Abstract
The progressing discovery of genetic variants associated with drug-related adverse events has raised expectations for pharmacogenetic tests to improve drug efficacy and safety. To further the use of pharmacogenetics in health care, tests with sufficient potential to improve efficacy and safety, as reflected by good clinical validity and population impact, need to be identified. The potential benefit of pharmacogenetic tests is often concluded from the strength of the association between the variant and the adverse event; measures of clinical validity are generally not reported. This paper describes measures of clinical validity and potential population health impact that can be calculated from association studies. We explain how these measures are influenced by the strength of the association and by the frequencies of the variant and the adverse event. The measures are illustrated using examples of testing for HLA-B*5701 associated with abacavir-induced hypersensitivity and SLCO1B1 c.521T>C (*5) associated with simvastatin-induced adverse events.
Introduction
Genetic variants associated with drug response or drug-related adverse events can potentially be used to improve the efficacy and safety of drugs.1, 2, 3 Pharmacogenetic tests are generally thought to be useful when the association between the genetic variant and the drug response or adverse event is strong.4 However, the ability of a pharmacogenetic test to improve drug efficacy and safety depends on more than just the association between the genetic variant and drug response or the adverse event. For that reason, the assessment of pharmacogenetic tests goes beyond quantification of association alone. Reporting measures of clinical validity and population impact in addition to measures of association allows a more informative evaluation of pharmacogenetic tests.
The clinical validity of a pharmacogenetic test indicates the test’s ability to predict the occurrence of the adverse event of interest. Clinical validity is determined by the strength of the association between the genetic variant and the adverse event, but also by the frequencies of the genetic variant and the adverse event. Therefore, a strong association is essential but not a sufficient condition to ensure good clinical validity.
The clinical validity subsequently impacts the clinical utility of the test, which is the ability of the test to prevent adverse effects through differentiation in treatments based on the test results.5 The population impact indicates the potential benefit of a pharmacogenetic test and differentiation in treatments and can be expressed as the expected reduction in adverse events or the number of patients that need a different treatment.
Evaluations of pharmacogenetic tests often report measures of association without considering clinical validity and population impact.4, 6, 7 For example, measures of clinical validity are included in the Clinical Pharmacogenetics Implementation Consortium guidelines for drug/gene pairs when this information is available from empirical studies, which is the case for only 5 of the 35 drugs.8 The reasons for this are that pharmacogenetic studies frequently investigate intermediate continuous end points instead of adverse events, such as drug plasma concentrations and biochemical markers of toxicity. Also, studies that investigate adverse events are often observational studies with a case–control design, which by design have a different proportion of cases than in the population of interest. Because of that, case–control studies do allow calculation of the pharmacogenetic association (odds ratio (OR)) but not of all measures of clinical validity.
In this paper we explain how measures of clinical validity and potential population impact can be calculated in pharmacogenetic association studies. Additionally, we demonstrate how the measures are impacted by variations in ORs, adverse event frequency and variant frequency, and illustrate their use in the assessment of pharmacogenetic testing for HLA-B*5701, which is associated with abacavir-induced hypersensitivity9 and SLCO1B1 c.521T>C (*5) associated with simvastatin-induced adverse events.10, 11
Measures of clinical validity
Clinical validity refers to the ability of a test to correctly identify or predict an outcome of interest, which, in pharmacogenetics, indicates the ability of the test to predict adverse events such as toxicity or lack of treatment efficacy. Clinical validity is indicated by measures of discriminative accuracy and predictive value.
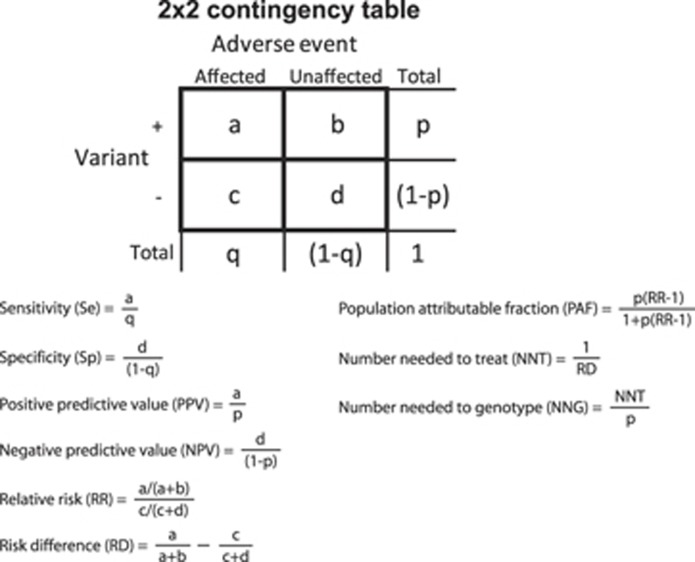
The discriminative accuracy refers to the ability of a test to discriminate between the presence and absence of adverse events and is indicated by the sensitivity and specificity. Sensitivity is the probability that the genetic variant associated with a higher adverse event risk (from here referred to as the genetic variant) is present in individuals with the adverse event while specificity is the probability that the genetic variant is absent in individuals without the adverse event (Figure 1). To indicate discriminative accuracy both sensitivity and specificity need to be reported. A pharmacogenetic test that has high sensitivity (97%) but low specificity (10%) will be able to predict 97% of the individuals who will develop an adverse event but it will misclassify 90% of the individuals who will not develop an adverse event. Measures of clinical validity can be calculated from a 2 × 2 contingency table that describes genetic test results by adverse events (Figure 1). In the abacavir examples (Box 1 and Table 1), the sensitivity and specificity were 48 and 97% for clinically diagnosed abacavir hypersensitivity and 100 and 96% for immunologically confirmed hypersensitivity. These values differ because immunologically confirmed hypersensitivity is a more accurate indicator of hypersensitivity reactions that are specifically caused by abacavir than clinically diagnosed hypersensitivity, which can also be caused by concomitant drug use. All immunologically confirmed hypersensitivities are clinically diagnosed hypersensitivities, but not vice versa.
Figure 1.
Calculation of clinical validity and potential population impact measures from 2 × 2 contingency tables reporting adverse event by genetic variant subgroups. Contingency tables can be constructed using empirical data or using hypothetical data calculated from summary statistics and association measures, such as odds ratios derived from observational studies with a case–control design in combination with the frequencies of the genetic variant and the adverse event (see Supplementary Information).
Examples.
Abacavir
Abacavir is a nucleoside reverse transcriptase inhibitor indicated for treatment of HIV infection. Treatment with abacavir is generally well tolerated, but 5–8% of the patients experience hypersensitivity reactions that can be life threatening and warrant immediate discontinuation of the drug. Hypersensitivity reactions can be clinically diagnosed based on symptoms only or be immunologically confirmed using patch testing. Multiple studies revealed a strong association between carriage of HLA-B*5701 and abacavir hypersensitivity in Caucasians and Hispanics.9, 44, 45, 46
Simvastatin
Simvastatin is among the most commonly prescribed generic statin formulations indicated to lower cholesterol levels. Its use is associated with muscle problems varying in severity, including myalgia (pain without evidence of muscle tissue damage), myopathy (pain with evidence of muscle tissue damage) and rhabdomyolysis (severe muscle tissue damage with (possible) complications). A genome-wide association study investigated simvastatin use and myopathy and identified SLCO1B1 as a gene of interest.10 Since then multiple association studies have linked SLCO1B1 c.521T>C (*5) (rs4149056 SNP) to lower plasma clearance of simvastatin and adverse events.10, 11, 14, 15
Table 1. Examples of calculating clinical validity and population impact.
| Example | Empirical data | Hypothetical data | ||
|---|---|---|---|---|
|
Abacavir |
Simvastatin |
|||
| Study | Mallal et al.9 | Mallal et al.9 | SEARCH collaborative group10 | Voora et al.11 |
| Variant | HLA-B*5701 | HLA-B*5701 | SLCO1B1 c.521T>C (*5) | SLCO1B1 c.521T>C (*5) |
| Risk genotype freq | 6.7% | 6.6% | 25% | 28% |
| Adverse event | Clinically diagnosed abacavir hypersensitivity | Immunologically confirmed abacavir hypersensitivity | Severe myopathy | Any side effect |
| Adverse event freq | 8.5% | 3.1% | 0.8% | 23% |
| ORa | 30 | 1176b | 8.5 | 2.8 |
| Sensitivity | 48% | 100% | 73% | 45% |
| Specificity | 97% | 96% | 76% | 77% |
| PPV | 60% | 47% | 2% | 37% |
| NPV | 95% | 100% | 99.7% | 83% |
| PAF | 44% | 100% | 64% | 24% |
| NNT | 2 | 3 | 49 | 6 |
| NNG | 27 | 33 | 195 | 19 |
Abbreviations: NNG, number needed to genotype; NNT, number needed to treat; NPV, negative predictive value; PAF, population attributable fraction; PPV, positive predictive value. Severe myopathy: muscle symptoms and creatine kinase level above 10 × upper limit of normal; any side effect: composite of any side effect, myalgia and/or creatine kinase level above 3 × upper limit of normal.
The odds ratios (OR) compare carriers versus non-carriers of HLA-B*5701 for abacavir and CC/CT versus TT genotype at SLCO1B1 rs4149056 for simvastatin.
The predictive value reflects the ability to predict adverse events from the presence or absence of the variant and is indicated by the positive and negative predictive value (PPV and NPV). PPV is the probability of an adverse event when the genetic variant is present, and NPV is the probability of no adverse event when the genetic variant is absent (Figure 1). Similarly as for discriminative accuracy, predictive ability is indicated by the combination of PPV and NPV, which implies that both need to be reported. Predictive value measures are sensitive to the prevalence of the adverse event and PPV remains generally low for rare adverse events even if the pharmacogenetic associations (ORs) are high. In the abacavir example, reported PPV and NPV were 60 and 95% for clinically diagnosed hypersensitivity and 47 and 100% for the immunologically confirmed hypersensitivity (Table 1). This means that 60 or 47% of the patients who carry the HLA-B*5701 variant will actually develop hypersensitivity from using abacavir, depending on which adverse events the test aims to predict, and that 95 or 100% of the patients who do not carry the variant will not develop hypersensitivity.
The different measures of clinical validity represent different perspectives. Sensitivity and specificity indicate the predictive performance from a population perspective, focusing on what proportions of patients with and without an adverse event are correctly predicted. PPV and NPV indicate the performance from an individual perspective as they quantify the adverse event risks for carriers and non-carriers of the genetic variant.
Population impact measures
Pharmacogenetic testing is performed to improve the effectiveness and efficiency by differentiating drug treatments between genotype groups. The potential impact of pharmacogenetic testing in terms of effectiveness can be indicated by the population attributable fraction (PAF). PAF is the proportion of events that is attributed to a risk factor or the proportion of events that would be eliminated from the population if exposure to the risk factor were eliminated (Figure 1). However, a pharmacogenetic variant cannot be eliminated, only a change in treatment can potentially prevent adverse events. Therefore, in pharmacogenetics, PAF indicates the proportion of adverse events that can potentially be eliminated if patients who carry the genetic variant receive different treatments. In the abacavir examples, PAF was 44 and 100% for clinically diagnosed and immunologically confirmed abacavir hypersensitivity, respectively. This means that 44 and 100% of the hypersensitivity is attributed to the effects of the HLA-B*5701 variant. This equals the maximum percentage of cases that can be prevented if individuals who test positive for HLA-B*5701 are not treated with abacavir but receive alternative treatment.
Pharmacogenetic testing may also increase the efficiency of treatment, which can be indicated by the number needed to treat (NNT). The NNT is generally defined as the number of individuals who would need to be treated to prevent one additional event and is calculated based on the event risks of individuals who receive or do not receive treatment. In pharmacogenetics, NNT is calculated by comparing adverse event risks in carriers and non-carriers of the genetic variant, which means that the NNT is interpreted as the number of patients who need treatment to prevent one patient from having an adverse event, with patients being the carriers of the genetic variant who need an alternative treatment (Figure 1). The number needed to genotype (NNG) is the number of patients that have to be genotyped to prevent one patient from having an adverse event. In the abacavir example, an NNG of 33 and an NNT of 3 means that for every 33 patients that are genotyped, 3 patients will learn that they test positive for HLA-B*5701 and need to receive alternative treatment to prevent hypersensitivity reactions in one.
Calculations in the absence of empirical data
Measures of clinical validity and potential population impact can be calculated from empirical cohort data when the adverse events are binary outcome variables. For studies with a case–control design, sensitivity and specificity can be calculated but PPV and NPV cannot because the proportions of cases and non-cases are generally not reflecting the actual proportions in the population of interest. In this case, or in the absence of empirical data, measures of clinical validity can be calculated from a 2 × 2 contingency table that can be constructed when the frequencies of the genetic variant and the adverse event as well as the OR are known (see Supplementary Information).12, 13 These data can be derived from published articles. For the simvastatin examples, we used reported summary statistics and association measures to calculate measures of clinical validity and population impact (Box 1 and Table 1).10, 11, 14, 15
Influence of OR, adverse event frequency and variant frequency
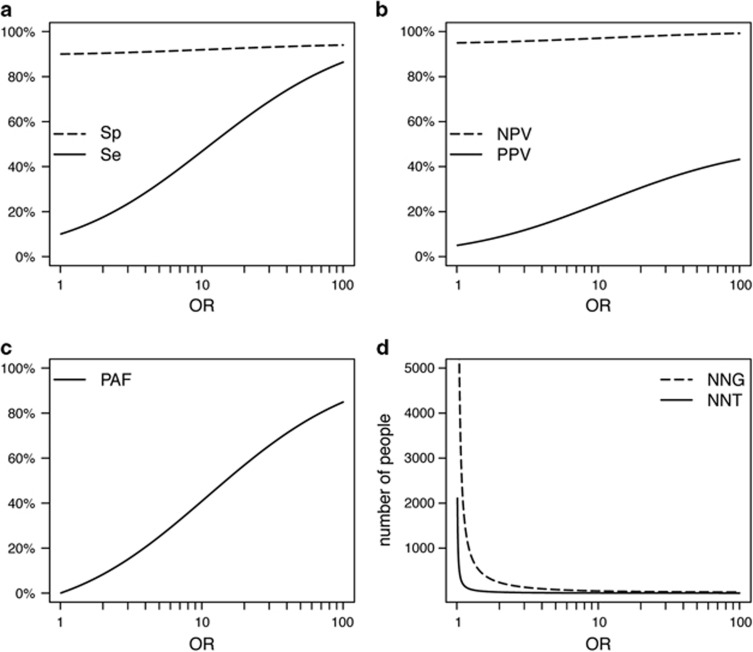
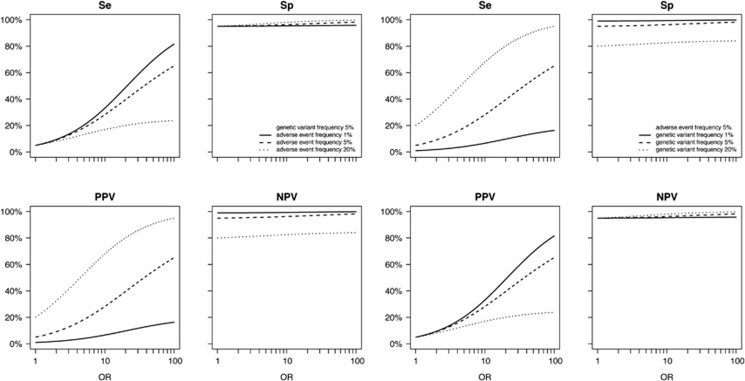
Measures of clinical validity and population impact evidently improve when the OR is higher (Figure 2), but a higher OR does not automatically result in higher values for all measures (Table 1). Clinical validity and potential population impact do not only depend on the OR, but also on the frequencies of the genetic variant and the adverse event (Figure 3 and Supplementary Figure S1). For example, in the simvastatin examples, the scenario in which the OR was higher did not have a higher treatment efficiency (Table 1); the lower adverse event frequency of 0.8% as compared with the other scenario (23%) led to a lower absolute risk difference and in turn to higher NNT and NNG.
Figure 2.
Effect of OR on measures of clinical validity and potential population impact. Top: Sensitivity (Se) and specificity (Sp) (a); positive predictive value (PPV) and negative predictive value (NPV) (b); bottom: Population attributable fraction (PAF) (c); number needed to genotype (NNG) and number needed to treat (NNT) (d). Adverse event frequency 5% and genetic variant frequency 10%.
Figure 3.
Effect of OR on measures of clinical validity when varying adverse event and genetic variant frequencies. NPV, negative predictive value; PPV, positive predictive value; Se, sensitivity; Sp, specificity.
The ratio between the frequency of the genetic variant and the frequency of the adverse event also influences clinical validity and potential population impact. In the abacavir example, the adverse event frequency of immunologically confirmed hypersensitivity (3.1%) was lower than the frequency of HLA-B*5701 carriers (6.6%). Despite the high OR (1176), sensitivity (100%), specificity (96%) and NPV (100%), the PPV was only 47%. When the variant frequency is higher than the adverse event frequency, by definition not all carriers will develop an adverse event. For the example of clinically diagnosed hypersensitivity, an OR of 30 was reported, while sensitivity was only 48% and PAF 44% (Table 1). This illustrates that when the frequency of the genetic variant is lower than the adverse event frequency, sensitivity and PAF can never reach 100% because only a proportion of the adverse events is attributed to the genetic variant.
Discussion
This article illustrates how the clinical validity and population impact of pharmacogenetic tests may vary with the population and setting in which tests are used. ORs, variant frequencies and adverse event frequencies often differ between (sub)populations, for example, according to ethnic background and gender, and therewith cause the clinical validity and potential population impact to vary between populations. Also, changes in the definition and measurement of the adverse event phenotype may impact the observed performance of genetic tests, as the different classification of individuals with and without the adverse event may lead to different adverse event frequencies and ORs.16
The calculation of the potential population impact comes with assumptions that impact its interpretation. PAF, NNT and NNG assume that changing treatment for patients who carry the genetic variant will lower their adverse event risk to the same level as the risk in the group who do not carry this variant. This assumption may be more realistic for some interventions than for others. The assumptions for PAF, NNT and NNG may hold in case the adverse event is rare. In this case, the adverse event risk after changing treatment will approximate the adverse event risk in patients who do not carry the genetic variant. A similar scenario is observed when the adverse event also has other causes than the drug treatment. For simvastatin, several placebo-controlled trials using statins have demonstrated that adverse event rates in the placebo group were comparable to the group treated with statins.17 For abacavir, the rate of clinically diagnosed hypersensitivity reactions that cannot be immunologically confirmed was similar to the rates of hypersensitivity reactions (2–7%) among patients not receiving abacavir in double-blind comparative-treatment studies.18, 19, 20 We therefore assumed that for both examples in this article the assumptions for PAF, NNT and NNG were reasonable.
A previous study has suggested that the strength of the association can be used as a single indicator of the diagnostic test performance21 because low ORs will never result in highly predictive diagnostic tests. This is true, but not sufficient. Clinical validity depends on the OR but the OR alone does not determine clinical validity and impact.22, 23, 24 We showed that the impact of the frequencies of the variant and the adverse event on clinical validity and population impact cannot be inferred from the strength of the association alone. Knowledge about the frequencies of the genetic variant and the adverse event in the target population and their influence on clinical validity and potential population impact can aid in identifying groups with increased or decreased drug efficacy or adverse event risks or aid in the selection of subpopulations that should be genotyped with priority.
The OR and the frequencies of the genetic variant and adverse event determine the clinical validity, but whether the clinical validity is high enough for the pharmacogenetic test to have clinical utility is determined by the intended use. The clinical utility of a genetic test is the ability of the test to prevent or ameliorate adverse health outcomes.5 Whether a test is worth implementing is a tradeoff of the benefits and costs that accrue from both positive and negative test results. For example, for the prevention of life-threatening adverse events, such as abacavir hypersensitivity, lower specificity may be acceptable to obtain the high sensitivity that is needed to prevent the vast majority of adverse events. However, for the prevention of milder adverse events, such as myalgia that often occurs during simvastatin therapy, a lower specificity may not outweigh the higher cost that come with targeting drug therapy. The potential implementation of a pharmacogenetic test is also determined by the alterative treatments available. If there were no alternative treatments to replace abacavir, the benefits of therapy could be considered to outweigh the risk of hypersensitivity reactions.
Insight in the clinical validity and population impact of pharmacogenetic tests helps understanding why some tests are widely used and others are not. The effectiveness and costs of abacavir and availability of alternative treatment explain why HLA-B*5701 testing is widely used before starting abacavir treatment, even though the PPV is lower than 50%.25, 26 In contrast, SLCO1B1 genotyping before prescribing simvastatin is not widely practiced, which also is supported by the calculations in this paper. While severe myopathy is an adverse outcome that is worth preventing and carriers of SLCO1B1 c.521T>C (*5) are at eightfold increased risk, their absolute risk (PPV) of severe myopathy was ‘only’ 2%. Forty-nine people would need to receive alternative treatment to prevent the adverse event in one (Table 1). The NPV was high (99.7%) but should be valued in comparison with the probability of no adverse event without testing, which in this case was 99.2%. Carriers had a higher absolute risk for mild adverse events, such as myalgia, but there is no evidence for clinical utility for SLCO1B1 genotyping.27 Also, statins are usually not discontinued when patients can tolerate mild muscle pain.28
The examples in this paper can also be generalized to other gene/drug examples. When the prevalence of the genetic variant is common and the adverse event is rare, by definition, most carriers will not develop the adverse event and PPV will be low. This is the case for carbamazepine and HLA-B*1502 associated with Stevens–Johnson syndrome and toxic epidermal necrolysis; despite a very strong association (OR=113) in Asian patients, the PPV of carriers of HLA-B*1502 is only 1.8%.29, 30 When the adverse event is common and the genetic variant is rare, non-carriers are still at substantial risk of developing adverse events, resulting in low sensitivity. This is the case for 5-fluorouracil and DPYD variants associated with toxicity where the sensitivity was only 31% despite an OR of 22.31, 32
Measures of clinical validity can be calculated in association studies when the end point of the study is a binary variable, such as the occurrence of an adverse event. In some instances, the end point is a time variable, namely the time to achieve certain therapeutic levels of the drug such as for warfarin and CYP2C9 and VKORC1. In these cases, measures of clinical validity can be calculated for cutoff values of time, which creates a binary end point as the percentage of patients that reached a therapeutic level of the drug within a clinically relevant period of time after initiating treatment.
The scarcity of resources available for translational research asks for approaches that can fill the current evidence gaps in available empirical pharmacogenetics data, but there is no consensus on the amount and type of evidence required to determine clinical validity. Randomized controlled trials are preferred in the evaluation of pharmacogenetic tests but often not available.33 The use of data from observational studies and modeling are an attractive alternative as these can provide important indications of clinical validity that can inform evaluations.2, 12 When using association data, it is important to verify that the study population is representative for the population in which the pharmacogenetic test is going to be used, as differences in the composition of the population might impact the risk of adverse events and the strength of the association. Also meta-analysis of existing data can be useful when differences in adverse event phenotypes are adequately taken into account.
The need to fill in evidence gaps in translational pharmacogenetics research is heightened by the increasing interest in preemptive (pre-prescription) genotyping. Preemptive pharmacogenetic testing has now been adopted in a few early-adopter health system settings,34 even though there remains great controversy about the clinical utility.35, 36, 37 The fact that pharmacogenetic data for variants associated with adverse events are ‘readily and freely’ available do not guarantee clinical utility. Measures of clinical validity are needed to inform whether and how to consider pharmacogenetic test results. Also, the pharmacogenetic data may be available, but its use is not ‘free’. Even though point-of-care testing is no longer required, other expenses such as costs of the retrieval and interpretation of the test results, counseling and alternative treatments still have to be considered.34, 36 Clinical validity and population impact measures are also important to determine the cost-effectiveness of pharmacogenetic testing.38, 39 Overall, evidence of clinical utility and cost-effectiveness is important for the adoption of pharmacogenetic testing in health-care practice.36
Finally, while the primary focus of pharmacogenetic testing has been improving drug selection and dosing, a potential benefit of testing may be increased personal utility and adherence.40 Testing may enhance the patient’s confidence in the efficacy of targeted therapy, which may increase medication adherence and improve clinical outcomes. Even though increased personal utility can have desirable benefits, pharmacogenetic tests without clinical validity are essentially placebo tests and should not be used because this may undermine confidence in medicine.
Conclusions
Implementing pharmacogenetics in clinical practice is new to many health-care providers. It is important to remember that there are many reasons for variability in drug responses between patients (e.g., age, liver and kidney function, pathology, lifestyle), which determine the proper application of pharmacogenetic testing.41 Pharmacogenetic tests should be adopted based on their clinical utility and cost-effectiveness. However, evidence gaps in available empirical pharmacogenetics data and scarcity of resources available for translational research can hinder current assessment of pharmacogenetic tests. The use of understandable and easily applied measures for clinical validity and potential population impact in addition to measures of association when reporting observational data will facilitate the identification of promising pharmacogenetic applications.
Acknowledgments
This work was supported by a consolidator grant from the European Research Council (GENOMICMEDICINE).
Footnotes
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website (http://www.nature.com/tpj)
The authors declare no conflict of interest.
Supplementary Material
References
- Abul-Husn NS, Owusu Obeng A, Sanderson SC, Gottesman O, Scott SA. Implementation and utilization of genetic testing in personalized medicine. Pharmacogenomics Pers Med 2014; 7: 227–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank PC, Swen JJ, Guchelaar HJ. Pharmacogenetic biomarkers for predicting drug response. Expert Rev Mol Diagn 2014; 14: 723–735. [DOI] [PubMed] [Google Scholar]
- Swen JJ, Nijenhuis M, de Boer A, Grandia L, Maitland-van der Zee AH, Mulder H et al. Pharmacogenetics: from bench to byte—an update of guidelines. Clin Pharmacol Ther 2011; 89: 662–673. [DOI] [PubMed] [Google Scholar]
- Gharani N, Keller MA, Stack CB, Hodges LM, Schmidlen TJ, Lynch DE et al. The Coriell personalized medicine collaborative pharmacogenomics appraisal, evidence scoring and interpretation system. Genome Med 2013; 5: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury MJ. Genetics and genomics in practice: the continuum from genetic disease to genetic information in health and disease. Genet Med 2003; 5: 261–268. [DOI] [PubMed] [Google Scholar]
- Ramsey LB, Johnson SG, Caudle KE, Haidar CE, Voora D, Wilke RA et al. The clinical pharmacogenetics implementation consortium guideline for SLCO1B1 and simvastatin-induced myopathy: 2014 update. Clin Pharmacol Ther 2014; 96: 423–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke RA, Ramsey LB, Johnson SG, Maxwell WD, McLeod HL, Voora D et al. The clinical pharmacogenomics implementation consortium: CPIC guideline for SLCO1B1 and simvastatin-induced myopathy. Clin Pharmacol Ther 2012; 92: 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical Pharmacogenetics Implementation Consortium guidelines. http://www.pharmgkb.org (last accessed on 1 December 2015).
- Mallal S, Phillips E, Carosi G, Molina JM, Workman C, Tomazic J et al. HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med 2008; 358: 568–579. [DOI] [PubMed] [Google Scholar]
- Link E, Parish S, Armitage J, Bowman L, Heath S, Matsuda F et al. SLCO1B1 variants and statin-induced myopathy—a genomewide study. N Engl J Med 2008; 359: 789–799. [DOI] [PubMed] [Google Scholar]
- Voora D, Shah SH, Spasojevic I, Ali S, Reed CR, Salisbury BA et al. The SLCO1B1*5 genetic variant is associated with statin-induced side effects. J Am Coll Cardiol 2009; 54: 1609–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens AC, Aulchenko YS, Elefante S, Borsboom GJ, Steyerberg EW, van Duijn CM. Predictive testing for complex diseases using multiple genes: fact or fiction? Genet Med 2006; 8: 395–400. [DOI] [PubMed] [Google Scholar]
- Kalf RR, Bakker R, Janssens AC. Predictive ability of direct-to-consumer pharmacogenetic testing: when is lack of evidence really lack of evidence? Pharmacogenomics 2013; 14: 341–344. [DOI] [PubMed] [Google Scholar]
- Brunham LR, Lansberg PJ, Zhang L, Miao F, Carter C, Hovingh GK et al. Differential effect of the rs4149056 variant in SLCO1B1 on myopathy associated with simvastatin and atorvastatin. Pharmacogenomics J 2012; 12: 233–237. [DOI] [PubMed] [Google Scholar]
- Carr DF, O'Meara H, Jorgensen AL, Campbell J, Hobbs M, McCann G et al. SLCO1B1 genetic variant associated with statin-induced myopathy: a proof-of-concept study using the clinical practice research datalink. Clin Pharmacol Ther 2013; 94: 695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurwitz D, Pirmohamed M. Pharmacogenomics: the importance of accurate phenotypes. Pharmacogenomics 2010; 11: 469–470. [DOI] [PubMed] [Google Scholar]
- Finegold JA, Manisty CH, Goldacre B, Barron AJ, Francis DP. What proportion of symptomatic side effects in patients taking statins are genuinely caused by the drug? Systematic review of randomized placebo-controlled trials to aid individual patient choice. Eur J Prev Cardiol 2014; 21: 464–474. [DOI] [PubMed] [Google Scholar]
- DeJesus E, Herrera G, Teofilo E, Gerstoft J, Buendia CB, Brand JD et al. Abacavir versus zidovudine combined with lamivudine and efavirenz, for the treatment of antiretroviral-naive HIV-infected adults. Clin Infect Dis 2004; 39: 1038–1046. [DOI] [PubMed] [Google Scholar]
- Gulick RM, Ribaudo HJ, Shikuma CM, Lalama C, Schackman BR, Meyer WA III et al. Three- vs four-drug antiretroviral regimens for the initial treatment of HIV-1 infection: a randomized controlled trial. JAMA 2006; 296: 769–781. [DOI] [PubMed] [Google Scholar]
- Staszewski S, Keiser P, Montaner J, Raffi F, Gathe J, Brotas V et al. Abacavir-lamivudine-zidovudine vs indinavir-lamivudine-zidovudine in antiretroviral-naive HIV-infected adults: a randomized equivalence trial. JAMA 2001; 285: 1155–1163. [DOI] [PubMed] [Google Scholar]
- Glas AS, Lijmer JG, Prins MH, Bonsel GJ, Bossuyt PM. The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol 2003; 56: 1129–1135. [DOI] [PubMed] [Google Scholar]
- Baldessarini RJ, Finklestein S, Arana GW. The predictive power of diagnostic tests and the effect of prevalence of illness. Arch Gen Psychiatry 1983; 40: 569–573. [DOI] [PubMed] [Google Scholar]
- Khoury MJ, Newill CA, Chase GA. Epidemiologic evaluation of screening for risk factors: application to genetic screening. Am J Public Health 1985; 75: 1204–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaescu R, Moonesinghe R, Khoury MJ, Janssens AC. Predictive genetic testing for the identification of high-risk groups: a simulation study on the impact of predictive ability. Genome Med 2011; 3: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JD, Lee KC, Kuo GM. HLA-B*5701 testing to predict abacavir hypersensitivity. PLoS Curr 2010; 2: RRN1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schackman BR, Scott CA, Walensky RP, Losina E, Freedberg KA, Sax PE. The cost-effectiveness of HLA-B*5701 genetic screening to guide initial antiretroviral therapy for HIV. Aids 2008; 22: 2025–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A. SLCO1B1 polymorphisms and statin-induced myopathy. PLoS Curr 2013; 4: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfirevic A, Neely D, Armitage J, Chinoy H, Cooper RG, Laaksonen R et al. Phenotype standardization for statin-induced myotoxicity. Clin Pharmacol Ther 2014; 96: 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip VL, Marson AG, Jorgensen AL, Pirmohamed M, Alfirevic A. HLA genotype and carbamazepine-induced cutaneous adverse drug reactions: a systematic review. Clin Pharmacol Ther 2012; 92: 757–765. [DOI] [PubMed] [Google Scholar]
- Leckband SG, Kelsoe JR, Dunnenberger HM, George AL Jr, Tran E, Berger R et al. Clinical Pharmacogenetics Implementation Consortium guidelines for HLA-B genotype and carbamazepine dosing. Clin Pharmacol Ther 2013; 94: 324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudle KE, Thorn CF, Klein TE, Swen JJ, McLeod HL, Diasio RB et al. Clinical Pharmacogenetics Implementation Consortium guidelines for dihydropyrimidine dehydrogenase genotype and fluoropyrimidine dosing. Clin Pharmacol Ther 2013; 94: 640–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel A, Boisdron-Celle M, Fey L, Soulie P, Craipeau MC, Traore S et al. Clinical relevance of different dihydropyrimidine dehydrogenase gene single nucleotide polymorphisms on 5-fluorouracil tolerance. Mol Cancer Ther 2006; 5: 2895–2904. [DOI] [PubMed] [Google Scholar]
- Caudle KE, Klein TE, Hoffman JM, Muller DJ, Whirl-Carrillo M, Gong L et al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr Drug Metab 2014; 15: 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnenberger HM, Crews KR, Hoffman JM, Caudle KE, Broeckel U, Howard SC et al. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu Rev Pharmacol Toxicol 2015; 55: 89–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis NK, Innocenti F. Evidence required to demonstrate clinical utility of pharmacogenetic testing: the debate continues. Clin Pharmacol Ther 2014; 96: 655–657. [DOI] [PubMed] [Google Scholar]
- Janssens AC, Deverka PA. Useless until proven effective: the clinical utility of preemptive pharmacogenetic testing. Clin Pharmacol Ther 2014; 96: 652–654. [DOI] [PubMed] [Google Scholar]
- Ratain MJ, Johnson JA. Meaningful use of pharmacogenetics. Clin Pharmacol Ther 2014; 96: 650–652. [DOI] [PubMed] [Google Scholar]
- Annemans L, Redekop K, Payne K. Current methodological issues in the economic assessment of personalized medicine. Value Health 2013; 16 (Suppl): S20–S26. [DOI] [PubMed] [Google Scholar]
- Deverka PA, Vernon J, McLeod HL. Economic opportunities and challenges for pharmacogenomics. Annu Rev Pharmacol Toxicol 2010; 50: 423–437. [DOI] [PubMed] [Google Scholar]
- Haga SB, LaPointe NM. The potential impact of pharmacogenetic testing on medication adherence. Pharmacogenomics J 2013; 13: 481–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson WT, Formea CM. Clinical Perspective on the Clinical Pharmacogenetics Implementation Consortium Updated 2014 Guidelines for CYP2D6 and Codeine. Clin Chem 2015; 61: 319–321. [DOI] [PubMed] [Google Scholar]
- Deeks JJ, Higgins J. Statistical Algorithms in Review Manager 5, 2010. http://ims.cochrane.org/revman/documentation/Statistical-methods-in-RevMan-5.pdf (last accessed on 1 December 2015).
- Pagano M, Gauvreau K. Principles of Biostatistics. 2nd edn Brooks/Cole: Belmont, CA, 2000. [Google Scholar]
- Hetherington S, Hughes AR, Mosteller M, Shortino D, Baker KL, Spreen W et al. Genetic variations in HLA-B region and hypersensitivity reactions to abacavir. Lancet 2002; 359: 1121–1122. [DOI] [PubMed] [Google Scholar]
- Mallal S, Nolan D, Witt C, Masel G, Martin AM, Moore C et al. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet 2002; 359: 727–732. [DOI] [PubMed] [Google Scholar]
- Martin AM, Nolan D, Gaudieri S, Almeida CA, Nolan R, James I et al. Predisposition to abacavir hypersensitivity conferred by HLA-B*5701 and a haplotypic Hsp70-Hom variant. Proc Natl Acad Sci USA 2004; 101: 4180–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.