Abstract
MicroRNAs (miRNAs) are crucial regulators of gene expression in tumorigenesis and are of great interest to researchers, but miRNA profiles are often inconsistent between studies. The aim of this study was to confirm candidate miRNA biomarkers for esophageal cancer from integrated‐miRNA expression profiling data and TCGA (The Cancer Genome Atlas) data in tissues. Here, we identify five significant miRNAs by a comprehensive analysis in esophageal cancer, and two of them (hsa‐miR‐100‐5p and hsa‐miR‐133b) show better prognoses with significant difference for both 3‐year and 5‐year survival. Additionally, they participate in esophageal cancer occurrence and development according to KEGG and Panther enrichment analyses. Therefore, these five miRNAs may serve as miRNA biomarkers in esophageal cancer. Analysis of differential expression for target genes of these miRNAs may also provide new therapeutic alternatives in esophageal cancer.
Keywords: Enrichment analysis, esophageal cancer, prognosis
Introduction
Esophageal cancers, including esophageal adenocarcinoma and esophageal squamous cell carcinoma, were the fifth highest in cancer incidence in China, accounting for 50% of all new cases with liver cancer and 49% of cancer‐related deaths worldwide according to the 2014 World Cancer Report 1. At present, the best strategy to improve the prognosis of esophageal cancer is early detection, diagnosis, and treatment. However, accurate diagnosis with an effective treatment strategy is still a challenge. Several miRNAs have been identified to play fundamental roles in the occurrence and development of cancer, and show altered expression in human various cancers 2, which motivated us to further explore the potential application of miRNAs in esophageal cancer diagnosis and therapy.
miRNA is a single‐stranded noncoding RNA consisting of 20–24 nucleotides, which functions in suppressing target gene expression by binding to complementary sequences in the 3__‐untranslated region of mRNAs, leading to degradation of mRNA and inhibition of their translation 3. Complex interactions existing among miRNAs, target genes, and phenotypes have emerged as valuable biomarkers of diagnosis and prognosis associated with various phenotypes in diseases. Since the first report of a direct link between miRNAs and human cancer, in which miR‐15 and miR‐16 were found to be absent or downregulated in B‐cell chronic lymphocytic leukemia, many studies have evaluated their biological roles and associations with disease 4. Owing to innovations in biotechnology, large datasets have emerged, providing a substantial amount of valuable miRNA data. However, miRNA profiling efforts are often inconsistent between studies because of small sample size, different technological platforms, and different methods for processing and analysis. To overcome these problems, a more advanced and powerful strategy is necessary to handle these comprehensive and complex high‐throughput data. These specific miRNA biomarkers could be applied in early diagnosis and therapy of esophageal cancer in the future.
Methods
Data collection and processing
We search for articles in PubMed published from 1999 to 2016 (last accessed on 20 January 2016), by means of the combination of the following key terms: ((esophageal and (cancer* or carcinoma or tumour* or tumor*)) and (microRNA* or miRNA* or miR‐*) and profil*. In total, we obtain 302 relevant articles, and 217 of them could be downloaded with full text and miRNA profiles from English‐language journals. Further screening filters out 48 papers in which the patient samples are cancer tissues and paired adjacent noncancerous tissues, and not serum samples. To maintain the strategic accuracy and standardize these data as much as possible, the fold change in each miRNA expression is regarded as rule for further screening, and then 12 articles are retained finally.
We download and extract the TCGA data including miRNA expression data, gene expression data and patients' clinical information from the TCGA Data Portal (https://tcga-data.nci.nih.gov/tcga/; last release, March 2016) 5. Gene expression profiles (Reads per kilobase per million, RPKM) and miRNA expression profiles (reads per million, RPM) are log2‐transformed and used for subsequent analysis. In total, 13 pairs of samples are applied to evaluate the expression levels of miRNAs by paired‐samples t‐test and their target genes by the DESeq2 program 6 between solid tumors and adjacent noncancerous tissues. The clinical record in 185 esophageal cancer samples is for prognosis analysis.
Standardization of miRNA data
Frequently, a miRNA exists as various precursors in primary data. Additionally, the mature form of miRNA always plays crucial roles in regulation of target gene expression. To analyze these data with greater precision and high efficiency, a unified standardization strategy is applied such that the precursors or alias of each miRNA are converted to the corresponding mature form according to the miRBase database (http://www.mirbase.org/; last release, Release 21 in June 2014) 7.
RRA analysis
The novel RRA method 8, based on the leave‐one‐out cross‐validation and Bonferroni correction, assigns a P‐value to each miRNA in the last aggregated list. The P‐value for each miRNA indicates how much better it is ranked compared with a null model, expecting random ordering. With the RRA method, we analyze the ranking miRNA lists based on their fold change in expression level from 12 studies to obtain meta‐signature miRNA. Here, the P‐value for each miRNA can indicate whether its expression in esophageal cancer tissues is statistically significant or not, compared with paired normal tissue. The RRA approach is openly available in Comprehensive R Archive Network (http://cran.r-project.org/).
Prediction, integration, and verification of target genes
Target genes of these five miRNAs in esophageal cancer are predicted with the miRDB database (http://mirdb.org/miRDB/) 9, TargetScan database (http://www.targetscan.org/) 10, microT‐CDS database (http://www.microrna.gr/microT-CDS) 11, and RNA22 database (http://cm.jefferson.edu/rna22/) 12. Additionally, the experimentally validated target genes of these five miRNAs are searched with the DIANA‐TarBase v7.0 database (http://www.microrna.gr/tarbase) 13 and the MiRTarBase database (http://mirtarbase.mbc.nctu.edu.tw/) 14. Then, the target gene lists for each miRNA are generated based on the intersection of the four predicted miRNA target gene databases (miRDB, TargetScan, microT‐CDS, and RNA22) and the union of the two experimentally validated miRNA targeted‐gene databases (DIANA‐TarBase v7.0 and MiRTarBase). Finally, the expression levels of these target genes are tested between 13 paired esophageal cancer and adjacent noncancerous tissues from the TCGA data to obtain significantly differently expressed genes. In theory, upregulation of miRNA expression would lead to downregulation of its target genes and vice versa. Thus, the target genes are confirmed with a further filter (downregulated miRNA: P ≤ 0.05 and log2 [fold change] >0; upregulated miRNA: P ≤ 0.05 and log2 [fold change] <0).
Enrichment analysis
To elucidate the biological function of these miRNAs, the candidate target genes are subjected to functional enrichment analyses individually with GeneCodis3 software. The GeneCodis3 (http://genecodis.cnb.csic.es/), a web‐based application for singular and modular enrichment analysis, integrates information for various types of data (functional, regulatory, and structural) by searching for frequent patterns among annotations and evaluating statistical relevance 15. This new approach, superior to DAVID, Onto‐Express, ProfCom or FATIGO+ by overcoming the lack of term–term relationships in these analyses, profiles different sides of the same information and offers a more accurate interpretation of the data.
GeneCodis3 is applied to the functional enrichment analyses (KEGG and Panther pathways) of the significant target genes for each miRNA with a hyp‐c ≤0.05. Here, the hyp‐c, the P‐value for the hypergeometric test with multiple hypothesis corrections, represents the significance of the association between each enrichment pathway and the input list of genes.
Prognosis for differentially expressed miRNAs
The association between miRNA expression and survival for esophageal cancer patients is explored by separating the cases from each cohort into a group with a high expression level and another with a low expression level. The data‐driven approach 16, a novel computational method for the identification of miRNAs with a significant influence on survival and patient grouping, estimates the optimal threshold expression level for each miRNA for grouping of patients by maximizing the separation of the survival curves related to the risks of the disease. The log‐rank test, based on Kaplan–Meier plots, determines the differences among survival curves with respect to the miRNA expression levels. The univariate HR value, based on the Wald's test, determines statistical significance along with 95% CIs in the Cox proportional hazards model.
Results
Confirmation of five significantly differentially expressed miRNAs in esophageal cancer
The general strategy of our study is described in detail in Figure 1A. We obtained 12 relevant articles 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 from the 302 articles with the screening criteria (Fig. 1B; Table 1). The miRNAs data from these 12 articles were then extracted and analyzed with the novel robust rank aggregation (RRA) method 29 to identify miRNAs with statistically significant differences in expression. Overall, 217 miRNAs were upregulated by >2‐fold and 97 miRNAs were downregulated by <0.5‐fold. Among them, 14 miRNAs showed integrated significance with the RRA method, including four upregulated miRNAs and 10 downregulated miRNAs (Table 2). To validate these 14 miRNAs, their expression levels in 13 paired esophageal cancer and adjacent noncancerous tissue samples from TCGA were reanalyzed. Ultimately, five of them showed significantly different expression, including three upregulated miRNAs (hsa‐miR‐155‐5p, hsa‐miR‐21‐5p, and hsa‐miR‐223‐3p) and two downregulated miRNAs (hsa‐miR‐100‐5p and hsa‐miR‐133b), with P ≤ 0.05 (Table 2). Notably, previously published quantitative real‐time PCR analyses (RT‐PCR) further supported our results. Hsa‐mir‐155 was upregulated in esophageal squamous cell carcinoma tissues compared to paired adjacent noncancerous tissues 30. At least two independent studies found that hsa‐mir‐21 was significantly upregulated in tumors compared to normal tissues 31, 32. Expression of hsa‐mir‐223 was also markedly increased 33, 34. Hsa‐mir‐100 35 and hsa‐mir‐133b 36, 37 were notably downregulated in tumor tissues compared to normal tissues. Collectively, these results conclusively demonstrate that compared with adjacent noncancerous tissues, these five miRNAs show significantly different expression in esophageal cancer tissues.
Figure 1.
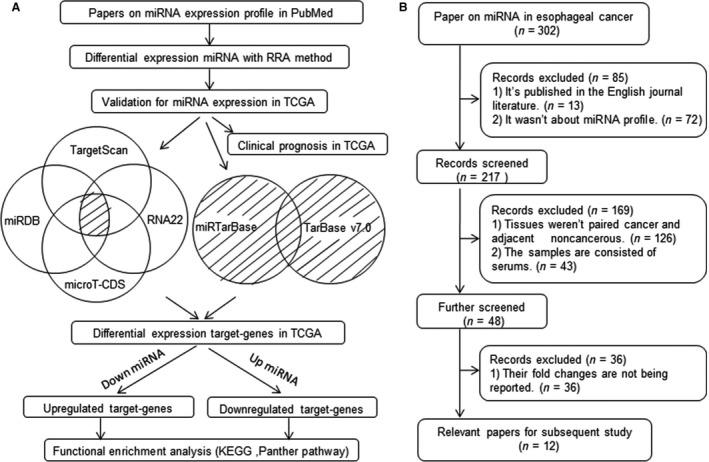
Flowchart for the study. (A) General strategy of the study. The RRA method (novel robust rank aggregation) was applied to obtaining significant miRNAs. Six databases including TargetScan, miRDB, microT‐CDS, RNA22, miRTarBase, and TarBase v7.0 were applied to obtaining the target genes of the miRNA. The dashed area in A represents the target genes we need. TCGA: The Cancer Genome Atlas. (B) Screening rules for articles on the miRNA expression profile in PubMed.
Table 1.
Characteristics of the analyzed articles
| Reference | Country | Cancer type | No. of tissue samples (cancer/normal) | Technique | No. of miRNAs in show |
|---|---|---|---|---|---|
| Feber (2008) | USA | EAC | 20 (10/10) | Microarray | 14 |
| Wijnhoven (2009) | Australia | EAC | 14 (7/7) | Microarray | 44 |
| Fu (2013) | China | ESCC | 68 (34/34) | Microarray | 12 |
| Kong (2011) | China | ESCC | 10 (5/5) | Microarray | 22 |
| Hong (2010) | China | ESCC | 20 (10/10) | Microarray | 12 |
| Kano (2010) | Japan | ESCC | 20 (10/10) | Microarray | 15 |
| Yang (2013) | China | ESCC | 6 (3/3) | Microarray | 15 |
| Fu (2013) | China | ESCC | 18 (9/9) | Microarray | 18 |
| Saad (2013) | USA | EAC | 68 (34/34) | Microarray | 21 |
| Liu (2013) | China | EAC | 6 (3/3) | Microarray | 60 |
| Zang (2013) | China | ESCC | 6 (3/3) | Microarray | 65 |
| Wu (2013) | USA | EAC | 70 (35/35) | Microarray | 138 |
EAC, esophageal adenocarcinoma; ESCC, esophageal squamous cell carcinoma.
Table 2.
miRNA expression of esophageal cancer samples in published articles and the TCGA
| miRNA | Stem‐loop | miRNA expression in article | miRNA expression in TCGA | ||
|---|---|---|---|---|---|
| Tumor versus noncancerous | P‐value | log2 (fold change) | P‐value | ||
| hsa‐miR‐100‐5p | hsa‐mir‐100 | Downregulated | 0.00002 | −1.695 | 0.04018 |
| hsa‐miR‐133b | hsa‐mir‐133b | Downregulated | 0.02949 | −2.097 | 0.00451 |
| hsa‐miR‐155‐5p | hsa‐mir‐155 | Upregulated | 0.00580 | 0.906 | 0.00136 |
| hsa‐miR‐21‐5p | hsa‐mir‐21 | Upregulated | 0.00020 | 1.138 | 0.00008 |
| hsa‐miR‐223‐3p | hsa‐mir‐223 | Upregulated | 0.00516 | 0.949 | 0.00197 |
| hsa‐miR‐424‐5p | hsa‐mir‐424 | Upregulated | 0.02595 | −0.10022 | 0.09163 |
| hsa‐miR‐375 | hsa‐mir‐375 | Downregulated | 0.00027 | −1.82663 | 0.13509 |
| hsa‐miR‐205‐5p | hsa‐mir‐205 | Downregulated | 0.00131 | 1.47158 | 0.41074 |
| hsa‐miR‐143‐3p | hsa‐mir‐143 | Downregulated | 0.00280 | −1.72661 | 0.10537 |
| hsa‐miR‐203a‐3p | hsa‐mir‐203a | Downregulated | 0.00699 | −1.09907 | 0.60401 |
| hsa‐miR‐192‐5p | hsa‐mir‐192 | Downregulated | 0.01563 | 0.27979 | 0.20536 |
| hsa‐miR‐27b‐3p | hsa‐mir‐27b | Downregulated | 0.02094 | −0.79117 | 0.15092 |
| hsa‐miR‐194‐5p | hsa‐mir‐194‐1 | Downregulated | 0.03672 | 0.07901 | 0.29485 |
| hsa‐let‐7c | hsa‐let‐7c | Downregulated | 0.04733 | −1.24124 | 0.13345 |
miRNA expression in this article is tested with the novel RRA method. miRNA expression in TCGA is tested with paired t‐test (two‐sided). The miRNA with bold text is coincident and significant between published article data and TCGA data.
Predicting, integrating, and verifying target genes
To further explore the function and mechanism of these five miRNAs in esophageal cancer, we identified the predicted target genes for each miRNA in six databases with the given integration strategy. The expression levels of these target genes were analyzed in 13 paired esophageal cancer and adjacent noncancerous tissue samples. In total, 1969 target genes were predicted and 686 of these target genes are verified as having significantly different expression between cancer and non‐cancer tissues (P ≤ 0.05). Then, 384 target genes were confirmed with a further filter (hsa‐miR‐100‐5p and hsa‐miR‐133b: log2 [fold change] >0; hsa‐miR‐155‐5p, hsa‐miR‐21‐5p, and hsa‐miR‐223‐3p: log2 [fold change] <0) (Table 3).
Table 3.
Significant miRNA target genes in TCGA
| miRNA | Significant target genes (P ≤ 0.05 and the log2 [fold change]) |
|---|---|
| miR‐100‐5p | CDK6, EIF5A, PSMA5, BAZ1B, LSM5, ATP1B3, RNFT1, CLCN5, COL4A1, RAB15, NKX3‐1, RB1, DDX21, EWSR1, YWHAE, NMT1, TTC30A, LIN28B, AP1AR, ABCB6, GNA13, GRB2, WDR4, ORC5, CCZ1B, CTDSPL2 |
| miR‐133b | SMARCD1, MET, ZNF131, FOXC1, COL5A3, SLC39A1 |
| miR‐155‐5p | CAB39L, METTL7A, INA, C3orf18, TTC37, PGRMC2, MITF, CD36, ATPAF1, PCDH9, MUT, TBC1D14, GABARAPL1, PDCD4, KIAA0430, KRCC1, ANAPC16, SETMAR, CDH2, EEF1A2, ZNF652, ERGIC1, RAB11FIP2, SAP30L, ZIC3, PHF17, GPT2, AGTR1, SKIV2L2, GHITM, DMD, ATP6V1H, PNPLA4, STIM1, NOVA1, TAB 2, JUN, SACM1L, CBR4, PCYOX1, ZNF493, GPM6B, OXCT1, AKR1C3, PALLD, CFL2, CAT, HSD17B12, IPO8, LTN1, PAK7, RIOK2, EIF3L, MPP2, UAP1, CYP2U1, FLNA, KANK2, TMBIM6, PIK3R1, MRS2, FOXO3, FGF2, SLC9A3R2, MPP5, MARCH2, PACSIN2, PRKAR2A, ARL6IP5, OSBPL9, ALDH1A2, PEBP1, ALDH3A2, NSA2, SNAP29, AGL, DDX17, ECI1, FAR1, AP1G1, INTS6, CRAT, MAN1A2, GLIPR2, PSIP1, XPC, MLH1, NR3C1, WRB, TNKS1BP1, RBPJ, TMED7‐TICAM2, CLIC4, SMAD4, SOX1, CSNK1A1, C16orf62, UGDH, SPECC1, NCKAP1, LONP2, TSHZ3, ZNF561, TRAK1, CTNNA1, RAB27B, ADH5, RAB6C, CLGN, CHD9, MEF2A, SNTB2, PKIA, LARS |
| miR‐21‐5p | FOXN3, FBXL17, LONRF2, CPEB3, LIFR, EPM2A, PAIP2B, PGRMC2, TGFBR3, LYRM7, LIMCH1, PDCD4, SASH1, ECI2, GPD1L, FBXL5, ARHGAP24, PURA, NFIA, FMOD, DCAF8, BTG2, CCNG1, ACAT1, KLF9, RNF11, UTRN, NBEA, ZBTB47, RAB11FIP2, DOCK3, SLAIN2, PRKAB2, FBXO3, ELAVL4, C20orf194, ISCU, PPM1L, PHF17, NEK1, RECK, COBLL1, MOAP1, GNAQ, DMD, ZBTB20, PDGFD, DDR2, FKBP5, PBX1, BDH2, FNBP1, MYO9A, TACC1, TIMP3, USP47, CYBRD1, MEGF9, BOC, SACM1L, DYNC1LI2, ATRX, FILIP1L, GOLGA4, ZBTB8A, TPRG1L, PALLD, TCF21, MYEF2, ZNF667, SOD3, PPAP2A, RHOB, WNK3, MKNK2, RUFY3, CALD1, TMX4, PRICKLE2, WFS1, SERPINI1, TUBGCP5, PHACTR2, SLMAP, PIK3R1, ARID4A, FERMT2, FAM63B, RNF38, IPP, SATB1, PER3, WDR47, TGFBR2, WDR7, PIGN, KLHL3, TSC1, MPP5, MON2, SEC63, CDS2, ARHGEF12, REV3L, SYBU, APPL1, AGGF1, JMY, BTBD3, WNK1, SESN1, DAAM1, TMEM56, PRKCE, SERAC1, ZYG11B, APOLD1, SPG11, FIGN, ANKRD46, PTGFR, PKD2, TM9SF3, MPDZ, RXRA, PTEN, CEP104, ITSN2, EIF4EBP2, CLOCK, ATP2B4, DCAF10, HECTD1, EIF2AK3, ZADH2, CSNK1A1, ESYT2, OSR1, CAPN2, DOCK8, NR2C2, MYCBP2, TEK, PTX3, VCL, ZRANB1, CYP4V2, FAM46A, TSHZ3, GPR64, SREK1, NIN, TPM1, PPP1R3B, EIF4A2, ARIH2, RAB6C, MLXIP, MTMR12, ABCD3, PPARA, MEF2A, NEK7, TOPORS, LARS |
| miR‐223‐3p | SLC2A4, ERO1LB, NFIA, FBXO8, RHOB, NFIX, ZFHX3, FBXW7, EPB41L3 |
The targeted genes participating in pathways (KEGG and Panther enrichment pathways) are tested in the TCGA database, and then screening with the rules for hsa‐miR‐100‐5p and hsa‐miR‐133b: P ≤ 0.05, and log2 (fold change) >0; hsa‐miR‐155‐5p, hsa‐miR‐21‐5p and hsa‐miR‐223‐3p: P ≤ 0.05, and log2 (fold change) <0.
Functional enrichment of target genes for miRNA
Overall, 38 KEGG pathways (Kyoto Encyclopedia of Genes and Genomes), and 22 Panther pathways were enriched for the target genes of hsa‐miR‐100‐5p, hsa‐miR‐133b, hsa‐miR‐155‐5p, hsa‐miR‐21‐5p, and hsa‐miR‐223‐3p.
In the KEGG pathways (Table 4), the target genes for hsa‐miR‐100‐5p were involved in cell growth and death, the immune system, the digestive system, and various cancers. Hsa‐miR‐133b functions as a signaling molecule and in cell‐cell interaction, cell communication, the digestive system, and cancer. Hsa‐miR‐155‐5p functions in cell communication, cell motility, transport and catabolism, signal transduction, cancer, and the immune system. Hsa‐miR‐223 functions in cancer protein folding, sorting, and degradation.
Table 4.
KEGG pathway of the target genes for these five miRNAs
| miRNA | Classification | KEGG pathway | Hyp‐c | Gene in pathway |
|---|---|---|---|---|
| miR‐155‐5p | Cell communication | Focal adhesion | 0.036 | FLNA, PIK3R1, JUN, PAK7 |
| Cell motility | Regulation of actin cytoskeleton | 0.016 | FGF2, CFL2, NCKAP1, PIK3R1, PAK7 | |
| Transport and catabolism | Peroxisome | 0.023 | CAT, CRAT, FAR1 | |
| Signal transduction | ErbB signaling pathway | 0.030 | PIK3R1, JUN, PAK7 | |
| Cancers | Pathways in cancer | 0.004 | FGF2, CTNNA1, PIK3R1, JUN, MITF, SMAD4, MLH1 | |
| Immune system | T‐cell receptor signaling pathway | 0.036 | PIK3R1, JUN, PAK7 | |
| Toll‐like receptor signaling pathway | 0.037 | PIK3R1, JUN, TAB 2 | ||
| miR‐21‐5p | Cell communication | Focal adhesion | 0.011 | CAPN2, PDGFD, PIK3R1, PTEN, VCL |
| Tight junction | 0.010 | MPP5, PTEN, MPDZ, PRKCE | ||
| Cell growth and death | p53 pathway feedback loops 2 | 0.005 | PIK3R1, PTEN, CCNG1 | |
| p53 signaling pathway | 0.009 | SESN1, PTEN, CCNG1 | ||
| Signal transduction | Phosphatidylinositol signaling system | 0.012 | PIK3R1, CDS2, PTEN | |
| Wnt signaling pathway | 0.041 | PRICKLE2, CSNK1A1, DAAM1 | ||
| Folding, sorting, and degradation | Protein processing in endoplasmic reticulum | 0.012 | CAPN2, EIF2AK3, SEC63, WFS1 | |
| Cell growth | Angiogenesis | 0.006 | TEK, PDGFD, RHOB, PIK3R1, PRKCE | |
| Cancers | Pathways in cancer | 0.025 | RXRA, APPL1, TGFBR2, PIK3R1, PTEN | |
| Endocrine system | Adipocytokine signaling pathway | 0.009 | RXRA, PRKAB2, PPARA | |
| Insulin signaling pathway | 0.007 | TSC1, MKNK2, PRKAB2, PPP1R3B, PIK3R1 | ||
| Insulin pathway‐protein kinase B signaling cascade | 0.005 | TSC1, PIK3R1, PTEN | ||
| Immune system | Fc gamma R‐mediated phagocytosis | 0.014 | PPAP2A, PIK3R1, PRKCE | |
| miR‐223‐3p | Folding, sorting, and degradation | Protein processing in endoplasmic reticulum | 0.041 | ERO1LB |
| Ubiquitin‐mediated proteolysis | 0.042 | FBXW7 | ||
| miR‐100‐5p | Cell growth and death | Cell cycle | 0.000 | CDK6, ANAPC1, YWHAE, ORC2, ORC5, PLK1, RB1 |
| Oocyte meiosis | 0.017 | ANAPC1, YWHAE, PLK1 | ||
| Replication and repair | Base excision repair | 0.018 | APEX1, HMGB1 | |
| Cancer | Pathways in cancer | 0.011 | CDK6, NKX3‐1, GRB2, COL4A1, RB1 | |
| Digestive system | Protein digestion and absorption | 0.012 | ACE2, COL4A1, ATP1B3 | |
| Immune system | Chemokine signaling pathway | 0.012 | CCL7, GRB2, CXCL16, CXCL13 | |
| miR‐133b | Cell communication | Adherens junction | 0.025 | MET |
| Focal adhesion | 0.007 | MET, COL5A3 | ||
| Signaling molecules and interaction | Cytokine‐cytokine receptor interaction | 0.048 | MET | |
| ECM‐receptor interaction | 0.023 | COL5A3 | ||
| Cancers | Melanoma | 0.029 | MET | |
| Renal cell carcinoma | 0.029 | MET | ||
| Digestive system | Protein digestion and absorption | 0.024 | COL5A3 |
In the Panther pathway analysis (Table 5), the target genes for hsa‐miR‐133b were enriched in signal transduction. Hsa‐miR‐155‐5p mainly functions in cancer development and signal transduction. Hsa‐miR‐21‐5p mainly functions in cell growth and death, cancer development, the endocrine system, and signal transduction. Hsa‐miR‐223‐3p functions in cell growth and death, cancer development, and signal transduction. However, no Panther pathway was enriched for target genes of hsa‐miR‐100‐5p.
Table 5.
Panther pathway of the target genes for hsa‐miR‐100‐5p, hsa‐miR‐133b, hsa‐miR‐155‐5p, hsa‐miR‐21‐5p, and hsa‐miR‐223‐3p
| miRNA | Classification | Panther pathway | Hyp‐c | Target genes in pathway |
|---|---|---|---|---|
| miR‐133b | Signal transduction | Wnt signaling pathway | 0.048 | SMARCD1 |
| miR‐155‐5p | Development | Angiogenesis | 0.048 | PIK3R1, FGF2, RBPJ, JUN |
| Signal transduction | Hedgehog signaling pathway | 0.033 | PRKAR2A, CSNK1A1 | |
| Wnt signaling pathway | 0.035 | SMAD4, CDH2, CSNK1A1, PCDH9, CTNNA1 | ||
| miR‐21‐5p | Cell growth and death | p53 pathway | 0.028 | CCNG1, PIK3R1, PTEN |
| p53 pathway by glucose deprivation | 0.023 | TSC1, PRKAB2 | ||
| p53 pathway feedback loops 2 | 0.015 | CCNG1, PIK3R1, PTEN | ||
| PDGF signaling pathway | 0.023 | PIK3R1, MKNK2, RHOB, NIN | ||
| Development | Angiogenesis | 0.012 | PIK3R1, PRKCE, PDGFD, RHOB, TEK | |
| Endothelin signaling pathway | 0.025 | PIK3R1, PRKCE, GNAQ | ||
| Endocrine system | Insulin/IGF pathway‐protein kinase B signaling cascade | 0.021 | PIK3R1, TSC1, PTEN | |
| Signal transduction | Hypoxia response via HIF activation | 0.027 | PIK3R1, PTEN | |
| PI3 kinase pathway | 0.015 | PIK3R1, GNAQ, PTEN | ||
| miR‐223‐3p | Cell growth and death | PDGF signaling pathway | 0.047 | RHOB |
| Development | Angiogenesis | 0.046 | RHOB | |
| Axon guidance mediated by Slit/Robo | 0.028 | RHOB | ||
| Cytoskeletal regulation by Rho GTPase | 0.032 | RHOB | ||
| Ras Pathway | 0.042 | RHOB | ||
| Signal transduction | Integrin signaling pathway | 0.041 | RHOB | |
| Notch signaling pathway | 0.033 | FBXW7 |
No Panther pathway is enriched for target genes of hsa‐miR‐100‐5p.
Effects of differentially expressed miRNAs on prognosis
To evaluate the prognostic impact of these five candidate miRNAs, survival and Cox regression analyses were applied to determine the risk of death and disease progression related to miRNAs in 185 esophageal cancer patient samples (Table 6). Owing to the complexity of miRNA metabolism, we selected the stem‐loop of mature miRNAs to standardize the analysis. The survival analysis results (Fig. 2) show that hsa‐mir‐100 and hsa‐mir‐133b were significantly associated with 3‐year and 5‐year survival, respectively. However, the other three miRNAs showed no significant difference in both 3‐year and 5‐year survival. Additionally, high hsa‐mir‐223 level was associated with a better prognosis in cancer, in contrast to the other miRNAs.
Table 6.
Characteristics of the 185 esophageal cancer patients in TCGA
| Characteristics | Frequency (No.) |
|---|---|
| Age | |
| <40 | 1.6% (3) |
| 40–49 | 11.9% (20) |
| 50–59 | 30.3% (56) |
| 60–69 | 18.9% (35) |
| 70+ | 31.4% (58) |
| Unknown | 7.0% (13) |
| Sex | |
| Male | 80.5% (149) |
| Female | 12.4% (23) |
| Unknown | 7.0% (13) |
| Alcohol history | |
| No | 27.0% (50) |
| Yes | 54.1% (100) |
| Unknown | 8.1% (15) |
| Smoking history | |
| Nonsmoker | 27.0% (50) |
| Current smoker | 19.5% (36) |
| ≤15 years | 18.9% (35) |
| >15 years | 18.4% (34) |
| Unknown | 16.2% (30) |
| Neoplasm histologic grade | |
| G1 | 9.7% (18) |
| G2 | 38.9% (72) |
| G3 | 23.2% (43) |
| GX | 21.1% (39) |
| Unknown | 7.0% (13) |
| Clinical stage | |
| Stage I | 1.6% (3) |
| Stage II | 14.6% (27) |
| Stage III | 11.4% (21) |
| Stage IV | 5.4% (10) |
| Unknown | 67.0% (124) |
| Radiation therapy | |
| No treatment | 66.5% (123) |
| Treatment | 16.8% (31) |
| Unknown | 16.8% (31) |
| Treatment prior to surgery | |
| No treatment | 26.5% (49) |
| Radiation and chemotherapy | 1.1% (2) |
| Unknown | 74.6% (138) |
| Additional pharmaceutical therapy | |
| No | 3.2% (6) |
| Yes | 5.4% (10) |
| Unknown | 91.4% (169) |
Figure 2.
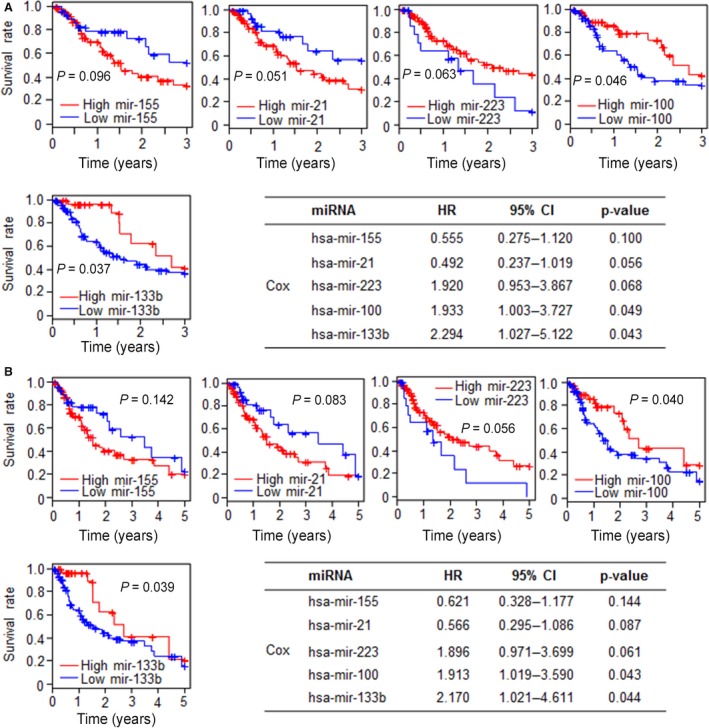
Prognostic value of miRNA in esophageal cancer. The survival rate analysis and Cox regression analysis are applied to hsa‐miR‐100‐5p, hsa‐miR‐133b, hsa‐miR‐155‐5p, hsa‐miR‐21‐5p, and hsa‐miR‐223‐3p for determination of prognosis in esophageal cancer. Here, the P value for survival analysis describes the significance of the log‐rank. (A) 3‐year‐survival rate analysis and 3‐year‐Cox regression analysis. (B) 5‐year‐survival rate analysis and 5‐year‐Cox regression analysis. HR, hazard ratio; CI, confidence interval.
Discussion
In general, the individual risk grade and decision of treatment largely depend on pathological and clinical factors, which show great variation between individuals, thereby influencing the predictive accuracy in cancer. A recent study demonstrates that differential expression of miRNAs can reflect tissue‐specific expression signatures through promotion or suppression of tumor development and progression 38. The application of miRNA‐based biomarkers for diagnosis thus provides a promising alternative.
In our study, we identified five differentially expressed miRNAs in esophageal cancer with comprehensive analyses of reported miRNA‐microarray sequencing data, miRNA‐generated sequencing data from the TCGA database, and RT‐PCR data from published studies. Here, we used paired cancer and adjacent noncancerous tissue samples to test miRNA and gene expression, which is more representative of the physiological status in the body than cell or serum samples. These five miRNAs are mainly involving in regulating esophageal cancer occurrence and development, according to comprehensive considerations of KEGG and Panther enrichment analyses. Additionally, high expressions of hsa‐mir‐100 and hsa‐mir‐133b, and low expressions of hsa‐mir‐155 and hsa‐mir‐21 tended to show a better prognosis, especially for hsa‐mir‐100 and hsa‐mir‐133b, which suggested high clinical value for esophageal cancer. Notably, hsa‐mir‐223 levels were increased in cancer tissue compared to normal tissue, but a good prognosis was also associated with a high expression level. In our results, hsa‐miR‐223‐3p mainly participates in protein folding, sorting, and degradation (Tables 4 and 5), which may underlie the anti‐tumor effects. Some studies have regarded the expression levels of different miRNAs as prognostic biomarkers in various cancers such as lung cancer 39, gastric cancer 40, and colorectal cancer 41. In our work, hsa‐miR‐100‐5p, hsa‐miR‐133b, hsa‐miR‐155‐5p, and hsa‐miR‐21‐5p were identified as differentially regulated, and associated with good prognosis; therefore, they can be used as miRNA biomarkers to increase the predictive ability in esophageal cancer, which will provide more choices in the treatment of esophageal cancer with further study. Hsa‐miR‐223‐3p also showed different expression in esophageal cancer tissue. Two or more of these miRNAs as testing indices could further improve the diagnostic accuracy of esophageal cancer.
Some significant target genes in our analysis have been verified experimentally in previous studies, for example, SMARCD 42 and PTEN 43, which play important roles in esophageal cancer occurrence and development (Table 5). Study of the other genes in Tables 4 and 5 may provide more constructive suggestions for esophageal cancer prognosis and treatment.
Conflict of Interest
The authors declare no conflicts of interest.
Supporting information
Acknowledgments
This work is supported by the National Natural Science Foundation of China (grant nos. 81572780, 31271383 to K.F.T.). We appreciate Ke‐Qing Shi (Wenzhou Medical University, Wenzhou, China), and Peng Fang (Wenzhou Medical University, Wenzhou, China) for technical assistance.
Cancer Medicine 2017; 6(8):1893–1903
References
- 1. Stewart, B. W. , and C. Wild. International Agency for Research on Cancer & World Health Organization. World Cancer Report 2014.
- 2. Tong, L. , Yuan Y., and Wu S.. 2015. Therapeutic microRNAs targeting the NF‐kappa B signaling circuits of cancers. Adv. Drug Deliv. Rev. 81:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lin, S. , and Gregory R. I.. 2015. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 15:321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Calin, G. A. , Dumitru C. D., Shimizu M., Bichi R., Zupo S., Noch E., et al. 2002. Frequent deletions and down‐regulation of micro‐RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA 99:15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gao, J. J. , Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Love, M. I. , Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kozomara, A. , and Griffiths‐Jones S.. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42:D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kolde, R. , Laur S., Adler P., and Vilo J.. 2012. Robust rank aggregation for gene list integration and meta‐analysis. Bioinformatics 28:573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wong, N. , and Wang X. W.. 2015. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 43:D146–D152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Agarwal, V. , Bell G. W., Nam J. W., and Bartel D. P.. 2015. Predicting effective microRNA target sites in mammalian mRNAs. Elife 4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Paraskevopoulou, M. D. , Georgakilas G., Kostoulas N., Vlachos I. S., Vergoulis T., Reczko M., et al. 2013. DIANA‐microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 41:W169–W173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miranda, K. C. , Huynh T., Tay Y., Ang Y. S., Tam W. L., Thomson A. M., et al. 2006. A pattern‐based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell 126:1203–1217. [DOI] [PubMed] [Google Scholar]
- 13. Vlachos, I. S. , Paraskevopoulou M. D., Karagkouni D., Georgakilas G., Vergoulis T., Kanellos I., et al. 2015. DIANA‐TarBase v7.0: indexing more than half a million experimentally supported miRNA: mRNA interactions. Nucleic Acids Res. 43:D153–D159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hsu, S. D. , Tseng Y. T., Shrestha S., Lin Y. L., Khaleel A., Chou C. H., et al. 2014. miRTarBase update 2014: an information resource for experimentally validated miRNA‐target interactions. Nucleic Acids Res. 42:D78–D85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tabas‐Madrid, D. , Nogales‐Cadenas R., and Pascual‐Montano A.. 2012. GeneCodis3: a non‐redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 40:W478–W483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Motakis, E. , Ivshina A. V., and Kuznetsov V. A.. 2009. Data‐driven approach to predict survival of cancer patients estimation of microarray genes' prediction significance by cox proportional hazard regression model. IEEE Eng. Med. Biol. Mag. 28:58–66. [DOI] [PubMed] [Google Scholar]
- 17. Feber, A. , Xi L., Luketich J. D., Pennathur A., Landreneau R. J., Wu M., et al. 2008. MicroRNA expression profiles of esophageal cancer. J. Thorac. Cardiovasc. Surg. 135:255–260; discussion 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wijnhoven, B. P. L. , Hussey D. J., Watson D. I., Tsykin A., Smith C. M., Michael M. Z., et al. 2010. MicroRNA profiling of Barrett's oesophagus and oesophageal adenocarcinoma. Br. J. Surg. 97:853–861. [DOI] [PubMed] [Google Scholar]
- 19. Fu, H. L. , Wu D. P., Wang X. F., Wang J. G., Jiao F., Song L. L., et al. 2013. Altered miRNA expression is associated with differentiation, invasion, and metastasis of esophageal squamous cell carcinoma (ESCC) in patients from Huaian, China. Cell Biochem. Biophys. 67:657–668. [DOI] [PubMed] [Google Scholar]
- 20. Kong, K. L. , Kwong D. L. W., Chan T. H. M., Law S. Y. K., Chen L. L., Li Y., et al. 2012. MicroRNA‐375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin‐like growth factor 1 receptor. Gut 61:33–42. [DOI] [PubMed] [Google Scholar]
- 21. Hong, L. , Han Y., Zhang H. W., Li M. B., Gong T. Q., Sun L., et al. 2010. The prognostic and chemotherapeutic value of miR‐296 in esophageal squamous cell carcinoma. Ann. Surg. 251:1056–1063. [DOI] [PubMed] [Google Scholar]
- 22. Kano, M. , Seki N., Kikkawa N., Fujimura L., Hoshino I., Akutsu Y., et al. 2010. miR‐145, miR‐133a and miR‐133b: tumor‐suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int. J. Cancer 127:2804–2814. [DOI] [PubMed] [Google Scholar]
- 23. Yang, M. , Liu R., Sheng J. Y., Liao J., Wang Y., Pan E. C., et al. 2013. Differential expression profiles of microRNAs as potential biomarkers for the early diagnosis of esophageal squamous cell carcinoma. Oncol. Rep. 29:169–176. [DOI] [PubMed] [Google Scholar]
- 24. Fu, M. G. , Li S., Yu T. T., Qian L. J., Cao R. S., Zhu H., et al. 2013. Differential expression of miR‐195 in esophageal squamous cell carcinoma and miR‐195 expression inhibits tumor cell proliferation and invasion by targeting of Cdc42. FEBS Lett. 587:3471–3479. [DOI] [PubMed] [Google Scholar]
- 25. Saad, R. , Chen Z., Zhu S. M., Jia P. L., Zhao Z. M., Washington M. K., et al. 2013. Deciphering the unique MicroRNA signature in human esophageal adenocarcinoma. PLoS ONE 8:e64463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu, S. G. , Qin X. G., Zhao B. S., Qi B., Yao W. J., Wang T. Y., et al. 2013. Differential expression of miRNAs in esophageal cancer tissue. Oncol. Lett. 5:1639–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zang, W. Q. , Wang Y. Y., Du Y. W., Xuan X. Y., Wang T., Li M., et al. 2014. Differential expression profiling of microRNAs and their potential involvement in esophageal squamous cell carcinoma. Tumor Biol. 35:3295–3304. [DOI] [PubMed] [Google Scholar]
- 28. Wu, X. F. , Ajani J. A., Gu J., Chang D. W., Tan W. Q., Hildebrandt M. A. T., et al. 2013. MicroRNA expression signatures during malignant progression from Barrett's esophagus to esophageal adenocarcinoma. Cancer Prev. Res. 6:196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shi, K. Q. , Lin Z., Chen X. J., Song M., Wang Y. Q., Cai Y. J., et al. 2015. Hepatocellular carcinoma associated microRNA expression signature: integrated bioinformatics analysis, experimental validation and clinical significance. Oncotarget 6:25093–25108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang, J. , Cheng C., Yuan X., He J. T., Pan Q. H., and Sun F. Y.. 2014. microRNA‐155 acts as an oncogene by targeting the tumor protein 53‐induced nuclear protein 1 in esophageal squamous cell carcinoma. Inter. J. Clin. Exper. Pathol. 7:602–610. [PMC free article] [PubMed] [Google Scholar]
- 31. Mori, Y. , Ishiguro H., Kuwabara Y., Kimura M., Mitsui A., Ogawa R., et al. 2009. MicroRNA‐21 induces cell proliferation and invasion in esophageal squamous cell carcinoma. Mol. Med. Rep. 2:235–239. [DOI] [PubMed] [Google Scholar]
- 32. Nouraee, N. , van Roosbroeck K., Vasei M., Semnani S., Samaei N. M., Naghshvar F., et al. 2013. Expression, tissue distribution and function of miR‐21 in esophageal squamous cell carcinoma. PLoS ONE 8:e73009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu, C. , Li M., Hu C., and Duan H.. 2014. Clinical significance of serum miR‐223, miR‐25 and miR‐375 in patients with esophageal squamous cell carcinoma. Mol. Biol. Rep. 41:1257–1266. [DOI] [PubMed] [Google Scholar]
- 34. Warnecke‐Eberz, U. , Chon S. H., Holscher A. H., Drebber U., and Bollschweiler E.. 2015. Exosomal onco‐miRs from serum of patients with adenocarcinoma of the esophagus: comparison of miRNA profiles of exosomes and matching tumor. Tumour Biol. 36:4643–4653. [DOI] [PubMed] [Google Scholar]
- 35. Zhou, S. , Yang B., Zhao Y., Xu S., Zhang H., and Li Z.. 2014. Prognostic value of microRNA‐100 in esophageal squamous cell carcinoma. J. Surg. Res. 192:515–520. [DOI] [PubMed] [Google Scholar]
- 36. Chen, Z. , Saad R., Jia P., Peng D., Zhu S., Washington M. K., et al. 2013. Gastric adenocarcinoma has a unique microRNA signature not present in esophageal adenocarcinoma. Cancer 119:1985–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen, G. , Peng J., Zhu W., Tao G., Song Y., Zhou X., et al. 2014. Combined downregulation of microRNA‐133a and microRNA‐133b predicts chemosensitivity of patients with esophageal squamous cell carcinoma undergoing paclitaxel‐based chemotherapy. Med. Oncol. 31:263. [DOI] [PubMed] [Google Scholar]
- 38. Pichler, M. , and Calin G. A.. 2015. MicroRNAs in cancer: from developmental genes in worms to their clinical application in patients. Br. J. Cancer 113:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang, R. J. , Zheng Y. H., Wang P., and Zhang J. Z.. 2015. Serum miR‐125a‐5p, miR‐145 and miR‐146a as diagnostic biomarkers in non‐small cell lung cancer. Inter. J. Clin. Exper. Pathol. 8:765–771. [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou, X. , Zhu W., Li H., Wen W., Cheng W. F., Wang F., et al. 2015. Diagnostic value of a plasma microRNA signature in gastric cancer: a microRNA expression analysis. Sci. Rep. 5:11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li, J. L. , Liu Y., Wang C., Deng T., Liang H. W., Wang Y. F., et al. 2015. Serum miRNA expression profile as a prognostic biomarker of stage II/III colorectal adenocarcinoma. Sci. Rep. 5:12921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Streppel, M. M. , Pai S., Campbell N. R., Hu C., Yabuuchi S., Canto M. I., et al. 2013. MicroRNA 223 is upregulated in the multistep progression of Barrett's esophagus and modulates sensitivity to chemotherapy by targeting PARP1. Clin. Cancer Res. 19:4067–4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li, P. , Mao W. M., Zheng Z. G., Dong Z. M., and Ling Z. Q.. 2013. Down‐regulation of PTEN expression modulated by dysregulated miR‐21 contributes to the progression of esophageal cancer. Dig. Dis. Sci. 58:3483–3493. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


