Abstract
Esophageal cancer ranks as the sixth leading cause of cancer-related deaths worldwide. Cancer stemness is mainly considered to be the key factor for cancer recurrence particularly in esophageal cancer. It is important to identify cancer stem cell markers as targets in future therapies. The present study aimed to investigate the expression of putative cancer stem cell-related marker musashi1 (Msi1) and assess the correlation with clinicopathologcal status of esophageal squamous cell carcinoma (ESCC) cases. We then clarified the role of Msi1 in esophageal cancer cells during proliferation, apoptosis, sphere formation and migration. Finally, we investigated the relationship of Msi1 with the prognosis of ESCC patients. ESCC tissue samples from 93 patients and 20 paired histologically normal tissues were procured for immunohistochemical analysis. We analyzed the characteristics of Msi1, using sphere formation and anchorage independent growth. Moreover, using flow cytometry and Cell Counting Kit-8 (CCK-8) assay, we investigated the role of Msi1 in cancer cell proliferation and apoptosis. Furthermore, we clarified the role of Msi1 in the process of sphere formation and migration of ESCC cells through knockdown of Msi1 expression by siRNA in ESCC cell lines. The results revealed that there was a higher expression of Msi1 in ESCC specimens compared with normal tissues. In addition, Msi1 expression was significantly associated with clinical stage and lymph node metastasis. Most importantly, the increased immunocytochemical staining of Msi1 in spheroid cells revealed the stemness characteristics of Msi1 in ESCC. In addition, we found that silencing of Msi1 decreased cell proliferation, migration and induced apoptosis in TE-7 and KYSE70 cells. Furthermore, downregulation of Msi1 attenuated the sphere formation ability of ESCC cells. Patients with higher expression of Msi1 had a shorter survival. In conclusion, Msi1 acts as a stemness-associated gene in esophageal cancer cell lines and could serve as a prognostic marker in patients with ESCC.
Keywords: ESCC, cancer stem cell, musashi1
Introduction
Esophageal cancer is one of the most aggressive gastrointestinal cancers ranking as the sixth leading cause of cancer-related deaths worldwide (1). Approximately 70% of global esophageal cancer cases occur in China, and squamous cell carcinoma accounts for 90% of these histopathologically diagnosed cases (2). Early therapeutic intervention is often hampered due to the insidious advancement of the disease, and poor elucidation of its cellular and molecular mechanism underlying its initiation and progression.
Carcinogenesis is a complex multistep process, in which numerous studies propose that tumor-initiating cells (TICs) or cancer stem cells (CSCs) are significant for its development (3). CSCs were first identified in human hematopoietic malignancies and then in several other solid tumors such as cancers of the brain (4), breast (5), prostate (6), colon (7), pancreas (8) and lung (9). However, their presence and identity in esophageal squamous cell carcinoma (ESCC) is yet to be defined. Lack of appropriate biomarkers unusually expressed by this subset of cancer cells is the main challenge.
Despite the fact that there are various studies affirming the role of aberrant expression of cancer stem-related markers CD90 (10), ALDH1 (11) and p75NTR (12) in the progression of ESCC and the clinicopathological status of patients, the RNA-binding protein, musashi1 (Msi1) which was initially identified in Drosophila melanogaster by its ability to regulate asymmetric cell division of neural and epithelial progenitor cells, has yet to be studied in relation to this disease (13). In mammals, Msi1 mainly expressed in stem and progenitor cells can regulate memory (14). In recent years, the role of Msi1 in tumors has attracted increasing interest. Recently, it was recognized as candidate cancer stem cell marker in pulmonary (15), colorectal (16), intestinal (17,18), endometrial (19), breast (20), gallbladder (21) and cervical squamous cell carcinomas (22). In addition, the latest studies show that Msi1, as the upstream protein of oncogenic and epigenetic signals, promoted poor prognosis and chemoresistance through the activation of the Akt pathway and IL-6 secretion (23,24). Moreover, a recent study speculated that Msi1 may be correlated with Notch1 expression in esophageal cancer (25), but no experimental studies have verified its impact on the development of esophageal cancer. In the present study, we set out to investigate the expression and clinicopathological significance of the putative cancer stem cell marker Msi1 in ESCC clinical samples and determine whether Msi1 plays a significant role in the proliferation, apoptosis, sphere formation and migration of esophageal cancer cell lines.
Materials and methods
Ethical standard and informed consent
All procedures performed in the present study involving human participants were in accordance with the ethical standards of the Institutional and/or National Research Committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the present study.
Cell lines
The TE-7 and KYSE70 cell lines (donated by Professor Mingzhou Guo, General Hospital of the Chinese People's Liberation Army) as well as TE-1, EC109, EC9706 and EC1 cell lines (donated by Professor Qingxia Fan, Department of Oncology, The First Affiliated Hospital of Zhengzhou University) in esophageal cancer research were preserved in our laboratory and maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (both from HyClone, Logan, UT, USA), 100 U/ml of penicillin, and 100 µg/ml of streptomycin at 37°C and an atmosphere of 5% CO2.
Clinical samples for qPCR and immunohistochemistry
Sixty-nine paired ESCC and adjacent non-cancerous tissues were previously collected and stored (2012–2014) for qPCR. Tissues were provided by the Department of Thoracic Surgery, The First Affiliated Hospital of Zhengzhou University, with confirmed histopathological results. Informed consent was obtained from each patient, and the collection of the samples was approved by the local Ethics Committee. Information pertaining to clinicopathological parameters was also available. Thick (5-µm) formalin-fixed paraffinized tissue sections were prepared from carcinomas derived from 93 tumors and 20 matched adjacent normal tissues. Informed consent was obtained from the patients or their guardians. None of the patients received any radiotherapy or chemotherapy before surgery.
RNA extraction, cDNA synthesis and quantitative real-time PCR
Total RNA was extracted from the cell lines and clinical samples using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA). cDNA was synthesized from RNA using PrimeScript RT reagent kit with gDNA Eraser (Takara, Shiga, Japan). RT-PCR and qPCR were performed to detect the expression of Msi1 in ESCC cell lines and clinical samples. GAPDH was used as an internal control. With the ΔΔCq method, we compared the expression level of clinical samples and cell lines (26).
Western blotting
Protein was extracted from the cell lines using an Ultrasonic Cell Cracking Apparatus. After denaturation, the samples were separated using 10% SDS-PAGE and transferred to NC membranes (G&E) for 2 h. After incubation, the bands were exposed using Bio-Rad with enhanced chemiluminescence (ECL). The exposure time was ~1 min.
Immunohistochemistry
For subsequent immunohistochemical analysis, aliquots of tissue sections were brought to 65°C for 30 min, and then dewaxed in two consecutive xylene washes for 15 min each and rehydrated in a series of decreasing alcohol concentrations. Sections were washed with phosphate-buffered saline (PBS) (pH 7.4) twice for 2 min. Antigen retrieval was achieved by steaming slides for 20 min in citrate buffer. H2O2 (3%) was applied for 30 min to suppress endogenous peroxidase activity. Sections were stained with primary antibodies diluted with PBS (pH 7.4) according to the manufacturers instructions and incubated at 4°C overnight. The samples were brought to room temperature for 1 h and signal amplification and visualization were performed using an avidin-biotin complex (ABC) and the substrate 3,3-diaminobenzidine (DAB), respectively. Sections were counterstained with hematoxylin (DingGuo Biotechnology Co., Ltd., Beijing, China), washed in running water for 10 min and treated with increasing serial concentrations of alcohol and two consecutive xylene washes. Finally, the samples were mounted for microscopic examination, immunoreactivity was scored using Leica Application Suite Version 3 software, and microphotographs were captured using Leica inverted Microscopy Digital Camera (magnification, ×200).
Sphere formation assay
TE-7 and KYSE-70 cells were resuspended in Dulbeccos modified Eagles medium (DMEM)/F12 (Invitrogen Life Technologies) supplemented with 4 µg/ml heparin (Sigma-Aldrich, St. Louis, MO, USA), B27 (1:50; Gibco, Life Technologies, Carlsbad, CA, USA), 20 ng/ml epidermal growth factor (EGF) and 20 ng/ml basic fibroblast growth factor (FGF) (both from PeproTech, Rocky Hill, NJ, USA), penicillin 100 IU/ml and streptomycin 100 µg/ml, and then seeded into 6-well ultra-low cluster plates (Corning Costar, Corning, NY, USA). After being cultured for 6 days, we calculated the rate of sphere formation and the spheres were collected and digested for adherent culture and further analysis.
Immunocytochemistry
To validate the expression of Msi1 in adherent and spheroid cells, immunocytochemistry was performed in the TE-7 and KYSE70 cell lines. Collected spheroid and adherent cells were transferred into the unused wells of the ultra-low attachment 24-well plate, and cultured for 8 h in stem culture medium. The medium was carefully aspirated and the cells were gently rinsed with PBS twice at room temperature and fixed using 4% paraformaldehyde. Antibody staining and color visualization processes were similar with those of immunohistochemistry. Finally, the cells were maintained in PBS to avoid drying until microphotographs were captured using Leica inverted Microscopy Digital Camera (magnification, ×200).
Scoring and standardized IHC staining
A semi-quantitative scoring approach which had previously been used (27–29) was applied in the present study. Each tumor was graded according to the intensity and the percentage of stained cells. The intensity scores were as follows: 0, no staining; +1, weak; +2, moderate; and +3, strong staining and the percentage of stained cells was as follows: 0–5%, 0; 6–25%, +1; 26–50%, +2; 51–75%, +3; and 76–100%, +4. Finally, the scores were combined by multiplying the intensity and the percentage of positive cells. Using this method, the possible score range was 0–12. Accordingly, scores in the range of 0–4 (inclusive) were considered as low (negative) and values >4 (6, 8, 9 and 12) as high (positive).
Msi1 knockdown
Using Lipofectamine, siRNA was transfected in TE-7 and KYSE70 cells. Then, we determined the effectiveness of siRNA by qPCR.
Flow cytometric (FACS) analysis
To assess the apoptosis of TE-7 and KYSE70 cells after transfection with Msi1 siRNA or negative control we used flow cytometry. The cells were collected and incubated with Annexin V binding buffer and Annexin V antibody for half an hour at 4°C. After being washed with buffer, the cells were resuspended in 300 µl of buffer, stained with propidium iodide (PI) and analyzed using a flow cytometer and CellQuest software (BD Biosciences, San Diego, CA, USA).
CCK-8 analysis
To investigate the effect of Msi1 on the proliferation of TE-7 and KYSE70 cells, we collected and seeded cells into a 96-well plate. After adherence, we added Cell Counting Kit-8 (CCK-8) solution at 0, 24 and 48 h and detection followed using a microplate reader 1 h later.
Migration assay
To detect the role of Msi1 in the process of ESCC cell migration, we carried out a Transwell assay. In this experiment, we resuspended KYSE70 or TE-7 cells in culture medium without serum and placed them in the upper chamber. In addition, complete medium was added in the training system. After 24 h, with fixation and staining, we observed the number of cells that had migrated to the lower chamber.
Statistical analysis
All statistical analyses were performed using SPSS 17.0 software. With paired or unpaired t-tests, we analyzed the expression of Msi1 in cell lines and clinical samples, as well as the difference of proliferation and apoptosis between si-Msi1 cells and the negative control group. Overall survival rates were determined from the time of curative surgery to the last follow-up contact dates or reported dates of death and estimated using Kaplan-Meier analysis. P-values <0.05 were considered to indicate a statistically significant result.
Results
Msi1 expression is correlated with clinical stage and lymphatic metastasis in patients with ESCC
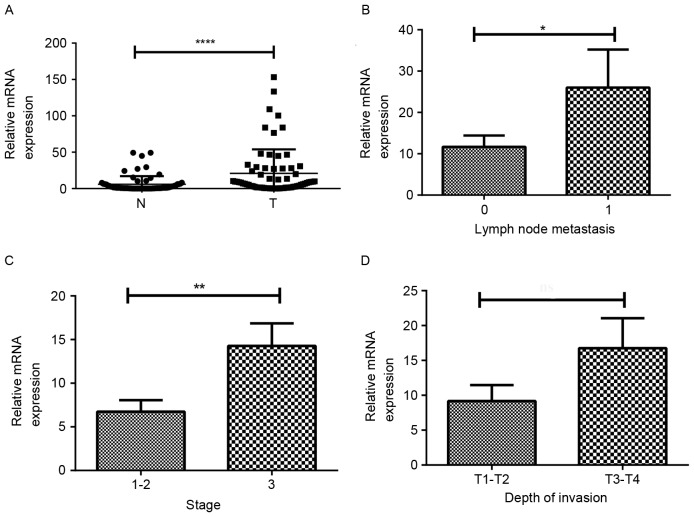
Msi1 is considered as a cancer stem cell marker in pulmonary, colorectal and breast cancer, and it regulates the tumorigenesis in tumor models. We compared the expression of Msi1 in tumor and adjacent non-cancerous tissues. As shown in Fig. 1A, Msi1 was found to be significantly upregulated in cancer tissues when compared with the corresponding non-tumor tissues (P<0.0001, n=67). We hypothesized that Msi1 plays a critical role in the progression of ESCC. In addition, we found that there was a higher expression of Msi1 in the samples from the patients suffering from lymph node metastasis or advanced-stage ESCC (Fig. 1B, P=0.04; Fig. 1C, P=0.005). However, there was no statistical significance in the depth of invasion (Fig. 1D). On the whole, the aforementioned results revealed that Msi1 may be an oncogene and act as a prognostic marker in ESCC patients.
Figure 1.
Expression of Msi1 is correlated with tumor stage and lymph node metastasis in human ESCC samples. (A) The qPCR analysis of Msi1 expression was investigated in paired human ESCC specimens (T) and adjacent non-cancerous tissues (N). The correlation of Msi1 expression with clinicopathological characteristics such as (B) stage, (C) lymphatic metastasis and (D) depth of invasion was analyzed in mRNA expression (*P<0.05, **P<0.01, ****P<0.0001). Msi1, musashi1; ESCC, esophageal squamous cell carcinoma.
Msi1 expression is enriched in esophageal cancer stem cells
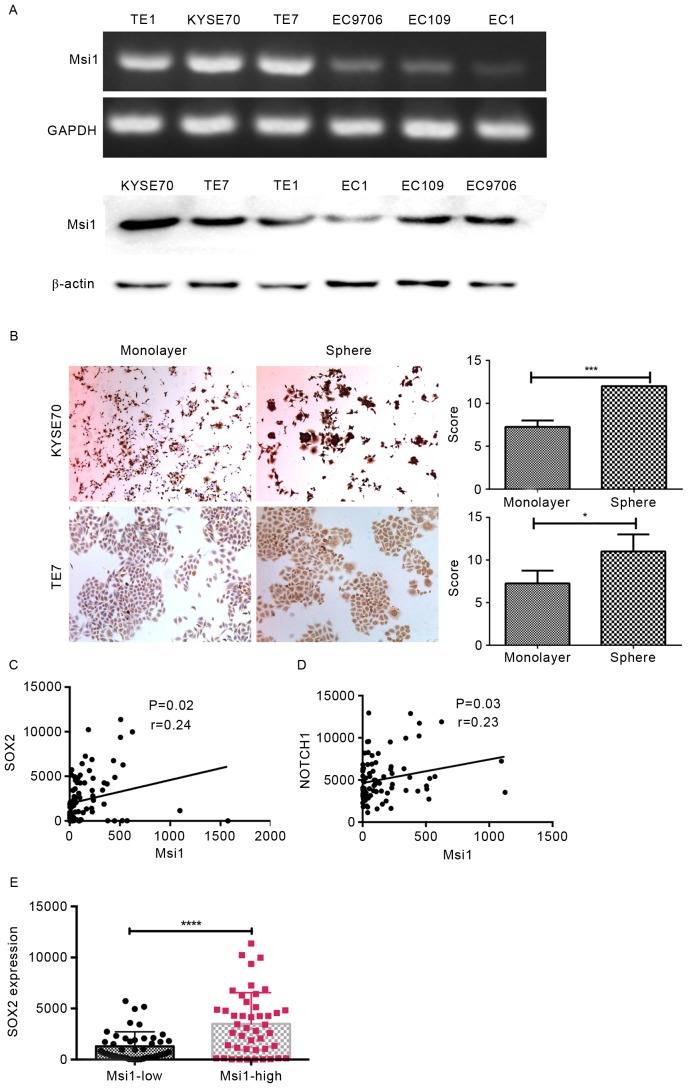
Msi1 had a higher expression in the poorly differentiated ESCC cell lines TE-7, TE-1 and KYSE70 than in the terminally differentiated cell lines EC109, EC9706 and EC-1 (Fig. 2A). Immunocytochemical staining revealed that Msi1 was strongly positive in spheroid cells, in TE-7 and KYSE70 cells when compared to the monolayer (Fig. 2B, P=0.03 and P=0.02, respectively). In addition, from TCGA analysis, we determined that Msi1 expression had a significant positive correlation with SOX2 and Notch1 (Fig. 2C, P=0.02; Fig. 2D, P=0.03) and that SOX2 had a significantly different expression in the Msi1 high-expression group compared with the low-expression group (Fig. 2E, P<0.0001). The latter two genes were the key factors regulating cancer cell stemness. Moreover, there was no similar phenomenon between the expression of Msi1 and Notch1 (data not shown). According to previous studies and the aforementioned results, we hypothesized that Msi1 plays a critical role in ESCC stemness and its characteristics could have an effect on the proliferation and apoptosis of cancer cells.
Figure 2.
Expression of Msi1 is enriched in esophageal cancer stem cells. (A) The incidence of Msi1 in different ESCC cell lines was detected through RT-PCR and western blotting. (B) Msi1 expression was investigated in the monolayer of KYSE70 and TE-7 cells and matched sphere cells by immunocytochemistry. (C and D) From the TCGA database, we analyzed the correlation between the expression of Msi1 and SOX2 and Notch1 in ESCC (n=91). (E) We divided the patients into two groups based on the expression of Msi1, and determined that SOX2 had a significantly different expression in the two groups (*P<0.05, ***P<0.01, ****P<0.0001). Msi1, musashi1; ESCC, esophageal squamous cell carcinoma.
Decrease of Msi1 expression inhibits esophageal cancer cell proliferation and promotes apoptosis
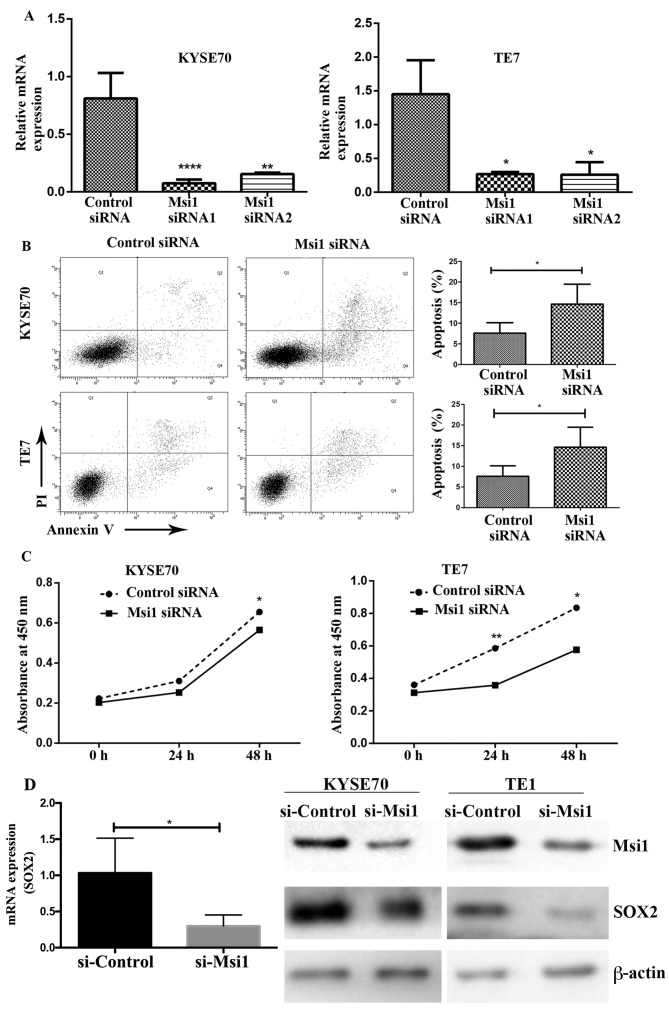
The role of Msi1 in the proliferation and apoptosis of TE-7 and KYSE70 cells was examined following the transfection of an Msi1 siRNA. RNA interference resulted in the decrease of Msi1 mRNA in TE-7 and KYSE70 cells compared to the control siRNA (Fig. 3A). After transfection for 48 h, we collected the cells and detected apoptosis using flow cytometry. Notably, we observed that knockdown of Msi1 increased the apoptosis rates in TE-7 and KYSE70 cells (Fig. 3B). In addition, the proliferation of TE-7 and KYSE70 cells transfected with Msi1-siRNA was observably lower than that of the control group (Fig. 3C). Using PCR and western blotting, we further determined that SOX2 expression was also decreased with Msi1 interference (Fig. 3D). Generally, Msi1 expression could promote the proliferation and decrease the apoptosis of esophageal cancer cell lines.
Figure 3.
Decrease of Msi1 promotes the apoptosis and inhibits the proliferation of esophageal cancer cells. (A) Msi1 mRNA expression was knocked down in KYSE70 and TE-7 cells by transfection with small interfering RNA-siRNA. The knockdown efficiency was determined by qPCR. (B) The apoptosis of KYSE70 and TE-7 cells before and after Msi1 knockdown was detected using flow cytometry. (C) The proliferation of KYSE70 and TE-7 cells was investigated by Cell Counting Kit-8 assay. (D) SOX2 expression was investigated with qPCR and western blotting under Msi1 interference (*P<0.05, **P<0.01, ****P<0.0001). Msi1, musashi1.
Capacity of sphere formation and migration is inhibited in Msi1 downregulated ESCC cells
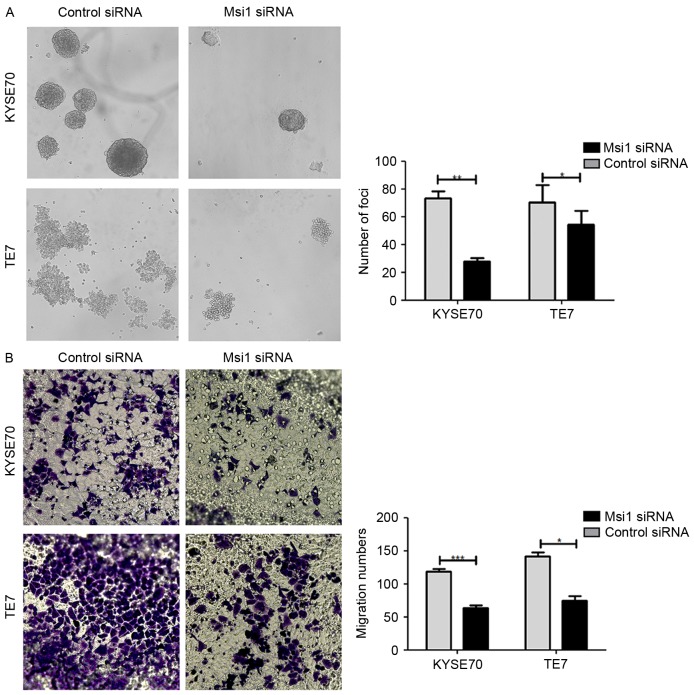
Using the same approach, we detected the influence of knocked down Msi1 expression on the capacity of sphere formation and migration in ESCC cells. We determined that in the si-Msi1 group the number of spheres formation was markedly decreased (Fig. 4A). Similarly, with Transwell assay, KYSE70 and TE-7 cells transfected with si-Msil lacked of the ability to migrate to the lower chamber (Fig. 4B). The data demonstrated that Msi1 had an effect on tumor cell stemness and migration capacity.
Figure 4.
Msi1 expression has a positive effect on the sphere formation and migration of ESCC cells. (A) Sphere formation assay was used to compare the stemness-associated phenotypes between wild-type Msi1 expression and si-Msi1 in ESCC cells. (B) Transwell assay was used to detect the migration capacity of KYSE70 and TE-7 cells (*P<0.05, **P<0.01, ***P<0.001). Msi1, musashi1; ESCC, esophageal squamous cell carcinoma.
Msi1 expression in ESCC tissues is negatively correlated with the overall survival
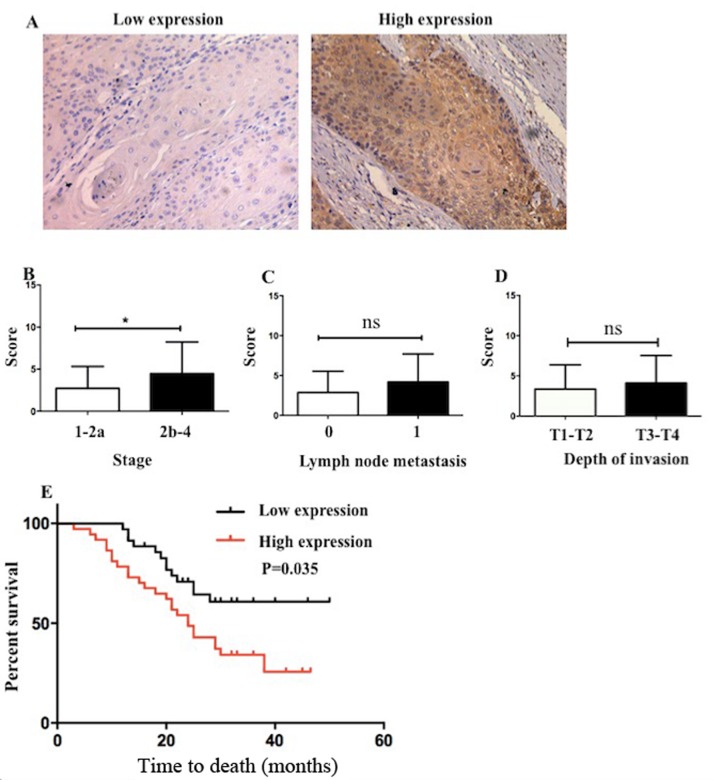
To further evaluate the relationship of Msi1 with the prognosis in ESCC, we investigated the protein expression of Msi1 in ESCC samples by immunohistochemistry. As shown in Fig. 5A, Msi1 had a varying expression, with patient tissues exhibiting high expression and low expression. At the protein level, we also found that Msi1 was correlated with late-stage ESCC. However, there was no statistical significance with different lymph node metastasis and invasion of depth (Fig. 5C-D). According to the expression of Msi1 in tumor tissues, we divided the 93 patients into two groups. There were 53 patients that had a high expression of the stemness-associated gene Msi1, and the rest (40 patients) had a low expression of Msi1 in tumor tissues. Moreover, compared with the low expression group, the high expression group had a significantly shorter overall survival (Fig. 5E; P=0.035).
Figure 5.
ESCC samples with a higher expression of Msi1 in tumor tissues have a worse clinical outcome. (A) Immunohistochemistry was used to detect the expression patterns of Msi1 in tumor tissues. The correlation of Msi1 expression with clinicopathological characteristics such as (B) stage, (C) lymphatic metastasis and (D) depth of invasion was analyzed at the protein level. (E) Patients with low expression of Msi1 had a better overall survival when cmpared to patients with high expression of Msi1 (*P<0.05). Msi1, musashi1; ESCC, esophageal squamous cell carcinoma. P=0.035.
Discussion
Despite the significant progress in ESCC diagnosis and treatment, the dismal 5-year survival rate of ESCC has not changed satisfyingly. Hence, the identification of important molecules to assist in an early diagnosis and treatment targets of ESCC are essential. Furthermore, with CSCs or TICs persisting in tumor development, cancer patients cannot acquire anticipatory prognosis after treatment with surgery, chemotherapy, radiotherapy or combination therapy. Thus, it is necessary to identify the pivotal point in ESCC development in order to target CSCs or TICs.
Msi1, a regulatory factor in the process of progenitor division, has also been identified as a regulator of memory. Both reported physiological functions are relevant in keeping the stemness of initial cells (14). Recently, an increasing number of results in research revealed that Msi1 plays a significant role in cancer development. In clinical samples, overexpression of Msi1 was correlated with tumor-node-metastasis (TNM) stage and lymph node metastasis. With Msi1 overexpression in tumor tissues, patients had a decreased progression-free survival and overall survival. Analogous results have been reported in lung, gastric and gallbladder cancer, and colorectal carcinoma. Moreover, in metastatic colorectal cancer cells it was demonstrated that NOTCH3 signaling regulated Msi1 expression, and played a key role in decreasing the activity of NUMB thus activating DLL4 or NOTCH1 and regulating of cancer cells (30). As an important upstream molecule of NOTCH, it has also been implicated in lung cancer. It has also been described as having a similar role to ALDH1, the widely accepted cancer stemness-associated gene in cancer development. In addition, a previous study demonstrated that Msi1 promotes tumor growth by targeting cell cycle checkpoint proteins p21, p27 and p53 (31).
Relevant studies revealed that CD271, CD133 and CD90 affected CSC development in ESCC, however, we are the first to propose that Msi1, a well known molecule regulating cell division, plays a significant role in ESCC development by promoting proliferation and decreasing apoptosis of cancer cells. Firstly, as revealed in other studies, Msi1 had a higher expression in ESCC tumor tissues compared with adjacent non-cancerous tissues and its distribution was also more diffuse (32,33). As suggested in gastric cancer (34), due to the difference in proliferation patterns between cancer cells and normal cells, Msi1 had a different expression in cancer and normal matched tissues. Furthermore, we determined a negative correlation between Msi1 expression in tumor tissues and lymph node metastasis and TNM stage. At the mRNA level, the patients with advanced-stage ESCC had a higher expression of Msi1 than early-stage patients, and the clinical samples with lymph node metastasis also exhibited a higher expression of Msi1 compared with the non-metastatic samples. We further investigated the expression patterns of Msi1 using immunohistochemistry. As determined, tumor tissues exhibited a higher expression of Msi1 compared to adjacent non-cancerous tissues. Moreover advanced-stage ESCC tissues had an increased expression of Msi1 when compared to ealy-stage patient tissues. Patients with lymph node metastasis also had a higher expression of Msi1, but the difference in expression with the non-metastatic group was not statistically significant.
To further clarify the importance of Msi1 in tumor development, we conducted a series of experiments in vitro using various ESCC cell lines. Compared with TE-1, EC109, EC9706 and EC1, the poorly differentiated TE-7 and KYSE70 cell lines had a higher expression of Msi1. We suspected that Msi1 was related to cancer histological grade. Upon investigation this relationship was not substantiated in the clinical specimens we observed due to the diffuse expression patterns of Msi1, and therefore we may further probe the expression of Msi1 in more ESCC patients. Notably, the spheroid cells when compared with the monolayer ones, exhibited a marked higher expression of Msi1. To the best of our knowledge, sphere formation is an effective approach to acquire CSC-like cells. Therefore, we believe that Msi1 can maintain the stemness of ESCC cancer cells. Consistent with other studies, using TCGA database, we found that Msi1 had a positive correlation with SOX2 in esophageal cancer (35). With the interference of RNA, we knocked down the expression of Msi1 in KYSE70 and TE-7 cells effectively. Then, significant changes were observed in the cancer cells. There was a marked increase in apoptosis in the si-Msi1 group and a decrease in proliferation. In addition, si-Msi1 had an effect on sphere formation and migration capacity. These results were similar to other types of cancer investigated in previous studies (24,30,31). Thus in clinical application, Msi1 can be considered as a target in the prevention of the malignant biological behavior of tumor cells and the improvement of the prognosis of cancer patients.
Furthermore, in future we need to construct an animal model to demonstrate the impact of Msi1 on the development of ESCC. In relation to the underlying mechanisms, in addition to NOTCH signaling as previously aforementioned, we may investigate the relationship with other cancer stemness-associated genes. We may also probe the stemness-associated genes regulating the tumor microenvironment as KLF4 which was studied in breast cancer (36).
In conclusion, the present study illustrated that Msi1 regulated the proliferation, apoptosis, sphere formation and migration capacity of cancer cells, and had a higher expression in spheroid cells. All the experimental results support that Msi1 may act as a potential prognostic marker in patients with ESCC. Furthermore, it could be used as a diagnostic and prognostic marker in clinical conversions.
Table I.
Patient characteristics in the present study.
| Clinicopatho-logical characteristics | n=60 | Relative Msi1 expression | n=93 | Msi1 expression (score) |
|---|---|---|---|---|
| Sex | ||||
| Male | 42 | 25.38±5.553 | 63 | 3.651±0.3861 |
| Female | 18 | 11.37±4.841 | 30 | 3.6±0.5416 |
| Age (years) | ||||
| <60 | 27 | 16.31±4.055 | 34 | 2.941±0.4221 |
| ≥60 | 33 | 27.14±7.885 | 59 | 4.0341±0.4229 |
| Lymph node metastasis | ||||
| Positive | 20 | 26.13±8.362 | 22 | 4.182±0.752 |
| Negative | 40 | 11.67±2.757 | 45 | 2.867±0.3964 |
| Histological grade | ||||
| G1–2 | 36 | 17.97±5.12 | 36 | 3.694±0.5328 |
| G3 | 24 | 24.42±7.348 | 30 | 5±0.5969 |
| Invasion | ||||
| T1-T2 | 27 | 9.169±2.31 | 38 | 3.357±0.5709 |
| T3-T4 | 30 | 16.75±4.301 | 28 | 4.105±0.5528 |
| Stage | ||||
| I-IIA | 36 | 6.749±1.309 | 43 | 2.721±0.3956 |
| IIB-IV | 17 | 14.27±2.578 | 26 | 4.462±0.7364 |
Msi1, musashi1.
Acknowledgements
The present study was supported by grants from the China-US (NFSC-NIH) Program for Biomedical Collaborative Research (grant no. 812111102), and the National Natural Science Foundation of China (grant no. 81171986).
References
- 1.Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: Defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137–2150. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 2.Song Y, Li L, Ou Y, Gao Z, Li E, Li X, Zhang W, Wang J, Xu L, Zhou Y, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91–95. doi: 10.1038/nature13176. [DOI] [PubMed] [Google Scholar]
- 3.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 4.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 5.Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, Pilotti S, Pierotti MA, Daidone MG. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 6.Patrawala L, Calhoun T, Schneider-Broussard R, Li H, Bhatia B, Tang S, Reilly JG, Chandra D, Zhou J, Claypool K, et al. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene. 2006;25:1696–1708. doi: 10.1038/sj.onc.1209327. [DOI] [PubMed] [Google Scholar]
- 7.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 8.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 9.Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, Conticello C, Ruco L, Peschle C, De Maria R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504–514. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 10.Tang KH, Dai YD, Tong M, Chan YP, Kwan PS, Fu L, Qin YR, Tsao SW, Lung HL, Lung ML, et al. A CD90+ tumor-initiating cell population with an aggressive signature and metastatic capacity in esophageal cancer. Cancer Res. 2013;73:2322–2332. doi: 10.1158/0008-5472.CAN-12-2991. [DOI] [PubMed] [Google Scholar]
- 11.Li S, Yue D, Chen X, Wang L, Li J, Ping Y, Gao Q, Wang D, Zhang T, Li F, et al. Epigenetic regulation of CD271, a potential cancer stem cell marker associated with chemoresistance and metastatic capacity. Oncol Rep. 2015;33:425–432. doi: 10.3892/or.2014.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang SD, Yuan Y, Liu XH, Gong DJ, Bai CG, Wang F, Luo JH, Xu ZY. Self-renewal and chemotherapy resistance of p75NTR positive cells in esophageal squamous cell carcinomas. BMC Cancer. 2009;9:9. doi: 10.1186/1471-2407-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okano H, Kawahara H, Toriya M, Nakao K, Shibata S, Imai T. Function of RNA-binding protein Musashi-1 in stem cells. Exp Cell Res. 2005;306:349–356. doi: 10.1016/j.yexcr.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 14.Hadziselimovic N, Vukojevic V, Peter F, Milnik A, Fastenrath M, Fenyves BG, Hieber P, Demougin P, Vogler C, de Quervain DJ, et al. Forgetting is regulated via Musashi-mediated translational control of the Arp2/3 complex. Cell. 2014;156:1153–1166. doi: 10.1016/j.cell.2014.01.054. [DOI] [PubMed] [Google Scholar]
- 15.Wang XY, Yu H, Linnoila RI, Li L, Li D, Mo B, Okano H, Penalva LO, Glazer RI. Musashi1 as a potential therapeutic target and diagnostic marker for lung cancer. Oncotarget. 2013;4:739–750. doi: 10.18632/oncotarget.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simon E, Petke D, Böger C, Behrens HM, Warneke V, Ebert M, Röcken C. The spatial distribution of LGR5+ cells correlates with gastric cancer progression. PLoS One. 2012;7:e35486. doi: 10.1371/journal.pone.0035486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rezza A, Skah S, Roche C, Nadjar J, Samarut J, Plateroti M. The overexpression of the putative gut stem cell marker Musashi-1 induces tumorigenesis through Wnt and Notch activation. J Cell Sci. 2010;123:3256–3265. doi: 10.1242/jcs.065284. [DOI] [PubMed] [Google Scholar]
- 18.Li N, Yousefi M, Nakauka-Ddamba A, Li F, Vandivier L, Parada K, Woo DH, Wang S, Naqvi AS, Rao S, et al. The Msi family of RNA-binding proteins function redundantly as intestinal oncoproteins. Cell Reports. 2015;13:2440–2455. doi: 10.1016/j.celrep.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Götte M, Greve B, Kelsch R, Müller-Uthoff H, Weiss K, Masouleh B Kharabi, Sibrowski W, Kiesel L, Buchweitz O. The adult stem cell marker Musashi-1 modulates endometrial carcinoma cell cycle progression and apoptosis via Notch-1 and p21WAF1/CIP1. Int J Cancer. 2011;129:2042–2049. doi: 10.1002/ijc.25856. [DOI] [PubMed] [Google Scholar]
- 20.Wang XY, Penalva LO, Yuan H, Linnoila RI, Lu J, Okano H, Glazer RI. Musashi1 regulates breast tumor cell proliferation and is a prognostic indicator of poor survival. Mol Cancer. 2010;9:221. doi: 10.1186/1476-4598-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu DC, Yang ZL, Jiang S. Identification of musashi-1 and ALDH1 as carcinogenesis, progression, and poor-prognosis related biomarkers for gallbladder adenocarcinoma. Cancer Biomark. 2010;8:113–121. doi: 10.3233/DMA-2011-0812. 2011. [DOI] [PubMed] [Google Scholar]
- 22.Hou T, Zhang W, Tong C, Kazobinka G, Huang X, Huang Y, Zhang Y. Putative stem cell markers in cervical squamous cell carcinoma are correlated with poor clinical outcome. BMC Cancer. 2015;15:785. doi: 10.1186/s12885-015-1826-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fox RG, Lytle NK, Jaquish DV, Park FD, Ito T, Bajaj J, Koechlein CS, Zimdahl B, Yano M, Kopp JL, et al. Image-based detection and targeting of therapy resistance in pancreatic adenocarcinoma. Nature. 2016;534:407–411. doi: 10.1038/nature17988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen HY, Lin LT, Wang ML, Lee SH, Tsai ML, Tsai CC, Liu WH, Chen TC, Yang YP, Lee YY, et al. Musashi-1 regulates AKT-derived IL-6 autocrinal/paracrinal malignancy and chemoresistance in glioblastoma. Oncotarget. 2016;7:42485–42501. doi: 10.18632/oncotarget.9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moghbeli M, Rad A, Farshchian M, Taghehchian N, Gholamin M, Abbaszadegan MR. Correlation between Meis1 and Msi1 in esophageal squamous cell carcinoma. J Gastrointest Cancer. 2016;47:273–277. doi: 10.1007/s12029-016-9824-6. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.McDonald JW, Pilgram TK. Nuclear expression of p53, p21 and cyclin D1 is increased in bronchioloalveolar carcinoma. Histopathology. 1999;34:439–446. doi: 10.1046/j.1365-2559.1999.00632.x. [DOI] [PubMed] [Google Scholar]
- 28.Xue LY, Hu N, Song YM, Zou SM, Shou JZ, Qian LX, Ren LQ, Lin DM, Tong T, He ZG, et al. Tissue microarray analysis reveals a tight correlation between protein expression pattern and progression of esophageal squamous cell carcinoma. BMC Cancer. 2006;6:296. doi: 10.1186/1471-2407-6-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langan RC, Mullinax JE, Ray S, Raiji MT, Schaub N, Xin HW, Koizumi T, Steinberg SM, Anderson A, Wiegand G, et al. A pilot study assessing the potential role of non-CD133 colorectal cancer stem cells as biomarkers. J Cancer. 2012;3:231–240. doi: 10.7150/jca.4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pastò A, Serafin V, Pilotto G, Lago C, Bellio C, Trusolino L, Bertotti A, Hoey T, Plateroti M, Esposito G, et al. NOTCH3 signaling regulates MUSASHI-1 expression in metastatic colorectal cancer cells. Cancer Res. 2014;74:2106–2118. doi: 10.1158/0008-5472.CAN-13-2022. [DOI] [PubMed] [Google Scholar]
- 31.Liu X, Yang WT, Zheng PS. Msi1 promotes tumor growth and cell proliferation by targeting cell cycle checkpoint proteins p21, p27 and p53 in cervical carcinomas. Oncotarget. 2014;5:10870–10885. doi: 10.18632/oncotarget.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moghbeli M, Forghanifard MM, Sadrizadeh A, Mozaffari HM, Golmakani E, Abbaszadegan MR. Role of Msi1 and MAML1 in regulation of Notch signaling pathway in patients with esophageal squamous cell carcinoma. J Gastrointest Cancer. 2015;46:365–369. doi: 10.1007/s12029-015-9753-9. [DOI] [PubMed] [Google Scholar]
- 33.Moghbeli M, Sadrizadeh A, Forghanifard MM, Mozaffari HM, Golmakani E, Abbaszadegan MR. Role of Msi1 and PYGO2 in esophageal squamous cell carcinoma depth of invasion. J Cell Commun Signal. 2016;10:49–53. doi: 10.1007/s12079-015-0314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuang RG, Kuang Y, Luo QF, Zhou CJ, Ji R, Wang JW. Expression and significance of Musashi-1 in gastric cancer and precancerous lesions. World J Gastroenterol. 2013;19:6637–6644. doi: 10.3748/wjg.v19.i39.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ying M, Wang S, Sang Y, Sun P, Lal B, Goodwin CR, Guerrero-Cazares H, Quinones-Hinojosa A, Laterra J, Xia S. Regulation of glioblastoma stem cells by retinoic acid: Role for Notch pathway inhibition. Oncogene. 2011;30:3454–3467. doi: 10.1038/onc.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu F, Shi Y, Wang J, Li J, Fan D, Ai W. Deficiency of Kruppel-like factor KLF4 in mammary tumor cells inhibits tumor growth and pulmonary metastasis and is accompanied by compromised recruitment of myeloid-derived suppressor cells. Int J Cancer. 2013;133:2872–2883. doi: 10.1002/ijc.28302. [DOI] [PMC free article] [PubMed] [Google Scholar]