Abstract
The study was performed on a male European bison (Bison bonasus bonasus L.) foetus spontaneously aborted at the fourth or fifth month of pregnancy in the Białowieża Forest. Serum samples from the foetus and mother revealed the presence of antibodies against T. gondii (S/P% = 88% and 75%, respectively). Mobile extracellular tachyzoites were first observed in a Vero cell culture, 110 days following inoculation of brain homogenate. PCR amplification with TGR1E1 and TGR1E2 primers confirmed the presence of T. gondii DNA, which was classified as Type I by PCR-RFLP genotyping. The sequences of 18S ribosomal RNA (18S rRNA) and 5.8S ribosomal RNA (5.8S rRNA) genes; internal transcribed spacer 1 (ITS1) and internal transcribed spacer 2 (ITS2), obtained from T. gondii isolate, have been deposited in GenBank (accession number KX459518.1). This is the first in vitro isolation and molecular identification of T. gondii from an aborted European bison foetus. The origin of this protozoan isolate indicates that the species is a significant threat to the European bison conservation program implemented in the Białowieża Forest.
Keywords: Toxoplasma gondii, Isolation, Genetic characterization, European bison, Aborted foetus, Białowieża Forest
Introduction
Toxoplasma gondii is one of the most important parasites of man and animals. It is a widely prevalent coccidian parasite capable of both sexual and asexual reproduction, and it is this flexibility that enables it to infect any warm-blooded animals as intermediate hosts (Dubey 2010). Transmission of T. gondii occurs via the faecal-oral route, through the consumption of infected meat, and by congenital transmission from mother to foetus (Dubey 2010; Guo et al. 2015).
The European bison (Bison bonasus) is the largest herbivore, and the heaviest living wild land animal, in Europe. The species is protected both by international and national laws. The species has been assigned endangered status by the IUCN Red List, and has been selected as a priority species under the EU Habitat Directive. The bison population at the Białowieża Forest consists of a group of animals maintained in captivity, and a group of animals living in the wild, and these are conserved within the framework of a European program (Krasiński et al. 2011). The conservation and management of European bison is aimed at increasing the number of animals, continuing the reintroduction process, and preserving the genetic diversity of captive and free herds (Pucek et al. 2004). Such actions might include legal protection in every country, according to its current status on Red Lists or Red Data Books, and the creation of free-ranging populations within the territories of national parks or reserves. The European bison in the Białowieża Forest play an important role in the restitution program and protection of the species.
The European bison and the American bison [Bison bison] are related to domestic cattle (Bos taurus) and they can interbreed. Both species of bison and cattle are considered resistant to T. gondii infection (Dubey 2010). Although T. gondii can be transmitted transplacentally in cattle, there is no confirmed report of clinical toxoplasmosis in cattle (Dubey 2010). With respect to T. gondii infection in European bison, antibodies to T. gondii were found in 24 of 95 (23.5%) of free-ranging bison in Poland (Majewska et al. 2014) and all of four captive European bison living in a zoo in the Czech Republic (Sedlák and Bártová 2006). Very low seropositivity has been reported for the American bison: T. gondii antibodies have been found in three (3.1%) of 93 bison from Montana (Dubey 1985), and two (0.8%) of 241 from Alaska (Zarnke et al. 2000). Experimentally, a 1-day-old bison fed oocysts of virulent T. gondii became infected but remained asymptomatic (Dubey 1983).
The present paper describes the first isolation and genetic characterization of T. gondii from the foetus of a European bison.
Materials and methods
Animals and source of samples
During a routine investigation, a European bison was found to have spontaneously aborted a male foetus with a gestational age of 4 to 5 months in the Białowieża Forest, Poland in 2014. The foetus was necropsied in the field by the attending veterinarian, and fresh samples of brain and blood of the foetus, and blood from the cow were sent to our laboratory for diagnosis. After separation, the serum from the cow was stored at −20 °C until analysis for antibodies to T. gondii and N. caninum.
Serological examination
The serum sample was analysed for the presence of antibodies to T. gondii using a multi-species ID Screen Toxoplasmosis Indirect kit (IDvet, Montpellier) and for N. caninum using an ELISA kit (IDEXX Laboratories Inc., Westbrook, ME, USA). The analysis was performed according to the manufacturer’s instructions.
Parasite isolation
The foetus brain was homogenized in PBS and the homogenate incubated in PBS with 0.25% trypsin at 37 °C for 1 h, with constant shaking. The suspension was filtered through sterilized gauze and centrifuged at 400 g for 10 min. The supernatant was then discarded and the pellet washed four times with sterile PBS. The sediment was suspended in approximately 10 ml of PBS and seeded into monolayer Vero cell culture flasks.
The Vero cell monolayers were cultured at 37 °C with 5% CO2, in RPMI 1640 medium (Sigma) supplemented with 1% horse serum, 100 μg/ml penicillin, and 100 μl/ml streptomycin. The culture medium was changed each week. Every 3 or 4 weeks, the Vero cells were scraped from the primary flask and transferred to a new culture flask. After detection of the parasites, the cultivation of Vero cells was continued and the parasites were passaged every 2 weeks to new culture flask. The tachyzoites from the successfully grown cell culture were frozen at −80 °C for further investigation.
DNA extraction
Total DNA was isolated from the genomic DNA of the purified tachyzoites cultured in Vero cells (Macharey-Nagel, Germany) according to the manufacturer’s instructions. The DNA was eluted in 50 μl of distilled water, and its concentration was determined using a NanoDrop ND 1000 Spectrophotometer (NanoDrop Technologies, USA).
PCR amplification procedures
Amplification with TGR1E1 and TGR1E2 primers
Amplification of the isolated DNA was carried out using standard PCR targeted at the T. gondii TGR1E gene region in accordance with the protocol described by Lamoril et al. (1996) with some modifications by Kornacka et al. (2016). The following specific primers were used: TGR1E1 (sense): 5′-ATG GTC CGG CCG GTG TAT GAT ATG CGA T, and TGR1E2 (antisense): 5′-TCC CTA CGT GGT GCC GCA TTG CCT. The final positive PCR product was 191 bp in size.
Amplification with Np21 and Np6 primers
The primers Np21 and Np6, based on the NC-5 region specific for N. caninum, were used in this study (Yamage et al. 1996).
DNA from Nc-1 N. caninum and T. gondii (RH strain) tachyzoites were used as positive or negative controls, respectively for the PCR protocol. Amplification products were analysed by electrophoresis through a 1.5% agarose gel stained with Gel Red (Nucleic Acid Gel Stain, Biotium) and visualised under UV light using ChemiDoc (Biorad, USA).
DNA sequencing and analysis
The sequence analysis was based on a DNA fragment of approximately 1000 bp amplified with primers NC18S and NC28S. The reaction was performed according to Vitale et al. (2013).
The PCR products were purified using the Clean-up Product Purification Kit (A&A Biotechnology, Poland) according to the manufacturer’s instructions, and then ligated into an pGEM-T easy cloning vector (Promega). Escherichia coli strain XL-1 Blue MRF electrocompetent cells (Promega) were used for cloning. Positive clones were identified by colony PCR with primers directed against vector sequences outside the multi-cloning site. Clones containing inserts were used for further examination. Positive plasmids were purified using GeneAll Exprep Plasmid SV mini (GeneAll, Korea) according to the manufacturer’s instructions.
The concentration of DNA was measured using a NanoDrop® ND-1000 Spectrophotometer (NanoDrop Technologies, USA). The DNA was then sequenced (Genomed, Poland).
Vector NTI® Advance 10 software (Invitrogen, Scotland) was used to assemble the sequence information from each of the isolated clones. The complete sequences were checked for similarities with published sequences in GenBank using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
Genetic characterization
PCR-RFLP genotyping was performed using ten genetic markers: SAG1, SAG2: (5′-3′SAG2, and alt.SAG2), SAG3, BTUB, GRA6, c22-8, c29-2, L358, PK1 and Apico as described previously (Su et al. 2010). Appropriate positive and negative controls were included in all batches. PCR-RFLP genotyping was conducted by USDA, ARS, Beltsville Maryland, USA.
Result
Macroscopic examinations of the aborted foetus revealed swelling of the subcutaneous head and trunk tissues. Enlarged, fragile, pale brick-coloured fatty degeneration was observed. Bacterial cultures and attempts of virus isolation have not confirmed the presence of these pathogens.
Serum samples from the foetus and mother revealed the presence of antibodies against T. gondii (S/P% = 88% and 75%, respectively). Anti-N. caninum antibodies were not detected. Mobile extracellular tachyzoites were first observed in Vero cell culture 110 days following inoculation of the brain homogenate from the aborted foetus.
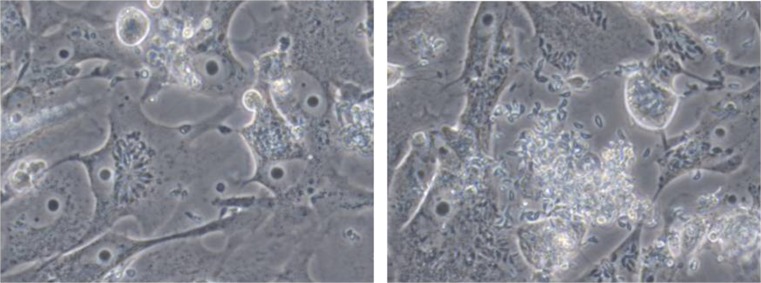
The tachyzoites were successfully propagated, and many were seen after several passages in cell cultures (Fig. 1).
Fig. 1.
Toxoplasma gondii tachyzoites successfully propagated after several passages in Vero cell cultures
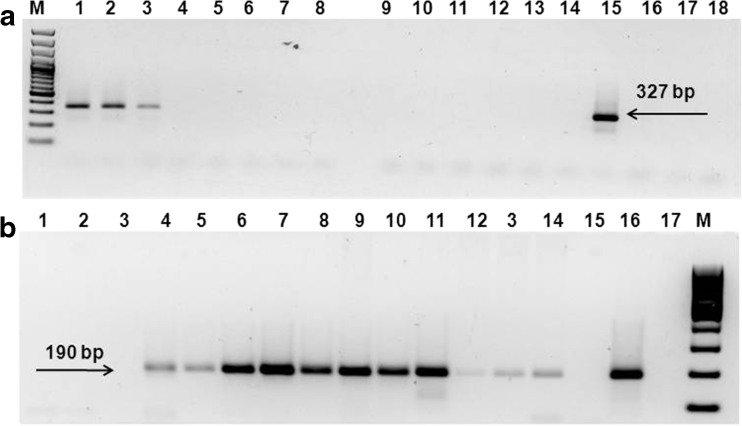
T. gondii DNA was confirmed in the tested samples by the presence of a specific 190 bp amplicon in the examined samples (Fig. 2); however, no 327 bp amplicon confirming the presence of N. caninum DNA was detected. PCR-RFLP analysis indicated a Type I genotype for samples 1 and 2 of the T. gondii tachyzoites propagated in cell culture (Table 1).
Fig. 2.
Specific amplicons with primers Np6/Np21 (a) and primers TGRE1/TGRE1-2 (b): M-molecular marker, lines 1–3—N. caninum DNA; lines 4–5—T. gondii RH DNA; lines 6–11 – tachyzoites from in vitro culture (Bison bonasus); lines 12–14—scrapings (Vero cells + tachyzoites); line 15—positive control—N. caninum DNA; line 16—positive control—DNA of T. gondii RH; lines 17–18—negative control
Table 1.
PCR-RFLP genotyping of T. gondii isolates from Bison bonasus
| Strain ID | ToxoDB PCR-RFLP Genotype# | Genetic markers | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SAG1 | (5′ + 3′) SAG2 | alt. SAG2 | SAG3 | BTUB | GRA6 | c22-8 | c29-2 | L358 | PK1 | Apico | ||
| GT-1 | #10 (Type I) | I | I | I | I | I | I | I | I | I | I | I |
| PTG | #1 (Type II) | II | II | II | II | II | II | II | II | II | II | II |
| CTG | #2 (Type III) | II/III | III | III | III | III | III | III | III | III | III | III |
| MAS | #17 | u-1 | I | II | III | III | III | u-1 | I | I | III | I |
| TgCgCa1 | #66 | I | II | II | III | II | II | II | u-1 | I | u-2 | I |
| TgCtBr5 | #19 | I | III | III | III | III | III | I | I | I | u-1 | I |
| TgCtBr64 | #111 | I | I | u-1 | III | III | III | u-1 | I | III | III | I |
| TgRsCr1 | #52 | u-1 | I | II | III | I | III | u-2 | I | I | III | I |
| Present study | ||||||||||||
| Sample 1 | #10 (Type I) | I | I | I | I | I | I | I | I | I | I | I or III |
| Sample 2 | #10 (Type I) | I | I | I | I | I | I | I | I | I | I | I or III |
The DNA sequences of the 18S ribosomal RNA (18S rRNA), 5.8S ribosomal RNA (5.8S rRNA), internal transcribed spacer 1 (ITS1) and internal transcribed spacer 2 (ITS2) genes obtained from T. gondii isolate Bb1 (isolate from Bison bonasus) have been deposited in GenBank with accession number KX459518.1.
A BLAST-N search using the T. gondii isolate Bb1 consensus sequence revealed 99% identity with other T. gondii sequences already published in GenBank: T. gondii strain YZ-2 (JQ235842.1), T. gondii strain YZ-1 (JQ235841.1), T. gondii RH (X75429.1) and 97% identity with T. gondii strain RH (L25635.1) and T. gondii isolate TG-CtPSU49 (KP895862.1).
Discussion
Toxoplasma gondii is a protozoan parasite of both medical and veterinary importance. The species is capable of causing infection and severe disease in both animals and humans. Clinical and subclinical toxoplasmosis has been reported in many host species (Dubey et al. 2007). However, natural T. gondii infection does not appear to cause clinical disease or abortion in cattle (Dubey 2010). Infected livestock and game animal species harbouring T. gondii tissue cysts represent a risk of infection to human consumers (Guo et al. 2015).
The isolation of T. gondii from the brain of the spontaneously aborted foetus confirms that transplacental transmission of the parasite is possible in Bison bonasus. However, as no histopathological examination was performed, it remains uncertain whether T. gondii was the cause of the abortion.
It has been documented that in Europe and North America the vast majority of T. gondii strains could be grouped into three lineages namely Types I, II, III (Howe and Sibley 1995; Dubey 2010). However, statistical analysis indicates significant difference among population in Africa, Asia, Central and South America (Shwab et al. 2014). It is significant that the T. gondii isolate in this study was found to be Type I, as these strains are rarely found in nature, perhaps because of their high virulence (Dubey 2010). Types I and II can cause congenital infection in humans, while types II and III are most prevalent in humans and animals in Europe. Interestingly, Type I strains have been isolated from a bovine foetus in Portugal (Canada et al. 2002) and a cow in the USA (Dubey 1992).
Studies have found that European bison are strict herbivores (Pucek et al. 2004; Krasiński et al. 2011). It is possible that the animal in question most likely became infected postnatally by ingesting food or water contaminated with oocysts excreted by felids. The only free-ranging felid in the Białowieża Forest is the Euroasian lynx (Lynx lynx), but its population density is not very high (Jędrzejewski et al. 2002). It has been documented that European bison feed on the meadows, pastures and cultivated fields located on the edges of the Białowieża Forest. It is possible that these environments may be contaminated with the faeces of the domestic and wild cats commonly present in this area. The high seroprevalence of T. gondii in European bison (25%) (Majewska et al. 2014) indicates the presence of environmental contamination with T. gondii oocysts shed by felids, combined with a lack of genetic variation and high level of inbreeding (Dubey 2010; Tokarska et al. 2011). Nothing is known of the prevalence of T. gondii in feral domestic and wild felids in Poland and needs investigation.
Toxoplasmosis and other such infections may represent a significant threat for the European bison conservation program (Pucek et al. 2004; Krasiński et al. 2011; Karbowiak et al. 2014). Due to the close relationship of cattle and bison, it is natural to assume that the two host species demonstrate similar clinical symptoms for this infection. Although congenital toxoplasmosis is rarely reported in cattle (Dubey 2010), some examples of reproductive disorders such as spontaneous abortion has been observed as a result of infection (Canada et al. 2002).
Conclusion
Our study describes the first molecular identification of a T. gondii isolate obtained from the aborted foetus of a European bison (Bison bonasus), and confirms that it had been acquired by transplacental transmission. Due to the origin of this protozoan isolate, we can assume that the species is a significant threat to the European bison conservation program implemented in the Białowieża Forest. It is important to note that both the aborted foetus and the carcasses of dead animals may represent potential sources of infection for many intermediate and definitive hosts living in this protected area.
Acknowledgements
We thank Drs. S. K. Verma and C. Su (United States Department of Agriculture, Agricultural Research Service, Beltsville Agricultural Research Center, Animal Parasitic Diseases Laboratory, Building 1001, Beltsville, MD 20705-2350, USA) for genotyping.
Authors’ contributions
BM conceived and designed the study area; AK, AC, JB, KR, KG performed experiments; MK collected samples; BM, JB, KR and WC analysed the data and wrote the manuscript. All authors read and approved the final manuscript.
Compliance with ethical standards
Competing interests
The authors declare that they have no competing interests.
References
- Canada N, Meireles CS, Rocha A, da Costa JM, Erickson MW, Dubey JP. Isolation of viable Toxoplasma gondii from naturally infected aborted bovine fetuses. J Parasitol. 2002;88(6):1247–1248. doi: 10.1645/0022-3395(2002)088[1247:IOVTGF]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Dubey JP. Experimental infection of a bison with Toxoplasma gondii oocysts. J Wildl Dis. 1983;19(2):148–149. doi: 10.7589/0090-3558-19.2.148. [DOI] [PubMed] [Google Scholar]
- Dubey JP. Serologic prevalence of toxoplasmosis in cattle, sheep, goats, pigs, bison, and elk in Montana. Am Vet Med Assoc. 1985;186(9):969–970. [PubMed] [Google Scholar]
- Dubey JP. Isolation of Toxoplasma gondii from a naturally infected beef cow. J Parasitol. 1992;78(1):151–153. doi: 10.2307/3283705. [DOI] [PubMed] [Google Scholar]
- Dubey JP. Toxoplasmosis of animals and humans. second. Boca Raton: CRC Press; 2010. [Google Scholar]
- Dubey JP, Sundar N, Nolden CA, Samuel MD, Velmurugan GV, Bandini LA, Kwok OC, Bodenstein B, Su C. Characterization of Toxoplasma gondii from raccoons (Procyon lotor), coyotes (Canis latrans), and striped skunks (Mephitis mephitis) in Wisconsin identified several atypical genotypes. J Parasitol. 2007;93(6):1524–1527. doi: 10.1645/GE-1245.1. [DOI] [PubMed] [Google Scholar]
- Guo M, Dubey JP, Hill D, Buchanan RL, Gamble HR, Jones JL, Pradhan AK. Prevalence and risk factors for Toxoplasma gondii infection in meat animals and meat products destined for human consumption. J Food Prot. 2015;78(2):457–476. doi: 10.4315/0362-028X.JFP-14-328. [DOI] [PubMed] [Google Scholar]
- Howe DK, Sibley DL. Toxoplasm gondii comprises three clonal lineages—correlation of parasite genotype with human disease. J Infect Dis. 1995;172(6):1561–1566. doi: 10.1093/infdis/172.6.1561. [DOI] [PubMed] [Google Scholar]
- Jędrzejewski W, Nowak S, Schmidt K, Jędrzejewska B. The wolf and the lynx in Poland—results of a census conducted in 2001. Kosmos. 2002;51(4):491–499. [Google Scholar]
- Karbowiak G, Demiaszkiewicz AW, Pyziel AM, Wita I, Moskwa B, Werszko J, Bień J, Goździk K, Lachowicz J, Cabaj W. The parasitic fauna of the European bison (Bison bonasus) (Linnaeus, 1758) and their impact on the conservation. Part 2. The structure and changes over time. Acta Parasitol. 2014;59(3):372–379. doi: 10.2478/s11686-014-0253-z. [DOI] [PubMed] [Google Scholar]
- Kornacka A, Cybulska A, Bień J, Goździk K, Moskwa B. The usefulness of direct agglutination test, enzyme-linked immunosorbent assay and polymerase chain reaction for the detection of Toxoplasma gondii in wild animals. Vet Parasitol. 2016;228(1):85–89. doi: 10.1016/j.vetpar.2016.08.010. [DOI] [PubMed] [Google Scholar]
- Krasiński ZA, Olech W, Perzanowski P, Bielecki W, Bereszyński A. Conservation plan for wisent Bison bonasus in Białowieski National Park. Eur Bison Conserv Newsl. 2011;4(1):101–116. [Google Scholar]
- Lamoril J, Molina JM, Gouvello AD, Garin YJ, Deybach JC, Modai J, Derouin F. Detection by PCR of Toxoplasma gondii in blood in the diagnosis of cerebral toxoplasmosis in patients with AIDS. J Clin Pathol. 1996;49(1):89–92. doi: 10.1136/jcp.49.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewska AC, Werner A, Cabaj W, Moskwa B. The first report of Toxoplasma gondii antibodies in free-living European bison (Bison bonasus bonasus Linnaeus) Folia Parasitol (Praha) 2014;61(1):18–20. [PubMed] [Google Scholar]
- Pucek Z, Belousova IP, Krasińska M, Krasiński ZA, Olech W. European bison. Status survey and conservation action plan. IUCN/SSC bison specialist group. Gland: IUCN; 2004. [Google Scholar]
- Sedlák K, Bártová E. Seroprevalences of antibodies to Neospora caninum and Toxoplasma gondii in zoo animals. Vet Parasitol. 2006;136(3–4):223–231. doi: 10.1016/j.vetpar.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Shwab EK, Zhu XQ, Majumdar D, Pena HF, Gennari SM, Dubey JP, Su C. Geographical patterns of Toxoplasma gondii genetic diversity revealed by multilocus PCR-RFLP genotyping. Parasitology. 2014;141(1):453–461. doi: 10.1017/S0031182013001844. [DOI] [PubMed] [Google Scholar]
- Su C, Shwab EK, Zhou P, Zhu XQ, Dubey JP. Moving towards an integrated approach to molecular detection and identification of Toxoplasma gondii. Parasitology. 2010;13(1):1–11. doi: 10.1017/S0031182009991065. [DOI] [PubMed] [Google Scholar]
- Tokarska M, Pertoldi C, Kowalczyk R, Perzanowski K. Genetic status of the European bison Bison bonasus after extinction in the wild and subsequent recovery. Mammal Rev. 2011;41(2):151–162. doi: 10.1111/j.1365-2907.2010.00178.x. [DOI] [Google Scholar]
- Vitale M, Galluzzo P, Currò V, Goździk K, Schillaci D, Presti VM. A high sensitive nested PCR for Toxoplasma gondii detection in animal and food samples. J Microb Biochem Technol. 2013;5:039–041. doi: 10.4172/1948-5948.1000097. [DOI] [Google Scholar]
- Yamage M, Flechtner O, Gottstein B. Neospora caninum: specific oligonucleotide primers for the detection of brain “cyst” DNA of experimentally infected nude mice by the polymerase chain reaction (PCR) J Parasitol. 1996;82(2):272–279. doi: 10.2307/3284160. [DOI] [PubMed] [Google Scholar]
- Zarnke RL, Dubey JP, Kwok OC, Ver Hoef JM. Serologic survey for Toxoplasma gondii in selected wildlife species from Alaska. J Wildl Dis. 2000;36(2):219–224. doi: 10.7589/0090-3558-36.2.219. [DOI] [PubMed] [Google Scholar]