Summary
Transcription initiation entails chromatin opening followed by pre-initiation complex formation and RNA polymerase II recruitment. Subsequent polymerase elongation requires additional signals, resulting in increased residence time downstream of the start site, a phenomenon referred to as pausing. Here, we harnessed single-molecule footprinting to quantify distinct steps of initiation in vivo throughout the Drosophila genome. This identifies the impact of promoter structure on initiation dynamics in relation to nucleosomal occupancy. Additionally, perturbation of transcriptional initiation reveals an unexpectedly high turnover of polymerases at paused promoters—an observation confirmed at the level of nascent RNAs. These observations argue that absence of elongation is largely caused by premature termination rather than by stable polymerase stalling. In support of this non-processive model, we observe that induction of the paused heat shock promoter depends on continuous initiation. Our study provides a framework to quantify protein binding at single-molecule resolution and refines concepts of transcriptional pausing.
Keywords: transcription, genomics, transcriptional pausing, DNA footprinting, single molecule, GTF, TBP
Graphical Abstract
Highlights
-
•
Genome-wide detection of protein-DNA contacts at single-molecule resolution
-
•
Simultaneous quantification of several transcription initiation intermediates
-
•
TATA box increases stability of the pre-initiation complex at promoters
-
•
High levels of polymerase turnover at the promoters of paused genes
Krebs et al. present a methodology to probe protein binding to the genome at the resolution of single DNA molecules, enabling disentanglement of binding heterogeneity at the promoter of genes, thereby revealing unexpected dynamics of RNA polymerase II at paused genes in Drosophila.
Introduction
In metazoans, launching transcription is a multistep process encompassing sequential binding of general transcription factors (GTFs) leading to the formation of a pre-initiation complex (PIC) and subsequent recruitment of RNA polymerase II (Pol II). PIC formation entails multiple protein-DNA contacts between GTFs (i.e., TATA binding protein [TBP]-TFIID) and conserved sequence elements of promoters (Louder et al., 2016). The loaded polymerase is activated by TFIIH-mediated phosphorylation allowing it to engage the DNA template and to transcribe a short RNA stretch followed by a transient pause 30–80 bp downstream of the transcription start site (TSS) (Jonkers and Lis, 2015). Subsequent transition of the engaged Pol II complex into a productive elongation complex requires 5′-capping of the RNA, which serves as an important checkpoint of the process.
In addition to reflect the time needed to assemble an elongation competent complex, pausing of Pol II allows post-initiation regulation of gene expression (Jonkers and Lis, 2015). It is, however, unclear how frequently pausing occurs and what weight it has in regulating genic output (Brannan et al., 2012, Ehrensberger et al., 2013, Henriques et al., 2013, Jonkers and Lis, 2015, Wagschal et al., 2012).
Direct probing of polymerase activity revealed imperfect correlation between the amount of Pol II loaded at promoters and the measured activity of the elongating polymerase (Core et al., 2012). This imbalance is particularly marked at some inducible genes including those that are activated upon heat shock in Drosophila melanogaster. These harbor high Pol II levels downstream of their TSS, yet display very low elongation activity (Jonkers and Lis, 2015). These genes are rapidly induced upon heat shock and show further strong activation if transcriptional elongation is enforced (Lis et al., 2000). Consequently, they serve as canonical examples for regulating genic output by controlling elongation, which in this particular case enables rapid induction during stress response (Guertin et al., 2010). This concept has been extrapolated to also function in control of gene induction during developmental and stimulus-responsive pathways (Gaertner and Zeitlinger, 2014, Henriques et al., 2013, Jonkers and Lis, 2015, Levine, 2011).
Current concept of transcriptional pausing favors a model where engaged Pol II is stabilized post-initiation, leading to the accumulation of transcriptionally competent polymerases downstream of the TSS. In support of this model, residence time of Pol II has recently been reported to be in the range of minutes at some genes (Buckley et al., 2014, Henriques et al., 2013, Shao and Zeitlinger, 2017). This paused state was proposed to be mediated through reinforced activity of negative elongation factor (NELF) and DRB sensitivity inducing factor (DSIF) possibly through the action of sequence-specific transcription factors (Jonkers and Lis, 2015). Alternatively, premature transcription termination and removal of Pol II could regulate transcriptional output post-initiation (Ehrensberger et al., 2013). Such termination would entail the combined action of exonucleases allowing decapping and degradation of the RNA protruding from the engaged polymerase (Brannan et al., 2012, Wagschal et al., 2012).
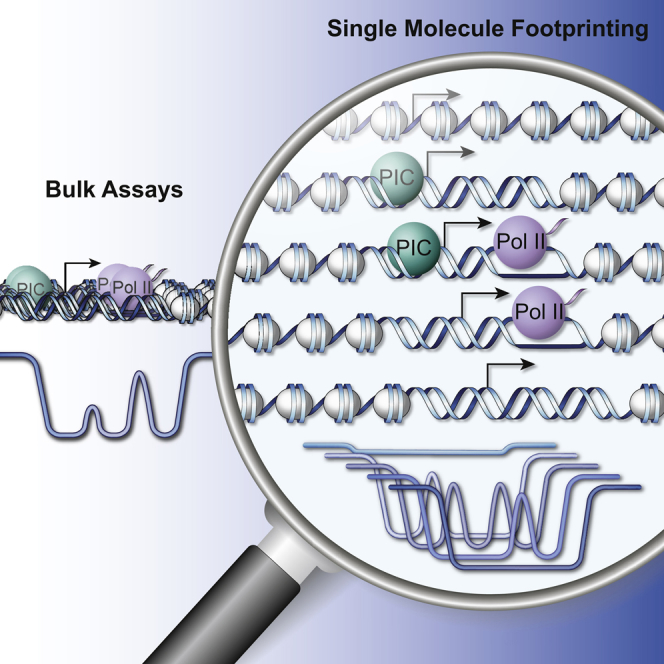
Our incomplete understanding of how early transcription events are regulated partly reflects the limited resolution of current approaches to probe binding and activity at promoters. Probing of Pol II and GTF binding has recently reached nucleotide resolution enabling to precisely locate binding of the transcription machinery in vivo (Pugh and Venters, 2016, Shao and Zeitlinger, 2017). Importantly, the above are bulk measurements that compile signals from many molecules. Due to this averaging, these are intrinsically unable to uncover the heterogeneity in protein-DNA interactions expected to result from complex binding events such as PIC formation and Pol II initiation (Louder et al., 2016).
To overcome these limitations, we establish a detection method that allows single-molecule footprinting of protein-DNA interactions genome-wide at high resolution. It simultaneously quantifies the occupancy of nucleosomes, the PIC, and Pol II at and downstream of the TSS. This permits to follow the changes in protein-DNA contacts associated with transcriptional activity. It identifies the effect of core promoter elements such as the TATA box on the dynamics of PIC formation. Furthermore, measuring changes in occupancy of polymerases upon chemically inhibiting initiation reveals that Pol II half-life at paused genes including the well-characterized Hsp70 promoter is equivalent to the one observed at normally elongating genes.
Results
Principles of Dual-Enzyme Single-Molecule Footprinting
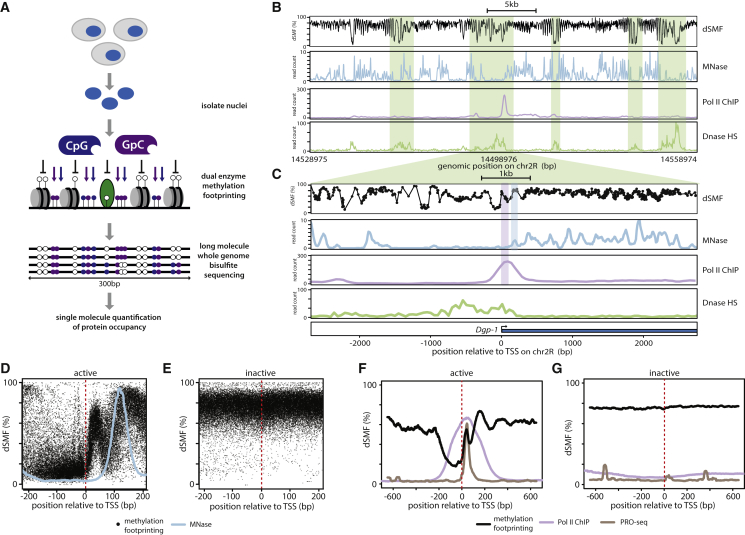
Detection and quantification of the multiple binding events that co-occur at promoters requires the ability to measure protein-DNA contacts at the level of single molecules. Using recombinant methyltranferases for footprinting combined with detection of their activity by bisulfite sequencing can probe nucleosome occupancy in intact nuclei at the resolution of single molecules (Kelly et al., 2012, Nabilsi et al., 2014). We thought to improve methyltransferase footprinting (Bell et al., 2010, Kelly et al., 2012, Nabilsi et al., 2014) in order to detect transcription initiation intermediates at the scale of the genome. To do so, we combine two recombinant methyl-transferases that target cytosines in different dinucleotide contexts (M.SssI targets CGs while M.CviPI methylates GCs) (Figure 1A). Both enzymes, when used separately or in combination, give remarkably similar footprinting profiles (R = ∼0.8, Figures S1A–S1C) arguing that the resulting signal reflects genuine DNA accessibility rather than individual biases of each enzyme. Importantly, their additive sequence specificity increases the median resolution to 7 bp (13 or 10 bp for M.SssI or M.CviPI alone), which is below the size of footprints created by transcription initiation factors. Bisulfite sequencing measures methylation at single cytosine resolution. Therefore, it can provide continuous information on neighboring cytosines as long as they reside within the same sequenced molecule (Figure 1A).
Figure 1.
High-Resolution Genomic Footprinting Detects Non-nucleosomal Binding Events
(A) Overview of dual-enzyme single-molecule footprinting (dSMF). Nuclei are isolated and incubated with recombinant cytosine methyl-transferases that target GpC (M.CviPI) and CpG (M.SssI). DNA bound by proteins is protected from methyltransferase activity (GpC, purple lollipops; CpG, blue lollipops), which results in distinct footprints of methylation (unmethylated, white lollipops). Combined treatment with both enzymes increases footprint resolution (∼7 bp). Resulting methylation is detected by whole-genome bisulfite sequencing at single-molecule and base-pair resolution. Sequencing is done with long reads (300 bp) to cover regions of interest within the same molecule.
(B) dSMF footprinting pattern of a 30-kb chromosomal region illustrating increased accessibility at regulatory regions. Shown is the inverse frequency of methylation (1-methylation (%)) (black line). Read counts for additional datasets are shown as smoothed signal (colored lines, datasets as indicated). Shaded green boxes indicate regions of increased accessibility as determined by DNase hypersensitivity (DHS).
(C) Zoom-in centered on the start site of the transcribed Dgp1 gene illustrating the presence of a non-nucleosomal footprint at the site of Pol II accumulation downstream of the TSS. Same tracks as in (B) except dSMF where the inverted methylation average for single cytosines is represented by black dots connected by a line. Position of the +1 nucleosome indicated by blue box, position of Pol II accumulation by purple box.
(D and E) Composite profile of footprinting signals at TSSs (dashed red line) of (D) active genes (top 10%, Pol II ChIP-seq) and (E) inactive genes (bottom 10%). Shown is the footprinting frequency (1-methylation [%]) of individual cytosines (black dots). The average MNase signal for the same regions is overlaid (light-blue line indicating MNase 5′ cut site). Note the footprint downstream of the TSSs of active genes that is not accounted for by nucleosomes.
(F and G) The non-nucleosomal footprint downstream of TSSs aligns with the site of Pol II accumulation. Shown are the averaged accessibility signal (black line), average Pol II occupancy (ChIP-seq, purple), and activity (PRO-seq, brown) for (F) active genes and (G) inactive genes as defined in (D) and (E).
We applied dual-enzyme single-molecule footprinting (dSMF) in Drosophila cells as an established model for studying early initiation events (Kwak et al., 2013, Lim et al., 2004, Nechaev et al., 2010). Components of the core transcription machinery are closely conserved from flies to mammals, and the absence of endogenous methylation in Drosophila prevents signal interference with footprinting. We generated high-coverage footprinting methylomes (∼40× median) in duplicate for two cell lines of divergent cellular origin with different transcriptional programs (Arnold et al., 2013) (Schneider-S2 and Ovarian Stem Cell [OSC]). Importantly, we optimized library preparation protocols to generate continuous footprinting information over long DNA stretches (∼300 bp).
An initial inspection of the resulting chromosomal profiles revealed that a large fraction of the genome is only weakly accessible to methyltransferase activity as expected for chromatinized DNA (Figure S1G). The distribution of footprints reveals a periodic structure highly reminiscent of nucleosomal phasing (Figures 1B and S1I). Similarly, we observe higher accessibility at linker regions, mirroring the activity of MNase (Figures 1B and S1I). In addition a subset of chromosomal regions show increased accessibility in a reproducible fashion (R > 0.89, Figures S1D, S1E, and S1I). A fraction of these regions show discrete differences between cell types (Figure S1F). In agreement with previous observations, these represent active regulatory regions that are free of nucleosomes (Figures 1B, 1C, and S1I) (Kelly et al., 2012, Nabilsi et al., 2014).
dSMF Detects Binding of Non-histone Proteins
While methylation footprinting is well documented to probe nucleosome organization (Kelly et al., 2012, Nabilsi et al., 2014), detection of binding by non-nucleosomal proteins such as transcription factors has also been shown to be possible in principle (Gal-Yam et al., 2006, Kelly et al., 2012, Levo et al., 2017). Consequently, we explored our dSMF profiles for such non-nucleosomal footprints. At promoters of highly expressed genes, we noticed the presence of a short (∼50 bp) footprint downstream of the TSSs and directly upstream of the highly phased +1 nucleosome (Figures 1C and 1D). This footprint is absent from inactive genes (Figure 1E). Comparison with MNase data indicates that this additional density does not reflect the presence of nucleosomes (Figure 1D). Instead, it nicely aligns with Pol II occupancy data suggesting that this footprint is caused by transcriptionally engaged RNA Pol II (Figures 1F, 1G, and S1H). Additionally, we noticed at a subset of promoters an additional density shortly upstream of the TSS at the same position where the PIC is reported to bind in vitro (−35:-22) (Figure 2A, upper panel). Together this suggests that high-resolution dSMF captures non-nucleosomal binding events at sites contacted by GTFs and Pol II.
Figure 2.
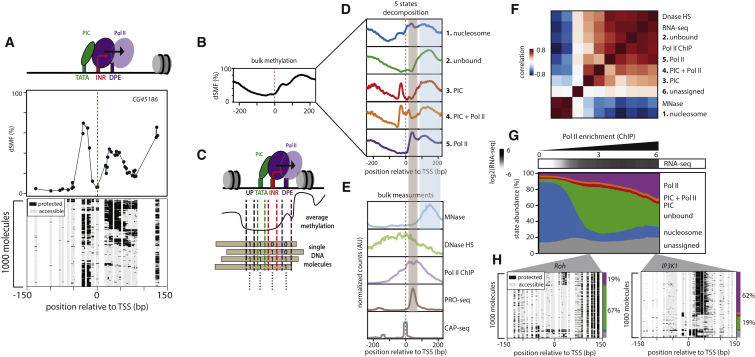
Pattern Decomposition into Discrete States of Transcriptional Initiation
(A) Single-molecule decomposition of footprinting signals over a single active promoter. Upper panel: graphic representation of promoter elements of the CG45186 gene. Middle panel: shown is the average footprint for single cytosines (black dots connected by a line, red line, TSS). Lower panel: stack of individual molecule reads illustrating the discrete distribution of accessible (methylated Cs, light gray) and protected regions (unmethylated Cs, black). DNA molecules measured by targeted amplicon bisulfite sequencing are organized by hierarchical clustering. Distribution of footprints illustrates the heterogeneity in protein binding at a single promoter within a population of cells.
(B) Average footprinting patterns centered on the TSS of the 10% most active promoters.
(C) Illustration of the strategy used to assign discrete states from dSMF datasets. Four bins anchored relative to core promoter elements were used to extract methylation from every sequenced molecule. Methylation was binarized within each bin creating a 4-bit vector leading to 16 possible combinations describing the state of each molecule. Five footprinting patterns were selected and defined as states (see STAR Methods).
(D) Average footprinting patterns of the five states (colored lines) centered on the TSS of active promoters computed after single-molecule separation. Sequencing reads were classified into five pre-defined states, and the average footprinting frequency was calculated for each covered position across the set of active promoters.
(E) Spatial alignment of footprinting signal with polymerase downstream of the TSSs (brown box) or positioned nucleosomes (blue box) across the same set of promoters. Average read count signal across active promoters is displayed for independent measurements.
(F) Global relation between state frequencies at promoters and independent bulk measurements of Pol II and nucleosomes depicted in form of a heatmap of similarity (Pearson correlation). States separate in two groups that either correlate with occupancy by the transcription machinery (Pol II ChIP-seq) or nucleosomes (MNase-seq) illustrating accurate state quantification.
(G) Distribution of state frequencies as a function of promoter activity. Cumulative bar plot depicting the distribution of state frequencies. Promoters were binned based on Pol II enrichment (log2 ChIP-seq), and the median frequency of each state was calculated within each bin. The frequency of each state is represented using the same color code as in (D). For comparison median expression (RNA-seq; reads per kilobase per million reads [RPKM]) is displayed (upper panel, white to black scale). A sharp decrease in nucleosome occupancy is observed between silent and lowly expressed genes, concomitant with a mirroring increase in unoccupied molecules. Promoters with increasing transcriptional activity display a reduction of unbound molecules concomitant with increasing Pol-II-bound molecules at a constant nucleosomal occupancy rate (∼10%).
(H) Single-locus examples of a weakly active promoter (left panel, Roh-Pol II fold enrichment 9×) versus a highly active promoter (right panel, IP3K1-Pol II fold enrichment 36×). Shown are stacks of individual molecules measured by targeted amplicon bisulfite sequencing, sorted into five states using the classification algorithm (methylated Cs, accessible, light gray; unmethylated Cs, protected, black). The vertical side bar depicts the frequency of each state using the same color code as 2D. The percentages of molecules in the Pol II or unbound state are indicated at the right side of the plot.
We next sought to investigate whether the sequence of protein-DNA binding events that occur during transcription initiation would create distinct patterns when analyzed at the single-molecule level. Indeed, such heterogeneity is evident when we cluster individual reads amplified from a specific promoter (Figure 2A, bottom panel). An active promoter displays fully accessible molecules, molecules with very large or discrete footprints at various positions around the TSS (exemplified in Figure 2A). While large footprints could reflect nucleosome occupancy, shorter ones (<50 bp) are unlikely to be able to accommodate nucleosomes. Instead, these probably represent PIC and Pol II occupancy since they spatially fit the expected binding regions for these protein complexes (Figure S2A).
Simultaneous Quantification of Multiple Transcription Initiation Intermediates
If these heterogeneous patterns represent distinct steps of the transcription initiation process, we should be able to link these patterns with occupancy of the protein complexes expected to create the footprints. In order to quantify pattern frequencies, we designed a molecular classifier that sorts molecules based on their footprints (Figure 2C). The separation was anchored around TSSs, and four collection bins were designed that cover regions contacted by the PIC and Pol II (Figures 2C, S2A, and S2B). For each individual molecule, the algorithm collects binarized methylation within the four bins creating 16 (24) possible combinations. We applied this method to the entire genome and quantified single-molecule footprints for ∼99% of those TSS that contain informative GCs or CG dinucleotides within all four collection bins. These represent 9,062 out of the 21,726 uniquely annotated TSS. Their almost complete experimental recovery argues for saturation in genome coverage.
Among the 16 theoretical patterns that were used for the classification, only the ones representing footprints occurring at promoters in vivo should be recurrently observed. Indeed, inspection of the frequencies of individual patterns across promoters identifies a subset of footprints that are frequently observed in vivo (Figure S2F). Interestingly, the frequencies of these abundant states tend to show correlation with promoter activity as defined by Pol II occupancy (Figure S2G). In opposition, the less abundant states tend to show no significant correlations with the tested features (Figure S2G). In order to facilitate data interpretation, we constructed a simplified set of five states (Figure S2C). This condenses the information of the most frequent ten states that show evidence of correlations with bulk data (Figures S2C and S2G). The remaining unassigned reads were compiled into a separate “unassigned” category (Figure S2F). Importantly, we observed high reproducibility between biological replicates in the frequency of each of the five states (R > 0.85 Figure S2I).
To delineate the nature of the observed footprints, we contrasted their spatial distribution with those of nucleosomes or Pol II as measured by bulk methodologies (Figures 2D and 2E). State 1 is characterized by a broad footprint that spans the entire promoter region compatible with presence of nucleosomes at these molecules. States 2–5 show a consistent footprint at ∼100 bp downstream of the TSS that aligns well with the phased +1 nucleosome as mapped by MNase sequencing (MNase-seq) (Figures 2D and 2E). Besides this nucleosomal footprint, state 2 is completely accessible at the promoter region. In contrast, states 3 and 4 harbor a short footprint upstream of the TSS (Figure 2D), indicative of binding of the PIC due to similarities with previous in vitro footprint patterns of TFIID-TFIIA (Cianfrocco et al., 2013). An additional peak downstream of the TSS (+30:+80 bp) characterizes state 4, a feature that is shared with state 5. Importantly, this short peak downstream of the TSS precisely aligns with the summit of Pol II density as measured by chromatin immunoprecipitation ChIP or precision nuclear run-on sequencing (PRO-seq) (Figures 2D and 2E).
If our single-molecule quantification strategy is accurate, the abundance of states at a given promoter should correlate with orthogonal bulk measures for chromatin and polymerase. To perform such comparison, we used molecule counts as a measure of state occurrences at individual promoters (Figure 2F). This revealed that all states but state 1 positively correlate with measures reflecting gene activity. Importantly, the strongest correlation for state 5 is observed with Pol II ChIP sequencing (ChIP-seq) (R = 0.69), confirming that abundance of this state largely recapitulates enrichments of Pol II. Additionally, we observe that state 1 is highly correlated with the abundance of nucleosomes at TSSs as measured by MNase-seq (R = 0.74, Figure 2F). The differences of states between cell types mirror those detected with other measures of accessibility or Pol II binding (Figure S2J). Based on these similarities, we assigned the states to those factors that explain best the observed footprint (Figures 2D–2F).
To validate the ability of our methodology to accurately reflect the binding frequency of the PIC (state 3 and 4) and Pol II (state 4 and 5), we determined state frequencies after depletion of either TBP using double-stranded RNA (dsRNA) (Figure S2K) or Pol II using α-amanitin-mediated degradation (Figure S2L). We observe that TBP depletion leads to selective reduction in the frequency of those states assigned to the PIC (state 3 and 4) (Figures S2M and S2R). In contrast, depletion of Pol II specifically reduces the frequency of state 5 (Figures S2N and S2S). As a control, we repeated this analysis using the 16-state classification (Figures S2O and S2P), confirming that most of the experimental variations were already captured by our simplified five-state classification (Figures S2M–S2P). We conclude that dSMF footprinting generates quantitative measures for multiple occupancy events at promoters genome-wide and at single-molecule resolution.
Next, we investigated how frequencies of these five states relate to each other. We ranked promoters according to their activity as defined by Pol II ChIP-seq (Figure 2G). While the frequency of the Pol II-bound state increases linearly, the frequency of the “nucleosome-occupied” state shows a bimodal pattern (Figure 2G). In other words, promoters of inactive genes are occupied by nucleosomes ∼80% of the time. This frequency drops rapidly to ∼10% for promoters of even lowly active genes (Figure 2G). This loss coincides with a simultaneous increase in the number of “unbound” molecules, which contain neither nucleosome nor polymerase. With increasing promoter activity the pool of Pol-II-bound molecules grows linearly at the expense of unbound molecules. (Figure 2G). Remarkably, this also illustrates that Pol II occupancy can reach up to 65% at a small set of very active promoters (Figures 2H and S2E). These results have several implications. First, they illustrate that within a population of dividing cells one-tenth of DNA molecules of even active TSSs remain occupied by nucleosomes. Second, it suggests that nucleosomal presence is equally low at weakly (Figure 2H, left panel) and highly transcribed promoters (Figure 2H, right panel). Thus a high number of molecules remain unoccupied at weakly active promoters (Figures 2G and 2H), arguing that increased output of promoters does not involve additional nucleosome removal but solely depend on increased Pol II recruitment. To directly test this hypothesis, we monitored state distributions at a hormonal-responsive promoter (Hr46) upon stimulation with ecdysone. This promoter displays low occupancy and basal transcription in untreated cells (Figure S2T). Upon hormonal induction it is upregulated as measured by RNA sequencing (RNA-seq) and displays a coinciding increase in the frequency of Pol II footprints and decrease in number of unoccupied molecules (Figure S2T). This directly illustrates how transcriptional upregulation can occur through increased recruitment of Pol II in absence of changes in nucleosome occupancy.
The TATA Box Stabilizes Binding of the PIC
Formation of the PIC is initiated through interactions of TFIID subunits with the TATA box as well elements downstream of the TSS (Louder et al., 2016). Presence of core promoter elements such as the TATA box influence TFIID binding in vitro (Cianfrocco et al., 2013). In contrast, in vivo binding of TBP-TFIID was reported to not be strongly enhanced at TATA-containing promoters when measured by ChIP-seq (Pugh and Venters, 2016). Given this apparent discrepancy, we wondered whether single-molecule quantification of DNA-protein contacts would reveal differences between promoters depending on the presence of a TATA box.
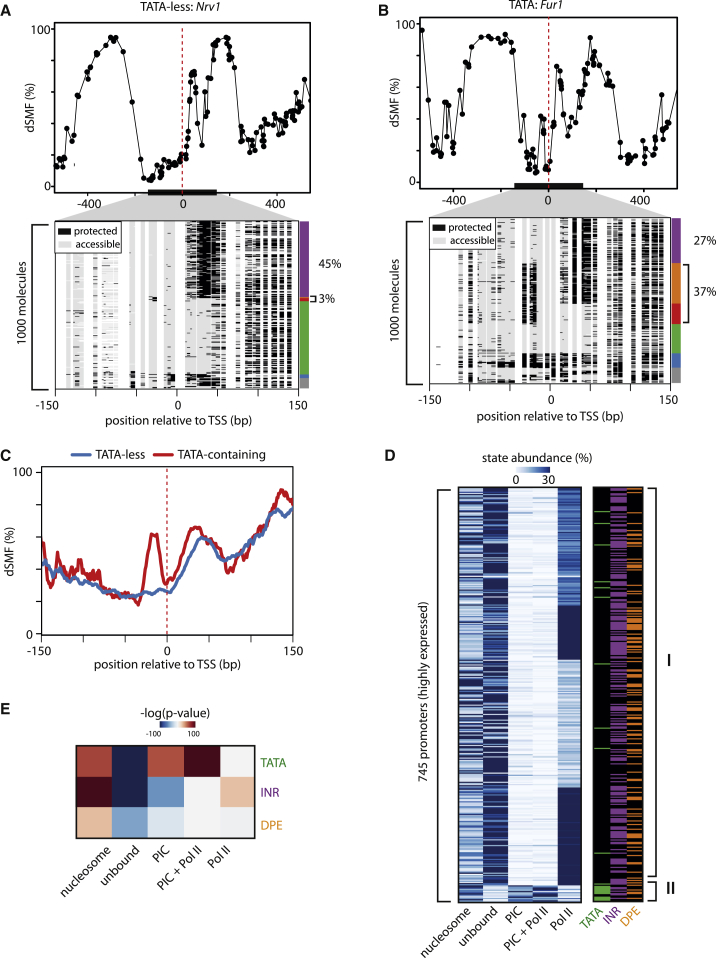
A comparison of active promoters with or without a TATA box revealed striking differences in their frequency of footprints upstream of the TSS (Figures 3A and 3B). These occurred much more often at a TATA-containing promoter such as Fur1 (37% of molecules, Figure 3B), compared to a TATA-less promoters such as Nrv1 (3% of molecules, Figure 3A). Inspection of the composite footprints for highly active promoters (Figure 3C) confirms these observations, since we rarely detect PIC footprints at TATA-less promoters.
Figure 3.
TATA Box Stabilizes Binding of the PIC
(A and B) Single-locus examples of TATA-less (A) versus TATA-containing (B) promoters. Shown are average methylation levels (top panel, black dots connected by a black line) and single-molecule stacks measured by targeted amplicon bisulfite sequencing, sorted into five states using the classification algorithm (methylated Cs, accessible, light gray; unmethylated Cs, protected, black). The vertical side bar depicts the frequency of each state using the same color code as in 2D. Red dashed line depicts position of the TSSs. The percentages of molecules in PIC- or Pol-II-bound states are indicated on the right side of the plot.
(C) Composite footprinting profiles around TATA-containing or TATA-less highly active promoters (red and blue lines, respectively) showing strong differences in PIC footprints between the categories.
(D) Heatmap showing state frequencies for transcriptionally active promoters. k-means clustering was used to organize the heatmap. Side bars indicate presence (colored) or absence (black) of core promoter elements, revealing that TATA-containing promoters cluster due to the increased abundance of PIC footprint.
(E) TATA-box presence alters state distribution, depleting accessible molecules and increasing the number of molecules bound by PIC (with or without Pol II binding). Shown are p values (–log) of a Wilcoxon rank-sum test multiplied by the sign of the statistic parameter to account for directionality of the differences.
To test whether this is generally the case, we clustered active promoters according to state frequencies (Figure 3D). This revealed that a fraction of active promoters (∼7%) show a high enrichment of PIC-bound molecules (Figure 3D, II) and indeed a large majority of these contain a TATA box (78%, Figure 3D). Furthermore, TATA-containing promoters display a strongly increased frequency of PIC-bound molecules, which coincides with a reduction of the unbound state (Figure 3E, p value <10−20) but which is independent of their transcriptional activity (Figure S3B). Importantly, this enrichment in PIC molecules at TATA-containing promoters appears as an intrinsic feature as we find it reproducible across cell types (Figure S2J) with no obvious cell-type-dependent redistribution of states (Figure S3A).
Interestingly, both PIC and engaged Pol II footprints frequently co-occur on the same molecule suggesting that the PIC is not necessarily evicted from DNA after initiation (Figure 3B). This observation is compatible with the concept that some GTFs of the PIC can form a stable scaffold able to promote re-initiation of transcription (Yudkovsky et al., 2000). Since no stable protein-DNA interactions are observed upstream of the TSS at TATA-less promoters (Figure 3A), PIC formation appears to be either transient and/or restricted to TFIID interactions with downstream promoter elements. In summary, these data suggest that PIC formation occurs by distinct protein/DNA interactions and with striking different dynamics in vivo at TATA-containing versus TATA-less promoters.
Polymerase Turnover at Paused Genes
The accumulation of Pol II in a paused state is considered not only to be a step of RNA quality control, but also to act as a regulatory switch at many developmental and stimulus-responsive genes. In order to directly probe the stability of the engaged Pol II, we inhibited transcription initiation by treating cells with Triptolide using previously established conditions (Figure 4A) (Henriques et al., 2013).
Figure 4.
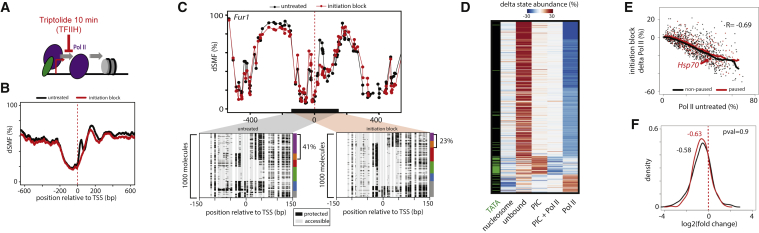
Blocking Transcriptional Initiation Changes Pol II Occupancy
(A) Schematic representation of the initiation process and its block through chemical inhibition of XPB-TFIIH with Triptolide.
(B) Composite footprinting profile around the top 10% expressed genes (as in Figure 1D) before and after Triptolide treatment for 10 min (black and red lines, respectively). Note the decrease of footprint downstream of TSS.
(C) Effect of inhibition of transcription initiation exemplified at the TATA-containing Fur1 promoter. Displayed are average methylation levels before and after inhibition (top panel, black dots [untreated]; red dots [initiation inhibition] connected by a line). The single-molecule stacks for untreated (left panel) versus 10-min treatment (right panel) display footprints measured by targeted amplicon bisulfite sequencing, sorted into five states using the classification algorithm (methylated Cs, accessible, light gray; unmethylated Cs, protected, black). The vertical side bar depicts the frequency of each state using the same color code as in 2D. Red dashed line depicts position of the TSSs. The percentages of molecules harboring footprints for the engaged Pol II are indicated on the right side of the plot.
(D) Inhibition of transcription initiation leads to a genome-wide reduction of engaged polymerases and an increase of the frequency of PIC and unbound states. Shown is a heatmap displaying the promoters showing the highest differences in state abundance (n = 468) after a 10-min inhibition of initiation (Triptolide). Left side bar depicts presence (green) or absence (black) of a TATA box at promoters. Promoters were organized by k-means clustering.
(E) Loss of Pol II at paused promoters upon inhibiting initiation is of similar amplitude than at non-paused promoters. Plotted is the starting amount of engaged Pol II at non-paused against the amount of Pol II lost after 10 min of initiation inhibition for two promoter categories (non-paused genes, black dots; paused genes, red dots). The running median for each starting Pol II amount is depicted for each category (non-paused genes, black; paused genes, red thick line).
(F) A similar fraction of Pol-II-bound molecules is lost at paused and non-paused promoters. Distribution of the fold change in Pol-II-bound amounts (log2) for both promoter categories (non-paused, black; paused, red) illustrating that all promoters loose a comparable fraction (∼65%) of Pol II irrespective of their category or the amount of Pol II before Triptolide treatment.
Blocking transcriptional initiation for 10 min resulted in a global reduction of Pol II footprints at active promoters (Figure 4B). These changes are directly evident when focusing at individual promoters such as the TATA-containing Fur1 (Figure 4C), where Pol II frequency drops but frequency of the PIC only state increases (Figure 4C). A genome-wide analysis of the effect of initiation inhibition identified reproducible changes in state frequencies (Figures 4D, S4A, and S4B). This reveals that most active genes lose engaged Pol II upon inhibition. Depending on the promoter type, this loss coincides with increased frequency of the unbound or the PIC-only state. These observations further support the model that the TATA box stabilizes the PIC at a stage preceding TFIIH activity. Notably, promoters that lose Pol II upon treatment do not gain nucleosomes (Figures 4C and 4D). Then within the tested time interval, engaged Pol II does not significantly contribute to the maintenance of open chromatin at promoters as previously proposed (Gilchrist et al., 2010).
Next, we asked for all promoters how the change in Pol II occupancy relates to the amount of polymerase present before treatment (Figure 4E). This reveals a strong negative correlation (R = −0.69) indicating that on average promoters lose a constant fraction of engaged Pol II regardless of their activity level. We then isolated a set of paused genes based on their “pausing index” as defined by the imbalance between Pol II at promoters compared to gene body (Figure S4C) (Core et al., 2012). Surprisingly, the loss of Pol II footprint observed at these genes is overall of comparable amplitude than the one observed at non-paused genes (Figures 4E and 4F). Importantly, this is similarly evident at the promoter of the Hsp70 gene (Figure 4E), which is the best-characterized example of a promoter whose output is indeed regulated by a switch to productive elongation (Guertin et al., 2010).
To show that the observed rapid loss of Pol II at paused genes can be observed with other detection methods, we performed ChIP-seq against Pol II under identical conditions (Figures S4D–S4G) and quantified the relative Pol II changes genome-wide. This confirmed that inhibiting initiation leads to a global decrease of Pol II levels (Figures S4D–S4G). Taken together these results suggest that the turnover rate of Pol II at paused promoters could be similar to that observed at actively transcribing genes, where polymerases continuously move into productive elongation.
Rapid Pol II Turnover at Paused Genes
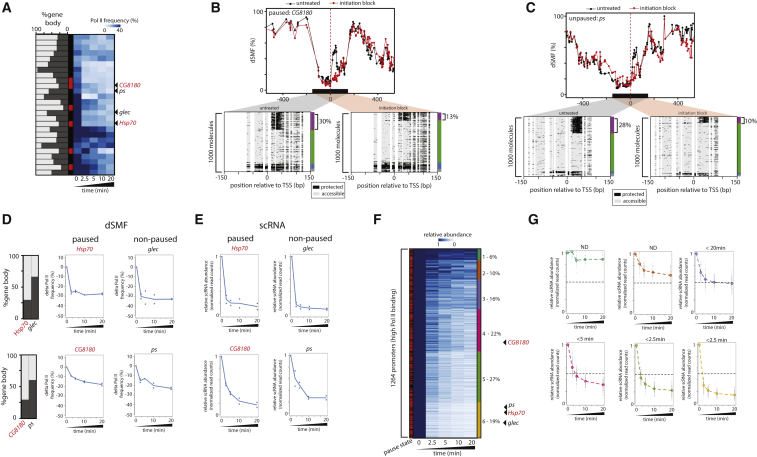
In order to monitor this process in more detail, we measured changes in Pol II occupancy during a time course of inhibition with Triptolide. For these experiments, we acquired high-coverage dSMF data from a focused set of 52 promoters sampled to have a broad range of transcriptional activities and pausing indices (Figure 5A, left panel). We observed a significant spread in Pol II turnover kinetics between tested genes (Figure 5A). While this spread is comparable to previous reports (Henriques et al., 2013), we did not observe an increased residence time for Pol II at promoters of paused genes (Figure 5A, red label). In fact, genes with comparable Pol II turnover differ significantly in their pausing index (Figure 5A). For instance, the genes CG8180 (Figure 5B) and ps (Figure 5C) have both ∼30% of their molecules occupied by Pol II even though they differ largely in their pausing index (Figures 5A and 5D). Upon inhibition of initiation, both lose two-thirds of their Pol II footprint (17% and 18%, respectively) within 10 min of inhibition (Figures 5B and 5C) and at similar kinetics (Figure 5D). Interestingly, one of the fastest turnover is detected at the paused Hsp70 promoter (Figure 5D). The observed half-life is well below 2.5 min (our earliest time point of measurement) and thus faster than previously reported for this gene (Buckley et al., 2014). It is as rapid as observed at glec, one of the most actively transcribed promoters within our set (Figure 5D).
Figure 5.
High Levels of Pol II Turnover at Paused Promoters
(A) Inhibition of transcription initiation leads to a rapid reduction of engaged polymerases at promoters also at paused genes. Heatmap depicting the changes in absolute levels of engaged Pol II after different time of inhibition of transcription initiation (time as indicated). Left side bar depicts the promoter category (non-paused, black; paused, red). Left bar plot depicts the amount of GRO-seq signal in gene body (dark gray) relative to promoter (light gray). Promoters were organized by hierarchical clustering.
(B and C) Single-locus examples of the effect of inhibition of transcription initiation at the (B) CG8180 paused promoter and the (C) ps non-paused promoter. Shown are average methylation levels before and after inhibition (top panel, black dots [untreated] and red dots [initiation inhibition] connected by a line). The single-molecule stacks for untreated (left panel) and incubated 10 min with initiation inhibitor (right panel) display footprints measured by targeted amplicon bisulfite sequencing, sorted into five states using the classification algorithm (methylated Cs, accessible, light gray; unmethylated Cs, protected, black). The vertical side bar depicts the frequency of each state using the same color code as Figure 2D. Red dashed line depicts position of the TSSs. The percentages of molecules harboring footprints for engaged Pol II are indicated on the right side of the plot.
(D) Examples of promoters that display different elongation rates yet comparable decay kinetics of engaged Pol II after inhibition of initiation. Bar plot depicting the amount of GRO-seq signal in gene body (dark gray) relative to promoter (light gray) for each example. Time-course data are displayed for the paused Hsp70 promoter versus the non-paused glec promoter and the CG8180 paused promoter versus the non-paused ps promoter. Blue dots represent the relative amount of engaged Pol II lost at each time point relative to the starting amount (two independent biological measurements are plotted for each time point). Blue line depicts average of replicates. Note the rapid loss at both the glec and Hsp70 promoter already at the first measurement point 2.5 min following inhibition.
(E) scRNAs decay with similar kinetics as Pol II footprints after inhibition of initiation. Relative scRNA abundance over a time course of inhibition at the same set of TSSs. Blue dots represent the amount of scRNA at each time point relative to the starting amount (two independent biological measurements are plotted for each time point). Blue line depicts the average signal.
(F) Inhibition of initiation leads to a global reduction of scRNA levels with various kinetics genome-wide. Heatmap depicting the relative changes in scRNA abundance after Triptolide inhibition at a highly active promoters (top 10%, Pol II ChIP-seq). The heatmap was organized using k-means clustering into six different clusters (right side bar). Left side bar depicts the promoter category (non-paused, black; paused, red). Shown are average values over two biological replicates.
(G) scRNA decay kinetics for each group of TSS (color as in A). Shown is the distribution of scRNA levels for each TSS (black dots) and the median abundance at each time point (colored lines and dots). The first time point where the median of the relative scRNA amount reached 50% of its original value was used to approximate the half-life for each cluster.
Paused promoters such as Hsp70 experience basal elongation levels. To test how much of the loss of Pol II at paused promoters could be explained by elongation, we repeated the inhibition of initiation in presence of the p-TEFb inhibitor Flavopridol that blocks entry into elongation (Figure S5A). This simultaneous inhibition indeed partially buffers the loss of Pol II at a fraction of non-paused promoters (Figures S5B and S5D). However, paused promoters including Hsp70 show a similar loss regardless of the additional inhibition of elongation (Figures S5C and S5E). We conclude that the loss of Pol II observed at paused promoters occurs independent of elongation.
Taken together, these results suggest that engaged polymerases are not particularly stable at paused promoters compared to promoters experiencing frequent elongation. This would in turn imply that a large fraction of Pol II engaged at paused promoters is rapidly lost independently of elongation, most probably through premature termination.
Widespread Rapid scRNA Turnover at Paused Genes
Abundance of short capped RNA (scRNA) at the 5′ of genes has been shown to indirectly reflect amounts of Pol II engaged at a particular promoter (Nechaev et al., 2010). To independently validate our above observations, we sequenced scRNA during the same time course of inhibition. Importantly, the generated dataset compares well with previous measures of scRNA decay kinetics made at the single-gene level (Figure S5F). We then compared the kinetics of Pol II loss as measured by SMF (Figure 5D), with that of scRNA decay (Figure 5E) for our selected set of TSSs. Both paused and non-paused genes show a very good agreement between the two measures, including the rapid decay of scRNA signal at the promoter of Hsp70 (Figure 5E). This independent readout further argues that engaged Pol II and its associated scRNA can be short lived at some paused promoters such as the Hsp70 promoter. We then asked generally how Pol II turnover relates to Pol II accumulation downstream of the TSS at paused genes. To address this question, we clustered all highly active genes (Figure S5G) according to their scRNA decay profile (Figure 5F), grouped them by similarity and estimated the approximate half-life of their scRNA (Figure 5G). We found that for a majority of promoters (68%) scRNA have a short half-life comprised between 0 and 5 min (Figures 5F and 5G, clusters 4–6), with a substantial fraction shorter than 2.5 min. When we compared the pausing index (Figures 5F, side bar, S5H), or the global run-on sequencing (GRO-seq) signal at the gene body (Figures S5I and S5J), we observe equivalent amount of paused or non-paused genes with half-life <2.5 min. Importantly, the half-life of Hsp70 scRNA ranks again among the fastest detected genome-wide (< 2.5 min, Figures 5E–5G). In fact none of the gene groups that differ significantly in their half-life display any particular enrichment for either category (Figures S5H–S5K, chi-square test, p value >0.01). Together these results further support a model where many paused genes have a short Pol II half-life. More generally our data suggest that stability of Pol II at promoters is not correlated with the efficiency of entry into elongation. Indirectly, these results imply that many paused genes including Hsp70 experience frequent premature termination rapidly after initiation.
Continuous Transcription Initiation Is Required for Induction of the Paused Heat Shock Gene
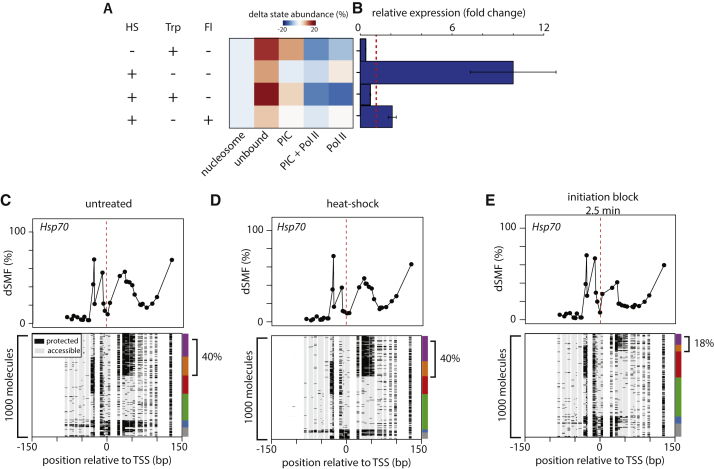
To functionally test such model of abundant non-processive pausing, we used the paradigm of heat shock response. We reasoned that if the large pool of polymerases engaged at the Hsp70 promoter (∼40%, Figure 6C) is stable as postulated in the classical model of “pausing,” the transcriptional response upon heat shock should be insensitive to a preceding short inhibition of transcription initiation. If Pol II is indeed unstable as we observed in our time-course experiment (Figures 5A, 5D, and 5E), a short inhibition of initiation should alter heat shock-induced transcriptional activation. To discriminate between these two models, we performed heat shock induction after short incubation with the same set of inhibitors (Figures 6A and 6B). In absence of Triptolide, we observed a rapid and over 10-fold increase in Hsp70 transcript upon 1 min of high temperature (Figure 6B) as previously reported (Guertin et al., 2010, Wirbelauer et al., 2005). During the strong transcriptional response upon normal heat shock, we observed that the amount of engaged Pol II is similar to the one prior to induction (Figures 6C and 6D). This shows that the initiation rate at Hsp70 is sufficient to directly replenish the pool of engaged Pol II that are going into elongation upon induction. This is in good agreement with the initiation and elongation rate measured at this promoter upon induction (Buckley et al., 2014). As expected, this fast induction is almost completely abolished upon chemical inhibition of elongation, in line with the established model that the engaged polymerase at the uninduced Hsp70 is controlled at the elongation step. Importantly, however, a short (2.5 min) chemical inhibition of initiation alone similarly abolishes the transcriptional response of Hsp70 (Figure 6B). This lack of transcriptional response fully agrees with the footprinting patterns that showed a rapid loss of the pool of engaged Pol II upon short initiation inhibition (Figures 5D, 6A, and 6E). This result indicates that the rapid transcriptional response observed upon heat shock is not mediated by a pool of polymerases that are stably associated with the DNA template. Instead, it suggests that this response relies on continuous initiation. We conclude that promoter-engaged Pol II is indeed unstable at the Hsp70 promoter, the canonical example of output regulation by pausing. We propose that the observed accumulation of engaged Pol II at these promoters is the result of continuous cycles of initiation followed by rapid premature termination. The fact that we observe this behavior throughout the genome argues that non-processive pausing is a common phenomenon. Activation of such paused genes appears not to be mediated through release of stably associated Pol II but rather through switching from termination to processive elongation.
Figure 6.
Heat Shock Response Requires Continuous Transcriptional Initiation
(A) Effect of short inhibition of initiation or elongation on heat shock response. Heatmap displays the redistribution of states upon various treatments (left panel) (HS, heat shock 1 min; Trp, Triptolide 2.5 min, initiation block; Fl, Flavopiridol 2.5 min, elongation block).
(B) Bar graph displaying the relative expression change with the same treatments as measured by qRT-PCR (fold change calculated using the comparative Ct method). Error bars represent SD from three biological replicates.
(C–E) Heat shock induction does not affect frequencies of early transcription intermediates. Examples of Hsp70 promoter footprints before (C) and after heat-shock (D) or after Triptolide inhibition (E). Shown are average footprinting levels (top panel) (black dots and line) and single-molecule stacks measured by targeted amplicon bisulfite sequencing, sorted into five states using the classification algorithm. The vertical side bar depicts the frequency of each state using the same color code as 2D. The percentages of molecules harboring footprints for engaged Pol II are indicated on the right side of the plot.
Discussion
This study establishes a methodological and computational framework for genome-wide SMF and applies it to quantify binding events that co-occur at promoters throughout the fly genome. By monitoring formation of various intermediates in the transcription initiation process this uncovers new insights about chromatin opening, PIC formation, and Pol II initiation dynamics leading to a revised model for polymerase pausing.
We find promoters of inactive genes to be covered by nucleosomes at a high frequency of 80%, while active genes show an expected lower occupancy. Surprisingly, however, active promoters display a rather homogeneous rate of ∼10% nucleosomal occupancy irrespective of their very different activity. Since our data were derived from an unsynchronized population of cells, it is tempting to speculate that the remaining 10% of occupied molecules originate from cells in a phase of the cell cycle where the transcriptional machinery is evicted from chromatin such as mitosis (Liang et al., 2015).
Our genome-wide promoter analysis further reveals that presence of core elements significantly alters footprinting patterns. This is most prominent in case of the TATA box, which displays frequent PIC footprints. While any footprinting assay is agnostic to the identity of the complexes binding, our data are in direct support of models previously derived from structural and biochemical data (Cianfrocco et al., 2013). These postulate that at TATA-containing promoters, TFIID binds through TAFs-MTE/DPE and TBP-TATA, while contacts will be limited to the downstream interaction for TATA-less sequences. Moreover the co-occurrence of this stabilized PIC and engaged Pol II suggests that this structure could allow re-initiation cycles as proposed by biochemical studies (Yudkovsky et al., 2000).
Quantitative monitoring of promoter footprints and their changes upon inhibition of initiation or elongation revealed surprisingly high turnover rates of Pol II at many paused promoters. Affected genes include the heat shock-responsive promoter Hsp70 that displayed one of the fastest Pol II turnover rates. This is not unique to our study and experimental approach since very fast turnover at Hsp70 was also very recently reported as part of a high-resolution genome-wide dataset of Pol II in Drosophila (not discussed by the authors, relevant data part of their Table S1 [Shao and Zeitlinger, 2017]). This is in contrast to a previous report using live tracking of photo-activated Pol II on polytene chromosomes, which estimated a significantly longer half-life than the one we derive from measuring Pol II occupancy (Buckley et al., 2014). This discrepancy might be explained by local recycling of the terminated polymerases, which could lead to extended fluorescence in a microscopical assay. In turn, this could result in an overestimation of residence time as already pointed out by the authors (Buckley et al., 2014).
Together our data revise the concept of promoter proximal pausing defined as the accumulation of a stable pool of polymerase to enable rapid gene activation upon stimuli (Guertin et al., 2010). Instead, our data argue for a non-processive model for pausing where a large fraction of initiated polymerases rapidly and prematurely terminates upon elongation block. In this scenario, rapid induction would not be mediated by release of a pool of polymerases pre-loaded on the DNA template. Instead, initiation at these genes leads to continuous replenishing of Pol II, which can be rapidly switched to elongation upon stimulus. Such regulation might enable higher and potentially sustained amplitude of expression, as induction does not rely solely on a single Pol II release but on continuous waves of initiation.
While mechanistically different from the prevalent concept of polymerase pausing, the model proposed here is nevertheless compatible with the idea that genes requiring rapid activation upon stimuli acquire a specific non-processive activation state. We confirm that in contrast to truly inactive promoters, this promoter state is characterized by extensive chromatin remodeling (>80% of molecules), high levels of PIC formation and transcription initiation. However, compared to truly active promoters, this state does not appear to involve extended stability of the engaged Pol II pool, but rather its continuous and rapid turnover. This turnover mechanism could in principle take place at promoters of developmental genes that have been suggested to be regulated at the level of elongation. If and how this precisely takes place during the course of early development remains to be determined.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Pol II CTD | (Wirbelauer et al., 2005) | 8WG16 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| M.CviPI | New England Biolabs | M0227L |
| M.SssI | New England Biolabs | M0226L |
| Triptolide | Sigma-Aldrich | T3652 |
| Flavopiridol | Sigma-Aldrich | F3055 |
| α-amanitin | Sigma-Aldrich | A2263 |
| Ecdysone | Sigma-Aldrich | H5142 |
| 5′ RNA phosphatase | Epicenter | RP8092H |
| 5′ terminator exonuclease | Epicenter | TER51020 |
| Cap-Clip Acid Pyrophosphatase | Cellscript | C-CC15011H |
| Critical Commercial Assays | ||
| NEBNext Chip Sample Prep Kit | New England Biolabs | E6240 |
| Truseq DNA Sample Prep Kit | Illumina | 15025064 |
| Megascript kit | LifeTech | AM1626 |
| TruSeq small RNA library | Illumina | RS-200 |
| RNA clean and concentrator | Zymo | R1016 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE77369 |
| Pol II - Rpb1 ChIP-seq | (Hu et al., 2013) | GEO: GSE47938 |
| DNase Hypersensitivity | (Arnold et al., 2013) | GSE40739 |
| RNA-seq | (Arnold et al., 2013) | GSE40739 |
| CAP-seq | (Kwak et al., 2013) | GSE42117 |
| PRO-seq | (Kwak et al., 2013) | GSE42117 |
| KMnO4 footprinting | (Lee et al., 2008) | N/A |
| short RNA | (Nechaev et al., 2010) | GSE18643 |
| GRO-seq | (Core et al., 2012) | GSE23544 |
| CAGE | (Roy et al., 2010) | modENCODE |
| MNase-seq | (Gilchrist et al., 2010) | GSE20472 |
| Experimental Models: Cell Lines | ||
| Schneider S2 cells | (Wirbelauer et al., 2005) | N/A |
| Ovarian somatic cells (OSC) | (Saito et al., 2009) | N/A |
| Oligonucleotides | ||
| dsRNA against dTBP fw:ACATGATGCCCATGAGTGAG; rv:AACCGAGCTTTTGGATGATG | This paper | N/A |
| Software and Algorithms | ||
| dsRNA design | N/A | http://www.flyrnai.org/snapdragon |
| QuasR | (Gaidatzis et al., 2015) | http://bioconductor.org/packages/release/bioc/html/QuasR.html |
| Other | ||
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dirk Schübeler (dirk.schubeler@fmi.ch).
Experimental Model and Subject Details
Schneider-S2 cells were grown at 25°C in Schneider’s Drosophila medium (LifeTech: 21720-001) supplemented with 10% FBS. Ovarian Somatic Cells (OSC) (Saito et al., 2009) were grown at 25°C in Shields and Sang M3 Insect Medium (Sigma, S8398) supplemented with 1% insulin, 1% glutathione, 10% heat-inactivated FBS, and 10% fly extract.
Method Details
Cell culture and treatments
Heat-shock was performed in S2 cells as previously described (Wirbelauer et al., 2005). Temperature was quickly raised by mixing equivalent amounts of cell containing medium at 25°C with pre-warmed medium (48°C). The mix was incubated for 1 min at 37°C. After heat shock, cells were let to recover for 5 min. Transcription inhibition was performed as previously described (Henriques et al., 2013). Triptolide and/or Flavopiridol were added at a final concentration of 10 μM to the culture media for the indicated time. Pol II degradation was achieved by addition of 20 μg/mL of α-amanitin for 24h into the culture medium. For ecdysone induction, S2 cells were incubated with 41 μM Ecdysone (Sigma - H5142) for the indicated time.
Dual Enzyme Single Molecule Footpriniting
Footprinting protocol was adapted from (Bell et al., 2010, Kelly et al., 2012). 2.5 10ˆ6 Drosophila cells (S2 or OSC) were re-suspended in ice-cold lysis buffer (10mM Tris (pH = 7.4), 10mM NaCl, 3mM Mgcl2, 0.1mM EDTA, 0.5% NP40), incubated 10min on ice, span down. Nuclei were washed (10mM Tris (pH = 7.4), 10mM NaCl, 3mM Mgcl2, 0.1mM EDTA) and re-suspended in M.CviPI reaction buffer (50mM Tris (pH 8.5), 50mM NaCl, 10mM DTT). The nuclei were then incubated with 200U of M.CviPI (NEB-M0227L) at 30°C for 7.5 min (in presence of 0.6mM SAM, and 300mM Sucrose). The reaction was supplemented with 100U of M.CviPI and 128pmol of SAM before a second incubation round of 30°C for 7.5 min. 10mM MgCl2, 128pmol of SAM and 60U of M.SssI (NEB-M0226L) were added for a third incubation round of 30°C for 7.5 min. Reaction was stopped by adding a SDS containing buffer (20mM Tris, 600mM NaCl, 1%SDS 10mM EDTA), DNA was extracted after overnight Proteinase K (200 μg/ml) digestion at 55°C. For single treatments only one of the enzyme was used under the same conditions.
Whole genome bisulfite libraries were prepared using Illumina Truseq DNA sample preparation kit following manufacturer recommendation. 5μg of sheared DNA (covaris) was used as an input for end-repair and A-tailing. Methylated adaptors were ligated and large DNA fragments (400-600bp) were selected on a low melting agarose gel (BIO-RAD −161-3107). The extracted material was used as an input for bisulfite conversion (Quiagen - 59104). The converted DNA was amplified by 10 cycles of PCR using Pfu Cx HotStart (Agilent - 600410) using Illumina primers. PCR product was purified using AMPureXP beads (Beckman Coulter - A63880) and controlled on Bioanalyser High sensitivity (Agilent 5067-4626). The samples were run on an Illumina HiSeq2500 generating 150bp paired-end reads (rapid-run). Two biological replicates was sequenced for each condition.
Amplicon bisulfite sequencing
Primers were designed against the in silico converted templates using Primer3 with slight modifications and subsequent selection of 96 bisulphite primer pairs (product size: 250–500 bp). Primers were commercially synthesized on 96 well plate format. A 2 μg sample of RNaseA-treated genomic DNA was converted following standard Epitect bisulphite conversion kit protocol (QIAGEN). Bisulphite-converted DNA was amplified with specific primers with following cycling conditions: twenty touch down cycles from 52 to 48°C with 30 s at 95°C, 30 s at 52/48°C and 30 s at 72°C, followed by 36 cycles of 30 s at 95°C, 30 s at 48°C and 30 s at 72°C. Then samples of 5 μL per reaction were pooled and bead purified (Agencourt - AMPureXP). Bisulfite amplicons libraries were prepared using NEBNext Chip (NEB - E6240) multiplexing up to 10 libraries (NEB - E7335) on a Miseq instrument generating 250bp paired-end reads. All targeted experiments have been performed in at least triplicates except for the time course data that have been performed in duplicates.
Short capped RNA preparation (scRNA)
For each condition 20 10ˆ6 of Triptolide treated Drosophila cells were spiked with 2 10ˆ6 untreated mouse embryonic stem cells. scRNA protocol was adapted from (Nechaev et al., 2010). Cells were re-suspended in ice-cold lysis buffer (10mM Tris (pH = 7.4), 10mM NaCl, 3mM Mgcl2, 0.1mM EDTA, 0.5% NP40), incubated 10min on ice, span down. Nuclei were washed with ice cold (10mM Tris (pH = 7.4), 10mM NaCl, 3mM Mgcl2, 0.1mM EDTA) and nuclear pellets were dissolved in Trizol (Thermofisher). RNA was size selected (17-200bp) using a two-step column purification strategy (RNA clean and concentrator, Zymo-R1016). 10 μg of purified RNA was successively treated by 5′ dephosphorylation - 20U at 37°C for 30min (Epicenter - RP8092H); 5′ terminator exonuclease - 1U in Buffer A at 30°C for 60min (Epicenter - TER51020); cap-clip decapping enzyme – 5U at 37°C for 90min. After each reaction, short RNA was column purified (RNA clean and concentrator, Zymo-R1016). The resulting RNA was used for library preparation using TruSeq small RNA library (Illumina). Libraries were purified on 6% TBE gels (150-300bp – Novex - EC6265BOX). Size distribution of the libraries were controlled on Bioanalyser High sensitivity (Agilent 5067-4626). Two biologically independent inhibition time courses were performed. The samples were run on an Illumina NextSeq generating 38bp paired-end reads.
dsRNA mediated RNA interference
Primers were designed using the snapdragon online tool. A two-step PCR was used to amplify the target sequence from the fly genome and add the full length T7 promoter. In vitro transcription was performed using the Megascript kit (LifeTech – AM1626). 30 μg of purified dsRNA was directly soaked into the medium with 1.5 10ˆ6 of S2 cells for 72 hr. Efficiency of downregulation was probed using RT-qPCR. Experiments were performed in biological duplicates.
Chomatin Immuno-precipitation (ChIP)
20 10ˆ6 S2 cells were cross-linked in S2 medium containing 1,1% formaldehyde for 10 min at room temperature. Cross-link was stopped by addition of 0.125 M glycine 10min at 4°C. Cells were rinsed twice with PBS. Nuclei were extracted by successive incubation in (10mM Tris (pH = 8.0), 10mM EDTA, 0.5mM EGTA, 0.25% Triton X-100) and (10 mM Tris (pH = 8.0), 1 mM EDTA, 0.5 mM EGTA, 200 mM NaCl). Nuclei were resuspended in (50 mM HEPES/KOH (pH = 7.5), 500mM NaCl, 1mM EDTA, 1% Triton X-100, 0.1%DOC, 0.1%SDS, protease inhibitors) and sonicated (40 cycles - 30 s) (Diagenode Bioruptor). 70 μg of pre-cleared chromatin was incubated with ∼5ug of antibody overnight at 4°C. The protein-DNA complexes were immuno-precipitated by addition of protein A-Sepharose for 3 hr at 4°C. Beads were washed twice with 1mL lysis buffer and once with 1 mL DOC buffer (10 mM Tris (pH = 8.0), 0.25 M LiCl, 0.5% NP- 40, 0.5% deoxycholate, 1 mM EDTA). Chromatin was eluted in (1% SDS, 0.1 M NaHCO3). After RNase A treatment, cross-linking was reversed by overnight incubation at 65°C followed by proteinase K digestion. DNA was isolated by phenol-chloroform extraction followed by ethanol precipitation and resuspension in 50 mL TE buffer.
Quantification and Statistical Analysis
Alignment and data extraction
Data alignment
For dSMF data, raw sequence files were pre-processed using Trimmomatic to remove Illumina adaptor sequences, remove low quality reads and trim low quality bases. The trimmed reads were then aligned using QuasR (using Bowtie as an aligner) (Gaidatzis et al., 2015) against a bisulfite index of the Drosophila melanogaster genome (BSgenome.Dmelanogaster.UCSC.dm3).
For other datasets (ChIP-seq, RNA-seq, scRNA), reads were aligned using QuasR against the Drosophila melanogaster genome (BSgenome.Dmelanogaster.UCSC.dm3), and or mouse genome (BSgenome.Mmusculus.UCSC.mm9). To enable analysis at the multi-copy Hsp70 locus, we allowed multiple hit mapping (< 10 – QuasR option maxHits = 10). For multi-mapping reads, only one hit is randomly selected for each read, avoiding artificial signal enhancement.
Average methylation call
Context independent cytosine methylation call was performed using QuasR. Custom R functions were developed to determine context dependent (CG, GC) average methylation. Methylation has been called genome wide for Cs covered at least 10 times. Single dot are used to display single C nucleotide data while curves represent smoothed data obtained by averaging data using a sliding window over 10 Cs.
Single molecule methylation call
Single molecule C methylation extraction was performed using QuasR (Gaidatzis et al., 2015). Custom R functions have been developed to determine nucleotide context and sort the molecules according to their methylation pattern using a molecular classifier.
Single molecule footprint quantification
A set of unique reference transcription start site has been constructed using Refseq (dm3) precisely repositioned according to the main CAGE signal within 50bp (S2 cells CAGE data from Modencode; Roy et al., 2010). A molecular classifier has been developed (Figures 2A and S2A–S2G) that extract methylation for every read in 4 bins designed around the theoretical position of promoter elements where the transcription machinery (PIC, Pol II) has been observed to create footprints (Cianfrocco et al., 2013, Lee et al., 2008, Lim et al., 2004): upstream:[-58:-43]; TATA/BRE:[-36:-22]; INR:[-6:14]; DPE:[28:47] (Figure S2A). All reads were aligned relative to the set of TSSs and methylation was extracted for every molecule in each bin. The methylation was binarized in each bin, creating a 4 bit vector classifying the state of every molecule among 2ˆ4 = 16 theoretical possibilities. These include a fully accessible pattern (1111); a fully inaccessible pattern (0000) which may indicate the presence of nucleosomes; a pattern which indicates accessibility only at the position of the DPE (1110) which may be reflective of engaged Pol II; and several patterns that could potentially be associated with PIC formation (1011, 1010, 1001 and 1101) (Cianfrocco et al., 2013, Lee et al., 2008, Lim et al., 2004). To simplify the interpretation of the data, we aimed to focus our analysis on states that are recurrently observed in vivo. We excluded some states based on their low frequency of occurrence at promoters (Figure S2F). These include some of the possible PIC footprints (1001, 1101) that either do not occur in vivo or are too transient to be captured in our assay. Additionally, comparison of the frequency count with bulk datasets enabled to separate states that capture variations related to gene activity (Figure S2G), from states that do not show any correlation with external measurements (0101, 0110, 0011, 1100). We noted that many of these correspond to the states occurring at low frequency. These states were grouped to an ‘unassigned’ category. The remaining states contained many of the patterns showing similarity with footprints observed in vitro (Cianfrocco et al., 2013) as well as in high resolution structure of transcription initiation (Louder et al., 2016) (Figures S2A and S2C). From these, we aggregated states sharing high similarities in their pattern (i.e., 0000, 1000, 0100, 0010, 0001) (Figure S2C) and show high correlation (Figure S2G) (state 1 and 2). Coverage cutoffs of 30 and 100 reads have been used for genome wide analysis and amplicon bisulfite sequencing respectively.
Data analysis
All analyses were performed using R- Bioconductor. Ad hoc R scripts are available upon request.
Detection of core promoter motifs
Motif occurrence has been determined by searching for occurrence of the consensus motif (INR:’TCAKTY’; TATA: ‘TATAWAAR’; DPE:’RGWYV’ (Lim et al., 2004)) allowing for one mismatch at their theoretical position relative to TSS flexible by 5bp. The effect of promoter motifs on state distribution has been measured by performing a Wilcoxon ranked-sum test comparing the abundance of a given state as a function of the presence/absence of the motif. The statistics of the test was used to display the directionality of the differences.
Comparison with external datasets
Data collection was performed using the qCount function of QuasR (Gaidatzis et al., 2015). Different collection windows were adopted to collect reads in external reference datasets. For comparison at TSS, reads for all datasets were collected in a [-150:150] window surrounding the TSSs with the exception of MNase datasets. For MNase the window was restricted to [-40:30] to exclude the +1 and −1 phased nucleosomes that sit around ± 100bp and restrict the counts to nucleosomes occupying the TSSs. Correlations were calculated on log2 transformed data.
Definition of ‘pausing’ index
Pausing index was calculated genome wide as previously described (Core et al., 2012). For each genes read counts were collected in GRO-seq data around the TSS (+/− 150bp) and in the gene body (+300:+600). The pausing index was defined as the ratio of TSS reads over gene body reads. ‘paused’ genes were defined as active genes having a pausing index > 8, and low read counts in the gene body (log2(RPKM) < 3), to extract genes with high levels of engaged Pol II but transcriptionally inactive. ‘unpaused’ genes were defined as active genes having a pausing index < 8.
scRNA data analysis
After alignment against Drosophila and mouse genomes (see above for details). Reads were counted in a window around TSSs [-100:200] using the qCount function of QuasR (Gaidatzis et al., 2015). To reduce potential contamination with full-length transcripts we used the second read from the pair representing the 3′ of the transcript for counting. Pairs having insert length > 180bp were excluded. We used the mouse spike-ins to perform inter-sample normalization and enable quantification of the global effects expected to occur upon inhibition of transcription. Under the Assumption that read counts from the mouse should be constant across samples; reads counts from each fly sample were normalized by the sum of the reads detected at mouse promoters within the same sample. Abundance of scRNA at each time-point was calculated relative to the respective amount before inhibition. This relative level was calculated for each replicate separately to correct for batch variations. While we generally observed the signal to be restricted to the 5′ of the genes, we noted unusually high gene body signal at a top-expressed genes (including many ribosomal protein coding genes). We interpreted this as a contamination by mRNA degradation products for highly abundant transcripts and therefore discarded those from the analysis. To avoid variations related to gene activity, the analysis was restricted to highly active genes (top 10% ChIP-seq as defined in Figure 1D).
ChIP -seq analysis
Reads were counted in a window around TSSs [-200:100] using the qCount function of QuasR (Gaidatzis et al., 2015) and enrichment over input were derived.
Targeted amplicon bis-seq primer design
Within the set of promoters covered in the genome wide experiment, a subset of 96 promoters were targeted for amplicon bisulfite sequencing. Promoters were selected to have presence/absence of core promoter elements (i.e., TATA box, INR), a wide range of expression levels and pausing indices. Moreover specific cases as the canonical ‘paused’ Hsp70 promoter were targeted. Primers were designed using R functions wrapping Primer 3. The design was performed using a C > T converted genome excluding CG and GC containing regions. Primer list is available upon request.
Data and Software Availability
Software
Software from this study has been previously published as detailed under “QUANTIFICATION AND STATISTICAL ANALYSIS.”
Data Resources
The accession number for the sequencing data reported in this paper is GEO: GSE77369.
Author Contributions
A.R.K. and D.S. designed the study and wrote the manuscript. A.R.K. designed and performed the experiments with the help of D.I. and L.H. for Illumina sequencing developments and library preparation. A.R.K. designed and implemented the single-molecule analysis pipeline with the technical support of D.G. A.R.K. analyzed the data with technical support by L.B. All authors discussed the results and commented on the manuscript.
Acknowledgments
The authors are grateful to Michael Stadler, Christiane Wirbelauer, Sebastien Smallwood, Kirsten Jacobeit, Fabio Mohn, Sophie Dessus-Babus, Tim Roloff, and Altuna Akalin for technical advise. The authors would like to thank Grzegorz Sienski and Julius Brennecke for providing OSCs and medium. We thank James Kadonaga for providing dTBP antibodies. The authors would like to thank Jeffrey Chao, Luca Giorgetti, Marc Bühler, Nico Thomä, Lászlò Tora, Didier Devys, Mohamed-Amin Choukrallah, and members of the Schübeler laboratory for helpful discussions and/or comments on the manuscript. Research in the laboratory of D.S. is supported by the Novartis Research Foundation, the European Union (NoE “EpiGeneSys” FP7-257082 and the “Blueprint” consortium FP7-282510), the European Research Council (ERC 204264 “EpiGePlas”), and the Swiss National Science Foundation (31003A_156963). A.R.K. and D.I. are supported by a Swiss National Science Foundation Ambizione grant (PZOOP3_161493).
Published: July 20, 2017
Footnotes
Supplemental Information includes five figures and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2017.06.027.
Contributor Information
Arnaud R. Krebs, Email: arnaud.krebs@fmi.ch.
Dirk Schübeler, Email: dirk@fmi.ch.
Supplemental Information
References
- Arnold C.D., Gerlach D., Stelzer C., Boryń Ł.M., Rath M., Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- Bell O., Schwaiger M., Oakeley E.J., Lienert F., Beisel C., Stadler M.B., Schübeler D. Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation and replication timing. Nat. Struct. Mol. Biol. 2010;17:894–900. doi: 10.1038/nsmb.1825. [DOI] [PubMed] [Google Scholar]
- Brannan K., Kim H., Erickson B., Glover-Cutter K., Kim S., Fong N., Kiemele L., Hansen K., Davis R., Lykke-Andersen J., Bentley D.L. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol. Cell. 2012;46:311–324. doi: 10.1016/j.molcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley M.S., Kwak H., Zipfel W.R., Lis J.T. Kinetics of promoter Pol II on Hsp70 reveal stable pausing and key insights into its regulation. Genes Dev. 2014;28:14–19. doi: 10.1101/gad.231886.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfrocco M.A., Kassavetis G.A., Grob P., Fang J., Juven-Gershon T., Kadonaga J.T., Nogales E. Human TFIID binds to core promoter DNA in a reorganized structural state. Cell. 2013;152:120–131. doi: 10.1016/j.cell.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L.J., Waterfall J.J., Gilchrist D.A., Fargo D.C., Kwak H., Adelman K., Lis J.T. Defining the status of RNA polymerase at promoters. Cell Rep. 2012;2:1025–1035. doi: 10.1016/j.celrep.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrensberger A.H., Kelly G.P., Svejstrup J.Q. Mechanistic interpretation of promoter-proximal peaks and RNAPII density maps. Cell. 2013;154:713–715. doi: 10.1016/j.cell.2013.07.032. [DOI] [PubMed] [Google Scholar]
- Gaertner B., Zeitlinger J. RNA polymerase II pausing during development. Development. 2014;141:1179–1183. doi: 10.1242/dev.088492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidatzis D., Lerch A., Hahne F., Stadler M.B. QuasR: Quantification and annotation of short reads in R. Bioinformatics. 2015;31:1130–1132. doi: 10.1093/bioinformatics/btu781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Yam E.N., Jeong S., Tanay A., Egger G., Lee A.S., Jones P.A. Constitutive nucleosome depletion and ordered factor assembly at the GRP78 promoter revealed by single molecule footprinting. PLoS Genet. 2006;2:e160. doi: 10.1371/journal.pgen.0020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist D.A., Dos Santos G., Fargo D.C., Xie B., Gao Y., Li L., Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin M.J., Petesch S.J., Zobeck K.L., Min I.M., Lis J.T. Drosophila heat shock system as a general model to investigate transcriptional regulation. Cold Spring Harb. Symp. Quant. Biol. 2010;75:1–9. doi: 10.1101/sqb.2010.75.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques T., Gilchrist D.A., Nechaev S., Bern M., Muse G.W., Burkholder A., Fargo D.C., Adelman K. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol. Cell. 2013;52:517–528. doi: 10.1016/j.molcel.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D., Smith E.R., Garruss A.S., Mohaghegh N., Varberg J.M., Lin C., Jackson J., Gao X., Saraf A., Florens L. The little elongation complex functions at initiation and elongation phases of snRNA gene transcription. Mol. Cell. 2013;51:493–505. doi: 10.1016/j.molcel.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I., Lis J.T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T.K., Liu Y., Lay F.D., Liang G., Berman B.P., Jones P.A. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 2012;22:2497–2506. doi: 10.1101/gr.143008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H., Fuda N.J., Core L.J., Lis J.T. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Li X., Hechmer A., Eisen M., Biggin M.D., Venters B.J., Jiang C., Li J., Pugh B.F., Gilmour D.S. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol. Cell. Biol. 2008;28:3290–3300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levo M., Avnit-Sagi T., Lotan-Pompan M., Kalma Y., Weinberger A., Yakhini Z., Segal E. Systematic investigation of transcription factor activity in the context of chromatin using massively parallel binding and expression assays. Mol. Cell. 2017;65:604–617. doi: 10.1016/j.molcel.2017.01.007. [DOI] [PubMed] [Google Scholar]
- Liang K., Woodfin A.R., Slaughter B.D., Haug J.S., Jaspersen S.L., Shilatifard A. Mitotic transcriptional activation: Clearance of actively engaged Pol II via transcriptional elongation control in mitosis. Mol. Cell. 2015;60:1–11. doi: 10.1016/j.molcel.2015.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C.Y., Santoso B., Boulay T., Dong E., Ohler U., Kadonaga J.T. The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 2004;18:1606–1617. doi: 10.1101/gad.1193404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis J.T., Mason P., Peng J., Price D.H., Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- Louder R.K., He Y., López-Blanco J.R., Fang J., Chacón P., Nogales E. Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature. 2016;531:604–609. doi: 10.1038/nature17394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabilsi N.H., Deleyrolle L.P., Darst R.P., Riva A., Reynolds B.A., Kladde M.P. Multiplex mapping of chromatin accessibility and DNA methylation within targeted single molecules identifies epigenetic heterogeneity in neural stem cells and glioblastoma. Genome Res. 2014;24:329–339. doi: 10.1101/gr.161737.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S., Fargo D.C., dos Santos G., Liu L., Gao Y., Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh B.F., Venters B.J. Genomic organization of human transcription initiation complexes. PLoS ONE. 2016 doi: 10.1371/journal.pone.0149339. Published online February 11, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Ernst J., Kharchenko P.V., Kheradpour P., Negre N., Eaton M.L., Landolin J.M., Bristow C.A., Ma L., Lin M.F., modENCODE Consortium Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Inagaki S., Mituyama T., Kawamura Y., Ono Y., Sakota E., Kotani H., Asai K., Siomi H., Siomi M.C. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- Shao W., Zeitlinger J. Paused RNA polymerase II inhibits new transcriptional initiation. Nat. Genet. 2017 doi: 10.1038/ng.3867. [DOI] [PubMed] [Google Scholar]
- Wagschal A., Rousset E., Basavarajaiah P., Contreras X., Harwig A., Laurent-Chabalier S., Nakamura M., Chen X., Zhang K., Meziane O. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell. 2012;150:1147–1157. doi: 10.1016/j.cell.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirbelauer C., Bell O., Schübeler D. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev. 2005;19:1761–1766. doi: 10.1101/gad.347705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudkovsky N., Ranish J.A., Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.