Abstract
Aldehyde dehydrogenase 1 family member L1 (ALDH1L1) is downregulated in hepatocellular carcinoma (HCC) tumors, and its decreased expression is associated with the poor prognosis of HCC patients. We, therefore, evaluated the effect of single nucleotide polymorphisms (SNPs) of ALDH1L1, and its mRNA expression on the survival of hepatitis B virus (HBV)-related HCC patients and the association with tumor protein p53 (TP53) expression. ALDH1L1 SNPs in 415 HBV-related HCC patients were genotyped via direct sequencing. Expression profile chip datasets and survival information were obtained from GSE14520. The C allele (CT/CC) carriers of rs2276724 were significantly associated with a favorable prognosis [adjusted P=0.040; adjusted hazard ratio (HR)=0.725; 95% confidence interval (CI)=0.533–0.986]. Joint-effect analyses suggested that the CT/CC genotype of rs2276724 in TP53-negative patients was significantly associated with a decreased risk of death, compared to the TT genotype of rs2276724 in TP53-positive patients (adjusted P=0.037; adjusted HR=0.621; 95% CI=0.396–0.973). Furthermore, low expression of ALDH1L1 predicted a poor prognosis for the HBV-related HCC patients (adjusted P=0.04 for disease-free survival; adjusted P=0.001 for overall survival). Patients with high ALDH1L1 expression and low TP53 expression were significantly associated with a decreased risk of recurrence and death, and patients with a high TP53 expression were also significantly associated with a decreased risk of death in HBV-related HCC, compared with low ALDH1L1 and low TP53 expression. Our results suggest that ALDH1L1 may be a biomarker for predicting postoperative clinical outcomes. Moreover, ALDH1L1-rs2276724 and mRNA expression were associated with TP53 expression in HBV-related HCC patients.
Keywords: prognosis, hepatocellular carcinoma, hepatitis B virus, ALDH1L1, TP53
Introduction
Liver cancer is the second leading cause of cancer-related deaths in males worldwide. More than half of these liver cancer-related deaths occurred in China during 2012 (1). A recent study estimated that approximately 422,100 Chinese patients died from liver cancer in 2015, which will make it the third leading cause of cancer-related death in China (2). A population-based study of 138,852 cancer cases reported that liver cancer is associated with poor survival with an age-standardized 5-year relative survival of 10.1% in China (3). Liver cancer death rates in Guangxi Province were the highest in China for both males and females (4). Hepatocellular carcinoma (HCC) is the most common type of liver cancer (85–90%) (5). The most prominent parameters associated with HCC in China include hepatitis B virus (HBV) and C viral infection, alcoholic liver disease, and aflatoxin-B1-contaminated food (6). Previous studies of the Guangxi population reported that high HBV infection and aflatoxin B1 (AFB1) exposure resulted in a higher HCC morbidity and mortality in this province than in other provinces in China (7,8).
Tumor protein p53 (TP53) is a tumor-suppressor protein involved in transcriptional activation, DNA binding, and oligomerization domains. TP53 wild-type protein can induce cell cycle arrest, apoptosis, senescence, DNA repair, and changes in metabolism (9,10). Wild-type TP53 is an important tumor-suppressor gene in many types of cancers, especially in HCC, and its mutation is regarded as oncogenic (10,11), and affects HCC tumorigenesis and cancer progression (12–16). AFB1 has been strongly associated with TP53 mutations at codon 249 in exon 7, and HCC patients in Guangxi have a high rate (34%) of TP53 mutations at codon 249 in exon 7 (8,17). Thus, the population in this region presents a unique opportunity to investigate the relationship of HBV infection, AFB1 exposure, and TP53 gene mutations with HCC. Recently, meta-analyses have reported that immunohistochemical characterization of TP53 expression is associated with a poor prognosis of HCC (18).
Aldehyde dehydrogenase 1 family member L1 (ALDH1L1), also known as 10-formyltetrahydrofolate dehydrogenase (FDH), is a folate metabolism enzyme with tumor suppressor-like properties and is involved in the regulation of cell proliferation. A previous study conducted by Oleinik and Krupenko demonstrated that the antiproliferative effects of FDH in human lung cancer cell line A549 induced G1 arrest and apoptosis, accompanied by an increase in TP53 and p21 (19). In addition, subsequent research by this group also demonstrated that FDH-induced tumor-suppressor effects were strictly TP53-dependent in A549 cells and the TP53 pathway was a downstream mechanism in response to induction of FDH expression (20). Another study of HCC patients in Guangxi reported that low ALDH1L1 protein expression was a new and potential prognostic marker for the survival of HCC patients (21). Using bioinformatic analyses, we found that expression of ALDH1L1 in the liver was the highest in various human normal tissues, and was significantly downregulated in HCC tumor tissues compared to tissues adjacent to the tumor. Our previous genome-wide association study also reported that single nucleotide polymorphisms (SNPs) were associated with positive immunohistochemical characterization of TP53 expression in Guangxi patients with HBV-related HCC (22). In the present study, we determined the association between ALDH1L1 genetic variations and mRNA expression and the postoperative prognosis in Chinese HBV-related HCC patients, and its interaction with TP53.
Materials and methods
Study population
This study was approved by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University (Guangxi, China) with approval number KY-E-032. Fresh specimens of 415 cases of HCC were collected from 2001 to 2013 at the First Affiliated Hospital of Guangxi Medical University and were confirmed by pathology. All the patients were positive for serum HBV surface antigen inspection. The TP53 expression status in the cancer tissues was detected by immunohistochemistry. The cancer tissues were collected during surgery and immediately stored at −80°C for further use. The tumor status was classified using the Barcelona Clinic Liver Cancer (BCLC) staging system, and the liver reserve function was determined using the Child-Pugh classification. Portal vein tumor thrombus (PVTT) was classified according to a previous study (23).
SNP selection and genotyping
ALDH1L1 tagged SNPs and non-synonymous SNPs were selected by using an SNP info Web Server (http://snpinfo.niehs.nih.gov/. accessed 20 October 2016). Evaluation of SNP non-synonymous mutations caused by changes in protein amino acids were determined using SIFT (http://sift.jcvi.org/. accessed October 20, 2016) (24) and PolPhen 2 (http://genetics.bwh.harvard.edu/pph2/index.shtml. accessed October 20, 2016) (25). Bioinformatic analyses of ALDH1L1 SNPs with the tagged SNP located in the exon region showed that rs2276724 was a non-synonymous SNP. The influence of SNP non-synonymous mutation analyses by SIFT also showed that rs2276724 S481G was deleterious to protein coding. Consistent results from PolyPhen2 also showed that rs2276724 S481G was possibly deleterious to protein coding. Thus, rs2276724 was further studied. The transcriptional regulation of rs2276724 S481G was detected by F-SNP database (http://compbio.cs.queensu.ca/F-SNP/. accessed February 7, 2017) and the prediction tool Golden Path suggested that non-synonymous mutation of rs2276724 S481G caused transcriptional regulation change (26).
Genomic DNA was extracted from surgical tumor samples using the TIANamp Genomic DNA kit (Tiangen Biotech, Beijing, China). All samples were genotyped by DNA sequencing using an ABI Prism 3100 (Applied Biosystems, Shanghai Sangon Biological Engineering Technology and Services, Shanghai, China) with the following primers: forward, 5′-GCCCTGTCTTCCCTTCCTGTG-3′ and reverse, 5′-CCTGAGCCCACTCTGCTGAAAT-3′ for rs2276724. The sequencing results were analyzed using Chromas software (http://technelysium.com.au/wp/chromas/ accessed October 20, 2016) with a signal/noise >98%.
GEO data and bioinformatic analysis
Based on the predictive result of F-SNP, non-synonymous mutation of rs2276724 S481G may affect gene transcriptional regulation. We hypothesized that ALDH1L1 mRNA expression may contribute to prognostic prediction of HBV-related HCC. To test this hypothesis, we further analyzed the association of ALDH1L1 and TP53 at the transcriptional level to evaluate the effects of ALDH1L1 mRNA expression and the interactions with TP53 on HCC prognosis after hepatectomy. The profile chip dataset of Chinese HBV-related HCC from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/. accessed October 20, 2016) was analyzed and Spearman's correlation coefficient was used to assess its correlations. The GEO data selection criteria were set as follows: i), expression profiling chip; ii), Chinese HBV-related HCC; iii), corresponding survival profiles was available; and iv), patients undergoing hepatectomy. By searching the GEO database, we found that only the data of GSE14520 met the criteria above. Then, the samples were divided into two groups according to the ALDH1L1 expression in tumors. The high ALDH1L1 group was composed of samples with ALDH1L1 expression levels above the median value, and the low ALDH1L1 group was composed of the remaining samples. TP53 expression was grouped in the same manner. Both disease-free survival (DFS) and overall survival (OS) were analyzed in the different ALDH1L1 expression groups and used for the joint-effect survival analyses of the TP53 groups. We also stratified the analyses of associations between different ALDH1L1 expression levels and clinical features, both for OS and DFS. A gene interaction analysis web site (GeneMANIA: http://www.genemania.org/ accessed October 20, 2016) was used for correlation analyses between genes. Online analysis tool was used to analyze the ALDH1L1 expression in multiple human normal tissues (http://www.gtexportal.org/home/ accessed October 20, 2016) and in differences of expression of HCC tumor and adjacent tumor tissues (MERAV, Metabolic gEne Rapid Visualizer: http://merav.wi.mit.edu/. accessed October 20, 2016).
Statistical analysis
Hardy-Weinberg equilibrium (HWE) of the selected SNPs was estimated using a goodness-of-fit χ2-test. The binary logistic regression model was used to analyze the genetic model of ALDH1L1 genotypes for the status of different TP53 expression levels and for the association between clinical risk factors with ALDH1L1 genotypes. Survival analyses were performed using the Kaplan-Meier method with the log-rank test for different clinical factors and different genotypes. Cox proportion haphazard regression analyses were used to calculate the crude or adjusted hazard ratio (HR) and the 95% confidence interval (CI) in univariate analyses and multivariate analyses, adjusted for those variables with P<0.1 in later multivariate analyses. A value of P<0.05 was considered statistically significant. All statistical analyses were conducted using SPSS Statistical Software for Windows, version 20.0 (SPSS, Chicago, IL, USA).
Results
Clinical features and outcomes
Patients were followed up after surgery until the final follow-up or until death. The final follow-up was conducted in September 2014. A total of 415 patients successfully completed the follow-up, with 6.7% of the patients lost in the follow-up. The duration of the follow-up ranged from 12–125 months, with an overall median survival time (MST) of 48 months. At the time of analyses, 192 (46.3%) of the patients had died. A total of 162 patients were negative for TP53 expression and 253 patients were positive for TP53 expression. Clinical features of all patients and the association with the OS are shown in Table I. Using Kaplan-Meier analyses, the biological characteristics of tumor size, tumor number, BCLC stage, and portal vein tumor thrombus (PVTT) were significantly associated with the OS (log-rank test, P<0.001) and increased risk of death. A Child-Pugh classification score for 356 (85.8%) patients was associated with the OS (log-rank P=0.005). Radical resection was conducted in 231 patients (55.7%), and was associated with the OS (log-rank P=0.052), and patients without cirrhosis had a better prognosis (log-rank P=0.027). Adjuvant antiviral treatment in 143 (34.5%) patients was significantly associated with the OS (log-rank P=0.019), compared with those without treatment. The other clinical parameters were not associated with the OS.
Table I.
Clinical features of the patients with HBV-related HCC.
| Variables | Patients (N=415) | No. of events (%) | MST (months) | HR (95% CI) | Log-rank p-value |
|---|---|---|---|---|---|
| Age (years) | 0.149 | ||||
| ≤60 | 367 | 169 (46.0) | 51 | 1 | |
| >60 | 48 | 23 (47.9) | 41 | 1.375 (0.888–2.128) | |
| Sex | 0.479 | ||||
| Male | 375 | 177 (47.2) | 48 | 1 | |
| Female | 40 | 15 (37.5) | 42 | 0.828 (0.488–1.404) | |
| Ethnicity | 0.989 | ||||
| Han | 260 | 122 (46.9) | 47 | 1 | |
| Minority | 155 | 70 (45..2) | 50 | 0.998 (0.743–1.341) | |
| BMI | 0.745 | ||||
| ≤25 | 328 | 150 (45.7) | 45 | 1 | |
| >25 | 87 | 42 (48.3) | 51 | 0.945 (0.6670–1.333) | |
| Smoking status | 0.107 | ||||
| None | 263 | 118 (44.9) | 51 | 1 | |
| Ever | 152 | 74 (48.7) | 39 | 1.269 (0.947–1.702) | |
| Drinking status | 0.084 | ||||
| None | 246 | 107 (43.5) | 51 | 1 | |
| Ever | 169 | 85 (50.3) | 40 | 1.284 (0.964–1.710) | |
| Child-Pugh score | 0.005 | ||||
| A | 356 | 153 (43.0) | 51 | 1 | |
| B | 59 | 39 (66.1) | 31 | 1.689 (1.159–2.460) | |
| Cirrhosis | 0.027 | ||||
| No | 46 | 16 (34.8) | NA | 1 | |
| Yes | 369 | 176 (47.7) | 44 | 1.769 (1.058–2.958) | |
| Radical resectiona | 0.052 | ||||
| Yes | 231 | 97 (42.0) | 71 | 1 | |
| None | 172 | 89 (51.7) | 40 | 1.330 (0.997–1.774) | |
| Portal hypertensionb | 0.243 | ||||
| No | 208 | 100 (48.1) | 52 | 1 | |
| Yes | 172 | 75 (43.6) | 42 | 1.197 (0.883–1.623) | |
| Pathological diagnosisc | 0.622 | ||||
| Well differentiated | 24 | 9 (37.5) | 79 | 1 | |
| Moderately differentiated | 341 | 159 (46.6) | 44 | 1.378 (0.703–2.699) | |
| Poorly differentiated | 11 | 4 (36.4) | NA | 1.200 (0.369–3.898) | |
| Adjuvant antiviral treatment | 0.019 | ||||
| Yes | 143 | 43 (30.0) | NA | 1 | |
| No | 272 | 149 (54.8) | 41 | 1.501 (1.065–2.116) | |
| AFP leveld | 0.233 | ||||
| <400 | 210 | 89 (42.4) | 51 | 1 | |
| ≥400 | 175 | 85 (48.6) | 42 | 1.197 (0.889–1.612) | |
| Tumor behavior | |||||
| Tumor size (cm) | <0.001 | ||||
| ≤5 | 158 | 59 (37.3) | 75 | 1 | |
| >5 | 229 | 133 (58.1) | 36 | 1.802 (1.326–2.450) | |
| Tumor number | <0.001 | ||||
| Single | 302 | 127 (42.0) | 58 | 1 | |
| Multiple | 113 | 65 (57.5) | 28 | 1.792 (1.326–2.420) | |
| Regional invasion | 0.156 | ||||
| Absence | 353 | 162 (45.9) | 51 | 1 | |
| Presence | 62 | 30 (48.4) | 35 | 1.323 (0.895–1.958) | |
| BCLC stage | <0.001 | ||||
| A | 236 | 81 (34.3) | 95 | 1 | |
| B | 68 | 37 (54.4) | 36 | 2.055 (1.391–3.035) | |
| C | 111 | 74 (66.7) | 24 | 2.741 (1.994–3.767) | |
| PVTT | <0.001 | ||||
| No | 342 | 139 (40.6) | 73 | 1 | |
| Yes | 73 | 53 (72.6) | 18 | 2.801 (2.032–3.861) |
Information regarding radical resection was unavailable for 12 patients
information regarding portal hypertension was unavailable for 35 patients
information regarding pathological diagnosis was unavailable for 39 patients
information regarding AFP level was unavailable for 30 patients. HBV, hepatitis B virus; HCC, hepatocellular carcinoma; BMI, body mass index; AFP, α-fetoprotein; BCLC, Barcelona Clinic Liver Cancer; PVTT, portal vein tumor thrombus; MST, median survival time; HR, hazard ratio; CI, confidence interval.
Gene expression analysis
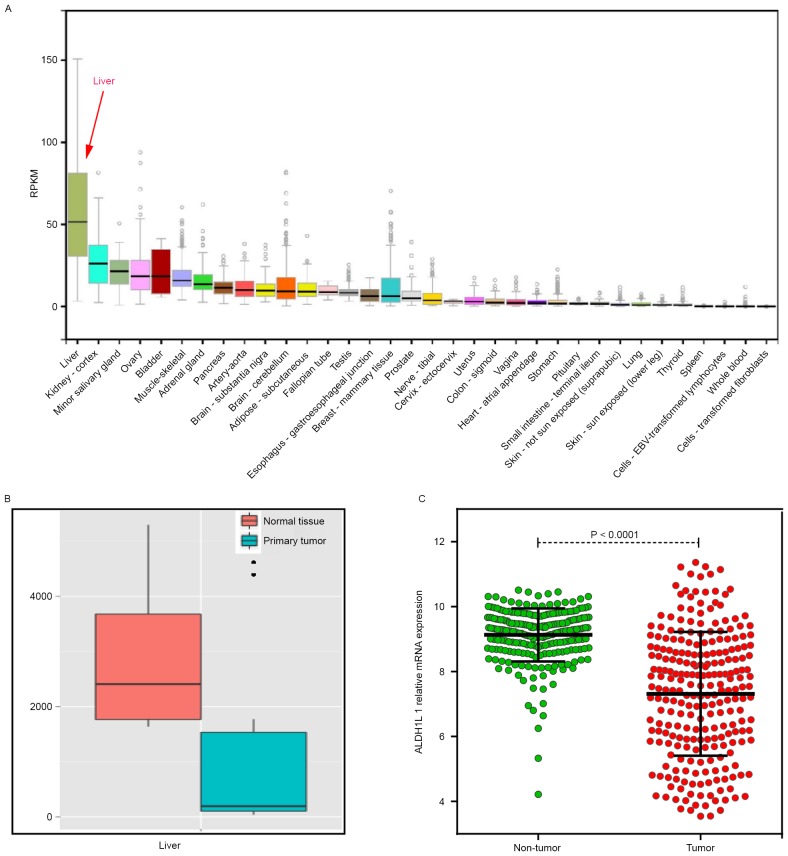
Bioinformatic analysis of ALDH1L1 gene expression in multiple human normal tissues showed that ALDH1L1 was the highest expression in normal liver tissue (Fig. 1A). ALDH1L1 expression was significantly downregulated in HCC tumor tissue, as determined for MERAV (Fig. 1B) and GSE14520 (Fig. 1C).
Figure 1.
(A) ALDH1L1 gene expression in multiple normal tissues. (B) Comparison of ALDH1L1 expression between HCC and normal tissues by MERAV. (C) Comparison of ALDH1L1 expression between HCC and non-tumor tissues by GSE14520. ALDH1L1, aldehyde dehydrogenase 1 family member L1; HCC, hepatocellular carcinoma.
Genetic model analysis of rs2276724
The success of genotyping for rs2276724 was 100%. The genotype frequencies met Hardy-Weinberg equilibrium as shown by the goodness-of-fit χ2-test (rs2276724, χ2=0.236; P=0.627). The genotype distribution of rs2276724 in patients with different TP53 expression is shown in Table II. The binary logistic regression model was used for adjustment for alcohol consumption, the Child-Pugh score, tumor size, tumor number, BCLC stage, radical resection, cirrhosis, adjuvant antiviral treatment, and PVTT. Using a co-dominant genetic model, the CT genotype of rs2276724 was significantly reduced with TP53 expression in HBV-related HCC patients (adjusted P=0.045; adjusted OR=0.644; 95% CI=0.418–0.990), compared with the TT genotype. The genotype distributions of rs2276724 in different types of TP53 expression were similar to the four genetic models.
Table II.
Genotype distribution of rs2276724 in HBV-related HCC patients with different TP53 expression statuses (genetic model).
| SNP | TP53- negative (n=162) | TP53-positive (n=253) | Crude OR (95% CI) | Crude p-value | Adjusted OR (95% CI) | Adjusted P-valuea |
|---|---|---|---|---|---|---|
| rs2276724 | ||||||
| Allele | ||||||
| T | 246 | 394 | 1 | 1 | ||
| C | 78 | 112 | 0.894 (0.640–1.249) | 0.512 | 0.878 (0.623–1.238) | 0.458 |
| Co-dominant | ||||||
| TT | 89 | 156 | 1 | 1 | ||
| CT | 68 | 82 | 0.688 (0.455–1.040) | 0.076 | 0.644 (0.418–0.990) | 0.045 |
| CC | 5 | 15 | 1.712 (0.602–4.867) | 0.314 | 1.838 (0.629–5.365) | 0.266 |
| Dominant | ||||||
| TT | 89 | 156 | 1 | 1 | ||
| CT+CC | 73 | 97 | 0.758 (0.508–1.131) | 0.175 | 0.725 (0.479–1.097) | 0.128 |
| Recessive | ||||||
| CT+TT | 157 | 238 | 1 | 1 | ||
| CC | 5 | 15 | 1.979 (0.705–5.554) | 0.195 | 2.153 (0.747–6.207) | 0.156 |
Adjustment for drinking status, Child-Pugh score, tumor size, tumor number, BCLC stage, radical resection, cirrhosis, adjuvant antiviral treatment, PVTT in logistic regression model. HBV, hepatitis B virus; HCC, hepatocellular carcinoma; TP53, tumor protein p53; SNP, single nucleotide polymorphism; AFP, α-fetoprotein; OR, odds ratio; CI, confidence interval.
Genetic polymorphisms in the HCC risk factors and in stratified analysis
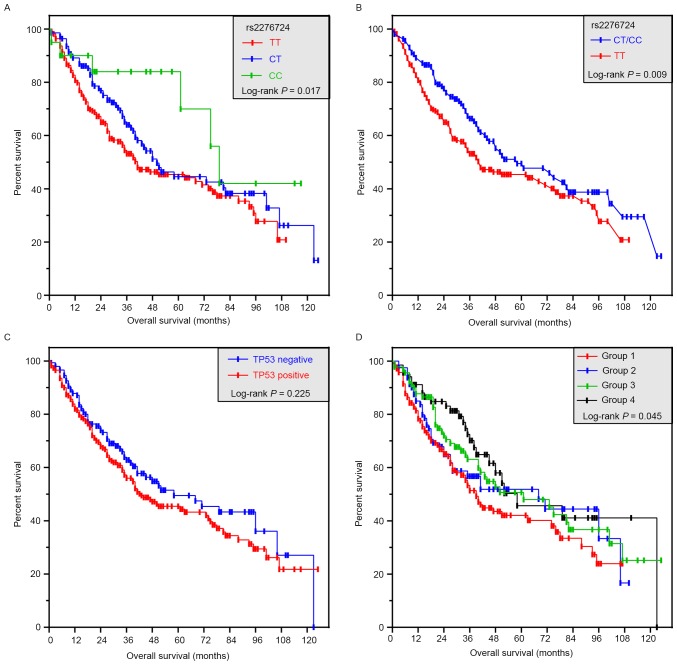
Association between risk factors and rs2276724 genotypes are summarized in Table III. None of the selected risk factors was associated with rs2276724 genotypes in the present study. Stratified analyses of the rs2276724 genotype and different strata of the OS of selected risk factors are shown in Fig. 2. In the favorable strata, the CT/CC genotype of rs2276724 significantly decreased the risk of death among patients with tumor sizes ≤5 cm, Child-Pugh score A, and without a PVTT. Regarding the invasion in the adverse strata, we also observed a similar effect that the CT/CC genotype of rs2276724 significantly decreased the risk of death among patients with BCLC stage B/C, non-radical resection, and the presence of regional invasion. The genotype distributions of rs2276724 in other strata showed no difference.
Table III.
Association between risk factors and rs2276724 in HBV-related HCC patients.
| Variables | TT | CT+CC | OR (95% CI) | P-value |
|---|---|---|---|---|
| Tumor size (cm) | ||||
| ≤5 | 98 | 70 | 1 | |
| >5 | 147 | 100 | 0.952 (0.640–1.418) | 0.810 |
| Tumor number | ||||
| Single | 176 | 126 | 1 | |
| Multiple | 69 | 44 | 0.891 (0.573–1.386) | 0.608 |
| Child-Pugh score | ||||
| A | 211 | 145 | 1 | |
| B | 34 | 25 | 1.070 (0.612–1.869) | 0.812 |
| BCLC stage | ||||
| A | 136 | 100 | 1 | |
| B | 43 | 25 | 0.791 (0.453–1.379) | 0.408 |
| C | 66 | 45 | 0.927 (0.586–1.467) | 0.747 |
| Radical resectiona | ||||
| Yes | 133 | 98 | 1 | |
| None | 103 | 69 | 0.909 (0.609–1.358) | 0.642 |
| AFP levelb | ||||
| <400 | 122 | 88 | 1 | |
| ≥400 | 109 | 66 | 0.839 (0.557–1.266) | 0.403 |
| Regional invasion | ||||
| Absence | 209 | 144 | 1 | |
| Presence | 36 | 26 | 1.048 (0.606–1.812) | 0.866 |
| PVTT | ||||
| No | 199 | 143 | 1 | |
| Yes | 46 | 27 | 0.817 (0.485–1.376) | 0.447 |
| Pathological diagnosisc | ||||
| Well differentiated | 13 | 11 | 1 | |
| Moderately differentiated | 198 | 143 | 0.854 (0.372–1.960) | 0.709 |
| Poorly differentiated | 7 | 4 | 0.675 (0.156–2.930) | 0.600 |
Information regarding radical resection was unavailable for 12 patients
information regarding pathological diagnosis was unavailable for 39 patients
information regarding AFP level was unavailable for 30 patients. HBV, hepatitis B virus; HCC, hepatocellular carcinoma; OR, odds ratio; CI, confidence interval; BCLC stage, Barcelona Clinic Liver Cancer stage; PVTT, portal vein tumor thrombus; AFP, α-fetoprotein
Figure 2.
Stratified analyses of associations of rs2276724 with the OS in the patients with HBV-related HCC. All variables were stratified by favorable and adverse strata. HBV, hepatitis B virus; HCC, hepatocellular carcinoma; OS, overall survival.
Relationship of rs2276724 and TP53 status with the OS
Using a dominant genetic model, patients with the TT genotype had a shorter MST compared to those with CT or CC genotypes of rs9275572 (TT vs. CT vs. CC; 39 vs. 50 vs. 79 months; log-rank P=0.017; Fig. 3A). However, the difference was similar after adjustment for alcohol consumption, Child-Pugh score, tumor size, tumor number, BCLC stage, radical resection, cirrhosis, adjuvant antiviral treatment, and a PVTT. In the dominant genetic model, patients with the TT genotype had a significantly smaller MST than the C allele carriers (TT vs.CT/CC; 39 vs. 58 months; log-rank P=0.009; Fig. 3B), and the CT/CC genotype of rs2276724 had a significantly decreased risk of death (adjusted P=0.040; adjusted HR=0.725; 95% CI=0.533–0.986; Table IV). Haplotype analysis showed that the C allele was associated with a significantly decreased risk of death (adjusted P=0.032; adjusted HR=0.747; 95% CI=0.572–0.976; Table IV), compared with the T allele. The prognosis for a different status of TP53 expression was similar in the patients (adjusted P=0.280; adjusted HR=1.183; 95% CI=0.872–1.605; Table IV and Fig. 3C).
Figure 3.
Survival curves of patients with rs2276724 and joint-effect analyses with different levels of TP53 expression. (A) Kaplan-Meier survival curves for patients with TT, CT, and CC genotypes. (B) Kaplan-Meier survival curves for patients with TT and CT/CC genotypes. (C) Kaplan-Meier survival curves for patients with different levels of TP53 expression. (D) Kaplan-Meier survival curves for joint-effect analyses of patients with different rs2276724 genotypes and different levels of TP53 expression. TP53, tumor protein p53.
Table IV.
Survival analysis of HBV-related HCC patients according to rs2276724 and TP53 status.
| Variable | Patients (n=415) | No. of events (%) | MST (months) | Crude HR (95% CI) | Crude p-value | Adjusted HR (95% CI) | Adjusted p-valuea |
|---|---|---|---|---|---|---|---|
| rs2276724 | |||||||
| Allele | |||||||
| T | 640 | 309 (48.3) | 41 | 1 | 1 | ||
| C | 190 | 75 (39.5) | 73 | 0.692 (0.535–0.893) | 0.005 | 0.747 (0.572–0.976) | 0.032 |
| Genotype | |||||||
| TT | 245 | 123 (50.2) | 39 | 1 | 1 | ||
| CT | 150 | 63 (42.0) | 50 | 0.716 (0.527–0.972) | 0.032 | 0.749 (0.545–1.029) | 0.074 |
| CC | 20 | 6 (30.0) | 79 | 0.421 (0.185–0.958) | 0.039 | 0.554 (0.240–1.278) | 0.166 |
| CT+CC | 170 | 69 (40.6) | 58 | 0.675 (0.502–0.909) | 0.010 | 0.725 (0.533–0.986) | 0.040 |
| TP53 status | |||||||
| Negative | 162 | 68 (42.0) | 58 | 1 | 1 | ||
| Positive | 253 | 124 (49.0) | 41 | 1.199 (0.892–1.612) | 0.229 | 1.183 (0.872–1.605) | 0.280 |
Adjustment for drinking status, Child-Pugh score, tumor size, tumor number, BCLC stage, radical resection, cirrhosis, adjuvant antiviral treatment, PVTT in Cox proportion haphazard regression model. HBV, hepatitis B virus; HCC, hepatocellular carcinoma; TP53, tumor protein p53; MST, median survival time; HR, hazard ratio; CI, confidence interval.
Joint-effect analysis
We further analyzed the TP53 status and rs2276724 genotypes with the mutual association with OS of the HBV-related HCC patients. TP53-positive patients with CT/CC genotypes had a significantly longer MST (Table V and Fig. 3D) as compared to the TP53-positive patients with TT genotype. After adjustment for alcohol consumption, Child-Pugh score, tumor size, tumor number, BCLC stage, radical resection, cirrhosis, adjuvant antiviral treatment, PVTT in the Cox proportion haphazard regression model, the TP53-negative patients with CT/CC genotypes showed a significantly decreased risk of death (adjusted P=0.037; adjusted HR=0.621; 95% CI=0.396–0.973; Table V).
Table V.
Joint effect survival analysis of rs2276724 and different TP53 expression statuses in HBV-related HCC patients.
| Group | Genotype | TP53 status | Patients (n=421) | No. of events (%) | MST (months) | Crude HR (95% CI) | Crude p-value | Adjusted HR (95% CI) | Adjusted P-valuea |
|---|---|---|---|---|---|---|---|---|---|
| 1 | TT | Positive | 156 | 81 (51.9) | 36 | 1 | 1 | ||
| 2 | TT | Negative | 89 | 42 (47.2) | 41 | 0.868 (0.598–1.260) | 0.457 | 0.903 (0.610–1.336) | 0.610 |
| 3 | CT+CC | Positive | 97 | 43 (44.3) | 61 | 0.695 (0.479–1.008) | 0.055 | 0.763 (0.518–1.126) | 0.173 |
| 4 | CT+CC | Negative | 73 | 26 (35.6) | 58 | 0.570 (0.366–0.888) | 0.013 | 0.621 (0.396–0.973) | 0.037 |
Adjustment for drinking status, Child-Pugh score, tumor size, tumor number, BCLC stage, radical resection, cirrhosis, adjuvant antiviral treatment, PVTT in Cox proportion hazard regression model. HBV, hepatitis B virus; HCC, hepatocellular carcinoma; MST, median survival time; HR, hazard ratio; CI, confidence interval.
GEO data and gene interaction analysis
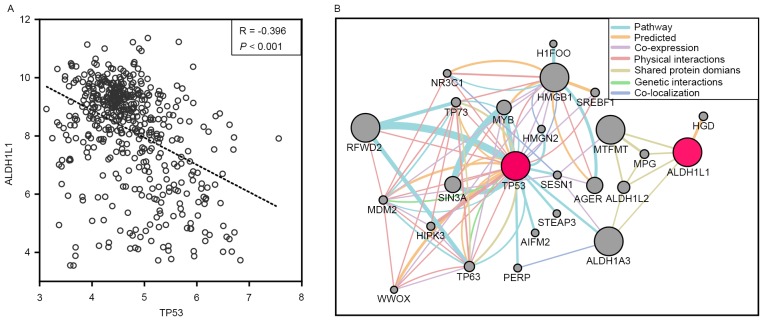
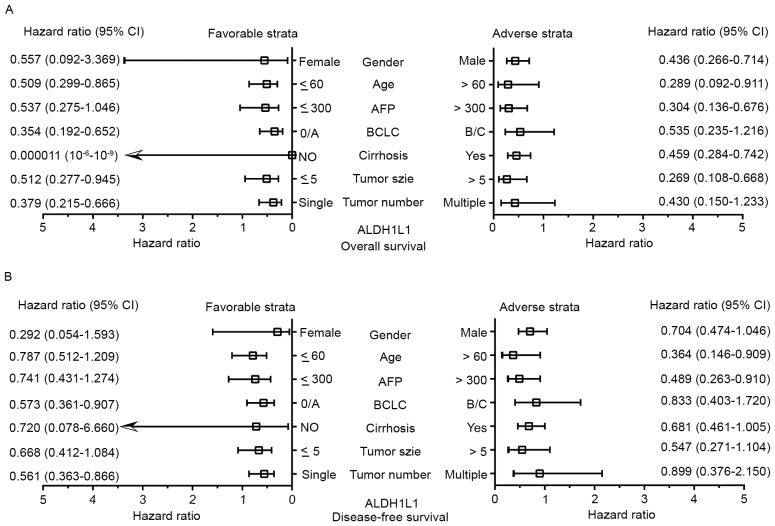
In order to determine the relationship of ALDH1L1 with TP53 at the transcriptional level, the GSE14520 database (including 218 Chinese HBV-related HCC patients with clinical information and prognosis) was used to correlate the ALDH1L1 and TP53 mRNA expression in HBV-related HCC patients. The results showed that ALDH1L1 had a weak negative correlation with TP53 (r=−0.396; P<0.001; Fig. 4A). Gene interaction analyses through GenMANIA also showed that ALDH1L1 shared protein domains with ALDH1A3, that affected the TP53 pathway (Fig. 4B). Stratified analyses for the DFS showed that high ALDH1L1 expression significantly decreased the risk of recurrence among patients >60 years of age, with a single tumor, BCLC stage 0/A, and AFP >300 ng/ml (Fig. 5A). Regarding the OS, high ALDH1L1 expression significantly decreased the risk of death among patients in both male age groups, both tumor size groups with a single tumor, who were characterized with cirrhosis, BCLC stage 0/A, and AFP >300 ng/ml (Fig. 5B). We further analyzed the effects of ALDH1L1 expression on the DFS and OS of GEO14520 HBV-related HCC patients by adjusting for age, sex, cirrhosis, BCLC stage and serum AFP level. The results showed that high ALDH1L1 expression was significantly associated with a favorable prognosis for both the DFS and OS (adjusted P=0.04; adjusted HR=0.669; 95% CI=0.456–0.981 for DFS; adjusted P=0.001; adjusted HR=0.446; 95% CI=0.277–0.719 for OS; Table VI and Fig. 6A and B).
Figure 4.
(A) Correlations between ALDH1L1 and TP53 mRNA expression using GSE14520. (B) A gene interaction diagram of the ALDH1L1 and TP53 genes using GeneMANIA. ALDH1L1, aldehyde dehydrogenase 1 family member L1; TP53, tumor protein p53.
Figure 5.
Stratified analyses of the associations of different ALDH1L1 mRNA expression levels with the GSE14520 prognosis of HCC patients. All variables were stratified by favorable and adverse strata. (A) Stratified analysis between ALDH1L1 and DFS. (B) Stratified analysis between ALDH1L1 and the OS. ALDH1L1, aldehyde dehydrogenase 1 family member L1; HCC, hepatocellular carcinoma; DFS, disease-free survival; OS, overall survival.
Table VI.
Survival analysis between ALDH1L1 and TP53 mRNA expression in GSE14520 HBV-related HCC patients.
| OS | DFS | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene expression | Patients (n=218) | No. of events (%) | MST (months) | Adjusted HR (95% CI) | Adjusted P-valuea | No. of events | MST (months) | Adjusted HR (95% CI) | Adjusted P-valuea |
| ALDH1L1 | |||||||||
| Low | 109 | 66 (60.6) | 28 | 1 | 54 (49.5) | 51 | 1 | ||
| High | 109 | 55 (50.5) | 53 | 0.669 (0.456–0.981) | 0.040 | 30 (27.5) | NA | 0.446 (0.277–0.719) | 0.001 |
| TP53 | |||||||||
| Low | 109 | 58 (53.2) | 50 | 1 | 37 (33.9) | NA | 1 | ||
| High | 109 | 63 (57.8) | 35 | 1.054 (0.726–1.5229) | 0.783 | 47 (43.1) | NA | 1.137 (0.722–1.791) | 0.580 |
Adjustment for age, sex, cirrhosis, BCLC stage, serum AFP level. ALDH1L1, aldehyde dehydrogenase 1 family member L1; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; DFS, disease-free survival; OS, overall survival; MST, median survival time; HR, hazard ratio; CI, confidence interval.
Figure 6.
Survival curves for the GSE14520 analyses of HCC patients with different ALDH1L1 mRNA expression levels, and the joint-effect analyses with TP53 mRNA expression levels. (A) Kaplan-Meier survival curves for DFS for different ALDH1L1 expression levels. (B) Kaplan-Meier survival curves for the OS analyses of different ALDH1L1 expression levels. (C) Kaplan-Meier survival curves for the joint-effect analyses for different ALDH1L1 and TP53 mRNA expression levels; analysis for DFS. (D) Kaplan-Meier survival curves for the joint-effect analyses for different ALDH1L1 and TP53 mRNA expression levels; analysis for OS. HCC, hepatocellular carcinoma; TP53, tumor protein p53; ALDH1L1, aldehyde dehydrogenase 1 family member L1; DFS, disease-free survival; OS, overall survival.
Joint-effect analyses among different ALDH1L1 and TP53 expression groups showed that patients with a high ALDH1L1 and low TP53 expression were significantly associated with a favorable prognosis, when compared with patients with a low ALDH1L1 and TP53 expression for both the DFS and OS (adjusted P=0.005; adjusted HR=0.460; 95% CI=0.266–0.795 for DFS; adjusted P=0.000011; adjusted HR=0.211; 95% CI=0.105–0.422 for OS; Table VII and Fig. 6C and D). Patients in groups b and d also had a reduced risk of death, compared with patients with low ALDH1L1 expression in the low TP53 expression group (adjusted P=0.023; adjusted HR=0.524; 95% CI=0.300–0.914 for group b; adjusted P=0.014; adjusted HR=0.434; 95% CI=0.222–0.846 for group d; Table VII and Fig. 6D).
Table VII.
Joint effect survival analysis between ALDH1L1 and TP53 mRNA expression level in GSE14520 HBV-related HCC patients.
| Group | ALDH1L1 expression | TP53 expression | Patients (n=218) | No. of events (%) | MST (months) | Adjusted HR (95% CI) | Adjusted P-valuea |
|---|---|---|---|---|---|---|---|
| DFS | |||||||
| i | Low | Low | 38 | 25 (65.8) | 23 | 1 | |
| ii | Low | High | 71 | 41 (57.7) | 35 | 0.675 (0.406–1.122) | 0.129 |
| iii | High | Low | 71 | 33 (46.5) | 57 | 0.460 (0.266–0.795) | 0.005 |
| iiii | High | High | 38 | 22 (57.9) | 36 | 0.614 (0.342–1.101) | 0.102 |
| OS | |||||||
| a | Low | Low | 38 | 22 (57.9) | 30 | 1 | |
| b | Low | High | 71 | 32 (45.1) | NA | 0.524 (0.300–0.914) | 0.023 |
| c | High | Low | 71 | 15 (21.1) | NA | 0.211 (0.105–0.422) | 0.000011 |
| d | High | High | 38 | 15 (39.5) | NA | 0.434 (0.222–0.846) | 0.014 |
Adjustment for age, sex, cirrhosis, BCLC stage, serum AFP level. ALDH1L1, aldehyde dehydrogenase 1 family member L1; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; TP53, tumor protein p53; DFS, disease-free survival; OS, overall survival; MST, median survival time; HR, hazard ratio; CI, confidence interval.
Discussion
ALDH1L1 has been widely accepted as an astroglial marker in the brain (27,28) and is also expressed in neural stem cells (29), but is not a suitable marker for enteric glial cells (30). Due to the cell-specificity of ALDH1L1 in nerve cells, ALDH1L1 polymorphisms have also been associated with neurological diseases, such as neural tube defects (31) and ischemic stroke (32), but ALDH1L1 polymorphisms have not been investigated in spina bifida (33). It's upregulation is involved in central nervous system development and reduced proliferation (34). ALDH1L1 is mainly expressed in human liver (35), implying that ALDH1L1 has an important function in this organ. Consistent with our bioinformatic analyses, ALDH1L1 is significantly downregulated in various human malignant tumors and cancer cell lines, including HCC (36). A similar result for ALDH1L1 downregulation in different tumor tissues was confirmed by ONCOMINE analyses (37) and other studies (21,38,39), but in non-small cell lung cancer (NSCLC), ALDH1L1 expression was upregulated (40). Our bioinformatic analyses also showed that ALDH1L1 was downregulated in HBV-related HCC tumor tissues. ALDH1L1 is upregulated in the presence of high concentrations of folate, and depletion of folate leads to the absence of ALDH1L1, resulting in cofilin dephosphorylation and inhibition of motility by protein phosphatase 1 (PP1) and protein phosphatase 2A (PP2A) in several cell lines. These results suggested that folate promotes a malignant phenotype in cancer (41). However, a study of oral cancer reported that folate supplementation decreased the risk of oral cancer even with alcohol abuse, thus, ALDH1L1 may play a causal role in oral cancer occurrence (42). In spite of the conflicting results of these studies, underexpressed ALDH1L1 was associated with an aggressive histology and/or biological behavior in renal cell carcinomas and pilocytic astrocytomas (43,44). ALDH1L1 knockdown in lung cancer cell lines showed that inhibition of ALDH1L1 expression reduced adenosine triphosphate (ATP) production by decreasing nicotinamide adenine dinucleotide (NADH) levels, resulting in cell death (40). Recent studies also reported that high expression of ALDH1L1 is correlated with better survival in HCC (21), neuroblastoma (45) and breast cancer (BC) (46). However, survival analyses of gastric cancer showed the opposite result that high expression of ALDH1L1 was associated with a worse prognosis (37,47). Furthermore, no significant relationship was observed between ALDH1L1 mRNA expression and OS in NSCLC (48). As previously mentioned, ALDH1L1 may play a different role as a tumor-suppressor during oncogenesis. Genetic variation analyses have reported that rs2276731 and rs2002287 of ALDH1L1 can affect the risk of BC morbidity (n=1007) (49), but this conclusion was not found for the risk of prostate cancer (n=2288) including other ALDH1L1 SNPs (50). A study of HCC and lung cancer reported that ALDH1L1 mRNA and protein levels correlated with the methylation status of the CpG island, and modicum ALDH1L1 CpG island methylation was sufficient to significantly decrease ALDH1L1 expression, suggesting that the mechanism of action of ALDH1L1 involves downregulation in cancers (51). A follow-up study in Chinese Kazakh patients with esophageal squamous cell carcinoma also showed that ALDH1L1 is involved in a one carbon metabolic process that plays a key role in DNA methylation (39). A study carried out by Oleinik and Krupenko also reported that inducible ALDH1L1 expression in A549 cells induced G1 cell cycle arrest and apoptosis. These anti-proliferative and apoptotic effects result in activation of TP53, followed by the TP53-mediated transcriptional activation of a downstream target of p21 (19), to function as a potent cyclin-dependent kinase inhibitor. Further studies of the relationship between ALDH1L1 and TP53 showed that expression of ALDH1L1 induced suppressor effects that were p53-dependent, and a TP53 deficit resulted in suppressor effects (20). This ALDH1L1-induced p53-dependent apoptosis also responded to folate stress, resulting in upregulation of ceramide synthesis (52).
A previous study of Guangxi HCC patients reported that ALDH1L1 expression was associated with the prognosis of HCC (21). HBV-related HCC patients in Guangxi were associated with a high morbidity of HBV-infection (53). The present study characterized ALDH1L1 polymorphism in HBV-related HCC patients and its association with TP53 expression. Bioinformatic analyses showed that rs2276724 S481G in ALDH1L1 affected gene expression and was possibly deleterious to patients. We further analyzed the distribution of rs2276724 genotypes in different TP53 expression groups and its possible association with the prognosis of HBV-related HCC patients. The results suggested that the occurrence of rs2276724 was similar between different TP53 expression groups when analyzed in different genetic models. Survival analyses showed that the C allele was associated with a decreased risk of death in HBV-related HCC patients, when compared to the T allele. Through stratified analyses, the C allele carriers of rs2276724 had significantly decreased risk of death among patients with a tumor size ≤5 cm, a Child-Pugh score, and without a PVTT, BCLC stage B/C, non-radical resection, and the presence of regional invasion. Joint-effect analyses showed that the CT/CC of rs2276724 in TP53-negative patients was associated with a significantly decreased risk of mortality, compared to the TT of rs2276724 with TP53-positive patients. We then used the Chinese HBV-HCC mRNA expression profiling chip from the GSE14520 dataset to evaluate the prognosis of ALDH1L1 expression in Chinese HBV-related HCC patients, and found that low ALDH1L1 expression predicted a poor prognosis for Chinese HBV-related HCC patients with low expression of HBV-related HCC tumor tissues. High ALDH1L1 expression significantly decreased the risk of HCC recurrence among patients with an age >60 years, a single tumor, BCLC stage 0/A, AFP >300 ng/ml, and showed a decreased risk of mortality among the male HCC patients in both age groups, both tumor size groups, a single tumor with cirrhosis, with BCLC stage 0/A, and an AFP >300 ng/ml. Gene interaction analyses showed that GeneMANIA ALDH1L1 and TP53 expression mRNA levels were negatively correlated in Chinese HBV-related HCC patients, and further showed that ALDH1L1 shared a protein domain with ALDH1A3 that was involved in the TP53 pathway. We combined the analyses of ALDH1L1 and TP53 expression in HBV-related HCC patients to show that high ALDH1L1 with low TP53 expression was associated with a significantly decreased risk of HBV-related HCC recurrence and mortality when compared with low ALDH1L1 and low TP53 expression. Patients with high TP53 expression also had a significantly decreased risk of HBV-related HCC death, compared with low ALDH1L1 and low TP53-expressing patients.
In conclusion, the present study showed, for the first time, that prognosis can be predicted for the rs2276724 genotypes of ALDH1L1 in HBV-related HCC patients and their associations with TP53 expression. The CT/TT genotype of rs2276724 may have a protective survival value and may be a potential prognostic marker in patients with HBV-related HCC receiving hepatic resection. We also confirmed that a decrease in ALDH1L1 expression predicts a poor prognosis for patients with HBV-related HCC. The expression of ALDH1L1 and genotypes of rs2276724 may therefore play a role in TP53 expression in HBV-related HCC of Chinese hepatic resection patients. Due to the limitations of the relatively small sample sizes and the long period of specimen collection, we did not analyze the association among rs2276724 genotypes and mRNA expression. Further well-designed, comprehensive, and large sample size studies are therefore needed to confirm our results.
Acknowledgements
This study was supported in part by the National Nature Science Foundation of China (nos. 81560535, 81072321, 30760243, 30460143 and 30560133), 2009 Program for New Century Excellent Talents in University (NCET), Guangxi Nature Sciences Foundation (no. GuiKeGong 1104003A-7), Guangxi Health Ministry Medicine Grant (Key-Scientific Research-Grant Z201018) and Self-raised Scientific Research Projects of the Guangxi Zhuang Autonomous Region Health and Family Planning Commission (Z2016318). The authors thank Professor Xiao Qin, Xigang Chen, Bin Chen, Zhixiong Su, Ming Su, Zhang Wen, Jingning Lu, Ning Peng, Hai Zhu who provided HCC samples for this study, and who are from the Department of Hepatobiliary Surgery, the First Affiliated Hospital of Guangxi Medical University. Thanks also go to researchers Jiaquan Li and Ying Gui from Guangxi Medical University for their contribution to specimen management and Professor Jiahong Dong from Beijing Tsinghua Changgung Hospital for his contribution to thesis guidance. In addition, we also would like to acknowledge the helpful comments on this paper received from our reviewers, and the contributors of GSE14520 for sharing their dataset on open access.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Zeng H, Zheng R, Guo Y, Zhang S, Zou X, Wang N, Zhang L, Tang J, Chen J, Wei K, et al. Cancer survival in China, 2003–2005: A population-based study. Int J Cancer. 2015;136:1921–1930. doi: 10.1002/ijc.29227. [DOI] [PubMed] [Google Scholar]
- 4.Zhou M, Wang H, Zhu J, Chen W, Wang L, Liu S, Li Y, Wang L, Liu Y, Yin P, et al. Cause-specific mortality for 240 causes in China during 1990–2013: A systematic subnational analysis for the Global Burden of Disease Study 2013. Lancet. 2016;387:251–272. doi: 10.1016/S0140-6736(15)00551-6. [DOI] [PubMed] [Google Scholar]
- 5.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 6.Wang FS, Fan JG, Zhang Z, Gao B, Wang HY. The global burden of liver disease: The major impact of China. Hepatology. 2014;60:2099–2108. doi: 10.1002/hep.27406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu L, Qian G, Tang L, Su J, Wang JS. Genetic variations of hepatitis B virus and serum aflatoxin-lysine adduct on high risk of hepatocellular carcinoma in Southern Guangxi, China. J Hepatol. 2010;53:671–676. doi: 10.1016/j.jhep.2010.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qi LN, Bai T, Chen ZS, Wu FX, Chen YY, De Xiang B, Peng T, Han ZG, Li LQ. The p53 mutation spectrum in hepatocellular carcinoma from Guangxi, China: Role of chronic hepatitis B virus infection and aflatoxin B1 exposure. Liver Int. 2015;35:999–1009. doi: 10.1111/liv.12460. [DOI] [PubMed] [Google Scholar]
- 9.Yin Y, Stephen CW, Luciani MG, Fåhraeus R. p53 stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat Cell Biol. 2002;4:462–467. doi: 10.1038/ncb801. [DOI] [PubMed] [Google Scholar]
- 10.Hainaut P, Wiman KG. 30 years and a long way into p53 research. Lancet Oncol. 2009;10:913–919. doi: 10.1016/S1470-2045(09)70198-6. [DOI] [PubMed] [Google Scholar]
- 11.Soussi T, Wiman KG. TP53: An oncogene in disguise. Cell Death Differ. 2015;22:1239–1249. doi: 10.1038/cdd.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: Functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26:2157–2165. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Ma Q, Zhang M, Wang X, Zhang D, Li W, Wang F, Wu E. Alterations of TP53 are associated with a poor outcome for patients with hepatocellular carcinoma: Evidence from a systematic review and meta-analysis. Eur J Cancer. 2012;48:2328–2338. doi: 10.1016/j.ejca.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Staib F, Hussain SP, Hofseth LJ, Wang XW, Harris CC. TP53 and liver carcinogenesis. Hum Mutat. 2003;21:201–216. doi: 10.1002/humu.10176. [DOI] [PubMed] [Google Scholar]
- 15.Hussain SP, Schwank J, Staib F, Wang XW, Harris CC. TP53 mutations and hepatocellular carcinoma: Insights into the etiology and pathogenesis of liver cancer. Oncogene. 2007;26:2166–2176. doi: 10.1038/sj.onc.1210279. [DOI] [PubMed] [Google Scholar]
- 16.Villanueva A, Hoshida Y. Depicting the role of TP53 in hepatocellular carcinoma progression. J Hepatol. 2011;55:724–725. doi: 10.1016/j.jhep.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 17.Gouas D, Shi H, Hainaut P. The aflatoxin-induced TP53 mutation at codon 249 (R249S): Biomarker of exposure, early detection and target for therapy. Cancer Lett. 2009;286:29–37. doi: 10.1016/j.canlet.2009.02.057. [DOI] [PubMed] [Google Scholar]
- 18.Ji YN, Wang Q, Xue J. TP53 immunohistochemical expression is associated with the poor outcome for hepatocellular carcinoma: Evidence from a meta-analysis. Tumour Biol. 2014;35:1653–1659. doi: 10.1007/s13277-013-1228-7. [DOI] [PubMed] [Google Scholar]
- 19.Oleinik NV, Krupenko SA. Ectopic expression of 10-formyltetrahydrofolate dehydrogenase in A549 cells induces G1 cell cycle arrest and apoptosis. Mol Cancer Res. 2003;1:577–588. [PubMed] [Google Scholar]
- 20.Oleinik NV, Krupenko NI, Priest DG, Krupenko SA. Cancer cells activate p53 in response to 10-formyltetrahydrofolate dehydrogenase expression. Biochem J. 2005;391:503–511. doi: 10.1042/BJ20050533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen XQ, He JR, Wang HY. Decreased expression of ALDH1L1 is associated with a poor prognosis in hepatocellular carcinoma. Med Oncol. 2012;29:1843–1849. doi: 10.1007/s12032-011-0075-x. [DOI] [PubMed] [Google Scholar]
- 22.Liao X, Han C, Qin W, Liu X, Yu L, Lu S, Chen Z, Zhu G, Su H, Mo Z, et al. Genome-wide association study identified PLCE1- rs2797992 and EGFR- rs6950826 were associated with TP53 expression in the HBV-related hepatocellular carcinoma of Chinese patients in Guangxi. Am J Transl Res. 2016;8:1799–1812. [PMC free article] [PubMed] [Google Scholar]
- 23.Kondo K, Chijiiwa K, Kai M, Otani K, Nagaike K, Ohuchida J, Hiyoshi M, Nagano M. Surgical strategy for hepatocellular carcinoma patients with portal vein tumor thrombus based on prognostic factors. J Gastrointest Surg. 2009;13:1078–1083. doi: 10.1007/s11605-009-0854-2. [DOI] [PubMed] [Google Scholar]
- 24.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 25.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee PH, Shatkay H. An integrative scoring system for ranking SNPs by their potential deleterious effects. Bioinformatics. 2009;25:1048–1055. doi: 10.1093/bioinformatics/btp103. [DOI] [PubMed] [Google Scholar]
- 27.Yang Y, Vidensky S, Jin L, Jie C, Lorenzini I, Frankl M, Rothstein JD. Molecular comparison of GLT1+ and ALDH1L1+ astrocytes in vivo in astroglial reporter mice. Glia. 2011;59:200–207. doi: 10.1002/glia.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feresten AH, Barakauskas V, Ypsilanti A, Barr AM, Beasley CL. Increased expression of glial fibrillary acidic protein in prefrontal cortex in psychotic illness. Schizophr Res. 2013;150:252–257. doi: 10.1016/j.schres.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 29.Foo LC, Dougherty JD. Aldh1L1 is expressed by postnatal neural stem cells in vivo. Glia. 2013;61:1533–1541. doi: 10.1002/glia.22539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boesmans W, Rocha NP, Reis HJ, Holt M, Vanden Berghe P. The astrocyte marker Aldh1L1 does not reliably label enteric glial cells. Neurosci Lett. 2014;566:102–105. doi: 10.1016/j.neulet.2014.02.042. [DOI] [PubMed] [Google Scholar]
- 31.Wu L, Lu X, Guo J, Zhang T, Wang F, Bao Y. Association between ALDH1L1 gene polymorphism and neural tube defects in the Chinese Han population. Neurol Sci. 2016;37:1049–1054. doi: 10.1007/s10072-016-2527-8. [DOI] [PubMed] [Google Scholar]
- 32.Williams SR, Yang Q, Chen F, Liu X, Keene KL, Jacques P, Chen WM, Weinstein G, Hsu FC, Beiser A, et al. Genomics and Randomized Trials Network; Framingham Heart Study: Genome-wide meta-analysis of homocysteine and methionine metabolism identifies five one carbon metabolism loci and a novel association of ALDH1L1 with ischemic stroke. PLoS Genet. 2014;10:e1004214. doi: 10.1371/journal.pgen.1004214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franke B, Vermeulen SH, Steegers-Theunissen RP, Coenen MJ, Schijvenaars MM, Scheffer H, den Heijer M, Blom HJ. An association study of 45 folate-related genes in spina bifida: Involvement of cubilin (CUBN) and tRNA aspartic acid methyltransferase 1 (TRDMT1) Birth Defects Res A Clin Mol Teratol. 2009;85:216–226. doi: 10.1002/bdra.20556. [DOI] [PubMed] [Google Scholar]
- 34.Anthony TE, Heintz N. The folate metabolic enzyme ALDH1L1 is restricted to the midline of the early CNS, suggesting a role in human neural tube defects. J Comp Neurol. 2007;500:368–383. doi: 10.1002/cne.21179. [DOI] [PubMed] [Google Scholar]
- 35.Hong M, Lee Y, Kim JW, Lim JS, Chang SY, Lee KS, Paik SG, Choe IS. Isolation and characterization of cDNA clone for human liver 10-formyltetrahydrofolate dehydrogenase. Biochem Mol Biol Int. 1999;47:407–415. doi: 10.1080/15216549900201433. [DOI] [PubMed] [Google Scholar]
- 36.Krupenko SA, Oleinik NV. 10-formyltetrahydrofolate dehydrogenase, one of the major folate enzymes, is down-regulated in tumor tissues and possesses suppressor effects on cancer cells. Cell Growth Differ. 2002;13:227–236. [PubMed] [Google Scholar]
- 37.Shen JX, Liu J, Li GW, Huang YT, Wu HT. Mining distinct aldehyde dehydrogenase 1 (ALDH1) isoenzymes in gastric cancer. Oncotarget. 2016;7:25340–25349. doi: 10.18632/oncotarget.8294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dmitriev AA, Kashuba VI, Haraldson K, Senchenko VN, Pavlova TV, Kudryavtseva AV, Anedchenko EA, Krasnov GS, Pronina IV, Loginov VI, et al. Genetic and epigenetic analysis of non-small cell lung cancer with NotI-microarrays. Epigenetics. 2012;7:502–513. doi: 10.4161/epi.19801. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y, Yin D, Li L, Deng YC, Tian W. Screening aberrant methylation profile in esophageal squamous cell carcinoma for Kazakhs in Xinjiang area of China. Mol Biol Rep. 2015;42:457–464. doi: 10.1007/s11033-014-3788-z. [DOI] [PubMed] [Google Scholar]
- 40.Kang JH, Lee SH, Lee JS, Nam B, Seong TW, Son J, Jang H, Hong KM, Lee C, Kim SY. Aldehyde dehydrogenase inhibition combined with phenformin treatment reversed NSCLC through ATP depletion. Oncotarget. 2016;7:49397–49410. doi: 10.18632/oncotarget.10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oleinik NV, Krupenko NI, Krupenko SA. ALDH1L1 inhibits cell motility via dephosphorylation of cofilin by PP1 and PP2A. Oncogene. 2010;29:6233–6244. doi: 10.1038/onc.2010.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang PH, Lian L, Zavras AI. Alcohol intake and folate antagonism via CYP2E1 and ALDH1: Effects on oral carcinogenesis. Med Hypotheses. 2012;78:197–202. doi: 10.1016/j.mehy.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dmitriev AA, Rudenko EE, Kudryavtseva AV, Krasnov GS, Gordiyuk VV, Melnikova NV, Stakhovsky EO, Kononenko OA, Pavlova LS, Kondratieva TT, et al. Epigenetic alterations of chromosome 3 revealed by NotI-microarrays in clear cell renal cell carcinoma. Biomed Res Int. 2014;2014:735292. doi: 10.1155/2014/735292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez FJ, Giannini C, Asmann YW, Sharma MK, Perry A, Tibbetts KM, Jenkins RB, Scheithauer BW, Anant S, Jenkins S, et al. Gene expression profiling of NF-1-associated and sporadic pilocytic astrocytoma identifies aldehyde dehydrogenase 1 family member L1 (ALDH1L1) as an underexpressed candidate biomarker in aggressive subtypes. J Neuropathol Exp Neurol. 2008;67:1194–1204. doi: 10.1097/NEN.0b013e31818fbe1e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hartomo TB, Van Huyen Pham T, Yamamoto N, Hirase S, Hasegawa D, Kosaka Y, Matsuo M, Hayakawa A, Takeshima Y, Iijima K, et al. Involvement of aldehyde dehydrogenase 1A2 in the regulation of cancer stem cell properties in neuroblastoma. Int J Oncol. 2015;46:1089–1098. doi: 10.3892/ijo.2014.2801. [DOI] [PubMed] [Google Scholar]
- 46.Wu S, Xue W, Huang X, Yu X, Luo M, Huang Y, Liu Y, Bi Z, Qiu X, Bai S. Distinct prognostic values of ALDH1 isoenzymes in breast cancer. Tumour Biol. 2015;36:2421–2426. doi: 10.1007/s13277-014-2852-6. [DOI] [PubMed] [Google Scholar]
- 47.Li K, Guo X, Wang Z, Li X, Bu Y, Bai X, Zheng L, Huang Y. The prognostic roles of ALDH1 isoenzymes in gastric cancer. Onco Targets Ther. 2016;9:3405–3414. doi: 10.2147/OTT.S102314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.You Q, Guo H, Xu D. Distinct prognostic values and potential drug targets of ALDH1 isoenzymes in non-small-cell lung cancer. Drug Des Devel Ther. 2015;9:5087–5097. doi: 10.2147/DDDT.S87197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stevens VL, McCullough ML, Pavluck AL, Talbot JT, Feigelson HS, Thun MJ, Calle EE. Association of polymorphisms in one-carbon metabolism genes and postmenopausal breast cancer incidence. Cancer Epidemiol Biomarkers Prev. 2007;16:1140–1147. doi: 10.1158/1055-9965.EPI-06-1037. [DOI] [PubMed] [Google Scholar]
- 50.Stevens VL, Rodriguez C, Sun J, Talbot JT, Thun MJ, Calle EE. No association of single nucleotide polymorphisms in one-carbon metabolism genes with prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:3612–3614. doi: 10.1158/1055-9965.EPI-08-0789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oleinik NV, Krupenko NI, Krupenko SA. Epigenetic silencing of ALDH1L1, a metabolic regulator of cellular proliferation, in cancers. Genes Cancer. 2011;2:130–139. doi: 10.1177/1947601911405841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoeferlin LA, Fekry B, Ogretmen B, Krupenko SA, Krupenko NI. Folate stress induces apoptosis via p53-dependent de novo ceramide synthesis and up-regulation of ceramide synthase 6. J Biol Chem. 2013;288:12880–12890. doi: 10.1074/jbc.M113.461798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yeh FS, Yu MC, Mo CC, Luo S, Tong MJ, Henderson BE. Hepatitis B virus, aflatoxins, and hepatocellular carcinoma in southern Guangxi, China. Cancer Res. 1989;49:2506–2509. [PubMed] [Google Scholar]