curcumin modulates cellular AP-1, NF-kB, and HPV16 E6 proteins in oral cancer. ecancermedicalscience9, doi:10.3332/ecancer.2015.525 (2015). [DOI] [PMC free article] [PubMed]
- 14.Bian Y, et al. MEK inhibitor PD-0325901 overcomes resistance to CK2 inhibitor CX-4945 and exhibits anti-tumor activity in head and neck cancer. Int J Biol Sci. 2015;11:411–422. doi: 10.7150/ijbs.10745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. Journal of cell science. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 16.Shaulian, E. & Karin, M. AP-1 in cell proliferation and survival. Oncogene20 (2001). [DOI] [PubMed]
- 17.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nature cell biology. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen P, et al. The FGFR1 inhibitor PD173074 induces mesenchymal–epithelial transition through the transcription factor AP-1. British journal of cancer. 2013;109:2248–2258. doi: 10.1038/bjc.2013.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rebollo A, et al. Bcl-3 expression promotes cell survival following interleukin-4 deprivation and is controlled by AP1 and AP1-like transcription factors. Molecular and cellular biology. 2000;20:3407–3416. doi: 10.1128/MCB.20.10.3407-3416.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 21.Lin C-W, et al. Kaempferol reduces matrix metalloproteinase-2 expression by down-regulating ERK1/2 and the activator protein-1 signaling pathways in oral cancer cells. PLoS One. 2013;8:e80883. doi: 10.1371/journal.pone.0080883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matthews CP, Colburn NH, Young MR. AP-1 a target for cancer prevention. Current cancer drug targets. 2007;7:317–324. doi: 10.2174/156800907780809723. [DOI] [PubMed] [Google Scholar]
- 23.Zenz R, et al. c-Jun regulates eyelid closure and skin tumor development through EGFR signaling. Developmental cell. 2003;4:879–889. doi: 10.1016/S1534-5807(03)00161-8. [DOI] [PubMed] [Google Scholar]
- 24.Oliveira-Ferrer L, et al. Prognostic impact of transcription factor Fra-1 in ER-positive breast cancer: Contribution to a metastatic phenotype through modulation of tumor cell adhesive properties. Journal of cancer research and clinical oncology. 2015;141:1715–1726. doi: 10.1007/s00432-015-1925-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao C, et al. Genome-wide profiling of AP-1-regulated transcription provides insights into the invasiveness of triple-negative breast cancer. Cancer Res. 2014;74:3983–3994. doi: 10.1158/0008-5472.CAN-13-3396. [DOI] [PubMed] [Google Scholar]
- 26.Eferl R, et al. Liver tumor development: c-Jun antagonizes the proapoptotic activity of p53. Cell. 2003;112:181–192. doi: 10.1016/S0092-8674(03)00042-4. [DOI] [PubMed] [Google Scholar]
- 27.Chang W-C, et al. PTX3 gene activation in EGF-induced head and neck cancer cell metastasis. Oncotarget. 2015;6:7741. doi: 10.18632/oncotarget.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang, X., Wu, J., Luo, S., Lechler, T. & Zhang, J. Y. FRA1 promotes squamous cell carcinoma growth and metastasis through distinct AKT and c-Jun dependent mechanisms. Oncotarget (2016). [DOI] [PMC free article] [PubMed]
- 29.Wisdom R, Johnson RS, Moore C. c‐Jun regulates cell cycle progression and apoptosis by distinct mechanisms. The EMBO journal. 1999;18:188–197. doi: 10.1093/emboj/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bossy‐Wetzel E, Bakiri L, Yaniv M. Induction of apoptosis by the transcription factor c‐Jun. The EMBO journal. 1997;16:1695–1709. doi: 10.1093/emboj/16.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schreiber M, et al. Control of cell cycle progression by c-Jun is p53 dependent. Genes & development. 1999;13:607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wisdon R, Verma IM. Transformation by Fos proteins requires a C-terminal transactivation domain. Molecular and cellular biology. 1993;13:7429–7438. doi: 10.1128/MCB.13.12.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wenzel A, et al. Fra‐1 substitutes for c‐Fos in AP‐1‐mediated signal transduction in retinal apoptosis. Journal of neurochemistry. 2002;80:1089–1094. doi: 10.1046/j.0022-3042.2002.00807.x. [DOI] [PubMed] [Google Scholar]
- 34.Jochum W, et al. Increased bone formation and osteosclerosis in mice overexpressing the transcription factor Fra-1. Nature medicine. 2000;6:980–984. doi: 10.1038/79676. [DOI] [PubMed] [Google Scholar]
- 35.Boeckx C, et al. Establishment and characterization of cetuximab resistant head and neck squamous cell carcinoma cell lines: focus on the contribution of the AP-1 transcription factor. American journal of cancer research. 2015;5:1921. [PMC free article] [PubMed] [Google Scholar]
- 36.Bedal KB, et al. Collagen XVI induces expression of MMP9 via modulation of AP-1 transcription factors and facilitates invasion of oral squamous cell carcinoma. PLoS One. 2014;9:e86777. doi: 10.1371/journal.pone.0086777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mangone FR, et al. Overexpression of Fos‐related antigen‐1 in head and neck squamous cell carcinoma. International journal of experimental pathology. 2005;86:205–212. doi: 10.1111/j.0959-9673.2005.00423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Sousa SOM, Mesquita RA, Pinto DS, Gutkind S. Immunolocalization of c‐Fos and c‐Jun in human oral mucosa and in oral squamous cell carcinoma. Journal of oral pathology & medicine. 2002;31:78–81. doi: 10.1046/j.0904-2512.2001.10012.x. [DOI] [PubMed] [Google Scholar]
- 39.Deng T, Karin M. JunB differs from c-Jun in its DNA-binding and dimerization domains, and represses c-Jun by formation of inactive heterodimers. Genes & development. 1993;7:479–490. doi: 10.1101/gad.7.3.479. [DOI] [PubMed] [Google Scholar]
- 40.Sabatini E, Bisgaard K, Ascani S. The EnVisionTM+ system: a new immunohistochemical method for diagnostics and research. Critical comparison with the APAAP, ChemMateTM, CSA, LABC, and SABC techniques. J Clin Pathol. 1998;51:506–511. doi: 10.1136/jcp.51.7.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olson, A. H. Image analysis using the Aperio ScanScope. Technical manual. Aperio Technologies Inc (2006).
- 42.Fox, J. Cox proportional-hazards regression for survival data. An R and S-PLUS companion to applied regression 1–18 (2002).
- 43.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 44.DeLong, E. R., DeLong, D. M. & Clarke-Pearson, D. L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics, 837–845 (1988). [PubMed]
- 45.Gönen M, Heller G. Concordance probability and discriminatory power in proportional hazards regression. Biometrika. 2005;92:965–970. doi: 10.1093/biomet/92.4.965. [DOI] [Google Scholar]
- 46.Therneau, T. M. & Lumley, T. Package survival. 2–37 (2017).
- 47.Fawcett T. An introduction to ROC analysis. Pattern recognition letters. 2006;27:861–874. doi: 10.1016/j.patrec.2005.10.010. [DOI] [Google Scholar]
- 48.Kappelmann M, Kuphal S, Meister G, Vardimon L, Bosserhoff AK. MicroRNA miR-125b controls melanoma progression by direct regulation of c-Jun protein expression. Oncogene. 2012;32:2984–2991. doi: 10.1038/onc.2012.307. [DOI] [PubMed] [Google Scholar]
- 49.Li B, et al. Mitogen-and stress-activated Kinase 1 mediates Epstein-Barr virus latent membrane protein 1-promoted cell transformation in nasopharyngeal carcinoma through its induction of Fra-1 and c-Jun genes. BMC cancer. 2015;15:1. doi: 10.1186/1471-2407-15-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Motrich RD, Castro GM, Caputto BL. Old players with a newly defined function: Fra-1 and c-Fos support growth of human malignant breast tumors by activating membrane biogenesis at the cytoplasm. PLoS One. 2013;8:e53211. doi: 10.1371/journal.pone.0053211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Serewko MM, et al. Alterations in gene expression and activity during squamous cell carcinoma development. Cancer research. 2002;62:3759–3765. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


