Abstract
Human papillomavirus negative (HPV-) head and neck squamous cell carcinomas (HNSCC) are deadly and common cancers. Recent genomic studies implicate multiple genetic pathways including cell-signalling, cell-cycle and/or immune evasion in their development. Here, we analyze public datasets and uncover a previously unappreciated role of epigenome deregulation in the genesis of 13% HPV-HNSCCs. Specifically, we identify novel recurrent p.K36M mutations occurring in multiple histone H3 genes. We further validate their presence in multiple independent HNSCC datasets and show that along with previously described NSD1 mutations, they correspond to a specific DNA methylation cluster. H3K36M and NSD1 defects converge on altering H3K36 methylation, subsequently blocking cellular differentiation and promoting oncogenesis. Our data further indicate surprisingly limited redundancy for NSD family members in HPV-HNSCCs and suggest a potential role of impaired H3K36 methylation in their development. Further investigation of drugs targeting chromatin regulators is warranted in HPV-HNSCCs driven by aberrant H3K36 methylation.
Introduction
Head and neck squamous cell carcinomas (HNSCC) are a heterogeneous group of tumours that develop through chemical - as a result of tobacco and/or alcohol abuse - or viral-induced carcinogenesis following infection with high-risk human papillomaviruses (HPV+)1. They represent the seventh most frequent cancer worldwide with ~600,000 new cases per year2 and occur throughout the oral cavity, hypopharynx, oropharynx, nasopharynx, or larynx. Tumors are often locally advanced invading proximal structures at diagnosis or show spread to cervical lymph nodes or distant metastases. Despite several advances and innovations in multimodality treatment, survival rates of locally advanced HNSCC have not substantially improved, and the prognosis for relapsed or metastatic tumors remains dismal3.
Several studies, including a recent comprehensive genomic profiling of a vast number of tumors by The Cancer Genome Atlas Project (TCGA), have shown that HPV negative (HPV-) and HPV+ HNSCC represent distinct disease entities4–6. The latter, which mainly arises in the oropharynx (tonsils and base of tongue), is associated with a substantially improved outcome and harbors distinct molecular alterations3,6,7. The vast majority of HPV- tumors have loss-of-function TP53 mutations and CDKN2A inactivation. Integrated genomic and DNA methylation analysis indicated a high level of heterogeneity in this entity with the presence of several molecular clusters seemingly enriched for specific genetic alterations6. These studies also identified frequent alterations in pathways involving growth factor receptors, RAS and PI3K signalling, the cell cycle, cell death and differentiation, and/or oxidative stress, which have thus become the focus of current research efforts and targeted therapies in HNSCCs3–6,8–13. Interestingly, novel genetic alterations in an epigenetic modulator Nuclear receptor-binding SET Domain protein 1 (NSD1) were recently listed in HPV-HNSCCs but insights into whether or how these mutations could drive oncogenesis are lacking6,12.
NSD1 is a member of a family of mammalian histone methyltransferases (NSD1, NSD2/MMSET/WHSC1, NSD3/WHSC1L1) that are essential in development. NSD family enzymes act as mono- and di-methyltransferases for histone H3 lysine 36 (H3K36, reviewed in14), while SETD2 mediates trimethylation of H3K36, a chromatin mark associated with active transcription, gene splicing, and DNA damage repair. Notably, aberrant regulation of H3K36 methylation has been recently identified in several neoplasms and could potentially promote oncogenesis15–20. Germline hypomorphic NSD1 mutations lead to the SOTOS overgrowth syndrome, where patients are more prone to cancer21. Somatic translocation and gain-of-function mutations involving NSD2 have been identified in leukemia and multiple myeloma, while NSD1 loss-of-function mutations have been described in lung cancers22. In AML, the recurring t(5;11)(q35;p15.5) translocation fuses NSD1 to nucleoporin-98 (NUP98) and leads to leukemogenesis through aberrant establishment of H3K36 methylation at specific genomic loci16. SETD2 mutations have also been described in a wide range of cancers15,19,23. Therefore, we reasoned that an abnormal epigenetic landscape, through alterations in the H3K36 methylation pathway, might constitute a previously unappreciated mechanism in the development of HPV-HNSCCs. To this end, we performed an integrative analysis of existing genomic and DNA methylation datasets of HNSCCs to assess the functional impact of NSD1 alterations and investigated the presence of other genetic alterations affecting the post-translational modifications of histones including H3K36. Our results characterize an epigenetically distinct subgroup in HPV-HNSCCs, where the main genetic alterations converge to specifically disrupt H3K36 methylation, potentially driving oncogenesis.
Results
TCGA HNSCC dataset reanalysis
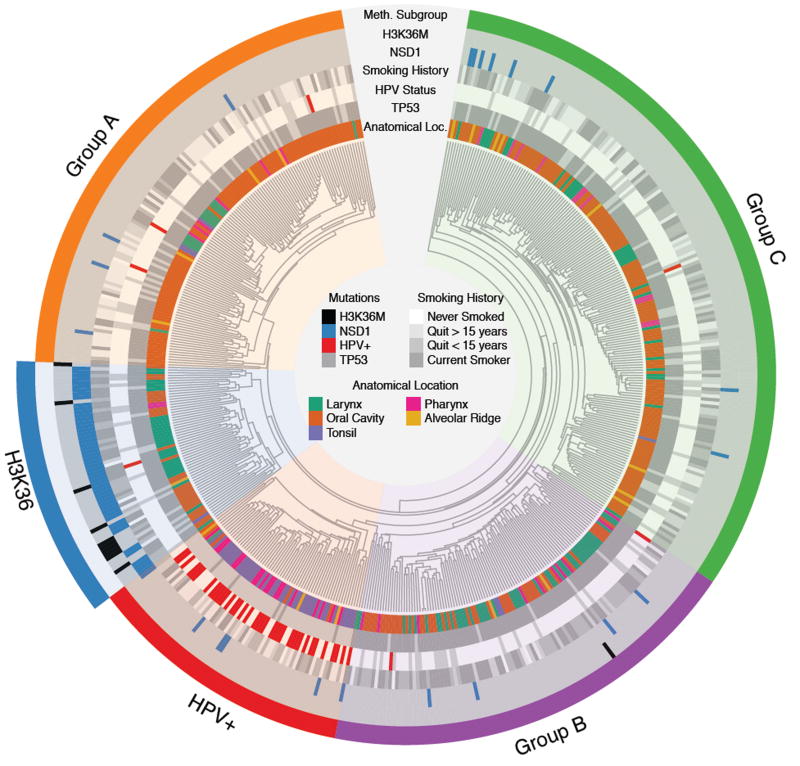
To assess the prevalence and nature of the genetic alterations affecting the epigenome in HNSCCs, we analyzed available HNSCC studies with reported results from next generation sequencing and DNA methylation profiling4–6,10,11. The TCGA dataset, the largest generated on HNSCCs, had DNA methylation data (450K arrays) available for 528 samples, whole-exome sequencing (WES) for 526 samples, and RNA sequencing for 520. A total of 518 samples were profiled on all three platforms (Supplementary Figure 1). We performed unsupervised hierarchical clustering of DNA methylation data (Figure 1) and identified a subgroup previously found to be enriched in NSD1 mutations. This subgroup was comprised of 61 samples, including 44 samples which carried damaging single nucleotide variants in NSD1 annotated in the initial TCGA analysis6 (Supplementary Figure 2C, Supplementary Table 1). Upon closer inspection of the primary data, we found that other tumors within this cluster had more complex genomic events including large chromosomal deletions encompassing this gene (n = 2), focal deletions within NSD1 (n = 2) or splicing defects (n = 1). Analysis of RNA-seq data showed absent NSD1 transcripts in two samples with non-interpretable WES data. In one sample, no conclusion could be drawn on NSD1 mutational status as only methylation data was available (Supplementary Table 2 & Supplementary Figure 3). Overall, the reanalysis indicated that 51/61 (84%) of samples within this methylation cluster carried NSD1 alterations. Notably, the vast majority of missense mutations in NSD1 in this cluster were located close to or within the functional SET domain and were frequently associated with additional truncating mutations in the same samples (Supplementary Figure 2C). In silico prediction of the functional impact of these mutations24 suggests that they likely compromise the methyltransferase activity of the enzyme (Supplementary Table 1). Two of these missense mutations were previously reported in SOTOS syndrome, a genetic disorder associated with germline NSD1 mutations21 including p.R2005Q, which has been shown to specifically ablate H3K36-methyltransferase activity25. The methylation-based clustering also clearly identified an HPV+/TP53 wild-type tumour subgroup, which is known to constitute a distinct entity within HNSCCs, along with three other clusters we labelled A, B and C that had no clear relationship to known alterations in HNSCC. NSD1 mutations/alterations were infrequent (4%), and were mostly missense and predicted to be less damaging in these clusters (Figure 1, Supplementary Table 3).
Figure 1. H3K36M and NSD1 mutations define a specific HNSCC subgroup.
Unsupervised hierarchical clustering of DNA methylation data identifies 5 HNSCC subgroups, including notably a subgroup comprised of NSD1 or H3K36M mutations (labeled the H3K36 cluster). The H3K36 cluster is enriched in HPV-, TP53 mutant, heavy smoking patients. Those tumors are found in the larynx and oral cavity. Another HPV+/TP53 wild-type subgroup is apparent (labeled the HPV+ cluster) as well as 3 other subgroups (labeled Groups A, B and C). Grey and white bars for clinical variables (HPV status, anatomical location) denote “No hit” or “Not available”, respectively.
Identification of H3K36M mutations in HNSCC
Given the high prevalence of NSD1 mutations in this DNA methylation cluster, we investigated the mutational status of other genes involved in H3K36 methylation. We identified 11 samples harboring p.K36M mutations in various histone H3 genes. The mutations occurred either in genes encoding the canonical histone H3.1/2 or the variant histone H3.3 (Supplementary Figure 2B, Supplementary Table 4). Remarkably, 10 of the H3K36M mutants occurred within the DNA methylation cluster predominantly defined by NSD1 defects (Figure 1, Supplementary Figure 2A–B). Taken together H3K36M and NSD1 mutations accounted for all samples with available genomic data (Supplementary Figure 2A) included in the DNA methylation derived cluster (thus labeled the H3K36 cluster) and jointly represent 13% of HPV-HNSCC. Only one sample with a H3K36M mutation was observed outside the H3K36 cluster (Figure 1). Other genes encoding potential H3K36 methyltransferases, including SETD2 and NSD2/WHSC1, were infrequently mutated in HNSCC and samples with these mutations were spread across all HNSCC methylation subgroups (Supplementary Figure 4).
Clinical and molecular features of the H3K36 HNSCC subgroup
A number of altered genes have been identified to be enriched in HPV-HNSCCs4–6,8–10,12,26–28, including TP53, EGFR, NOTCH1, PI3KCA and HRAS. We found that only the prevalence of TP53 and CASP8 significantly vary among HNSCC methylation subgroups. (Figure 1, Supplementary Figure 4). Similarly, there were no copy number alterations specific to the H3K36 cluster (data not shown). Consistent with the original TCGA report, we found tumors within this cluster to be strongly DNA hypomethylated (p < 1.99e-14, Student’s t-test) with no difference between H3K36M and NSD1 mutants (p = 0.36, Student’s t-test, Supplementary Figure 5), suggesting that the unique DNA methylation signature of the H3K36 cluster is driven by defects in the H3K36 methylation pathway. NSD1-mutated samples within the H3K36 cluster showed a lower NSD1 expression (Student’s t-test, p = 0.0007), global DNA hypomethylation (Student’s t-test, p = 1.41e-6) and more truncating mutations compared to NSD1-mutated samples outside the H3K36 cluster (Supplementary Figure 6).
The H3K36 cluster included tumors from heavy smokers or heavy smokers who had recently quit smoking (Figure 1). When assessing the mutational burden in a given cluster, NSD1 mutant tumors were hypermutated (p < 0.0004, Student’s t-test), while H3K36M mutant samples had similar number of mutations per sample compared to the other methylation clusters (Supplementary Figure 7). Notably, when we analyzed the somatic mutational pattern in relation to smoking, the signatures we obtained for each molecular entity showed that NSD1 but not H3K36M mutants had a mutational signature strongly associated with smoking (Supplementary Figure 8). Except for HPV+ cancers, which are associated with a favorable outcome, there were no differences in tumor grade or outcome among HPV-HNSCCs (Supplementary Figure 9). Moreover, we observed distinct anatomical locations for samples within the H3K36 cluster: all H3K36M tumors (100%, 10/10) occurred in the oral cavity while NSD1 mutant tumors localized more often in the larynx (29/51, 57%, p < 0.007, Chi-Square test) (Figure 2, Supplementary Figure 10). No other clinical feature reached a statistically significant enrichment (Supplementary Table 5). We also re-analyzed four other available WES datasets on HNSCCs. In combination (196 samples in total), these studies led us to identify 14 NSD1 mutations, most of which were located in the larynx, and one HIST3H3H p.K36M in the oral cavity4,5,29,30. Those 15 patients were HPV- and heavy smokers, consistent with our findings using the TCGA dataset (Figure 1, Supplementary Table 6).
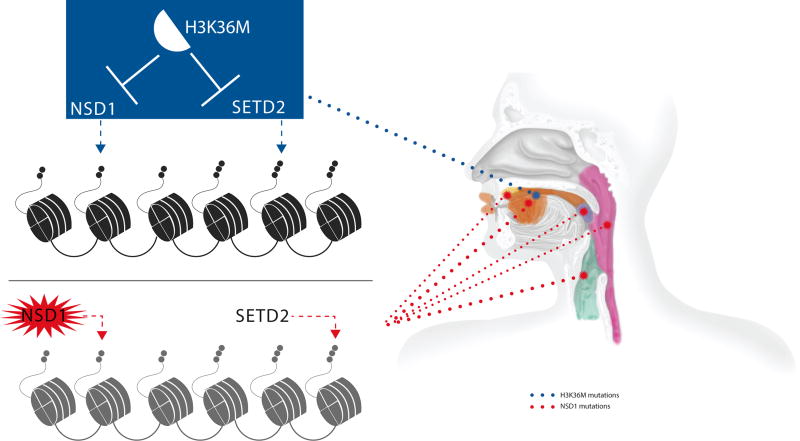
Figure 2. Schematic representation of the anatomical locations of NSD1 and H3K36M mutations in HNSCCs.
Tumors with H3K36M were mainly located in the oral cavity while NSD1 mutant tumors mainly occurred in the larynx and less frequently in other known locations of HNSCCs. The size of the dotted lines is proportional to the frequency of the mutation in a given anatomical location.
Effects of NSD1 and H3K36M mutations on epigenome and transcriptome
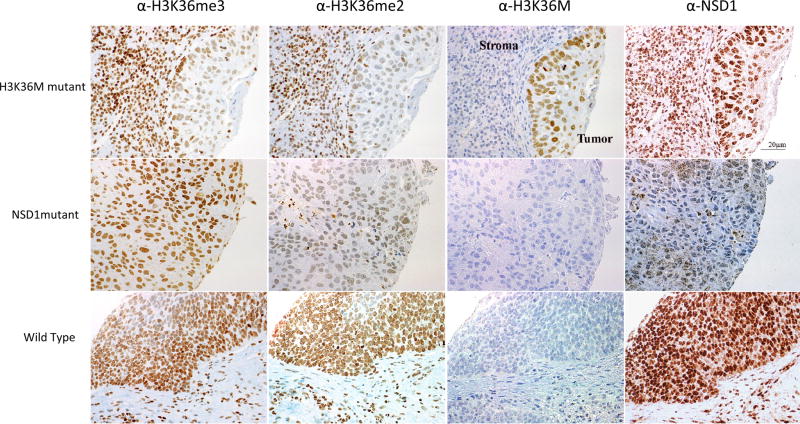
An independent cohort of 158 oropharyngeal HPV+ (n = 78) and HPV- (n = 80) HNSCCs with available tissue microarrays (TMA) was assembled to determine the prevalence and impact of H3K36M mutations on H3K36 methylation. Immunohistochemistry analysis identified three samples with typical nuclear staining of H3K36M, confirming the protein expression of the mutant histone in HNSCC tumor tissues in this independent cohort (Figure 3). We have previously shown that K-to-M substitutions at H3K36, although affecting only one of the 32 alleles encoding histone H3, act as dominant mutations and inhibit the respective SET-domain containing methyltransferases leading to decreased di- and tri-methylation of H3K3631,32. As expected, immunohistochemical staining of H3K36me2 and H3K36me3 of the TMAs showed that samples carrying H3K36M have a drastic decrease in their K36me2/me3 levels (Figure 3). In tumor cores lacking NSD1 staining, global levels of H3K36me3 were mostly unchanged while, as expected, H3K36me2 levels were markedly reduced, in keeping with the role of NSD1 as a di- but not tri-methyltransferase for H3K36 (Figure 3).
Figure 3. Representative immunohistochemical (IHC) staining of H3K36M, NSD1, H3K36me2 and H3K36me3 in H3K36M or NSD1 mutant HNSCCs and in a HNSCC sample wild-type for these genes.
Representative IHC of HNSCC samples using anti-H3K36M, anti-NSD1, anti-H3K36me3, or anti-H3K36me2 antibodies and counterstained with hematoxylin. H3K36M was expressed in 3/85 HPV-HNSCCs. H3K36M shows strong nuclear positivity in tumor cells but no staining in adjacent nuclei of stromal/inflammatory cells. Tumors wild-type for H3K36M and/or NSD1 show no nuclear staining with the anti-H3K36M antibody. Corresponding H3K36me3 and H3K36me2 staining on the same samples shows global decrease in the levels of these histone marks in H3K36M mutant tumors compared to tumors wild-type for this mutation. Notably, strong positivity for H3K36me3 and H3K36me2 was seen in the adjacent stromal cells not carrying H3K36M mutation, while uniform strong staining in tumor and stroma was observed for both marks in HNSCCs wild-type for H3K36M. Loss of NSD1 was associated with a marked decrease in global levels of H3K36me2 but not in H3K36me3. Pictures were taken under a 40X magnification and collated using Photoshop.
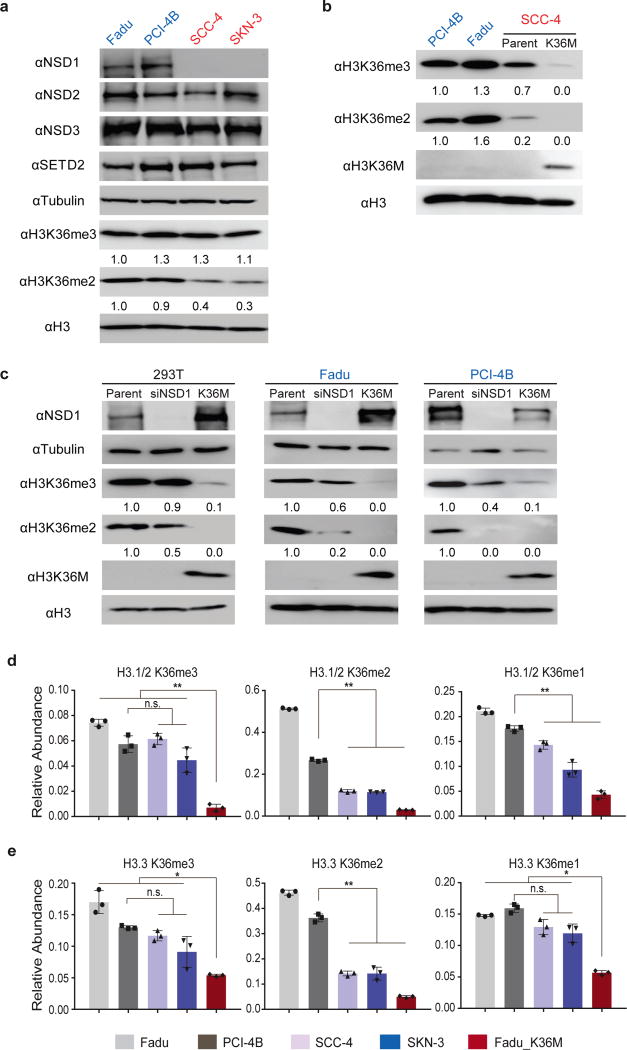
We also obtained two HNSCC cell lines carrying NSD1 truncating mutations (SCC-4 and SKN-3) and two cell lines wild-type for NSD1 (Fadu and PCI-4B). Immunoblotting and mass spectrometry analyses revealed a marked decrease in H3K36me2 levels and relatively preserved H3K36me3 levels in histone extracts from the NSD1 mutant cell lines (Figure 4A, D, E). Ectopic expression of H3K36M in HEK293 embryonic kidney cells, two HNSCC cell lines wild-type for H3 and NSD1, and an NSD1-deficient HNSCC cell line led to a significant decrease in H3K36me2/3 levels similar to that observed in HNSCC patient TMA samples, while siRNA knockdown of NSD1 specifically led to decreased H3K36me2 (Figure 4B–E). These findings suggest that both mutations may contribute to the progression of HNSCC through reducing H3K36me2. It is of interest that H3K36me2 levels were significantly altered despite a relatively normal expression of NSD2 in NSD1 mutant HNSCC samples and cell lines (p >0.22, Student’s t-test) (Figure 4A & Supplementary Figure 11). Furthermore, knockdown of NSD1 resulted in a more pronounced reduction in H3K36me2 in HNSCC cells compared to HEK293 cells (Figure 4C). These results highlight a major and non-redundant role of NSD1 in establishing H3K36me2 in HNSCC.
Figure 4. H3K36M and NSD1 mutations decrease levels of H3K36me2 in HNSCC.
(A) Immunoblots showing levels of NSD1/2/3, SETD2, H3K36me2 and H3K36me3 in NSD1 wild-type (Fadu and PCI-4B) and NSD1-deficient (SCC-4 and SKN-3) HNSCC cell lines. (B) Immunoblots showing levels of H3K36me2, H3K36me3 and H3K36M in NSD1 wild-type (Fadu and PCI-4B) and NSD1-deficient SCC-4 HNSCC cell lines, and SCC-4 cells stably expressing H3K36M mutant histone. (C) Immunoblots showing levels of NSD1, H3K36me2, H3K36me3 and H3K36M in 293T and NSD1 wild-type (Fadu and PCI-4B) HNSCC cell lines that were treated with NSD1 siRNA or stably express H3K36M mutant histone. (D) and (E) Mass spectrometry-based quantification of levels of H3K36 methylation in H3.1/2 (D) or H3.3 (E) in NSD1 wild-type (Fadu and PCI-4B) and NSD1-deficient (SCC-4 and SKN-3) HNSCC cell lines, and Fadu cells stably expressing H3K36M mutant histone. For (A) (B) and (C), blot images are cropped and representative of two independent experiments. Full-length blots are included in Supplementary Figure 14. For (D) and (E), error bars represent standard deviation from three independent cell cultures. Error bars represent standard deviation from biological triplicates. n.s., not significant; *, P<0.05; **, P<0.01 by Student’s t-test.
Finally, we analyzed transcriptome data available with the TCGA dataset and were able to identify a cluster that corresponds, to a large extent, to the DNA methylation-defined H3K36 subgroup (Supplementary Figure 12), suggesting that altered methylation at H3K36 affects both the epigenome and transcriptome in HNSCC patient samples. We next performed differential expression analysis focusing on genes that are differentially expressed between H3K36 and other HNSCC subgroups. Our results indicate that NSD1 and H3K36M mutants specifically suppressed the expression of genes involved in epidermal differentiation and keratinization processes (Supplementary Figure 13 & Supplementary Table 7). These findings mirror our recent data in mouse mesenchymal progenitor cells where overexpression of H3K36M resulted in a blockade in differentiation to the respective downstream lineages and induced undifferentiated sarcomas in mice32.
Discussion
In this study, we re-analyzed public genomic and epigenomic datasets from HNSCCs and identified a previously largely unappreciated role of epigenome deregulation in HNSCC tumorigenesis, specifically at lysine 36 of histone H3. Our unsupervised clustering approach using DNA methylation data segregated 61 HNSCC samples into a cluster we labeled H3K36. This DNA methylation cluster is exclusively comprised of samples carrying NSD1 alterations or H3K36M mutations reported here for the first time in HNSCC. Characterized by H3K36 alteration and DNA hypomethylation, this subgroup represents a significant subset of HPV-tumors (13%). Other clusters include a subgroup highly enriched in HPV+ samples along with Groups A B C, which do not harbor obviously defining clinico-pathological features or genetic alterations. Except for HPV+ cancers, we did not observe any statistically significant differences in event-free survival among methylation clusters. This may be due to the relatively short median follow-up time (401 days) of the TCGA HNSCC cohort. When using a similar unsupervised clustering approach on RNA-Seq data (Supplementary Figure 12), we are also able to recuperate a group of samples enriched in NSD1 or H3K36M mutations suggesting that the H3K36 cluster corresponds to a specific molecular entity.
In depth analysis of the NSD1 locus in samples included in the H3K36 subgroup allowed us to identify alterations such as splicing defects and focal deletion of a small set of exons that were not detectable using standard automated approaches (Supplementary Table 2). Moreover, a gene-based approach has previously failed to identify the enrichment of H3K36M substitutions as they are observed in a wide array of histone H3 genes (Supplementary Figure 2B & Supplementary Table 4). This is consistent with our previous work, which demonstrated that the effect of H3K36M mutation is dominant and independent of H3 isoforms31,32 (Figures 3 & 4). A complementary meta-analysis of earlier studies confirmed the prevalence of H3K36M and NSD1 alterations in HNSCC and their association with smoking and specific anatomical locations.
We replicated our genomic findings using an independent, previously unpublished cohort of HNSCC patient samples and showed that the mutant histone is expressed exclusively in tumor cells and leads to decreased H3K36me2 and me3 (Figure 3). In contrast, NSD1 loss leads to decreased H3K36me2 (Figures 3 & 4), consistent with previous biochemical characterization of the enzyme as a specific H3K36 mono- and di-methyltransferase. Given the converging effects of NSD1 and H3K36M mutations on DNA methylation and H3K36me2, it appears that these mutations may help drive HNSCC pathogenesis through a common mechanism involving epigenome deregulation. H3K36M mutations have been observed in chondroblastoma17 and more recently in pediatric soft-tissue tumors32. Our group also showed that when introduced into mesenchymal progenitor cells, H3K36M was sufficient to induce undifferentiated sarcomas in mice by blocking cellular differentiation32. Our transcriptome analysis reveals that genes specifically downregulated in the H3K36 subgroup are involved in cellular differentiation. These results support a model whereby an altered epigenetic landscape, mediated by the impaired H3K36 methylation subsequent to the genetic alterations, arrests keratinocytes in a progenitor state refractory to differentiation stimuli. This differentiation blockade in turn synergizes with signalling and cell cycle deregulation to facilitate HNSCC development.
Interestingly, no NSD2/WHSC1 mutation was found in the H3K36 subgroup and NSD1 deficiency was sufficient to decrease H3K36me2 in HNSCC cell lines despite the normal expression of NSD2. Moreover, a recent report showed overexpression of NSD2/WHSC1 and increased H3K36me2 in a set of HNSCC samples examined35. Therefore, it appears that NSD1 and NSD2 may play non-redundant roles in certain cell types and that their effect could vary based on the DNA methylation group. Further studies are warranted to investigate the distinct mechanisms by which gain or loss of H3K36me2 promotes HNSCC development.
The H3K36 cluster showed marked DNA hypomethylation compared to the other methylation subgroups. Interestingly, a recent study observed a DNA hypomethylation signature in SOTOS syndrome, a monogenic disorder defined by germline mutations in NSD136. Therefore, the DNA hypomethylation signature in the H3K36 subgroup is likely caused by genetic or K36M-mediated biochemical inactivation of NSD1. Several recent reports suggest a link between H3K36 methylation and the binding of de novo DNA methyltransferases DNMT3A/B37,38. The PWWP domain of DNMT3A, which is essential for its chromatin recruitment, specifically binds to H3K36me2/me3. In agreement, the gene-body localization of Dnmt3b in mouse embryonic stem cells depends on Setd2-mediated H3K36me3. Therefore the recruitment of DNMT3A/B could be potentially impaired in NSD1 or H3K36M mutant tumors, leading to the global DNA hypomethylation phenotype observed in the H3K36 cluster.
Large and comprehensive genomic and epigenomic surveys of HNSCCs have increased our understanding of the key driver mutations and heterogenic nature of this disease. Most of these studies emphasize the role of alterations affecting the cell cycle (TP53, CDKN2A) or signalling pathways including growth factor receptors (FGFR, EGFR), the RAS and PI3K pathways. Accordingly, a number of novel targeted therapies tailored to attenuate signalling defects are currently investigated in pre-clinical studies and clinical trials. However, recent studies in HNSCCs indicate that recurrent or metastatic tumors often fail to reproduce the mutational landscape of the primary tumor, specifically for the aforementioned targetable alterations10. In contrast, this evolutionary dynamic has not been observed in tumors carrying epigenetic drivers. Indeed, in the previously studied cancers that are promoted by mutations in epigenetic modulators, such as DNMT3A, IDH39 or histone H340, the epigenetic drivers are invariably present throughout the course of the disease. Our current study uncovers a previously unsuspected role of impaired H3K36 methylation in HNSCCs. This finding underscores the need for further investigations into the mechanisms of epigenome deregulation in HNSCC tumorigenesis, as targeting epigenetic modifiers may be, when further validated by additional in vitro and in vivo studies, of therapeutic benefit for HNSCC patients with altered H3K36 methylation.
Online Methods
DNA Methylation Clustering
We obtained the Level 1 (IDAT format) methylation data from the TCGA data portal for 528 HNSCC samples. We extracted raw beta values using the R package minfi41. We removed probes that contained or targeted SNPs as per the Illumina HumanMethylation450k Manifest. We performed hierarchical clustering using the 1,000 most variable sites. Distance was assessed using d = 1 − r, where r is the Pearson product-moment coefficient. Clustering was performed using average linkage (UPGMA). In order to establish robustness of our groups, we tested their predictive ability using a 10-fold cross-validation. We computed the sample-by-sample correlation between the test and training and averaged on the training set subgroups. The test set labels were predicted according to the highest average correlation among the training set subgroup.
Somatic Mutations
We used all the somatic mutation calls from Level 2 (MAF format) data from the TCGA data portal for 517 samples. We merged calls from the different sequencing centers, removed duplicates and converted MAF to VCF using a custom script. We annotated the resulting comprehensive mutation list using ANNOVAR42 to obtain the resulting amino-acid changes.
In order to address the prevalence of specific mutations in HPV-HNSCC methylation subgroups, we performed an ANOVA on the presence or absence of mutation, excluding the HPV+ subgroup. In order to control for the mutation count per sample, we divided the presence of mutation (1) by the total amount of mutations, thus asking “Does the subgroup of a sample explain the variance observed in the fraction of mutations in a given gene”. Every gene was treated independently and p-values were corrected for multiple testing using Bonferroni.
To predict the impact of NSD1 missense mutations on the protein function, we used mutation assessor (http://mutationassessor.org) which evaluates the functional impact of an amino-acid substitution based on the evolutionary conservation of amino-acid residues in a protein family multiple sequence alignment, assuming that mutations that affect conserved residues are more likely to be functional.
Mutations in the H3K36 cluster
Samples in the H3K36 cluster not carrying a somatic H3K36M or NSD1 mutations were manually assessed at the NSD1 locus. We identified large chromosomal deletions containing NSD1 (n = 2), focal deletion of a set of exons (n = 2) or splicing defects (n = 1). RNA-seq analysis showed absent NSD1 transcripts in two samples with non-interpretable WES data and one sample in the cluster lacked genomic data (Supplementary Table 2). Lolipot plots were obtained from the mutation mapper tool (http://www.cbioportal.org/mutation_mapper.jsp).
Clinical Annotation
Smoking: We tested for association with smoking using the 4-factor definition as per the CDR browser https://cdebrowser.nci.nih.gov/CDEBrowser/search?listValidValuesForDataElements=9&tabClicked=2&PageId=DataElementConceptDetails). To test for enrichment for smoking in H3K36 samples, we performed a Fisher’s Exact Test on those factors.
Anatomical Location: Some anatomical locations were grouped together (as per http://www.cancer.gov/types/head-and-neck/head-neck-fact-sheet#q1) in order to simplify the visual representation. Alveolar Ridge: {Alveolar Ridge}. Oral Cavity: {Buccal Mucosa, Floor of Mouth, Hard Palate, Oral Cavity, Oral Tongue}. Pharynx: {Base of Tongue, Hypopharynx, Oropharynx}. Larynx: {Larynx}. Tonsil: {Tonsil}
HPV Status: ISH and p16 tests were combined together so that sample flagged as HPV positive by either tests would be considered as HPV+ in our analysis. Vice versa for a negative diagnosis. Samples with no information were labeled as NA.
Copy Number Analysis
We obtained the Level 3 CNV calls from the TCGA data portal for 520 samples. We used the nocnv_hg19.seg.txt as input for GISTIC 2.0 to extract significant CNVs. We clustered samples based on CNV using the method previously described6. We considered NSD1 GISTIC value of −0.3 as sufficient evidence for LOH.
Mutation Signature
In order to infer the mutagenic processes in HNSCC subgroups, we used the somatic mutations obtained as described above. Samples from each methylation-defined subgroup were merged together because of the overall low amount of mutations from WES. We appended mutations from 8 TCGA cancers using the R package SomaticCancerAlterations. We extracted the mutational context (flanking nucleotides), extracted the somatic spectrum of each cancer and computed a 5 signature model using non-negative matrix decomposition using the R package SomaticSignatures. Hierarchical clustering was done on the somatic spectrum matrix using cosine distance.
RNA-Seq
We obtained raw bam files using gtdownload from cghub. Gene expression was quantified with featureCount43 using UCSC hg19 annotation with isoforms collapsed into a single entry per gene. We used the raw counts and normalized them for total library size using DESeq244. We performed hierarchical clustering using log2 transformed read count for the 1,000 most variable genes. Distance was assessed using d = 1 − r, where r is the Pearson product-moment coefficient. Clustering was performed using average linkage (UPGMA). In order to obtain differentially expressed genes that are specific to the H3K36 cluster, we performed two independent differential expression analyses. We used all HNSCC tumor samples and looked for genes differentially expressed specifically in the H3K36 cluster, compared to all other subgroups using anatomical location as a covariate. The threshold for establishing if a gene was differentially expressed was: Bonferroni adjusted p-value < 0.01, and an absolute value of log2 fold change > 2.
Cell culture, siRNA transfection and lentiviral infection
Fadu (ATCC), SKN-3 (JCRB cell bank) and PCI-4B (University of Pittsburgh) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) with 10% fetal bovine serum (FBS, CellGro). SCC-4 (ATCC) cells were cultured in DMEM:F12 Medium (Invitrogen) with 10% FBS and 400 ng/ml hydrocortisone (Sigma). These HNSCC cell lines have been demonstrated to be tumorigenic45,46,47,48 and the NSD1 mutational status was obtained through COSMIC or Cancer Cell Line Encyclopedia and verified by immunoblotting. For siRNA transfection, cells were reverse transfected with Lipofectamine® RNAiMAX (Life Technologies) and ON-TARGETplus SMARTpool siRNA against human NSD1 (GE Healthcare). After 72 h, cells were harvested for immunoblotting analysis. To generate cells stably expressing H3K36M mutant histone, cells were transduced with concentrated lentivirus (2 × 107 IFU) generated as previously described32. Transduced cells were grown under puromycin selection (2 µg/ml) 48 h after transduction and selected for a week before harvested for immunoblotting or mass spectrometry analysis.
Immunoblotting
For immunoblotting, acid-extracted histones or whole cell lysates were separated by SDS-PAGE, transferred to nitrocellulose membrane, blocked in 5% non-fat milk in PBS plus 0.5% Tween-20, probed with primary antibodies, and detected with horseradish peroxidase-conjugated α-rabbit or α-mouse secondary antibodies (GE Healthcare). Primary antibodies used include: αNSD1 (NeuroMab clone N312/10); αNSD2 (Millipore, MABE191); αNSD3 (Abcam, ab180500); αSETD2 (Abcam, ab31358); αH3K36me3 (Active Motif, 61021); αH3K36me2 (Cell Signaling Tech, 2901); αH3K36M (gift of Millipore); αH3 (Abcam, ab1791); αTubulin (Cell Signaling Tech, 2144).
Immunohistochemistry
All samples were obtained with informed consent after approval of the Institutional Review Board of the respective hospitals they were treated in, and were independently reviewed by pathologists with expertise in Head and Neck cancer (W.S., C.J.H.). Tissue microarrays (TMA) from a collection of 158 patients with HNSCCs were obtained following standard procedures. Consecutive sections from the TMAs were cut at 4 µm, placed on SuperFrost/Plus slides (Fisher) and dried overnight at 37°C, before IHC processing. The slides were then loaded onto the Discovery XT Autostainer (Ventana Medical System). All solutions used for automated IHC were from Ventana Medical System unless otherwise specified. Slides underwent deparaffinization, heat-induced epitope retrieval (CC1 prediluted solution Ref: 950–124, standard protocol). Immunostaining was performed online using a heat protocol. A negative control was performed by the omission of the primary antibody. Mouse monoclonal anti-H3K36me3 antibody (Diagenode Inc.) and anti-H3K36me2 (Cell Signaling Tech, 2901) were diluted at 1:500 in the antibody diluent (Ref: 251–018). Rabbit polyclonal anti-H3K36M antibody and anti-NSD1 (custom made by Millipore) were diluted at 1:22 and 1:20 in the antibody diluent for 32 min at 37°C and subsequently processed as previously described32. Slides were counterstained with hematoxylin for 4 min, blued with Bluing Reagent for 4 min, removed from the autostainer, washed in warm soapy water, dehydrated through graded alcohols, cleared in xylene and mounted with Eukitt Mounting Medium (EMS, Ref: 15320). Sections were scanned using the aperio system. Scoring was done on 100 tumor cells from 4 separate fields for all 4 antibodies. Staining was considered conclusive for H3K36M when strong specific nuclear staining was seen in tumor cells with no immunoreactivity in adjacent stroma; In the absence of genotype data, a tumor was considered as possibly carrying NSD1 mutation if more than 70% of tumor cells lacked nuclear staining while normal NSD1 nuclear staining could be seen in infiltrating inflammatory cells or adjacent stroma. H3K36me3 and H3K36me2 stains were similarly scored to NSD1.
Histone acid extraction, histone derivatization and PTM analysis by nano-LC-MS
Cells were lysed in hypotonic lysis buffer (10 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, protease inhibitors) for 1 hr at 4°C. H2SO4 was added to 0.2 N followed by overnight rotation at 4°C. After centrifugation, supernatants were collected, proteins were precipitated in 33% TCA, washed with acetone, and resuspended in deionized water.
Acid-extracted histones (15–25 µg) were resuspended in 100 mM ammonium bicarbonate (pH 8), derivatized using propionic anhydride, and digested with trypsin as previously described49. The resulting histone peptides were desalted using C18 Stage Tips, dried using a centrifugal evaporator, and reconstituted using 0.1% formic acid in preparation for nanoLC-MS analysis.
nanoLC was performed using a Thermo Scientific™ Easy nLC™ 1000 system equipped with a 75 µm × 20 cm in-house packed column using Reprosil-Pur C18-AQ (3 µm; Dr. Maisch GmbH, Germany). Buffer A was 0.1% formic acid and Buffer B was 0.1% formic acid in 100% acetonitrile. Peptides were resolved using a two-step gradient from 0% to 26% B over 45 min, then from 26% B to 98% B over 10 min at a flow rate of 300 nL/min. The HPLC was coupled online to an Orbitrap Elite mass spectrometer operating in the positive mode using a Nanospray Flex™ Ion Source (Thermo Scientific) at 2.40 kV. MS was performed using data-independent acquisition (DIA) as previously described50 with slight modifications. Briefly, two full MS scans (m/z 300–1100) were acquired in the orbitrap with a resolution of 120,000 (at 200 m/z) every 8 DIA MS/MS events, using isolation windows of 50 m/z each (e.g. 300–350, 350–400, etc.). The full MS scan was performed twice within the same duty cycle to allow for a more resolved definition of the precursor peak profile. MS/MS events were acquired in the ion trap operating in normal mode. Fragmentation was performed using collision-induced dissociation (CID) in the ion trap mass analyzer with a normalized collision energy of 35. AGC target and maximum injection time were 10e6 and 50 ms for the full MS scan, and 10e4 and 150 ms for the MS/MS scan, respectively.
Mass-to-charge ratios were calculated for each modified form of H3 peptide. The area under the curve was calculated from extracted ion chromatograms using Xcalibur Qual Browser for each histone peptide mass-to-charge ratio using a 10 ppm mass accuracy cutoff. Data from all detectable charge states were summed. The area for each modification state of a peptide was normalized against the sum of all peptides sharing the same sequence to give the relative abundance of each modified state. Three biological replicates were analyzed per condition, and the relative abundance of each peptide modification was averaged across the runs. Statistical significance was determined using Student’s t-test.
Supplementary Material
Acknowledgments
This research was supported by funding from NIH grants (P01CA196539 to N.J., J.M., P.W.L., B.A.G. and C.D.A.; K99CA212257 to C.L.; T32GM008275 to D.M.M,), the Rockefeller University (to C.D.A), startup provided by the Wisconsin Institute for Discovery (to P.W.L.), The Greater Milwaukee Foundation (to P.W.L.), Sidney Kimmel Foundation (Kimmel Scholar Award to P.W.L.), Canadian Institutes for Health Research (CIHR) grants (MOP 142491 to J.S.M. and A.C.N.; MOP 340674 to A.C.N.) and the Cedars Cancer Foundation. B.A.G. is funded by Leukemia and Lymphoma Society Dr. Robert Arceci Scholar Award. This work was performed within the context of the I-CHANGE consortium and supported by funding from Genome Canada, Genome Quebec, The Institute for Cancer Research of the CIHR, McGill University and the Montreal Children’s Hospital Foundation. C.L. is the Kandarian Family Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2195-14). N.J. is a member of the Penny Cole lab and the recipient of a Chercheur Boursier, Chaire de Recherche Award from the Fond de la Recherche du Québec en Santé. J.M. holds a Canada Research Chair (tier 2). S.P.C. is supported by a studentship from the Fond de la Recherche du Québec en Santé. D.B. and TG are supported by a fellowship from the T.D trust/Montreal Children’s Hospital Foundation and CIHR respectively. We thank EMD Millipore for its role in H3.3K36M antibody generation. We thank Andres Ponce de Leon for his help with figure formatting. We would like to thank the TCGA group for the wealth of data available and for their rapid and helpful answers to other enquiries we had on this dataset.
Footnotes
Data Availability
All sequencing and methylation data were obtained from the TCGA Genomic Data Commons (https://gdc.cancer.gov/).
Author Contributions:
S.P.C., C.L., J.M., N.J. conceived and designed the experiments, and analyzed data. C.L., T.G., L.G.M., D.B., C.K., J.K., O.M.D., S.D., W.S., C.J.H., J.W.B., D.M.M., B.A.G., performed experiments and analyzed data. L.A., I.W., D.G., J.S.M., P.W.L., A.C.N., C.D.A. contributed materials and analyzed data. All authors contributed to the written manuscript.
Conflict of Interest
The authors declare no conflicts of interest.
References
- 1.Gillison ML, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92:709–20. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Baxi S, Fury M, Ganly I, Rao S, Pfister DG. Ten years of progress in head and neck cancers. J Natl Compr Canc Netw. 2012;10:806–10. doi: 10.6004/jnccn.2012.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–7. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ang KK, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun W, Califano JA. Sequencing the head and neck cancer genome: implications for therapy. Ann N Y Acad Sci. 2014;1333:33–42. doi: 10.1111/nyas.12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaykalova DA, et al. Novel insight into mutational landscape of head and neck squamous cell carcinoma. PLoS One. 2014;9:e93102. doi: 10.1371/journal.pone.0093102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hedberg ML, et al. Genetic landscape of metastatic and recurrent head and neck squamous cell carcinoma. J Clin Invest. 2016;126:1606. doi: 10.1172/JCI86862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pickering CR, et al. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov. 2013;3:770–81. doi: 10.1158/2159-8290.CD-12-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seiwert TY, et al. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res. 2015;21:632–41. doi: 10.1158/1078-0432.CCR-13-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammerman PS, Hayes DN, Grandis JR. Therapeutic insights from genomic studies of head and neck squamous cell carcinomas. Cancer Discov. 2015;5:239–44. doi: 10.1158/2159-8290.CD-14-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115–26. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fontebasso AM, et al. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol. 2013;125:659–69. doi: 10.1007/s00401-013-1095-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9:804–12. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- 17.Behjati S, et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet. 2013;45:1479–82. doi: 10.1038/ng.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaffe JD, et al. Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nat Genet. 2013;45:1386–91. doi: 10.1038/ng.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–3. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–31. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 21.Douglas J, et al. NSD1 mutations are the major cause of Sotos syndrome and occur in some cases of Weaver syndrome but are rare in other overgrowth phenotypes. Am J Hum Genet. 2003;72:132–43. doi: 10.1086/345647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tonon G, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci U S A. 2005;102:9625–30. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–63. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiao Q, et al. The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation. J Biol Chem. 2011;286:8361–8. doi: 10.1074/jbc.M110.204115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun W, et al. Activation of the NOTCH pathway in head and neck cancer. Cancer Res. 2014;74:1091–104. doi: 10.1158/0008-5472.CAN-13-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayes DN, Van Waes C, Seiwert TY. Genetic Landscape of Human Papillomavirus-Associated Head and Neck Cancer and Comparison to Tobacco-Related Tumors. J Clin Oncol. 2015;33:3227–34. doi: 10.1200/JCO.2015.62.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vokes EE, Agrawal N, Seiwert TY. HPV-Associated Head and Neck Cancer. J Natl Cancer Inst. 2015;107:djv344. doi: 10.1093/jnci/djv344. [DOI] [PubMed] [Google Scholar]
- 29.India Project Team of the International Cancer Genome, C. Mutational landscape of gingivo-buccal oral squamous cell carcinoma reveals new recurrently-mutated genes and molecular subgroups. Nat Commun. 2013;4:2873. doi: 10.1038/ncomms3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pickering CR, et al. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin Cancer Res. 2014;20:6582–92. doi: 10.1158/1078-0432.CCR-14-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis PW, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–61. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu C, et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science. 2016;352:844–9. doi: 10.1126/science.aac7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walter V, et al. Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PLoS One. 2013;8:e56823. doi: 10.1371/journal.pone.0056823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Cecco L, et al. Head and neck cancer subtypes with biological and clinical relevance: Meta-analysis of gene-expression data. Oncotarget. 2015;6:9627–42. doi: 10.18632/oncotarget.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saloura V, et al. WHSC1 promotes oncogenesis through regulation of NIMA-related kinase-7 in squamous cell carcinoma of the head and neck. Mol Cancer Res. 2015;13:293–304. doi: 10.1158/1541-7786.MCR-14-0292-T. [DOI] [PubMed] [Google Scholar]
- 36.Choufani S, et al. NSD1 mutations generate a genome-wide DNA methylation signature. Nat Commun. 2015;6:10207. doi: 10.1038/ncomms10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhayalan A, et al. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem. 2010;285:26114–20. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baubec T, et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015;520:243–7. doi: 10.1038/nature14176. [DOI] [PubMed] [Google Scholar]
- 39.Johnson BE, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–93. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nikbakht H, et al. Spatial and temporal homogeneity of driver mutations in diffuse intrinsic pontine glioma. Nat Commun. 2016;7:11185. doi: 10.1038/ncomms11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aryee MJ, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–9. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic feature. Bioinformatics. 2014;30:923–30. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 44.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nii M, Kayada Y, Yoshiga K, Takada K, Yanagihara K. Expression of type IV collagen-degrading metalloproteinases and tissue inhibitors of metalloproteinases in newly established human oral malignant tumor lines. Jpn J Clin Oncol. 1996;26:117–23. doi: 10.1093/oxfordjournals.jjco.a023193. [DOI] [PubMed] [Google Scholar]
- 46.Heo DS, et al. Biology, cytogenetics, and sensitivity to immunological effector cells of new head and neck squamous cell carcinoma lines. Cancer Res. 1989;49:5167–75. [PubMed] [Google Scholar]
- 47.Zietman AL, Suit HD, Ramsay JR, Silobrcic V, Sedlacek RS. Quantitative studies on the transplantability of murine and human tumors into the brain and subcutaneous tissues of NCr/Sed nude mice. Cancer Res. 1988;48:6510–6. [PubMed] [Google Scholar]
- 48.Rheinwald JG, Beckett MA. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultured from human squamous cell carcinomas. Cancer Res. 1981;41:1657–63. [PubMed] [Google Scholar]
- 49.Sidoli S, Bhanu NV, Karch KR, Wang X, Garcia BA. Complete Workflow for Analysis of Histone Post-translational Modifications Using Bottom-up Mass Spectrometry: From Histone Extraction to Data Analysis. J Vis Exp. 2016 doi: 10.3791/54112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sidoli S, Simithy J, Karch KR, Kulej K, Garcia BA. Low Resolution Data-Independent Acquisition in an LTQ-Orbitrap Allows for Simplified and Fully Untargeted Analysis of Histone Modifications. Anal Chem. 2015;87:11448–54. doi: 10.1021/acs.analchem.5b03009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.