Abstract
Lung adenocarcinoma is a multifactorial disease. MicroRNA (miRNA) expression profiles are extensively used for discovering potential theranostic biomarkers of lung cancer. This work proposes an optimized support vector regression (SVR) method called SVR-LUAD to simultaneously identify a set of miRNAs referred to the miRNA signature for estimating the survival time of lung adenocarcinoma patients using their miRNA expression profiles. SVR-LUAD uses an inheritable bi-objective combinatorial genetic algorithm to identify a small set of informative miRNAs cooperating with SVR by maximizing estimation accuracy. SVR-LUAD identified 18 out of 332 miRNAs using 10-fold cross-validation and achieved a correlation coefficient of 0.88 ± 0.01 and mean absolute error of 0.56 ± 0.03 year between real and estimated survival time. SVR-LUAD performs well compared to some well-recognized regression methods. The miRNA signature consists of the 18 miRNAs which strongly correlates with lung adenocarcinoma: hsa-let-7f-1, hsa-miR-16-1, hsa-miR-152, hsa-miR-217, hsa-miR-18a, hsa-miR-193b, hsa-miR-3136, hsa-let-7g, hsa-miR-155, hsa-miR-3199-1, hsa-miR-219-2, hsa-miR-1254, hsa-miR-1291, hsa-miR-192, hsa-miR-3653, hsa-miR-3934, hsa-miR-342, and hsa-miR-141. Gene ontology annotation and pathway analysis of the miRNA signature revealed its biological significance in cancer and cellular pathways. This miRNA signature could aid in the development of novel therapeutic approaches to the treatment of lung adenocarcinoma.
Introduction
Lung cancer has consistently been one of the most lethal cancers. Lung carcinomas are classified into either small-cell lung carcinomas (SCLC) or non-small cell lung carcinomas (NSCLC)1. Lung adenocarcinoma is the most common sub-type of NSCLC. Despite advances in cancer therapy, the 5-year survival rate of lung cancer is only 17.4%2. Due to the limitation of tumor detection using bronchoscopy and computed tomography techniques3, 4, poor early stage detection of lung tumor is a major obstacle to recovery. Therefore, there is a great need of treatment options for NSCLC diagnosis. For accurate detection and potential diagnosis during the NSCLC’s early stage, it is necessary to identify the molecular signature associated with patient survival which may assist in the development of gene target based therapy.
Microarray methods for large-scale analysis of gene expression have helped to systematically identify the molecular biomarkers of cancers5, 6. Microarray technology is one of the leading methods for subtyping of cancers on the basis of characteristic expression profiles. Meyerson et al. determined the molecular network of lung carcinogenesis by systematically analysing the patient’s protein and mRNA expression profiles7. Several researchers focusing on genes and proteins have discovered valuable information such as RB/p16, PTEN, K-ras, FHIT, p53, and MYO18b gene alterations, which are all observed in lung carcinoma4, 8, 9. MicroRNAs (miRNAs) are small non-coding RNAs that regulate gene expression and are involved in several biological processes, including human carcinogenesis and embryonic development. Gene and miRNA expression profiles have revealed abundant information on the molecular signatures of various cancer types. MiRNA expression profiles have been used to classify cancers into various subtypes. Many studies reported the relationship between miRNA and cancers, such as colorectal cancer, lung cancer and human cell lymphomas10, 11.
Several studies investigated the miRNA expression associated with lung adenocarcinoma. Liu et al. distinguished lung adenocarcinoma patients from healthy subjects using miRNA expression profiles and identified seven significantly expressed miRNAs in lung cancer tissue12. Yanaihara et al. investigated the diagnostic role of miRNAs in lung cancer and reported 43 differently expressed miRNAs in lung cancer tissue when compared with non-cancerous lung tissue13. MiRNA expression associated with early stage detection and disease progression was reported in lung cancer14, 15. Patnaik et al. predicted the recurrence of early stage in 77 NSCLC cases using a support vector machine (SVM) classifier and identified the miRNA expression pattern differentiation in recurrence groups16. Yu et al. predicted the clinical outcome of 112 NSCLC patients using miRNA expression profiles, and identified five miRNAs that can predict relapse and survival in lung cancer17. Raponi et al. identified 20 miRNAs that can predict prognosis in 61 squamous cell carcinoma samples using statistical analysis18. Besides the well-known survival analysis methods, alternatively, there are linear regression models proposed for survival estimation using censored data19–21. Support vector regression methods have been used in medical survival data analysis based on censored data and obtained a significant improvement in accuracy when comparing with standard survival analysis methods22. Zhao et al. estimated mean survival time using a reinforcement Q-learning method, which is developed based on support vector regression. The reinforcement Q-learning method utilized censored data of patients with advanced metastatic stage IIIB/IV of non-small cell lung cancer. Estimated mean survival time is used as clinical outcome to initiate the second-line therapy in patients with NSCLC23.
Current treatment modalities often fail to successfully treat lung cancer though strenuous efforts have been made to find better therapeutics to cure this cancer. Most of these researchers develop microarray methods to identify tumors and cancer stages. MiRNAs exceptionally influence developmental and oncogenic pathways by regulating gene expression11, 24. Defects in the miRNA biogenesis mechanism cause oncogenesis in lung cancer. Kumar et al. reported that conditional deletion of Dicer 1 associated with the lung tumor development in a mouse model25. Reduced dicer expression is involved in the development of lung tumors and shows a significant prognostic impact on survival of lung cancer patients26. A collection of evidences shows that miRNAs are differently expressed in non-small cell lung cancers and act as tumor suppressor and oncogenes27. For example, hsa-let-7 family often acts as a tumor suppressor. Hsa-let-7 family was found to be frequently deleted in chromosomal regions of lung cancer cell lines28 and inhibition of let-7 expression leads to increased cell division in A549 lung cancer cell lines29. Additionally, let-7 family negatively regulates oncogenes such as MYC and RAS25, 30. MiRNAs such as miR-31 function as oncogenic miRNAs in lung cancer by suppressing the tumor suppressor genes PP2A regulatory subunit B alpha isoform and large tumor suppressor-231. MiR-17-92 cluster promotes cell proliferation in non-small cell lung cancer32. Furthermore, miRNAs such as miR-1244 are down-regulated in A549 cells and involved in the progress of cisplatin resistance in NSCLC33. MiR-630 controls the p-53 regulated pro-apoptotic pathway and is involved in chemo resistant determination in lung cancer cells33. These studies imply a significant role of miRNAs in the development and progression of lung cancers.
MiRNA expression profiles are helpful to identify survival-related variants of lung adenocarcinoma. The miRNA signature associated with patient survival is also helpful for the development and evaluation of gene target based therapy. However, few studies develop machine learning approaches to identify the miRNA signature of patient survival34. Accordingly, the aim of this work is to identify the miRNA signature that can predict patients’ survival time in lung adenocarcinoma. This work proposes a support vector regression (SVR) based predictor, SVR-LUAD, to identify the miRNA signature associated with the survival time of patients with lung adenocarcinoma. Estimating survival time is very important, especially in cancer studies, to evaluate the personalized treatment effects in individuals with cancer. Identification of miRNA signature associated with survival time will help to further understand the miRNA mechanism in lung cancer as well as develop the therapeutics.
The SVR-LUAD method uses an optimal feature selection method, an inheritable bi-objective combinatorial genetic algorithm (IBCGA)35 to identify a small set of informative miRNA cooperating with SVR by maximizing estimation accuracy of survival time. There were 102 lung adenocarcinoma patients’ miRNA expression profiles along with survival information extracted from the cancer genome atlas (TCGA) database. SVR-LUAD identified a set of 18 miRNAs from the expression profiles of 332 miRNAs. The 18-miRNA signature is highly associated with lung adenocarcinoma survival. SVR-LUAD achieved a correlation coefficient of 0.88 ± 0.01 and mean absolute error of 0.56 ± 0.03 year (mean and standard deviation) between the real and estimated survival time using 10-fold cross-validation (10-CV). We validated the SVR-LUAD method using an independent test cohort of 51 lung adenocarcinoma patients obtained from the TCGA database. Additionally, we analysed the 18-miRNA signature using gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. These findings may be helpful towards understanding the individual miRNA role in lung adenocarcinoma survival.
Results and Discussion
SVR-LUAD simultaneously identified the miRNA signature and estimated the survival time of lung adenocarcinoma patients using miRNA expression profiles. Based on the accurate estimation of survival time, we can further understand individual miRNAs of the signature. There were 102 and 51 patients along with expression profiles of 332 miRNAs for training and testing the prediction model.
Identifying the miRNA signature with survival time estimation
SVR-LUAD used an optimal feature selection algorithm IBCGA to identify a set of 18 informative miRNAs (referred to a miRNA signature) associated with the estimation of lung adenocarcinoma survival time. Since the optimal feature selection method IBCGA is a non-deterministic method, we employed 30 independent runs to select one robust feature set. The 30 runs and their corresponding appearance scores are shown in Supplementary Table S1. We compare the SVR-LUAD method with penalized regression methods, such as LASSO, Ridge regression, Elastic net, and Multiple linear regression. The Elastic net is a combination of both methods LASSO and Ridge regression. The comparison results of SVR-LUAD, Elastic net and Multiple linear regression methods are shown in Table 1.
Table 1.
Prediction performance comparison of SVR-LUAD.
| Method | MiRNAs selected | Correlation coefficient | Mean absolute error |
|---|---|---|---|
| Multiple linear regression | 5 | 0.53 | 0.99 |
| SVR-LUAD-5 | 5 | 0.66 | 0.94 |
| Elastic net | 8 | 0.55 | 1.02 |
| SVR-LUAD-8 | 8 | 0.72 | 0.81 |
| SVR-LUAD | 18 | 0.90 | 0.52 |
| SVR-LUAD-mean | 24.7 | 0.88 | 0.56 |
The highest appearance score of SVR-LUAD was 0.53 (=16.0/30) with 18 miRNAs indicating that each miRNA of the signature may be selected with the probability of 0.53 in a run of SVR-LUAD on average (Fig. S1). The estimation accuracy of SVR-LUAD was the correlation coefficient of 0.90 and mean absolute error of 0.52 year using 10-CV. SVR-LUAD-mean achieved a correlation coefficient of 0.88 ± 0.01 and mean absolute error of 0.56 ± 0.03 year on average. The LASSO method achieved a correlation coefficient and mean absolute error of 0.48 and 1.083 year, and Ridge regression achieved a correlation coefficient and mean absolute error of 0.51 and 1.086 year, respectively. The Elastic net method with 8 miRNAs achieved a correlation coefficient and mean absolute error of 0.55 and 1.02 year, and Multiple linear regression with 5 miRNAs obtained a correlation coefficient and mean absolute error of 0.53 and 0.99 year, respectively. To fairly compare SVR-LUAD with these methods, the feature numbers of SVR-LUAD were restricted to 5 and 8. SVR-LUAD-5 with 5 miRNAs obtained a correlation coefficient and mean absolute error of 0.66 and 0.94 year, respectively. SVR-LUAD-8 with 8 miRNAs obtained a correlation coefficient and mean absolute error of 0.72 and 0.81 year, respectively. The correlation plots of the real and estimated survival time for SVR-LUAD, Elastic net, and Multiple linear regression are shown in Fig. 1. The correlation plots for LASSO and Ridge regression are shown in Supplementary Fig. S2. SVR-LUAD is better than these compared methods.
Figure 1.
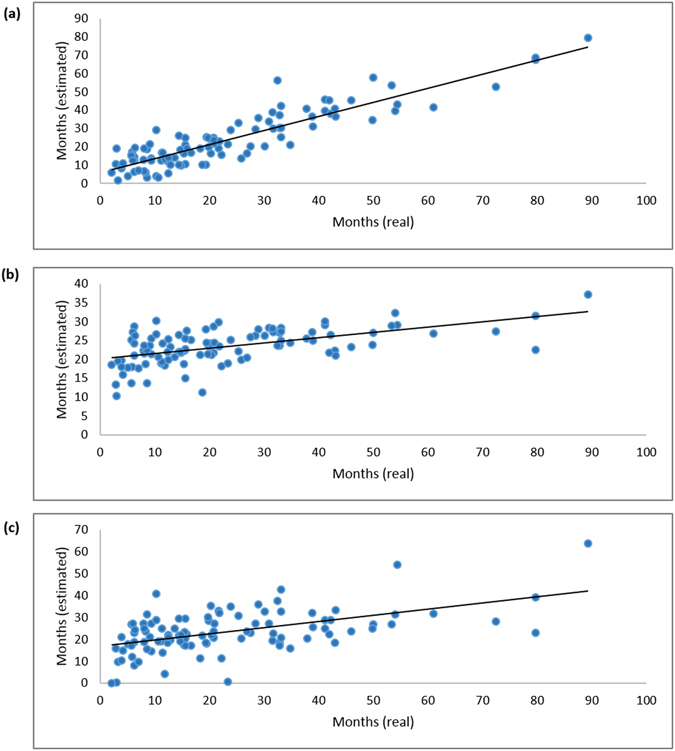
(a) Prediction performance of SVR-LUAD with a correlation coefficient of 0.90. (b) Prediction performance of Elastic net with a correlation coefficient of 0.55. (c) Prediction performance of Multiple linear regression with a correlation coefficient of 0.53. X-axis refers to real survival time and Y-axis refers to estimated survival time.
Evaluation on an independent test cohort
The 51 patients with follow-up time are alive and have tumors after therapy. The prediction result of SVR-LUAD for individual patients is shown in Fig. 2. The mean and standard deviation of the follow-up time are 25.27 ± 12.95 months. SVR-LUAD predicted and obtained the survival time of 38.92 ± 23.68 months. There were 38 of the 51 (75%) patients whose predicted survival time is larger than his/her follow-up time. The predicted survival time and follow-up time of the remaining 13 patients were 18.01 ± 11.52 and 34.57 ± 14.27 months on average, respectively. Comparing to the prediction error of 0.52 years (Table 1) and the mean difference, 16.56 (=34.57–18.01) months, of follow-up time and predicted survival time, it reveals that the pharmaceutical therapy may be promising.
Figure 2.
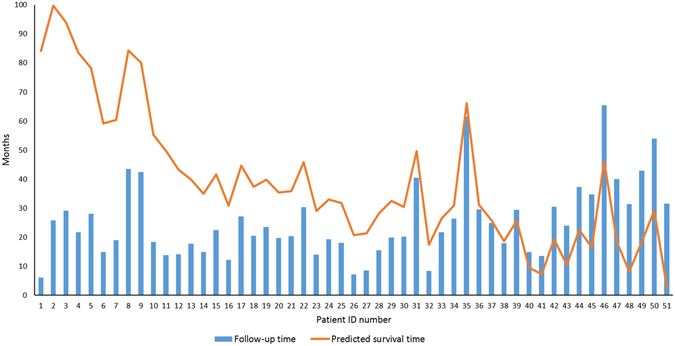
The validation of SVR-LUAD on an independent cohort of 51 patients with lung adenocarcinoma. Predicted survival time is larger than the follow-up time for the first 38 patients (1–38).
Contribution of individual miRNAs
SVR-LUAD with the 18-miRNA signature can achieve high estimation accuracy, but it doesn’t mean that the 18 miRNAs are the only informative miRNAs. For example, some miRNAs (e.g., hsa-let-7a-2, hsa-miR-192, hsa-miR-20b, hsa-miR-24-1, hsa-miR-25, hsa-miR-3187, hsa-miR-34b, hsa-miR-3617 and hsa-miR-1254, hsa-miR-1291, hsa-miR-194-2, hsa-miR-212, hsa-miR-3920) obtained from SVR-LUAD-8 and SVR-LUAD-5 don’t belong to the 18-miRNA signature. An increase of the patient cohort may be helpful to improve the prediction accuracy and identify the really informative miRNAs associated with survival time of patients with lung adenocarcinoma. We evaluated the contribution of individual miRNAs towards the estimation of survival time using two analysis methods, main effect difference and individual miRNA effect. The 10 top-ranked miRNAs according to the contribution of survival time estimation were further analyzed.
Main effect difference
We measure the individual effect of each miRNA in the 18-miRNA signature using an orthogonal array experimental design36. The larger the value of the main effect difference37, the larger the contribution of this miRNA toward to the survival time prediction using the signature is. The 10 top-ranked miRNAs are hsa-let-7f-1, hsa-miR-16-1, hsa-miR-152, hsa-miR-217, hsa-miR-18a, hsa-miR-193b, hsa-miR-3136, hsa-let-7g, hsa-miR-155 and hsa-miR-3199-1. Hereafter these will be referred to as top-10. All the 18 miRNAs and their corresponding main effect difference are shown in Table 2.
Table 2.
Contribution of individual miRNAs using MED and individual miRNA effect.
| Rank | MiRNA | MED | MiRNA effect | MAE (month) |
|---|---|---|---|---|
| value | Correlation coefficient | |||
| 1 | hsa-let-7f-1 | 1.790 | 0.25 | 12.95 |
| 2 | hsa-miR-16-1 | 1.575 | 0.60 | 9.43 |
| 3 | hsa-miR-152 | 0.966 | 0.07 | 13.24 |
| 4 | hsa-miR-217 | 0.955 | 0.35 | 12.05 |
| 5 | hsa-miR-18a | 0.952 | 0.42 | 11.57 |
| 6 | hsa-miR-193b | 0.921 | 0.61 | 8.74 |
| 7 | hsa-miR-3136 | 0.779 | 0.29 | 12.08 |
| 8 | hsa-let-7g | 0.775 | 0.57 | 9.63 |
| 9 | hsa-miR-155 | 0.622 | 0.35 | 12.13 |
| 10 | hsa-miR-3199-1 | 0.446 | 0.27 | 12.69 |
| 11 | hsa-miR-219-2 | 0.407 | 0.29 | 11.88 |
| 12 | hsa-miR-1254 | 0.398 | 0.49 | 10.75 |
| 13 | hsa-miR-1291 | 0.396 | 0.54 | 11.05 |
| 14 | hsa-miR-192 | 0.324 | 0.30 | 12.71 |
| 15 | hsa-miR-3653 | 0.274 | 0.47 | 11.30 |
| 16 | hsa-miR-3934 | 0.234 | 0.46 | 10.73 |
| 17 | hsa-miR-342 | 0.216 | 0.38 | 11.05 |
| 18 | hsa-miR-141 | 0.033 | 0.43 | 10.08 |
Individual miRNA effect
We assess the ability of an individual miRNA in estimating survival time of cancer patients using this miRNA only. We take one miRNA from the 18-miRNA signature and obtain the estimation performance of this miRNA in terms of correlation coefficient and mean absolute error, shown in Table 2. The results show that the three miRNAs, hsa-miR-193b, hsa-miR-16-1 and hsa-let-7g, have the highest correlation coefficients 0.61, 0.60 and 0.57 and mean absolute errors 8.74, 9.43 and 9.63 months, respectively. The ability of a miRNA in estimating survival time is slightly different between the roles in a signature and a single miRNA. Correlation plots for each miRNA are shown in Supplementary Fig. S3. The top-10 miRNAs are discussed below.
Among top-10 miRNAs, most of the miRNAs are involved in several major cancer types including lung cancer, bladder cancer, breast cancer, hepatocellular carcinoma, glioblastoma, ovarian and gastric cancers. We summarize the top-10 miRNAs and their involvement in various cancer types in Table 3.
Table 3.
The top-10 miRNAs involved in various cancers.
| MiRNA | Cancer | Regulation | Reference |
|---|---|---|---|
| hsa-let-7f-1 | Lung cancer | down | 25, 30, 38 |
| Breast cancer | down | 102 | |
| Colon cancer | down | 103 | |
| Hepatocellular carcinoma | down | 104 | |
| Neuroblastoma | down | 105 | |
| Pancreatic ductal adenocarcinoma | down | 106 | |
| hsa-miR-16-1 | Lung cancer | down | 42, 43 |
| Prostate cancer | down | 107 | |
| Neuroblastoma | up | 108 | |
| Chronic lymphocytic leukemia | down | 109 | |
| hsa-miR-152 | Lung cancer | down | 44, 48, 49 |
| Breast cancer | down | 49 | |
| Colorectal cancer | down | 110 | |
| Glioblastoma | down | 111 | |
| Ovarian cancer | down | 112 | |
| hsa-miR-217 | Lung cancer | down | 113 |
| Breast cancer | up | 114 | |
| Gastric cancer | down | 115 | |
| Hepatocellular carcinoma | up | 116 | |
| Pancreatic ductal adenocarcinoma | down | 117 | |
| hsa-miR-18a | Lung cancer | up | 32, 54 |
| Colorectal cancer | up | 118 | |
| Colon cancer | up | 119 | |
| Gastric cancer | up | 120 | |
| hsa-miR-193b | Lung cancer | down | 55–57 |
| Breast cancer | down | 121 | |
| Gastric cancer | down | 122 | |
| Hepatocellular carcinoma | down | 123 | |
| Ovarian cancer | down | 124 | |
| hsa-miR-3136 | Acute myeloid leukemia | — | 67 |
| Esophageal adenocarcinoma | — | 125 | |
| Breast cancer | — | 68 | |
| hsa-let-7g | Lung cancer | down | 58–60 |
| Breast cancer | down | 102 | |
| Colon cancer | down | 103 | |
| Esophageal squamous cell carcinoma | down | 126 | |
| Hepatocellular carcinoma | down | 104 | |
| hsa-miR-155 | Lung cancer | up | 50, 63, 64 |
| Acute myeloid leukemia | up | 37 | |
| Bladder cancer | up | 127 | |
| Breast cancer | up | 128 | |
| Nasopharyngeal carcinoma | up | 129 | |
| hsa-miR-3199–1 | Prostate cancer | up | 69 |
| Breast cancer | — | 130 |
Roles of the identified miRNAs
We analysed individual roles of the top-10 miRNAs in the 18-miRNA signature. Among the top-10 miRNAs, eight miRNAs are involved not only in lung cancer but also in other major cancer types. The two miRNAs hsa-miR-3136 and hsa-miR-3199-1 are involved in tumorigenesis but they are not reported as being involved in lung cancer.
(1) Hsa-let-7f-1: According to the MED analysis, hsa-let-7f-1 with rank 1 is the most effective in estimating survival time of lung adenocarcinoma patients. SVR-LUAD achieved a correlation coefficient of 0.25 and MAE of 12.95 months revealing that hsa-let-7f-1 is also very informative associated with the survival time. The hsa-let-7 family functionally inhibits oncogenes, such as c-Myc25, the Ras family30, and HMGA238. Hsa-let-7f-1 often acts as a tumor suppressor and is down-regulated in NSCLC due to its reduced expression39. The genes of the hsa-let-7 family are frequently located in the genomic regions of human cancers28. Takamizawa et al. investigated 143 lung cancer cases and reported that the reduced expression of the hsa-let-7 family is significantly associated with poor prognoses of lung cancer patients40. Real-time RT-PCR analysis on hsa-let-7a revealed that the expression of hsa-let-7 in lung cancer patients is significantly different (p = 0.000398) from the non-cancerous lung tissue; and altered expression of hsa-let-7a is associated with poor survival in lung adenocarcinoma patients13. Significance analysis on microarray studies reported that hsa-let-7f-1 is differently expressed in plasma of NSCLC patients (mean 0.53 fold change) when compared with healthy controls41. Altogether, these findings suggest that expression alteration of hsa-let-7f-1 has a prognostic impact on lung adenocarcinoma patients.
(2) Hsa-miR-16-1: Experimental investigation on NSCLC reported that hsa-miR-16-1 is mostly down-regulated in lung adenocarcinoma cell lines, Calu-1, H2009, H1299, H358 and A549, and this miRNA induced cell cycle arrest42. Normalized PCR experimental results of 70 NSCLC patients showed that hsa-miR-16-1 was down-regulated in tumor tissue (p < 0.001) when compared with normal tissue43. A clinical level study of 77 surgically-treated NSCLC patients found that hsa-miR-16-1 over expressed (2.650 fold change) in a recurrence group when compared with no-recurrence cases16.
(3) Hsa-miR-152: Chen et al. reported that hsa-miR-152 was up-regulated in NSCLC patients serum compared with control subjects’ serum44. Su et al. reported that hsa-miR-152 is down-regulated in NSCLC cells and acts as a tumor suppressor45. Hsa-miR-152 induces cell proliferation, migration, invasion and colony formation by targeting ADAM metallopeptidase domain 17 in NSCLC tissue45. Down-regulated expression of hsa-miR-152 targets the neuropilin-1 and regulates cancer metastasis in NSCLC A549 cell lines46. Hsa-miR-152 expression is significantly down-regulated in NSCLC patients when comparing with the healthy controls47. This miRNA was also majorly involved in cancer types such as epithelial ovarian48 and breast cancers49.
(4) Hsa-miR-217: Real-time PCR study on 100 patients revealed that over expression of hsa-miR-217 inhibited the cell proliferation, migration and promoted the apoptosis in lung cancer cells50. Hsa-miR-217 expression was significantly lower in lung cancer tissue when compared to the normal tissue50. Hsa-miR-217 has an emerging role in pancreatic ductal adenocarcinoma51, and its expression was up-regulated in B-cell lymphocytic leukemia52. Hsa-miR-217 targets phosphatase and tensin homologue, resulting in the activation of Akt kinase in diabetic nephropathy53.
(5) Hsa-miR-18a: Hayashita et al. reported that hsa-miR-18 is overexpressed in the amplified chromosomal region of lung cancer32. Its expression was significantly higher (20.25 fold change) in high-grade neuroendocrine pulmonary tumors of the lungs54.
(6) Hsa-miR-193b: Hsa-miR-193b was up-regulated and differentially expressed (1.50 fold change) in short survival versus long survival NSCLC patients55. A microarray study on 38 NSCLC patients reported that hsa-miR-193b is up-regulated in lung cancer tissue with a 6.8 fold change when compared with the normal tissue56. Quantitative RT-PCR analysis reported that hsa-miR-193b expression was lower in the NSCLC cell line A549 when compared with normal tissue, and that it modulates cell migration, invasion, and proliferation in NSCLC cells57.
(7) Hsa-let-7g: The hsa-let-7 family plays a key role in lung cancer. Hsa-let-7g inhibits tumor cell proliferation and promotes tumor cell death as observed in murine K-Ras expressing lung adenocarcinoma cells (LKR13)58. This miRNA can actively suppress the tumor formation in K-Ras mutant NSCLC cell lines58. Single nucleotide poly morphisms potentially modify the hsa-let-7g binding and target gene regulation, which eventually increases the risk of NSCLC cells59. Park et al. reported that hsa-let-7g is aberrantly expressed in NSCLC cells. Further-more, hsa-let-7g targets the genes HMGA2 and K-Ras resulting in the inhibition of A549 lung cancer cell migration60. This miRNA’s expression inhibits the nuclear-factor kappa B1 and plays a significant role in enhancing the ability of radiotherapy in lung cancer61. Joeng et al. investigated the role of hsa-let-7g in radio-sensitivity in lung cancer, and reported that over expression of this miRNA enhances radio-sensitivity in radio-resistant H1299 lung cancer cells, and also increases the response to ionizing radiation62.
(8) Hsa-miR-155: Yanaihara et al. investigated miRNA expression associated with the lung cancer patient survival and found that higher expression of hsa-miR-155 is associated with poor survival in lung adenocarcinoma patients. Its expression was significantly different in lung cancer tissue (p = 1.00e-07) when compared with non-lung cancer tissue13. A real time PCR study of 74 lung cancer patients reported that hsa-miR-155 expression was significantly higher in lung cancer tissue than control tissue (p < 0.001) and this miRNAs expression was used to discriminate the lung cancer cells from controls63. Volinia et al. reported higher expression of hsa-miR-155 in lung, breast and colon cancers64. Hsa-miR-155 was reported as a significant cancer regulator and it is under-expressed in lung cancer cells and other cancers50.
(9) Hsa-miR-3136 and hsa-miR-3199-1: He et al. reported that single nucleotide polymorphism occurs between hsa-miR-3136 and 3′ UTR of lung cancer-related inflammatory gene KSR1 with a mean allele frequency of 0.29365. Hsa-miR-3136 is involved in Klinefelter syndrome66 and childhood acute lymphoblastic leukemia67, and its over expression is observed in breast cancer68. Hsa-miR-3199-1 is involved in castration resistant prostate cancer69. According to the individual miRNA effects, the two miRNAs, hsa-miR-3136 and hsa-miR-3199-1, have correlation coefficients of 0.29 and 0.27, respectively, that means their contribution towards survival prediction is relatively high in lung adenocarcinoma.
Additionally, we investigated the miRNA expression differentiation between lung cancer tissue and normal tissue using the TCGA database, in which 473 cancer cases and 45 controls were used70. Hsa-miR-3136 expression is up-regulated in lung cancer when compared to the normal tissue with a fold change of 0.093 and a p-value of 0.009. Hsa-miR-3199-1 expression is down-regulated in lung cancer with a fold change of −0.12 and a p-value of 0.002 between cancer and normal tissue70. Hence, this work found that the two miRNAs have significant association with lung adenocarcinoma survival and should be further investigated for their roles in lung adenocarcinoma.
We summarize the top-10 miRNAs and their functions as either oncogenic or tumor-suppressor in Table 4. We constructed a miRNA-target interaction network using the CyTargetLinker application, supported by the Cytoscape71 to explore the regulatory interactions derived from interaction databases. The top-10 miRNAs are annotated with miRBase accession numbers, and identified 7654 predicted miRNA-target interactions. In the predicted network, only the miRNA-target interaction network in MicroCosm and TargetScan are visualized for the top-10 miRNAs, shown in Supplementary Fig. S4. The experimentally validated target genes of the top-10 miRNAs were reported in supplementary Table S2 from DIANA-TarBase72 and existing studies. We describe the top-10 miRNAs with some selected target genes using the validation methods including immunohistochemistry, western blot analysis, qPCR and immunoprecipitation below.
Table 4.
Role of the top-10 miRNAs in lung cancer.
| Rank | MiRNAs | Oncogenic/Tumor suppressor | Reference |
|---|---|---|---|
| 1 | hsa-let-7f-1 | Tumor-suppressor | 25, 30, 38, 39 |
| 2 | hsa-miR-16-1 | Tumor-suppressor | 42, 131 |
| 3 | hsa-miR-152 | Tumor-suppressor | 45, 46 |
| 4 | hsa-miR-217 | Tumor-suppressor | 132 |
| 5 | hsa-miR-18a | Oncogenic | 133 |
| 6 | hsa-miR-193b | Tumor-suppressor | 57 |
| 7 | hsa-miR-3136 | — | — |
| 8 | hsa-let-7g | Tumor-suppressor | 58–60 |
| 9 | hsa-miR-155 | Oncogenic | 13 |
| 10 | hsa-miR-3199-1 | — | — |
There were five miRNAs with validation using immunohistochemistry, western blot analysis and qPCR which are hsa-let-7f-1, hsa-miR-217, hsa-miR-18a, hsa-let-7g and hsa-miR-155. The hsa-let-7 family targets PRDM1 and regulates its function in a diffuse large B-cell lymphoma73. Hsa-miR-217 targets E2F3 which is positively associated with the hepatocellular carcinoma metastasis74. Hsa-miR-18a alters the PTEN protein expression in neural progenitor cells75. Hsa-let-7g is one of the hypoxia-responsive miRNAs targeting argonaute 1, which controls the miRNA induced silencing complex76. Hsa-miR-155 down-regulates the JMJD1A and BACH1 expression in nasopharyngeal cell lines77.
There were eight miRNAs with validation on their functional targets using western blot analysis and qPCR. Besides the five miRNAs mentioned above, the other three miRNAs were hsa-miR-16-1, hsa-miR-152 and hsa-miR-193b. For instance, hsa-miR-16-1 regulates the expression of CyclinD1 (CCDN1), which is an important regulator of cell-cycle progression78. Inhibition of miR-152 regulates DNA methylation via targeting DNA methyl transferase 1 in nickel sulfide transformed human bronchial epithelial cells79. Hsa-miR-193b directly targets the oncogenes CCD1 and ETS1 and regulates the invasion and migration in hepatoma cells80. The studies with immunoprecipitation experiments reported that hsa-miR-3136 targets SOCS5 with down-regulation in lymphoblastoid cell lines81 and hsa-miR-3199-1 targets CDK16 in 293 C cell lines82.
Aside from the top-10 miRNAs, other identified miRNAs such as hsa-miR-219-2, are down-regulated and differently expressed (p = 5.56e-05) in lung cancer cells when compared with non-lung cancer cells13. Foss et al. investigated differently expressed serum-based miRNAs in NSCLC, and found that hsa-miR-1254 expression was significantly higher in early stage NSCLC cells; they then used this miRNA to distinguish early stage NSCLC samples from controls83. Hsa-miR-192 is down-regulated in lung cancer cells and its expression was significantly higher (p = 0.000119) in lung cancer tissue when compared with non-lung cancer tissues13. Overexpression of hsa-miR-192 suppresses cell proliferation in NSCLC cells and it is usually down-regulated in NSCLC cells when compared with adjacent non-cancer cells84. It has been reported that hsa-miR-141 is differently expressed in primary lung tumors85.
Biological significance of the top-ranked miRNAs
We investigated whether the selected miRNAs have biological significance in cellular pathways and molecular functions using two analysis procedures, the KEGG pathway analysis and gene ontology terms using the Diana tool. First, we employed the KEGG pathway analysis for the top-10 miRNAs. In this analysis, the top-10 miRNAs are functionally enriched in a total of 60 cancer/non-cancer KEGG pathways, shown in Supplementary Table S3.
The pathway union analysis results showed that hsa-miR-7f-1, hsa-miR-16-1, hsa-miR-217, hsa-miR-18a, hsa-miR-193b, and hsa-miR-3199-1 are highly enriched in specific pathways such as extra cellular biosynthesis of unsaturated fatty acids, miRNAs in cancer pathway, adherence junction, in hippo and TGF-beta signaling pathways, ECM-receptor interaction in fatty acid biosynthesis, and fatty acid metabolism respectively. Additionally, all the top-10 miRNAs are involved in endometrial cancer, colorectal cancer, pathways in cancer, bladder cancer, proteoglycans in cancer, chronic myeloid leukemia, melanoma, hepatitis-B, and lysine degradation, to name a few. The pathway union enrichment analysis is shown in Fig. 3, and KEGG pathway analysis for all the 18 miRNAs are shown in Supplementary Fig. S5.
Figure 3.
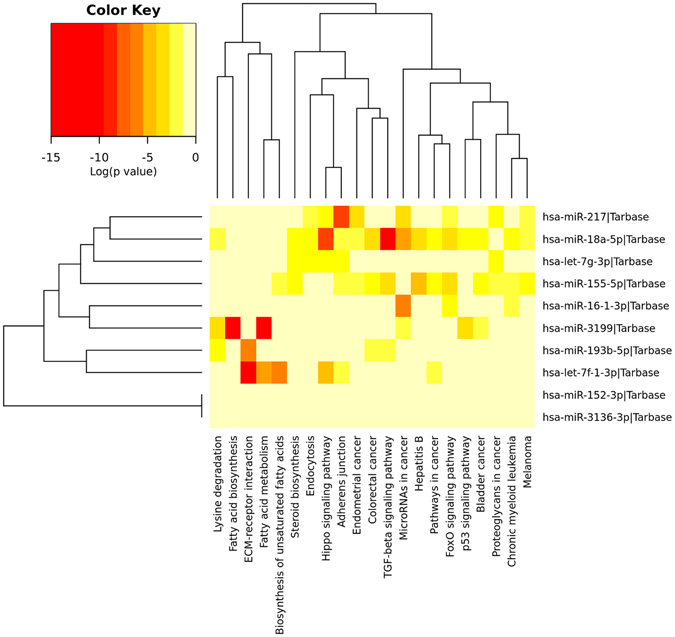
Heat map of the KEGG pathway. Top-10 miRNAs involved in cancer and non-cancer pathways.
Secondly, we employed a GO slim to provide a summary of gene ontology annotation for the identified miRNA signature. GO slim annotation results showed that hsa-miR-193, hsa-miR-155, hsa-miR-7g and hsa-miR-18a are highly enriched in specific molecular functions, biological processes and cell components, such as catabolic process, small molecule catabolic process, cellular protein modification process, cell death, cellular component, protein binding transcription factor activity, macro molecular complex assembly, protein complex assembly, cellular component assembly, enzyme binding, biosynthetic process, protein complex, cellular nitrogen compound metabolic process, cytosol, nucleoplasm, RNA binding, organelle, ion binding and molecular functions. Gene ontology enrichment of the top-10 miRNAs are depicted in Fig. 4. All the 18 miRNAs’ GO slim analysis is shown in Supplementary Fig. S6. Detailed process of gene ontology annotation results are shown in Supplementary Table S4.
Figure 4.
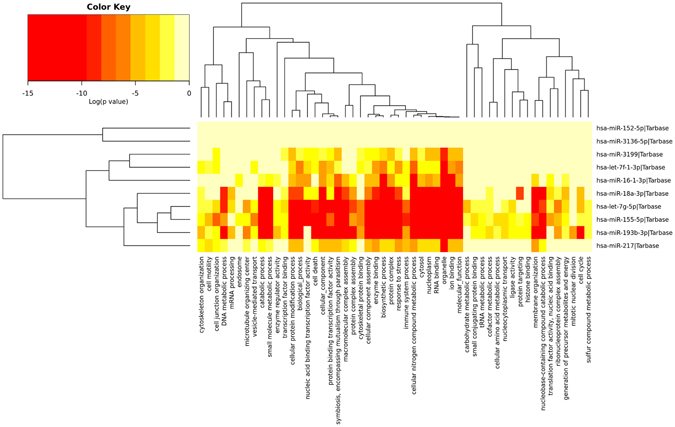
Heat map of the GO term analysis. Top-10 miRNAs involved in cellular component, molecular function and biological pathways in brief.
Conclusions
Due to the limitation in conventional therapies and diversified nature of diseases, multi-dimensional strategies are needed in cancer therapy. At present there are two strategies for implementing miRNAs as therapeutics in lung cancer. One is to inhibit the function of oncogenic miRNAs and the other is to restore the tumor-suppressor miRNA functions. Promising pre-clinical studies have shown that the therapeutic potentials of miRNAs in cancer treatment by restoring the miRNA functions. For instance, restoration of miR-34 inhibits the tumor growth in animal models86, 87. Let-7, miR-31 and miR-16 were proven to have anticancer effect in pre-clinical models88. Moreover, the company, miRNA therapeutics, developed a MRX34, a miRNA mimic, which was put into the clinical practice. Thus, identification of an effective miRNA signature can guide therapeutic decision and diagnosis in lung cancer.
Accordingly, we developed a method SVR-LUAD to identify the potential miRNA signature associated with survival time of lung adenocarcinoma patient. We first proposed a support vector regression based method cooperated with an optimal feature selection algorithm IBCGA to identify the miRNA signature associated with survival time in lung adenocarcinoma patients. The proposed survival time estimator identified the 18-miRNA signature out of 332 miRNAs strongly correlated with the survival time of lung cancer patients and obtained a correlation coefficient of 0.90 and mean absolute error of 0.52 year using 10-CV. Further, we ranked miRNAs based on the MED experiment and discussed the top-10 miRNA signatures’ characterization in lung cancer and other major cancers. Among the top-10 miRNA signatures, two miRNAs, hsa-miR-3136 and hsa-miR-3199-1, were previously unreported for the involvement in lung cancer. However, our method has found that these two miRNAs, like the other eight reported miRNAs, are strongly correlated with the survival of lung adenocarcinoma patients. Additionally, gene ontology enrichment annotations and KEGG pathway involvement of these top-10 miRNAs are discussed. The analysis suggests that these two miRNAs might be important subjects for further examination.
Validation of clinical applicability of the miRNA signature is necessary. We hope that the identified miRNA signature will assist in comprehensively understanding their pathway mechanism in lung cancer and improve the therapeutic strategies for the treatment of lung adenocarcinoma patients.
Materials and Methods
Dataset
There were 521 patients with lung adenocarcinoma in the TCGA database. We downloaded level-3 miRNA expression data from the TCGA portal that the miRNA profiling was implemented on the Illumina HiSeq. 2000 miRNA sequencing platform. We filtered out the used dataset using the following criteria. We included only the patients who have clinical data and survival information, and excluded the patients with a survival period of less than one month. All patients with their clinical data and survival periods were merged into a single dataset to eliminate duplicate entries. As a result, there were 102 patients with expression profiles of 332 miRNAs along with their clinical data including gender, age, and days until death (survival time).
Another set of 51 patients who are still alive with clinical data and follow-up time was used as an independent test cohort. The 51 patients with lung adenocarcinoma were selected by considering 1) who have tumors after pharmaceutical therapy, 2) vital status (alive), and 3) offer of follow-up days.
SVR-LUAD
The proposed method SVR-LUAD is an integration approach to combiningνsupport vector regression (ν-SVR) and feature selection algorithm IBCGA. SVR-LUAD is designed to simultaneously identify the miRNA signature and predict the survival time in order to discover the mechanism of the miRNA signature and develop effective therapies of lung adenocarcinoma patients.
Support vector regression
Support vector machine (SVM) is a state-of-the-art method for solving classification and regression problems. SVM has extensively been used in solving biological problems89. SVR is one of the practical modes of SVM. Due to the potential regression ability, SVR has been applied to a wide range of biological issues, such as estimation of survival time in glioblastoma cancer patients34, estimation of missing values of microarray data90, prediction of gene expression levels91, and prediction of siRNA efficacy92.
The ν-SVR is a new regression method of SVM which presents good performance depending on the number of support vectors and training error93. Given a set of N data points, {(x1, y1), (x2, y2), …, (xN, yN)}, where xi Rm is an input sample (patient) and yi R1 is a target label. In this study, yi is the survival time. The optimization problem of the ν-SVR can be described as follows.
| 1 |
Subject to
| 2 |
| 3 |
| 4 |
where 0 ≤ v ≤ 1. C is a regularization parameter and b is a constant. The ε-insensitive loss function means that if w T∅(xi) is in the range of y ± ε, no loss is considered. The y T is known as the soft margin where ν is an upper bound on the fraction of margin errors and a lower bound of the fraction of support vectors.
Fitness function
The fitness function of the IBCGA is the only guide to search for an optimal solution. In this study, the fitness function is to maximize the Pearson’s correlation coefficient (CC) of 10-CV as follows:
| 5 |
where y i and z i are real and predicted survival time of the i th patient, and and are their corresponding means. M is the total number of patients (M = 102 in this study). The mean absolute error (MAE) is also used for measuring prediction performance:
| 6 |
Inheritable bi-objective combinatorial genetic algorithm
SVR-LUAD used the optimal feature selection method IBCGA to identify a small set of m informative miRNAs from n = 332 miRNAs cooperating with ν-SVR by maximizing estimation accuracy of survival time. The IBCGA uses an intelligent evolutionary algorithm94 for solving the large combinatorial optimization problem C(n, m) to obtain an optimized ν-SVR where n is a given large constant and the best value of the variable m is not known beforehand. The intelligent evolutionary algorithm uses an orthogonal array crossover with a systematic reasoning ability to reproduce better offspring instead of random recombination in the crossover operation. The intelligent evolutionary algorithm can obtain a good solution S k to C(n, k) if k is a given constant. The IBCGA can obtain a set of solutions, S r, where r = r start, r start + 1, …, r end in a single run to efficiently search for a solution S r+1 to C(n, r + 1) by inheriting a good solution S r to C(n, r). The S m is the best solution among the solutions S r. In this work, the LibSVM package95 was used for implementation of ν-SVR.
The chromosome of the IBCGA consists of 332 genes for encoding the 332 miRNAs and three 4-bit genes for encoding the three variables γ, C, andνof theν-SVR. The parameter tuning of IBCGA was same with the previous study34, 35. The customized IBCGA for obtaining the m-miRNA signature where r start ≤ m≤ r end is described below.
Step 1) (Initialization) Randomly generate an initial population with N pop individuals. Each individual has r 1′s and n-r 0′s encoded into the n binary genes f i, where r = r start.
Step 2) (Evaluation) Evaluate all individuals in the population using the fitness function (2).
Step 3) (Selection) Use a tournament selection method that selects the winner from two randomly selected individuals to form a mating pool.
Step 4) (Crossover) Select P c · N pop parents from the mating pool to perform the orthogonal array crossover94, where P c is the crossover probability.
Step 5) (Mutation) A traditional mutation operator is applied to the randomly selected P m · N pop individuals except the best individual, where P m is the mutation probability.
Step 6) (Termination) If the stopping condition of performing G max generations is satisfied, output the best individual in the population as S r. Otherwise, go to Step 2.
Step 7) (Inheritance) If r < r end, randomly change one bit in the binary genes f i from 0 to 1 for each individual; increase the number r by one, and go to Step 2. Otherwise, stop the algorithm.
Step 8) (Output) Let m be equal to the value of r that S r is the best solution in the population. Output the m miRNAs and the corresponding ν-SVR model.
Appearance score
Since the IBCGA is a non-deterministic algorithm that the solutions of multiple runs are not always the same, selection of a robust solution is necessary. SVR-LUAD automatically identifies a robust solution (miRNA signature) from R (R = 30 in this study) independent runs for estimating the survival time of patients with lung adenocarcinoma. The robust set of features (miRNAs) has the highest appearance score obtained using the following procedure.
Step 1: Prepare the training dataset for 10-CV.
Step 2: Perform R independent runs of SVR-LUAD by maximizing CC of 10-CV for obtaining R miRNA signatures. There are features in the t-th signatures, t = 1, …, R.
Step 3: Appearance score is calculated as follows:
Calculate the appearance frequency f(p) for each feature p that ever presents in the R sets of miRNAs.
Calculate the score St, t = 1, …, R where pi is the i-th feature in the t-th solution:
| 7 |
Step 4: Output the t-th feature set with the highest appearance score St.
Multiple regression analysis
We employed the Multiple linear regression method96 to estimate the survival time in lung adenocarcinoma patients. The Multiple linear regression method is formulated as
| 8 |
where y i is a dependent variable (survival time of the i-th patient in this study); x i1, x i2, …, and x im are independent variables (miRNA expression); β 0 is a regression constant; β 1 , β 2, …, and β m are the regression coefficients; m is the number of terms in the model, and ε is the error term. In this study, m is the number of selected miRNAs. A stepwise feature addition method was used for feature selection97.
Elastic net
Elastic net is a regularization with an automatic feature selection technique98, which is a combination of ridge regression99 and least absolute shrinkage and selection operator (LASSO)100. The objective function of the Elastic net method using 10-CV is formulated as follows:
| 9 |
where y i is the sample response (survival time) at observation i (patient); x i Rm is the vector of m miRNA expression values for the i-th observation, λ is a regularization parameter, β 0 and β are regression coefficients, and M is the total number of observations.
KEGG pathway and Gene ontology annotation analysis
We used DIANA-mirpath web-based server to analyze the miRNA profiles101. The DIANA-Tarbase algorithm provided the predicted miRNAs targets for the pathway analysis. We used fisher’s exact test for enrichment analysis with a threshold p-value 0.05. In order to estimate the specificity of results, we performed the pathway analysis for all identified miRNAs.
We employed gene ontology annotations in order to identify miRNAs belonging to specific GO categories based on the experimental findings using the DIANA-mirpath webserver101. This webserver uses predicted miRNA targets obtained from the DIANA-microT-CDS algorithm. We used a hypergeometric distribution method for enrichment analysis.
Electronic supplementary material
Acknowledgements
This work was funded by the National Science Council of Taiwan under the contract numbers MOST-105-2627-M-009-008- and MOST-105-2221-E-009-138-MY2-, and “Center for Bioinformatics Research of Aiming for the Top University Program” of National Chiao Tung University and the Ministry of Education, Taiwan, R.O.C. for the project 105W962. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
Y.S.S. and S.Y.H. designed the system, participated in manuscript preparation, and carried out the detail study. All authors have read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-07739-y
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Travis, W. D. Pathology of lung cancer. Clin Chest Med23, 65–81, viii (2002). [DOI] [PubMed]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers–a different disease. Nat Rev Cancer. 2007;7:778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 4.Minna JD, Roth JA, Gazdar AF. Focus on lung cancer. Cancer Cell. 2002;1:49–52. doi: 10.1016/S1535-6108(02)00027-2. [DOI] [PubMed] [Google Scholar]
- 5.Lockhart DJ, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharjee A, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proceedings of the National Academy of Sciences. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyerson M, Carbone D. Genomic and proteomic profiling of lung cancers: lung cancer classification in the age of targeted therapy. J Clin Oncol. 2005;23:3219–3226. doi: 10.1200/JCO.2005.15.511. [DOI] [PubMed] [Google Scholar]
- 8.Yokota J, Kohno T. Molecular footprints of human lung cancer progression. Cancer Sci. 2004;95:197–204. doi: 10.1111/j.1349-7006.2004.tb02203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sekido Y, Fong KM, Minna JD. Molecular genetics of lung cancer. Annu Rev Med. 2003;54:73–87. doi: 10.1146/annurev.med.54.101601.152202. [DOI] [PubMed] [Google Scholar]
- 10.Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004;39:167–169. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 11.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu L, et al. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer. 2010;127:2870–2878. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 14.Saito M, et al. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin Cancer Res. 2011;17:1875–1882. doi: 10.1158/1078-0432.CCR-10-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donnem T, et al. Prognostic impact of MiR-155 in non-small cell lung cancer evaluated by in situ hybridization. J Transl Med. 2011;9:6. doi: 10.1186/1479-5876-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patnaik SK, Kannisto E, Knudsen S, Yendamuri S. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res. 2010;70:36–45. doi: 10.1158/0008-5472.CAN-09-3153. [DOI] [PubMed] [Google Scholar]
- 17.Yu SL, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 18.Raponi M, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776–5783. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 19.Ding, Y. & Nan, B. A sieve M-theorem for bundled parameters in semiparametric models, with application to the efficient estimation in a linear model for censored data. 3032–3061, doi:10.1214/11-AOS934 (2011). [PMC free article] [PubMed]
- 20.Jin Z, Lin DY, Wei LJ, Ying Z. Rank-based inference for the accelerated failure time model. Biometrika. 2003;90:341–353. doi: 10.1093/biomet/90.2.341. [DOI] [Google Scholar]
- 21.Wei LJ, Ying Z, Lin DY. Linear Regression Analysis of Censored Survival Data Based on Rank Tests. Biometrika. 1990;77:845–851. doi: 10.1093/biomet/77.4.845. [DOI] [Google Scholar]
- 22.Khan, F. M. & Zubek, V. B. In 2008 Eighth IEEE International Conference on Data Mining. 863–868.
- 23.Zhao Y, Zeng D, Socinski MA, Kosorok MR. Reinforcement Learning Strategies for Clinical Trials in Non-small Cell Lung Cancer. Biometrics. 2011;67:1422–1433. doi: 10.1111/j.1541-0420.2011.01572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 25.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 26.Karube Y, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calin GA, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson CD, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 30.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 31.Liu X, et al. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. The Journal of clinical investigation. 2010;120:1298–1309. doi: 10.1172/JCI39566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashita Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 33.Li W, et al. MiR-1244 sensitizes the resistance of non-small cell lung cancer A549 cell to cisplatin. Cancer Cell International. 2016;16:30. doi: 10.1186/s12935-016-0305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yerukala Sathipati S, Huang H-L, Ho S-Y. Estimating survival time of patients with glioblastoma multiforme and characterization of the identified microRNA signatures. BMC Genomics. 2016;17:1022. doi: 10.1186/s12864-016-3321-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho, S. Y., Chen, J. H. & Huang, M. H. Inheritable genetic algorithm for biobjective 0/1 combinatorial optimization problems and its applications. IEEE Trans Syst Man Cybern B Cybern34, 10.1109/tsmcb.2003.817090 (2004). [DOI] [PubMed]
- 36.Tung CW, Ho SY. Computational identification of ubiquitylation sites from protein sequences. BMC Bioinformatics. 2008;9:310. doi: 10.1186/1471-2105-9-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khalife J, et al. Pharmacological targeting of miR-155 via the NEDD8-activating enzyme inhibitor MLN4924 (Pevonedistat) in FLT3-ITD acute myeloid leukemia. Leukemia. 2015;29:1981–1992. doi: 10.1038/leu.2015.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pasquinelli AE, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 40.Takamizawa J, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 41.Silva J, et al. Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur Respir J. 2011;37:617–623. doi: 10.1183/09031936.00029610. [DOI] [PubMed] [Google Scholar]
- 42.Bandi N, et al. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009;69:5553–5559. doi: 10.1158/0008-5472.CAN-08-4277. [DOI] [PubMed] [Google Scholar]
- 43.Navarro A, et al. Prognostic implications of miR-16 expression levels in resected non-small-cell lung cancer. J Surg Oncol. 2011;103:411–415. doi: 10.1002/jso.21847. [DOI] [PubMed] [Google Scholar]
- 44.Chen X, et al. Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for nonsmall cell lung cancer diagnosis. Int J Cancer. 2012;130:1620–1628. doi: 10.1002/ijc.26177. [DOI] [PubMed] [Google Scholar]
- 45.Su Y, Wang Y, Zhou H, Lei L, Xu L. MicroRNA-152 targets ADAM17 to suppress NSCLC progression. FEBS Lett. 2014;588:1983–1988. doi: 10.1016/j.febslet.2014.04.022. [DOI] [PubMed] [Google Scholar]
- 46.Zhang YJ, Liu XC, Du J, Zhang YJ. MiR-152 regulates metastases of non-small cell lung cancer cells by targeting neuropilin-1. Int J Clin Exp Pathol. 2015;8:14235–14240. [PMC free article] [PubMed] [Google Scholar]
- 47.Yang JS, et al. Serum miR-152, miR-148a, miR-148b, and miR-21 as novel biomarkers in non-small cell lung cancer screening. Tumour Biol. 2015;36:3035–3042. doi: 10.1007/s13277-014-2938-1. [DOI] [PubMed] [Google Scholar]
- 48.Zhang L, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2008;105:7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Q, et al. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J Mol Cell Biol. 2013;5:3–13. doi: 10.1093/jmcb/mjs049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roa W, et al. Identification of a new microRNA expression profile as a potential cancer screening tool. Clin Invest Med. 2010;33:E124. doi: 10.25011/cim.v33i2.12351. [DOI] [PubMed] [Google Scholar]
- 51.Szafranska AE, et al. Analysis of microRNAs in pancreatic fine-needle aspirates can classify benign and malignant tissues. Clin Chem. 2008;54:1716–1724. doi: 10.1373/clinchem.2008.109603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calin GA, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kato M, et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol. 2009;11:881–889. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mairinger FD, et al. Different micro-RNA expression profiles distinguish subtypes of neuroendocrine tumors of the lung: results of a profiling study. Mod Pathol. 2014;27:1632–1640. doi: 10.1038/modpathol.2014.74. [DOI] [PubMed] [Google Scholar]
- 55.Donnem T, et al. MicroRNA signatures in tumor tissue related to angiogenesis in non-small cell lung cancer. PLoS One. 2012;7:e29671. doi: 10.1371/journal.pone.0029671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vosa U, et al. Identification of miR-374a as a prognostic marker for survival in patients with early-stage nonsmall cell lung cancer. Genes Chromosomes Cancer. 2011;50:812–822. doi: 10.1002/gcc.20902. [DOI] [PubMed] [Google Scholar]
- 57.Hu H, Li S, Liu J, Ni B. MicroRNA-193b modulates proliferation, migration, and invasion of non-small cell lung cancer cells. Acta Biochim Biophys Sin (Shanghai) 2012;44:424–430. doi: 10.1093/abbs/gms018. [DOI] [PubMed] [Google Scholar]
- 58.Kumar MS, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chin LJ, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park S, et al. Suppression of A549 lung cancer cell migration by precursor let-7g microRNA. Mol Med Rep. 2010;3:1007–1013. doi: 10.3892/mmr.2010.373. [DOI] [PubMed] [Google Scholar]
- 61.Arora H, Qureshi R, Jin S, Park AK, Park WY. miR-9 and let-7g enhance the sensitivity to ionizing radiation by suppression of NFkappaB1. Exp Mol Med. 2011;43:298–304. doi: 10.3858/emm.2011.43.5.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jeong SH, Wu HG, Park WY. LIN28B confers radio-resistance through the posttranscriptional control of KRAS. Exp Mol Med. 2009;41:912–918. doi: 10.3858/emm.2009.41.12.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng D, et al. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int J Clin Exp Pathol. 2011;4:575–586. [PMC free article] [PubMed] [Google Scholar]
- 64.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He F, et al. Inferring single nucleotide polymorphisms in microRNA binding sites of lung cancer-related inflammatory genes. Asian Pacific journal of cancer prevention: APJCP. 2014;15:3601–3606. doi: 10.7314/APJCP.2014.15.8.3601. [DOI] [PubMed] [Google Scholar]
- 66.Sui W, et al. microRNA expression profile of peripheral blood mononuclear cells of Klinefelter syndrome. Exp Ther Med. 2012;4:825–831. doi: 10.3892/etm.2012.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schotte D, et al. Discovery of new microRNAs by small RNAome deep sequencing in childhood acute lymphoblastic leukemia. Leukemia. 2011;25:1389–1399. doi: 10.1038/leu.2011.105. [DOI] [PubMed] [Google Scholar]
- 68.Godfrey AC, et al. Serum microRNA expression as an early marker for breast cancer risk in prospectively collected samples from the Sister Study cohort. Breast Cancer Res. 2013;15:R42. doi: 10.1186/bcr3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ottman R, Nguyen C, Lorch R, Chakrabarti R. MicroRNA expressions associated with progression of prostate cancer cells to antiandrogen therapy resistance. Mol Cancer. 2014;13:1. doi: 10.1186/1476-4598-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Z, et al. dbDEMC 2.0: updated database of differentially expressed miRNAs in human cancers. Nucleic Acids Research. 2017;45:D812–D818. doi: 10.1093/nar/gkw1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shannon P, et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vlachos IS, et al. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015;43:D153–159. doi: 10.1093/nar/gku1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leppert U, Henke W, Huang X, Muller JM, Dubiel W. Post-transcriptional fine-tuning of COP9 signalosome subunit biosynthesis is regulated by the c-Myc/Lin28B/let-7 pathway. Journal of molecular biology. 2011;409:710–721. doi: 10.1016/j.jmb.2011.04.041. [DOI] [PubMed] [Google Scholar]
- 74.Su J, Wang Q, Liu Y, Zhong M. miR-217 inhibits invasion of hepatocellular carcinoma cells through direct suppression of E2F3. Molecular and cellular biochemistry. 2014;392:289–296. doi: 10.1007/s11010-014-2039-x. [DOI] [PubMed] [Google Scholar]
- 75.Liu XS, et al. MicroRNA-17-92 cluster mediates the proliferation and survival of neural progenitor cells after stroke. The Journal of biological chemistry. 2013;288:12478–12488. doi: 10.1074/jbc.M112.449025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen Z, et al. Hypoxia-responsive miRNAs target argonaute 1 to promote angiogenesis. The Journal of clinical investigation. 2013;123:1057–1067. doi: 10.1172/JCI65344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Du Z-M, et al. Upregulation of MiR-155 in Nasopharyngeal Carcinoma is Partly Driven by LMP1 and LMP2A and Downregulates a Negative Prognostic Marker JMJD1A. PLOS ONE. 2011;6:e19137. doi: 10.1371/journal.pone.0019137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen RW, et al. Truncation in CCND1 mRNA alters miR-16-1 regulation in mantle cell lymphoma. Blood. 2008;112:822–829. doi: 10.1182/blood-2008-03-142182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ji W, et al. MicroRNA-152 targets DNA methyltransferase 1 in NiS-transformed cells via a feedback mechanism. Carcinogenesis. 2013;34:446–453. doi: 10.1093/carcin/bgs343. [DOI] [PubMed] [Google Scholar]
- 80.Xu C, et al. MicroRNA-193b regulates proliferation, migration and invasion in human hepatocellular carcinoma cells. European journal of cancer (Oxford, England: 1990) 2010;46:2828–2836. doi: 10.1016/j.ejca.2010.06.127. [DOI] [PubMed] [Google Scholar]
- 81.Skalsky RL, et al. The Viral and Cellular MicroRNA Targetome in Lymphoblastoid Cell Lines. PLOS Pathogens. 2012;8:e1002484. doi: 10.1371/journal.ppat.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Karginov FV, Hannon GJ. Remodeling of Ago2–mRNA interactions upon cellular stress reflects miRNA complementarity and correlates with altered translation rates. Genes & development. 2013;27:1624–1632. doi: 10.1101/gad.215939.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Foss KM, et al. miR-1254 and miR-574-5p: serum-based microRNA biomarkers for early-stage non-small cell lung cancer. J Thorac Oncol. 2011;6:482–488. doi: 10.1097/JTO.0b013e318208c785. [DOI] [PubMed] [Google Scholar]
- 84.Feng S, et al. MicroRNA-192 targeting retinoblastoma 1 inhibits cell proliferation and induces cell apoptosis in lung cancer cells. Nucleic Acids Res. 2011;39:6669–6678. doi: 10.1093/nar/gkr232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gibbons DL, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23:2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Craig VJ, et al. Systemic microRNA-34a delivery induces apoptosis and abrogates growth of diffuse large B-cell lymphoma in vivo. Leukemia. 2012;26:2421–2424. doi: 10.1038/leu.2012.110. [DOI] [PubMed] [Google Scholar]
- 87.Bader AG. miR-34 - a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim M, Kasinski AL, Slack FJ. MicroRNA therapeutics in preclinical cancer models. Lancet Oncol. 2011;12:319–321. doi: 10.1016/S1470-2045(11)70067-5. [DOI] [PubMed] [Google Scholar]
- 89.Vapnik VN. An overview of statistical learning theory. IEEE Transactions on Neural Networks. 1999;10:988–999. doi: 10.1109/72.788640. [DOI] [PubMed] [Google Scholar]
- 90.Wang X, Li A, Jiang Z, Feng H. Missing value estimation for DNA microarray gene expression data by Support Vector Regression imputation and orthogonal coding scheme. BMC Bioinformatics. 2006;7:32. doi: 10.1186/1471-2105-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Raghava GP, Han JH. Correlation and prediction of gene expression level from amino acid and dipeptide composition of its protein. BMC bioinformatics. 2005;6:1. doi: 10.1186/1471-2105-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Qiu S, Lane T. A framework for multiple kernel support vector regression and its applications to siRNA efficacy prediction. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2009;6:190–199. doi: 10.1109/TCBB.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chang C-C, Lin C-J. Training nu-support vector regression: theory and algorithms. Neural Computation. 2002;14:1959–1978. doi: 10.1162/089976602760128081. [DOI] [PubMed] [Google Scholar]
- 94.Ho S-Y, Shu L-S, Chen J-H. Intelligent evolutionary algorithms for large parameter optimization problems. Trans. Evol. Comp. 2004;8:522–541. doi: 10.1109/TEVC.2004.835176. [DOI] [Google Scholar]
- 95.Chang C-C, Lin C-J. LIBSVM: a library for support vector machines. ACM Transactions on Intelligent Systems and Technology (TIST) 2011;2:27. [Google Scholar]
- 96.Aiken, L. S., West, S. G. & Pitts, S. C. In Handbook of Psychology (John Wiley & Sons, Inc., 2003).
- 97.Hocking RR. A Biometrics Invited Paper. The Analysis and Selection of Variables in Linear Regression. Biometrics. 1976;32:1–49. doi: 10.2307/2529336. [DOI] [Google Scholar]
- 98.Zou H, Hastie T. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2005;67:301–320. doi: 10.1111/j.1467-9868.2005.00503.x. [DOI] [Google Scholar]
- 99.Hoerl AE, Kennard RW. Ridge regression: Biased estimation for nonorthogonal problems. Technometrics. 1970;12:55–67. doi: 10.1080/00401706.1970.10488634. [DOI] [Google Scholar]
- 100.Tibshirani, R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society. Series B (Methodological), 267–288 (1996).
- 101.Vlachos IS, et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Research. 2015;43:W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yan Y, Zhang F, Fan Q, Li X, Zhou K. Breast cancer-specific TRAIL expression mediated by miRNA response elements of let-7 and miR-122. Neoplasma. 2014;61:672–679. doi: 10.4149/neo_2014_082. [DOI] [PubMed] [Google Scholar]
- 103.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 104.Saito Y, et al. MicroRNAs in Hepatobiliary and Pancreatic Cancers. Front Genet. 2011;2:66. doi: 10.3389/fgene.2011.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Molenaar JJ, et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat Genet. 2012;44:1199–1206. doi: 10.1038/ng.2436. [DOI] [PubMed] [Google Scholar]
- 106.Torrisani J, et al. let-7 MicroRNA transfer in pancreatic cancer-derived cells inhibits in vitro cell proliferation but fails to alter tumor progression. Hum Gene Ther. 2009;20:831–844. doi: 10.1089/hum.2008.134. [DOI] [PubMed] [Google Scholar]
- 107.Bonci D, et al. A microRNA code for prostate cancer metastasis. Oncogene. 2016;35:1180–1192. doi: 10.1038/onc.2015.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gao SM, et al. miR-15a/16-1 enhances retinoic acid-mediated differentiation of leukemic cells and is up-regulated by retinoic acid. Leuk Lymphoma. 2011;52:2365–2371. doi: 10.3109/10428194.2011.601476. [DOI] [PubMed] [Google Scholar]
- 109.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li B, Xie Z, Li B. miR-152 functions as a tumor suppressor in colorectal cancer by targeting PIK3R3. Tumour Biol. 2016;37:10075–10084. doi: 10.1007/s13277-016-4888-2. [DOI] [PubMed] [Google Scholar]
- 111.Ma J, et al. MiR-152 functions as a tumor suppressor in glioblastoma stem cells by targeting Kruppel-like factor 4. Cancer Lett. 2014;355:85–95. doi: 10.1016/j.canlet.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 112.Zhou X, et al. Altered expression of miR-152 and miR-148a in ovarian cancer is related to cell proliferation. Oncol Rep. 2012;27:447–454. doi: 10.3892/or.2011.1482. [DOI] [PubMed] [Google Scholar]
- 113.Guo J, Feng Z, Huang Za, Wang H, Lu W. MicroRNA-217 Functions as a Tumour Suppressor Gene and Correlates with Cell Resistance to Cisplatin in Lung Cancer. Molecules and Cells. 2014;37:664–671. doi: 10.14348/molcells.2014.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang Q, Yuan Y, Cui J, Xiao T, Jiang D. MiR-217 Promotes Tumor Proliferation in Breast Cancer via Targeting DACH1. J Cancer. 2015;6:184–191. doi: 10.7150/jca.10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen DL, et al. microRNA-217 inhibits tumor progression and metastasis by downregulating EZH2 and predicts favorable prognosis in gastric cancer. Oncotarget. 2015;6:10868–10879. doi: 10.18632/oncotarget.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xia H, Ooi LL, Hui KM. MicroRNA-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer. Hepatology. 2013;58:629–641. doi: 10.1002/hep.26369. [DOI] [PubMed] [Google Scholar]
- 117.Zhao WG, et al. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis. 2010;31:1726–1733. doi: 10.1093/carcin/bgq160. [DOI] [PubMed] [Google Scholar]
- 118.Brunet Vega A, et al. microRNA expression profile in stage III colorectal cancer: circulating miR-18a and miR-29a as promising biomarkers. Oncol Rep. 2013;30:320–326. doi: 10.3892/or.2013.2475. [DOI] [PubMed] [Google Scholar]
- 119.Wu CW, et al. MicroRNA-18a attenuates DNA damage repair through suppressing the expression of ataxia telangiectasia mutated in colorectal cancer. PLoS One. 2013;8:e57036. doi: 10.1371/journal.pone.0057036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen YJ, et al. MicroRNA-18a modulates P53 expression by targeting IRF2 in gastric cancer patients. J Gastroenterol Hepatol. 2016;31:155–163. doi: 10.1111/jgh.13041. [DOI] [PubMed] [Google Scholar]
- 121.Li XF, Yan PJ, Shao ZM. Downregulation of miR-193b contributes to enhance urokinase-type plasminogen activator (uPA) expression and tumor progression and invasion in human breast cancer. Oncogene. 2009;28:3937–3948. doi: 10.1038/onc.2009.245. [DOI] [PubMed] [Google Scholar]
- 122.Wang L, et al. MicroRNA-193b inhibits the proliferation, migration and invasion of gastric cancer cells via targeting cyclin D1. Acta Histochem. 2016;118:323–330. doi: 10.1016/j.acthis.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 123.Yin W, Nie Y, Zhang Z, Xie L, He X. miR-193b acts as a cisplatin sensitizer via the caspase-3-dependent pathway in HCC chemotherapy. Oncol Rep. 2015;34:368–374. doi: 10.3892/or.2015.3996. [DOI] [PubMed] [Google Scholar]
- 124.Li H, Xu Y, Qiu W, Zhao D, Zhang Y. Tissue miR-193b as a Novel Biomarker for Patients with Ovarian Cancer. Med Sci Monit. 2015;21:3929–3934. doi: 10.12659/MSM.895407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang K, et al. Circulating miRNA profile in esophageal adenocarcinoma. Am J Cancer Res. 2016;6:2713–2721. [PMC free article] [PubMed] [Google Scholar]
- 126.Liu Q, et al. Role of microRNA let-7 and effect to HMGA2 in esophageal squamous cell carcinoma. Mol Biol Rep. 2012;39:1239–1246. doi: 10.1007/s11033-011-0854-7. [DOI] [PubMed] [Google Scholar]
- 127.Wang H, Men CP. Correlation of Increased Expression of MicroRNA-155 in Bladder Cancer and Prognosis. Lab Med. 2015;46:118–122. doi: 10.1309/LMWR9CEA2K2XVSOX. [DOI] [PubMed] [Google Scholar]
- 128.Chen J, Wang BC, Tang JH. Clinical significance of microRNA-155 expression in human breast cancer. J Surg Oncol. 2012;106:260–266. doi: 10.1002/jso.22153. [DOI] [PubMed] [Google Scholar]
- 129.Du ZM, et al. Upregulation of MiR-155 in nasopharyngeal carcinoma is partly driven by LMP1 and LMP2A and downregulates a negative prognostic marker JMJD1A. PLoS One. 2011;6:e19137. doi: 10.1371/journal.pone.0019137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cava C, et al. How interacting pathways are regulated by miRNAs in breast cancer subtypes. BMC Bioinformatics. 2016;17:348. doi: 10.1186/s12859-016-1196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bandi N, Vassella E. miR-34a and miR-15a/16 are co-regulated in non-small cell lung cancer and control cell cycle progression in a synergistic and Rb-dependent manner. Mol Cancer. 2011;10:55. doi: 10.1186/1476-4598-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Guo J, Feng Z, Huang Z, Wang H, Lu W. MicroRNA-217 functions as a tumour suppressor gene and correlates with cell resistance to cisplatin in lung cancer. Mol Cells. 2014;37:664–671. doi: 10.14348/molcells.2014.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Matsubara H, et al. Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene. 2007;26:6099–6105. doi: 10.1038/sj.onc.1210425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


