Abstract
Abnormalities of chromosome 17, recognised over two decades ago to be important in tumorigenesis, often occur in breast cancer. Changes of specific loci on chromosome 17 including ERBB2 amplification, P53 loss, BRCA1 loss, and TOP2A amplification or deletion are known to have important roles in breast-cancer pathophysiology. Numerical aberrations of chromosome 17 are linked to breast-cancer initiation and progression, and possibly to treatment response. However, the clinical importance of chromosome 17 anomalies, in particular the effect on ERBB2 protein expression, is unknown. Reports are conflicting regarding the association of copy gain of chromosome 17 (polysomy 17) with strong ERBB2 protein expression in the absence of true ERBB2 gene amplification. Copy-number anomalies in chromosome 17 seem to be common in tumours that show discrepant ERBB2 expression and in tumours with discordant ERBB2-protein and ERBB2 gene copy number measurements. The mechanisms of ERBB2 dosage changes—gene amplification versus chromosome gain and loss—probably differ in primary and metastatic disease; however, a correction for chromosome 17 copy-number is necessary to completely distinguish between these mechanisms. A better understanding of how polysomy 17 affects gene-copy number and protein expression will help to select patients who will respond to therapies targeting ERBB2 and other protein products of chromosome 17 loci.
Introduction
Changes in the number of individual whole chromosomes (aneusomy) seems to indicate genetic instability and was first proposed to cause tumorigenesis in 1902.1 Gains (polysomy) and losses (aneusomy) of specific chromosomes are common in breast cancer and are respectively associated with activation of oncogenes and inactivation of tumour-suppressor genes.2–4 Abnormalities in chromosome 17 are common in breast cancer,5 including whole chromosome and gene-copy-number anomalies, allelic losses, and structural rearrangements shown by conventional cytogenetic and molecular cytogenetic techniques.2,3,6,7 These chromosome abnormalities have been linked to mechanisms of breast-cancer pathophysiology including reduced apoptosis, unchecked proliferation, increased motility, and increased angiogenesis.4,8,9 Up to 93% of breast tumours have whole chromosome 17 copy-number changes.10 Analytical approaches with comparative genomic hybridisation and gene-expression profiling confirm the high proportion of whole and regional chromosome 17 changes in breast cancer.4,5,8,9 Distinct patterns of changes are associated with different clinicopathological features and gene-expression subtypes of breast cancer.11
Researchers debate the clinical importance of gain in copy number of chromosome 17 (polysomy 17) in breast cancer. The effect of polysomy 17 on expression of human epidermal-growth-factor receptor 2 (ERBB2) in ERBB2 non-amplified breast tumours is of particular interest (figure 1), as is its effect on treatment response to ERBB2-targeted therapies (eg, trastuzumab, lapatinib). Figures 1 and 2 show fluorescence in-situ (FISH) staining for ERBB2 (red signals) and chromosome 17 (green signals) in invasive breast cancer. The tumour in figure 1 is defined as ERBB2 non-amplified with chromosome 17 polysomy. A gain of signal is seen for the probe against the centromere of chromosome 17 (centromere enumerator probe 17 [CEP 17]). Although there seems to be a gain of signal for the probe against ERBB2, the ERBB2 signal is not increased with respect to the number of CEP 17 signals. About 97% of cells displayed three or more CEP17 signals with a resultant ERBB2:CEP17 ratio of 1·10. Figure 2 shows a gain of signal for ERBB2 and typically two signals per nuclei for chromosome 17. Thus, this figure shows a breast tumour with ERBB2 amplification and chromosome 17 disomy (about 5% of cells displayed 3 or more CEP17 signals with a resultant ERBB2:CEP17 ratio of 12·23).
Figure 1. Fluorescent in-situ hybridisation (FISH) detection of ERBB2 non-amplification and chromosome 17 polysomy in invasive breast cancer.
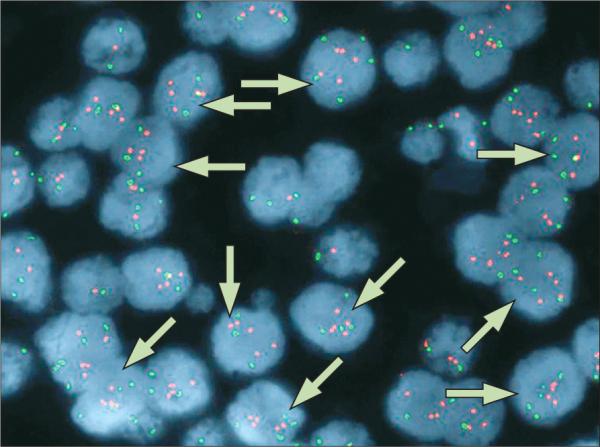
Red signals represent the detection of the ERBB2 gene and the green signals represent the detection of the centromere enumerator probe (CEP) for chromosome 17. Arrows indicate ERBB2 non-amplified, polysomic chromosome 17 nuclei.
Figure 2. FISH detection of ERBB2 amplification and chromosome 17 disomy in invasive breast cancer.
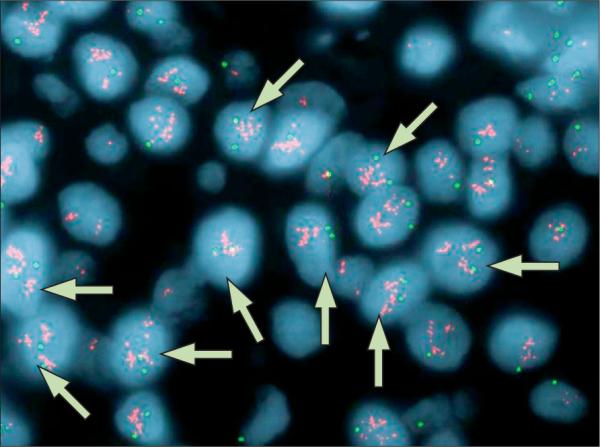
Red signals represent the detection of the ERBB2 gene and the green signals represent the detection of the centromere enumerator probe (CEP) for chromosome 17. Arrows indicate ERBB2 amplified, disomic chromosome 17 nuclei.
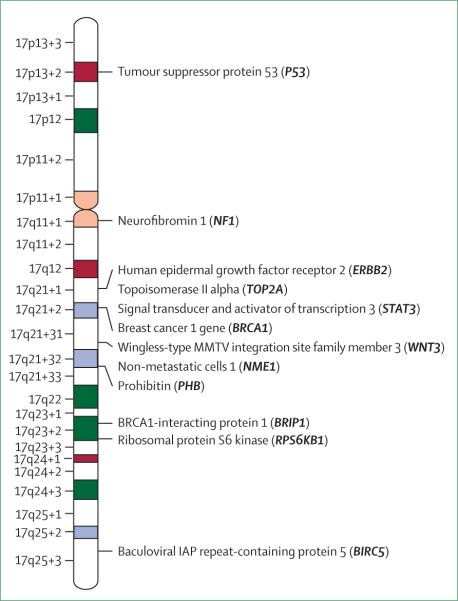
In addition to ERBB2, genes involved in breast-cancer pathophysiology that are located on chromosome 17 include tumour-suppressor genes P53 and BRCA1, and the gene for topoisomerase IIα (TOP2A; figure 3). These key genes are located in regions that are often deleted (P53, BRCA1, TOP2A) or amplified (ERBB2, TOP2A). Therefore, elucidating the effects of aneuploidy 17 copy-number changes will provide a better understanding of the role of genetic instability in breast cancer. For example, P53 is deleted in more than 50% of primary breast carcinomas,12–14 leading to inactivation of cellular tumour antigen P53 and genetic instability. Increased numbers of chromosome 17 and abnormal P53 expression have been observed simultaneously in breast-cancer cells, further evidence of an association between loss of P53 and genetic instability of chromosome 17.15 Additionally, about 40% of breast cancers have loss-of-heterozygosity (LOH) on the long arm of chromosome 17:12–14 the location of possible tumour-suppressor genes, including BRCA1, prohibition (PHB), non-metastatic cells 1 (NME1), and wingless-type MMTV integration site family member 3 (WNT3) (figure 3).
Figure 3. Ideogram of chromosome 17.
Genes important in breast cancer are indicated.
ERBB2 and TOP2A are in close proximity on chromosome 17 (figure 3) and copy number changes together in many tumours. TOP2A is either amplified or deleted, with equal probability, in nearly 90% of ERBB2-amplified primary breast tumours.16–18 By contrast, TOP2A copy-number anomalies are rare in ERBB2 non-amplified tumours (<7%).19,20 TOP2A deletion also affects tumours with polysomy 17, and so TOP2A deletion probably happens before polysomy.18,19 Furthermore, the presence of abnormalities in both ERBB2 and TOP2A might help to identify patients best suited to trastuzumab and anthracycline therapies.21–24 Preliminary findings suggest that polysomy 17 affects response to trastuzumab.25–28 Therefore, whole chromosome 17 copy-number anomalies might affect the clinical assessment and importance of ERBB2 and TOP2A amplification and protein expression.
Although polysomy in chromosome 17 is associated with several diseases and cancers, in this Review we focus on breast cancer. Because FISH is the prevailing technique to visualise and quantify chromosomal anomalies, we have primarily such data. FISH methods maintain tumour architecture and spatial relationships between cells, enabling investigation of the genetic heterogeneity of anomalies. We discuss the relevance of different cut-off points used to define chromosome 17 aneusomy and the implications of copy-number changes in breast carcinogenesis. Reports of the prevalence of aneusomy 17 and its association with prognostic factors in breast cancer are summarised. Finally, we discuss the emerging relevance of chromosome 17 aneusomy in response to anti-ERBB2 therapies.
Identification of aneusomy 17
Definitions of aneusomy 17 differ with the threshold criteria for monosomy and polysomy (tables 1 and 2).29–50 Setting of these thresholds is complicated not only by genomic heterogeneity and proliferative activity of tumours, but also by the substantial nuclear truncation resulting from tissue sectioning. Control specimens used to set these thresholds have included human lymphocytes,29,31,33,50 cholangiocarcinoma cell-line controls,31,51,52 benign breast-lesions or biopsies (eg, fibroadenomas, sclerosing adenosis),38,53 healthy breast tissue,30,32,36,40,54,55 and paired healthy breast tissue.36 Typically, 5–30% of cells displayed monosomy and less than 5% displayed polysomy in healthy (control) epithelium.10,56–58
Table 1.
Prevalence of chromosome 17 aneusomy in invasive breast cancer estimated by FISH analysis
| Breast material | Number of specimens | Number of nuclei counted | Disomy | Monosomy | Polysomy | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||
| Cutoff | % | Cutoff | % | Cutoff | % | Association with ERBB2 protein expression* | ||||
| Herrington et al (1995)29 | FNA | 49 | ≥100 | MS=2 | 40 | MS<2 | 5.0 | MS>2 | 55 | NR |
| Ichikawa et al (1996)30 | FNA | 80 | >200 | >80† | 48 | >15‡ | 14 | >20§ | 34 | NR |
| McManus et al (1999)31 | FNA | 69 | ≥100 | 2¶ | 26 | ≥20‡ | 0.0 | ≥10§ | 68 | NR |
| Tsukamoto et al (2001)32 | FNA | 113 | >100 | NR | 41 | >15‡ | 22 | >20§ | 37 | NR |
| Fehm et al (2002)33 | Touch prep | 74 | ≥100 | 2¶ | 41 | >15‡ | 11 | >6§ | 38 | NR |
| Nakopoulou et al (2002)34 | FFPE | 42 | 200–400 | >70† | 24 | >40‡ | 38 | >15§ | 38 | NR |
| Wang et al (2002)35 | FFPE | 189 | ≥60 | 1.76–2.25¶ | 49 | <1.76¶ | 2.6 | ≥2.26¶ | 49 | - |
| 2.26–3.75]∥ | 43 | - | ||||||||
| >3.76** | 5 | 3+†† | ||||||||
| Watters et al (2003)36 | FFPE | 214 | 60 | 1.35–1.85¶ | 46 | <1.35¶ | 7.5 | >1.86¶ | 47 | NR |
| Ma et al (2005)37 | FFPE | 893 | 60 | 1.5–2.25¶ | 49 | <1.5¶ | 8.9 | ≥2.26¶ | 42 | 3+ |
| 2.26–3.75∥ | 35 | - | ||||||||
| >3.76** | 7 | 3+ | ||||||||
| Merola et al (2006)38 | FFPE | 343 | 200 | 2¶ | 30 | 1¶ | 24 | >3¶ | 46 | 2+ |
| 2–4∥ | 40 | NR | ||||||||
| >4** | 5.8 | NR | ||||||||
| Takehisa et al (2007)39 | FNA | 40 | >100 | NR | 48 | >15‡ | 10 | >20§ | 43 | NR |
| Hofmann et al (2007)25 | FFPE | 95 | 60 | NR | NR | NR | NR | ≥3¶ | 27 | 3+ |
| Hyun et al (2008)40 | FFPE | 309 | ≥60 | 1.25–2.25¶ | NR | <1.25¶ | 1.3 | ≥2.26¶ | 32 | 3+ |
| 2.26–3.75∥ | 26 | NR | ||||||||
| >3.76** | 5.8 | NR | ||||||||
FNA=fine needle aspirate. FFPE=formalin-fixed paraffin-embedded. NR=not explicitly reported in reference. MS=mode chromosome 17 signal per nucleus. ..=no association found.
The association between polysomy 17 and ERBB2 expression (scored as 0–3+ staining intensity) in ERBB2 non-amplified tumours.
Percent of cells displaying 2 signals per nucleus.
Percent of nuclei with loss of centromeric region or entire chromosome (typically with 0 or 1 chromosome 17 signal per nucleus).
Percent of nuclei with gain of centromeric region or entire chromosome (typically >2 or ≥3 chromosome 17 signals per nucleus).
Average number of chromosome 17 signals per nucleus.
Low-level polysomy.
High-level polysomy.
Association was found between high polysomy (>3.76) and 10 tumours with ERBB2 immunohistochemical scores of 3+.
Table 2.
Prevalence of chromosome 17 aneusomy in invasive ERBB2 amplified* and non-amplified breast cancer estimated by FISH analysis
| Number of specimens | Disomy | Monosomy | Polysomy | Association with ERBB2 expression in ERBB2 non-amplified tumours | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||||||||
| Total | Amp* | Non-amp | Cutoff | Total N (%) | Amp N (%)* | Non-amp N (%) | Cutoff | Total N (%) | Amp N (%) | Non-amp N (%) | Cutoff | Total N (%) | Amp N (%) | Non-amp N (%) | ||
| Mezzelani et al (1998)41 | 58 | 24 | 28 | 2† | 20 (35) | 15 (63) | 5 (18) | 1† | 2 (3.4) | 1 (4.1) | 1 (3.6) | >2† | 5(8.6) | 4 (17) | 1 (3.6) | NR |
| Farabegoli et al (1999)42 | 79 | 19 | 60 | 2‡ | NR | NR | 15 (25) | 1‡ | NR | NR | 12 (20) | ≥3‡ | NR | NR | 33 (55) | NA |
| Jimenez et al (2000)43 | 34 | 10§ | 15§ | >80 | 14 (41) | 4 (40) | 9 (60) | >80∥ | 3 (8.8) | 1 (10) | 1 (6.7) | >80** | 5 (15) | 1 (10) | 1 (6.7) | NA |
| Bose et al (2001)44 | 74 | 8 | 66 | 2‡ | 46 (62) | 1 (13) | 45 (68) | 0–1‡ | 3 (4.1) | 0 (0) | 3(4.5) | 3–4‡ | 25 (34) | 7 (88) | 18 (27) | 3+ |
| Tubbs et al (2001)45 | 145 | 29 | 116 | NR | 92 (63)†† | 12 (41) | 80 (69) | NR | NR | NR | NR | NR | 53 (37) | 17 (59) | 36 (31) | NR |
| Varshney et al (2004)46 | 687 | 78 | 609 | NR | NR | NR | NR | NR | NR | NR | NR | ≥3‡ | 71 (10) | 30 (39) | 41 (6.7) | 3+ |
| Salido et al (2005)47 | 175 | 80†† | 95 | NR | 153 (87)†† | 72 (90) | 81 (85) | NR | 3 (1.7) | NR | NR | ≥3‡ | 22 (13) | 14 (18) | 8 (15) | 2–3+ |
| Downs-Kelly et al (2005)48 | 727 | 134 | 593 | NR | NR | NR | NR | NR | NR | NR | NR | >2.1‡ | 257 (35) | 54 (40) | 203 (34) | NA |
| Beser et al (2007)19 | 50 | 11 | 39 | 1.35–1.85‡ | 35 (70) | 9 (82) | 26 (67) | <1.35‡ | 6 (12) | 0 (0) | 6 (15) | >1.86‡ | 9 (18) | 2 (18) | 7 (18) | NR |
| Torrisi et al (2007)49 | 457 | 54 | 403 | NR | NR | NR | NR | NR | NR | NR | NR | ≥3‡ | 77 (17) | 9 (17) | 68 (17) | NR |
| Merola et al (2006)38 | 343 | 101 | 242 | 2‡ | 102 (30) | NR | NR | 1‡ | 83 (24) | 49 (49) | 34 (14) | >3‡ | 158 (46) | 24 (24) | NR | 2+ |
| Reinholz et al (2007)28 | 1888 | 1488 | 156 | Loss ≤60∥ | 625 (33) | 544 (37) | 81 (52) | >60∥ | 89 (4.7) | 79 (5.3) | 5 (3.2) | >30** | 935 (50) | 865 (58) | 70 (36) | NR |
| Gain ≤30** | ||||||||||||||||
| Vanden Bempt et al (2008)50 | 226 | 97§§ | 126 | NR | NR | NR | NR | NR | NR | NR | NR | >3‡ | 104 (46) | 42 (43) | 62 (49) | NA |
Amp=ERBB2 amplification. Non-amp=ERBB2 non-amplification. N=number of specimens. NR=not explicitly reported in reference. NA=ERBB2 expression was not associated with polysomy 17.
ERBB2 amplification defined as ERBB2/CEP17 ratio ≥2.0 unless otherwise indicated.
Disomy defined as 2 ERBB2 gene signals related to 2 chromosome 17 signals was found in all cells; monosomy defined as ERBB2 signals <chromosome 17 signals; polysomy defined as equal numbers of both signals and ratio >2.
Average number of chromosome 17 signals per nucleus.
ERBB2 amplification defined as: >80% with >5 ERBB2 signals per nucleus; ERBB2 non-amplification: >80% with ≤2 signals per nucleus. Percent of nuclei with 2 chromosome 17 signals per cell.
Percent of nuclei with loss of centromeric region or entire chromosome (typically with 0 or 1 chromosome 17 signal per cell).
Percent of nuclei with gain of centromeric region or entire chromosome (typically >2 or ≥3 chromosome 17 signals per cell).
Inferred from literature to be disomic.
‡‡ ERBB2 amplification defined by mode ERBB2 signal or mode chromosome 17 signal ≥2.
ERBB2 amplification defined as ERBB2/CEP17 ratio >2.2.
Chromosome 17 aneusomy is typically identified with one of two methods. The first method finds the mean number of centromere-17 signals per control cell and defines aneuploidy as more than 3 standard deviations (SD) from this value. In the second method, the abnormal range is determined by a pre-specified proportion of nuclei displaying a prespecified number of abnormal centromeric signal counts. For each method, different signal counts have been used as cut-offs, leading to large differences in the reported incidences of chromosome 17 aneusomy (tables 1 and 2).29–50
Because the definition of chromosome-17 aneusomy is controversial, we analysed FISH data from about 11000 breast tumours (7400 ERBB2 non-amplified and 3200 ERBB2 amplified specimens) from the clinical ERBB2 database maintained by the cytogenetics laboratory at Mayo Clinic, Rochester, MN, USA (Schroeder MJ, Jenkins RB, unpublished data). We classified specimens by either the proportion of nuclei with three or more centromere-17 signals (to validate our polysomy 17 cutoff) or with 1 centromere-17 signal (to validate our monosomy 17 cut-off). We compared the distribution of specimens with these classifications with the distribution of specimens classified by the average number of centromere-17 signals. For both ERBB2-non-amplified and ERBB2-amplified specimens, we observed two populations separated at threshold of either greater than 30% nuclei with three or more centromere-17 signals or an average of 2·2 centromere-17 copies per cell, thus, distinguishing normal and polysomic cases. We also observed that two populations were separated at thresholds of either greater than 60% nuclei with one or more centromere-17 signals or an average of 1·4 centromere-17 copies per cell, thus, distinguishing normal and monosomic cases. These cut-offs should be investigated and validated in other clinical trials with ERBB2-targeted therapies.
Aneusomy 17 and breast cancer
Several studies have examined the prevalence of changes in copy number of chromosome 17 in invasive breast cancer (tables 1 and 2).29–50 Monosomy 17 is observed less often than polysomy 17 (typically less than 15% and greater than 35%, respectively), but the prevalence for both types of aneusomy vary greatly between studies. Reported incidences range from 0–38% for monosomy 17 and 8–68% for polysomy 17 (tables 1 and 2).29–50 Several studies further divided polysomy 17 into low-level and high-level polysomy and observed that low-level polysomy 17 was the more common of the two (26–43% vs 5–7%; table 1).25,35–38,40,44 The range in prevalence values is a result of different types of material examined, different selection criteria (eg, ERBB2 immunohistochemical scores), and the varying methods used to define thresholds of disomy, monosomy, and polysomy as discussed above.
Aneusomy 17 in breast-cancer progression
The understanding that genetic instability can drive tumorigenesis prompted descriptive studies that investigated genetic changes associated with early breast-neoplasia progression. Both gains and losses of chromosome 17 happen in all stages of breast cancer, including non-invasive (proliferative lesions), preinvasive (ductal carcinoma in situ [DCIS] and lobular carcinoma in situ [LCIS]), and invasive breast disease (invasive ductal carcinomas [IDC]; table 3).43,51,55–58,60 Proliferative lesions were characterised mainly by borderline chromosome losses, whereas advanced lesions (LCIS, DCIS, and IDC) were characterised by unequivocal losses and gains. The role of aneusomy 17 in non-invasive disease is supported by a study that found copy-number changes of chromosome 17 in 25 of 32 women with non-proliferative epithelium or hyperplasia with no evidence of invasive disease.59
Table 3.
Prevalence of aneusomy 17 in breast-cancer progression estimated by FISH analysis
| Breast tissue | Number of specimens | Number of nuclei counted | Disomy | Monosomy | Polysomy | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||||
| Cut-off | % | N | Cut-off | % | N | Cut-off | % | N | ||||
| Micale et al (1994)56 | Proliferative lesion | 8 | 200–400 | Loss ≤45* | 50 | 4 | >45* | 50 | 4 | >10† | 0 | 0 |
| Ductal carcinoma in situ | 1 | Gain ≤10† | 0 | 0 | 0 | 0 | 100 | 1 | ||||
| Lobular carcinoma in situ | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Invasive ductal carcinoma | 1 | 0 | 0 | 0 | 0 | 100 | 1 | |||||
| Tubular carcinoma | 1 | 0 | 0 | 100 | 1 | 0 | 0 | |||||
| Murphy et al (1995)60 | Ductal carcinoma in situ | 3 | 200 | ≥60‡ | NR | <2§ | NR | >10† | 100 | 3 | ||
| Invasive ductal carcinoma | 3 | NR | NR | NR | ||||||||
| Visscher et al (1996)57 | Proliferative lesion | 6 | 200–300 | Loss ≤40* | NR | >40* | 17 | 1 | >10† | 0 | 0 | |
| Ductal carcinoma in situ | 10 | Gain ≤10† | NR | 50 | 5 | 20 | 2 | |||||
| Lobular carcinoma in situ | 9 | NR | 67 | 3 | 0 | 0 | ||||||
| Mendelin et al (1999)58 | Ductal carcinoma in situ | 12 | 200–300 | Loss ≤40* | 34¶ | 17¶ | >40* | 16¶ | 8¶ | >20† | 50¶ | 25¶ |
| Paired invasive componentg | 12 (50 hybridisations)¶ | Gain ≤10† | 12¶ | 6¶ | 10¶ | 5¶ | 78¶ | 39¶ | ||||
| Visscher et al (2000)51 | Ductal carcinoma in situ | 28 (143 hybridisations)¶ | 200–300 | Loss ≤40* | 35¶ | 50¶ | ≥40* | 19¶ | 27¶ | >20† | 46¶ | 66¶ |
| Gain ≤10† | ||||||||||||
| Jimenez et al (2000)43 | Ductal carcinoma in situ | 7 | 100–300 | >80‡ | 29 | 2 | >80* | 0 | 0 | >80‡ | 14 | 1 |
| 57∥ | 4∥ | |||||||||||
| Marinho et al (2000)55 | Proliferative lesion | 9 | ≥200 | NR | 100 | 9 | >37.5* | 0 | 0 | >10† | 0 | 0 |
| Ductal carcinoma in situ | 11 | NR | NR | 46 | 5 | 46 | ||||||
| Invasive ductal carcinoma | 16 | 38 | 6 | 50 | 8 | |||||||
N=number of specimens. NR=Not explicitly reported in reference.
Percent of nuclei with loss of centromeric region or entire chromosome (typically with 0 or 1 chromosome 17 signals per cell).
Percent of nuclei with gain of centromeric region or entire chromosome (typically >2 or ≥3 chromosome 17 signals per cell).
Percent of cells displaying 2 signals per nucleus.
Average number of chromosome 17 signals per nucleus.
Prevalence based on hybridisations.
Heterogeneous chromosome 17 copy number (tumour cells contained variable number of chromosome 17 copies per nucleus).
** Invading cells were compared to residual in-situ population in the 12 breast carcinomas studied.
The presence and extent of chromosome aneuploidy differed substantially between neoplastic cells from the invasive component of a breast carcinoma and cells of the residual preinvasive population.51,58 Furthermore, intraductal carcinomas associated with invasive neoplasms showed a greater extent of chromosomal aneusomies than did DCIS without an invasive component.51 Chromosomal instability might correlate (perhaps causally) with progression of DCIS to invasive growth, suggesting that genetic instability is a pattern that affects the likelihood of progression of early breast carcinoma.51 Paired DCIS and invasive specimens have common and unique genetic changes, suggesting clonal diversity within the same tumours.54,60 Indeed, distinctive, but overlapping patterns of genetic instability are found in primary breast-tumours and adjacent uninvolved parenchyma.10
Monosomy 17 seems to be more widespread than polysomy 17 in non-invasive and low grade in-situ carcinomas (tables 3 and 4).43,51,55–58,60 Loss of chromosome 17 has been observed in hyperplasia and malignant lesions but not in corresponding healthy tissue, suggesting that hyperplasia might be clinically relevant in breast-cancer development.56 Additionally, monosomy is more common than polysomy in LCIS, suggesting that subsets of preinvasive breast neoplasia have divergent patterns of genetic instability.56,57 Monosomy of chromosomes 7, 8, 16, and 17 is more common in grade I DCIS than in grade III DCIS tumours (29% [9/31 hybridisations] vs 4% [2/49 hybridisations]).51
Table 4.
Summary of aneusomy 17 in breast-cancer progression (combination of study results from table 3)
| Specimens (N)* | Disomy | Monosomy | Polysomy | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||
| NNR | N | %† | NNR | N | %† | NNR | N | %† | ||
| Proliferative lesion | 23 | 6 | 13 | 76 | 0 | 5 | 22 | 0 | 0 | 0 |
| Ductal carcinoma in situ | 32 | 24 | 2 | 25 | 3 | 10 | 34 | 0 | 16 | 50 |
| Lobular carcinoma in situ | 10 | 9 | 0 | 0 | 0 | 3 | 30 | 0 | 0 | 0 |
| Invasive carcinoma | 21 | 19 | 0 | 0 | 3 | 7 | 39 | 3 | 9 | 50 |
N=number of specimens. NR=not explicitly reported in reference.
Percent hybridisations from table 3 and associated tumours are not included.
Does not include NR specimens.
Polysomy 17 is often seen in DCIS (tables 3 and 4),43,51,54,57,58,60 and the pattern of chromosomal gain differs between healthy and DCIS tissues.60 Visscher and colleagues51 noted polysomy 17 in 88% (43/49 hybridisations) of grade III DCIS compared with none in grade I DCIS. However, neoplasms of grade II DCIS had varied chromosome aneuploidy: disomy in 38% (24/63 hybridisations), monosomy in 26% (16/63 hybridisations), and polysomy in 36% (19/63 hybridisations) of specimens. The presence of multiple aneuploidy patterns in DCIS supports the notion that diverse mechanisms of genetic alteration are involved in the development of breast cancer.
The identification of copy-number changes in lesions that are potentially premalignant supports the classification of these lesions as biologically premalignant. Moreover, aneusomy 17 might be an intermediate biomarker of breast tumorigenesis and help to detect patients at high risk who might gain from preventive action. The overall goal is to elucidate the multistep mechanism of breast carcinogenesis. In specimens of preinvasive and early invasive breast-cancer lesions, associating tumour subtype and allelotype with specific chromosome copy-number changes, gene mutations, and gene expression will help.
Aneusomy 17 in invasive breast cancer
Polysomy 17 and ERBB2 amplification
The development of trastuzumab, an ERBB2-targeted antibody, and findings that ERBB2 overexpression and gene amplification often predict its benefit, prompted numerous investigations of the relation between chromosome 17 monosomy and polysomy, ERBB2 amplification and non-amplification, and ERBB2 expression in invasive breast cancer (table 2).19,28,38,41–50 Reported prevalences for chromosome 17 monosomy were typically less than 15%, irrespective of ERBB2 amplification. Two studies did not find monosomy in any ERBB2 amplified tumours,19,44 whereas another group reported a prevalence of 49% (49/101).38 Chromosome 17 polysomy was usually more prevalent in tumours with ERBB2 amplification (10% [1/10]–88% [7/8]) than in tumours without ERBB2 amplification (3·6% [1/28]–55% [33/60]). In our N9831 clinical trial,28 we observed polysomy 17 in 58% (865/1488) of ERBB2 amplified tumours and in 36% (70/156) of ERBB2 non-amplified tumours.
Polysomy 17 and ERBB2 expression in the absence of ERBB2 amplification
Because ERBB2 overexpression without gene amplification has been observed in up to 10% of breast tumours, several studies assessed the association between chromosome 17 polysomy and ERBB2 expression in tumours without ERBB2 amplification (tables 1 and 2).29–50 Findings were contradictory. Many studies suggest that, at least in a subset of breast carcinoma, increases in ERBB2 copy number that result from polysomy 17 can lead to protein overexpression in the absence of ERBB2 amplification.37,38,40,44,46,47,61 Polysomy 17 is more common in non-amplified tumours with overexpression of ERBB2 (immunohistochemical [IHC] scores of 3+) than in tumours with no or low ERBB2 expression (IHC scores of 0–1+).37,46,61 In our N9831 study,28 among 156 patients with ERBB2 non-amplified tumours, there is an association between polysomy 17 and ERBB2 expression. Breast tumours scored as IHC 2–3+ were more likely to be polysomic for chromosome 17 than tumours scored as 0–1+ (p<0·001). High polysomy (four or more chromosome 17 signals per nucleus) seems to be more strongly associated between ERBB2 overexpression (immunohistochemical score of 3+) and chromosome 17 copy-number than low polysomy (fewer than four chromosome 17 signals per nucleus).35
Conversely, weak associations between ERBB2 expression and ERBB2 copy number have been observed in other studies of non-amplified tumours.62,63 Several studies have found that in the absence of ERBB2 gene amplification, high polysomy 17 was not associated with ERBB2 protein or mRNA overexpression.12,36,42,43,49,64 Molecular detection techniques (eg, reverse transcription PCR and isotopic in-situ hybridisation) showed that ERBB2 mRNA expression was not increased in nonamplified breast tumours with polysomy 17, and that amplification of ERBB2 resulted in increased ERBB2 expression, independent of chromosome 17 polysomy.48,50,64–66 Therefore, in the absence of amplification, polysomy 17 does not seems to result in increased expression of ERBB2 mRNA. In summary, whether chromosome 17 polysomy can cause of ERBB2 overexpression in the absence of true ERBB2 amplification is unclear.
Polysomy 17 might cause slight ERBB2 expression (IHC 2+) in instances of gene amplification with FISH ratio 2–4 or 4–6 ERBB2 copies.38,44,47 An additive effect on gene dosage and protein expression has been seen in scenarios of high polysomy 17 (≥4 chromosome 17 signals per nuclei) with gene duplication or modest gene amplification (ERBB2/CEP17 ratio 2–3).35,43 Furthermore, many patients with IHC 2+ did not have gene amplification but had chromosome 17 polysomy.35,67,68 Polysomy 17 has been reported in 41–86% of ERBB2 non-amplified tumours scored as IHC 2+ or 3+.37,38,40,42,44,46
Association of polysomy 17 with prognostic factors
Genomic aberrations recurrent in a specific type of cancer can be important prognostic markers for tumour progression. Because chromosome 17 copy-number alterations have been repeatedly identified in preinvasive and invasive lesions (table 3),43,51,55–58,60 aneusomy 17 is a predictor of cancer aggressiveness.
Table 5 lists studies of associations between common pathological characteristics and aneusomy of chromosome 17. Commonly examined characteristics were tumour grade, nodal metastasis, and hormone receptor status. In general, high tumour grade was associated with polysomy 17.10,12,31,36,39,40,51,69 However, other reports did not find this relation.29,33,34,47,50,64 Many investigations have shown a link between polysomy 17 and lymph node metastasis,32,47,69 although this is not always true.10,36,31,50 Monosomy 17 also has been associated with nodal metastasis.34 The association between aneusomy 17 and hormone-receptor status is controversial. Studies have shown that both monosomy and polysomy were associated with oestrogen-receptor negativity.32,34,36 Results are also inconsistent regarding links between polysomy 17 and oestrogen-receptor positivity12,39,47,50 and tumour size.12,29,31,33,50,64 Polysomy 17 was found more often in invasive ductal carcinomas than in invasive lobular carcinomas by use of chromogenic in-situ hybridisation.70 Overall, it seems that aberrations of chromosome 17 copy-number are associated with indicators of poor prognosis in certain groups of patients with breast cancer, and that these associations might be related to differences in the ERBB2 amplification status of the tumour.
Table 5.
Aneusomy 17 and association with clinicopathological characteristics
| Number of specimens | Nottingham prognostic index | Tumour grade | Tumour histology | Tumour size | Tumour nodal status | Oestrogen receptor | Progesterone receptor | Survival | |
|---|---|---|---|---|---|---|---|---|---|
| Herrington et al (1995)29 | 49 | NR | NA | NR | NA | CI | NR | NR | NR |
| Persons et al (1996)21 | 55 | NR | Y, p17 | NR | Y, p17 | NR | NA | NA | NR |
| Ichikawa et al (1996)30 | 106 | NR* | NR | NR | NR | Y, a17 | NR | NR | NR |
| Adeyinka et al (1999)69 | 16 | NR | Y† | NR | NR | Y† | NR | NR | NR |
| McManus et al (1999)31 | 69 | NR | Y, p17 | Y, p17 | NA | NA | NR | NR | NR |
| Botti et al (2000)10 | 28 | NR | Y, p17 | NR | NR | NA | NR | NR | NR |
| Visscher et al (2000)51 | 28 | NR | Y, a17 | NR | NR | NR | NR | NR | NR |
| Tsukamoto et al (2001)32 | 113 | NR | NR | NR | NR | Y, p17 | m17/ER- | p17/PR- | NR |
| Fehm et al (2002)33 | 74 | NR | NA | NR | Y, p17 | NA | NA | m17/PR- | NR |
| Nakopoulou et al (2002)34 | 42 | NR | NA | NA | Y | Y, m17 | m17/ER- | NR | Y, m17 |
| Watters et al (2003)36 | 214 | Y, p17 | Y, p17 | NR | NR | NA | p17/ER- | NR | NA |
| Salido et al (2005)47 | 175 | NR | NA | NR | NR | Y, p17 | NA | NA | NR |
| Dal Lago et al (2006)64 | 893 | NR | NA | NR | NA | NR | NA/ER+ | NR | NR |
| Takehisa et a (2007)39 | 42 | NR | Y, p17 | NR | NR | NR | Y, p17 | NR | NR |
| Hyun et al (2008)40 | 309 | NR | Y, p17 | NR | NR | NR | NR | NR | NR |
| Vanden Bempt et al (2008)50 | 226 | NA | NA | NA | NA | NA | NA | NA | Trend, p17 |
N=number of specimens. NR=not reported in reference. CI=chromosome imbalance. NA=characteristic examined in study but not associated with chromosome 17 copy number status. Y=association exists with chromosome 17 aneusomy (a17), polysomy (p17), or monosomy (m17) as indicated. ER−=oestrogen receptor-negativity. ER+=oestrogen receptor-positivity. PR−=progesterone receptor negativity. PR+=progesterone receptor positivity.
No correlation with tumour stage (using TNM tumour size, nodal status, metastasis staging criteria).
Associated with polysomy of multiple chromosomes.
Chromosome 17 copy-number correction
Clinicians are interested in aneusomy 17 because of its possible effect on classification of ERBB2 status (ie, interpretation of ERBB2 testing), especially for tumours with differing protein and gene measurements. Results from several studies show that polysomy 17 is regularly seen in tumours with discrepant ERBB2 protein and gene copy number measurements.28,35,45,46,50,61,62 Polysomy 17 (especially highly polysomic cases) seems to cause the inconsistency in ERBB2 amplification defined by gene-copy number versus amplification defined by the ratio of gene-copy number to chromosome-copy number.37,50 Furthermore, in most ERBB2-positive examples (positive by either ERBB2 protein overexpression [immunohistochemical score of 3+] or ERBB2 gene amplification [ERBB2/CEP17 ratio ≥2]), ERBB2 overexpression results from gene amplification independent of polysomy 17—although polysomy 17 is often found with ERBB2 amplification. However, in another class of ERBB2-positive tumours ERBB2 overexpression happens in the absence of ERBB2 amplification, and in this instance protein overexpression might result from deregulated gene transcription and not from extra copies of chromosome 17.50,68,71 Chromosome correction is necessary to accurately identify these rare tumours that likely have different biological characteristics than tumours with ERBB2 amplification.50
Reports indicate chromosome correction (chromosome copy number normalisation) as the best method to adjust for ERBB2/neu pseudoamplification due to chromosome 17 polysomy.45,50,71 Inclusion of the probe for the centromere gives reassurance that the chromosome bearing the aberrant gene is still being detected in truncated nuclei (during tissue sectioning), and the probe serves as an independent positive control for hybridisation reaction.45 Therefore, the copy number of chromosome 17 should be routinely examined to show technical validity and to help distinguish between low and high ERBB2 amplification. Chromosome correction will then help differentiate a subgroup of patients that probably have genetic and clinical differences.50,71 These differences will affect patient selection for ERBB2-targeted therapies and the efficacy of therapy.
Chromosome 17 and prediction of therapeutic response
In the last 3 years, investigations have begun on associations between aneusomy of chromosome 17 and trastuzumab benefit.25–28 Preliminary findings suggest that patients with metastatic breast cancer with ERBB2 amplification and chromosome 17 monosomy did not respond to trastuzumab.27 Our results from N9831 further suggest that patients with primary breast cancer with ERBB2/CEP17 ratios greater than 15, most of whom displayed monosomy 17, did not benefit from adjuvant trastuzumab (hazard ratio 1·01).28 This implies that aneusomy 17 might be more important in tumours with ERBB2 amplification (ratio greater than or equal to 2) and monosomy 17 (although rare), if gene dosage is the main mechanism of protein overexpression. For patients with high ERBB2/CEP17 ratios and monosomy 17, precautions should be taken and absolute gene copy number might be unimportant because ratio alone might not be a reliable indicator of ERBB2 status.
Conflicting results have come from studies that addressed the question: in tumours with ERBB2 overexpression but without ERBB2 amplification, does polysomy 17 predict trastuzumab responsiveness?25,26,28 Some researchers hypothesised that polysomy 17 might not have predictive value for trastuzumab therapy and only tumours with true gene amplification respond to trastuzumab therapy.71 Findings from two studies25,26 suggested that that polysomy of chromosome 17 was associated with ERBB2 overexpression in absence of ERBB2 amplification, indicating that polysomy 17 possibly can be used in clinical assessment of ERBB2 status and treatment prediction for anti-ERBB2 therapies. Three of seven patients with non-amplified tumours and ERBB2 IHC 3+ scores responded in the WO16229 trastuzumab trial,25 and two of these patients had polysomy 17. A subset analysis of the CALGB 9840 trial26 suggested that patients who were FISH-negative and had polysomy 17 (defined as greater than or equal to 2·2 centromere 17 signals) possibly responded to trastuzumab (p=0·048) but did not have significantly longer progression-free and overall survival. However, a preliminary report from EGF3000172 found that polysomy 17 did not predict response to lapatinib; the median progression-free survival was not significantly different between and within treatment arms based on polysomy 17.73 These results could be interpreted to mean that polysomy 17 does not predict anti-ERBB2 treatment response, or that polysomy 17 is clinically important in the metastatic setting, but not in the adjuvant setting. In our N9831 adjuvant trastuzumab trial,28 we reported a benefit from trastuzumab for patients with ERBB2 amplified tumours (ratio ≥2·0) with either polysomy 17 (hazard ratio 0·52) or normal chromosome 17 copy-number (0·37). Also, the 423 patients who received chemotherapy alone and had ERBB2 amplified and polysomy 17 tumours had a longer 5-year disease-free survival rate (78%) than the 282 patients who received chemotherapy alone and had ERBB2 amplified and disomy 17 tumours (68%; p=0.04). Furthermore, our exploratory analyses showed that 5-year disease-free survival rate for patients treated with trastuzumab with ERBB2 normal (non-amplified and IHC 0–2+) tumours was 66% for those with polysomy 17 and 84% for disomy backgrounds.28 Because there are so few patients with polysomy 17 in reported studies, the response of polysomic, ERBB2-non amplified but IHC-positive tumours to trastuzumab therapy needs further investigation. Gains and losses of chromosome 17 (and other chromosomes or regions) might be biologically and clinically relevant for reasons other than responsiveness to anti-ERBB2 therapy, warranting further subset studies of large trials. At this time, we do not recommend using polysomy 17 in treatment decisions for ERBB2-directed therapies.
Conclusion
Aneuploidy is an indication of genetic instability and might deregulate global gene transcription in cancer cells, either by driving or inhibiting tumorigenesis.74 Accumulation of genomic and epigenomic aberrations enables the development of breast cancer pathophysiology. Discovery of recurrent aberrations and the genes that are deregulated by these aberrations will aid in understanding the mechanisms of cancer formation and progression and guide improvements in cancer diagnosis and treatment.
Abnormalities of chromosome 17 result in key changes in genes including ERBB2, BRCA1, P53, and TOP2A. These changes are known to have an important role in breast-cancer pathophysiology. Whole chromosome 17 copy-number alterations are also common in breast cancer, but their clinical relevance is much less defined. This is partly because of the different criteria used for classifying aneusomy (monosomy and polysomy) of chromosome 17. Non-standardised criteria also explains the wide range of incidences of aneusomy 17 reported and why conflicting evidence exists for the role of polysomy 17 in ERBB2 expression in ERBB2 non-amplified breast tumours.
Aneusomy of chromosome 17 is observed in all stages of breast carcinogenesis and is an indicator of poor prognosis. Chromosome 17 monosomy is more common in non-invasive and preinvasive cancers than in invasive breast lesions. By contrast, chromosome 17 polysomy is more common in invasive than in non-invasive and preinvasive breast lesions. Polysomy of chromosome 17 is also common in cases with equivocal ERBB2 protein expression and ERBB2 gene amplification as well as in cases with discrepant ERBB2 protein and gene copy number measurements.
Furthermore, ERBB2 overexpression in invasive breast cancer typically results from ERBB2 amplification independent of polysomy 17. Polysomy 17 should be distinguished from true ERBB2 amplification by use of chromosome correction. Tumours with polysomy 17 seem to be more similar to ERBB2-negative than to ERBB2-positive tumours.50 Polysomy 17 in the absence of ERBB2 amplification has not been associated with clinical characteristics of ERBB2-positive breast cancer.50,71 Chromosome 17 aneusomy might have different roles in prediction of anti-ERBB2 treatment response for primary versus metastatic breast cancers. Identification of chromosome 17 polysomy might be important when planning treatment approaches targeting other amplicons or genes located on chromosome 17. Finally, it is essential to optimise staining, normalise for chromosome 17 copy-number, and standardise criteria for clinical assessment and interpretation of ERBB2 amplification. Investigators who do FISH should be aware of aneusomy and do careful studies to validate criteria for chromosome 17 gain and loss,75 including comparison with healthy breast specimens from the same sample if available. Wide ranges exist in ERBB2 expression using different analytes (ie, DNA, RNA, and protein), suggesting the importance of other factors such as transcription and translation regulation, tumour-cell heterogeneity, preanalytic variability, or assay variability. Overall, the integration of molecular cytogenetics, whole-genome screening, and gene-expression profiling will allow for more detailed investigation of the mechanisms of chromosome aneusomy in protein expression, gene amplification, and breast-cancer pathophysiology. This approach, combined with a review of existing clinical trial data, will lead to improved assessment of ERBB2 status, more accurate selection of patients for targeted therapy, and better outcomes.76
Search strategy and selection criteria.
Data for this Review were identified by searches of Medline, Current Contents, and PubMed using the search terms (alone and in combination): “polysomy 17”, “breast cancer”, “polysomy 17 and trastuzumab”, and “therapy response”. References from relevant articles, abstracts and reports from meetings were included only when they related directly to the scope of this Review. Articles published in English between January, 1980, and November, 2008, were included.
Acknowledgments
This work was supported by the National Cancer Institute grants, CA25224 and CA114740, awarded to the North Central Cancer Treatment Group.
Footnotes
Conflicts of interest The authors declared no conflicts of interest.
References
- 1.Boveri T. On multipolar mitosis as a means of analysis of the cell nucleus. Develop Biol. 1902;35:67–90. [Google Scholar]
- 2.Geleick D, Muller H, Matter A, Torhorst J, Regenass U. Cytogenetics of breast cancer. Cancer Genet Cytogenet. 1990;46:217–29. doi: 10.1016/0165-4608(90)90107-l. [DOI] [PubMed] [Google Scholar]
- 3.Thompson F, Emerson J, Dalton W, et al. Clonal chromosome abnormalities in human breast carcinomas. I. Twenty-eight cases with primary disease. Genes Chromosomes Cancer. 1993;7:185–93. doi: 10.1002/gcc.2870070402. [DOI] [PubMed] [Google Scholar]
- 4.Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–41. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Yao J, Weremowicz S, Feng B, et al. Combined cDNA array comparative genomic hybridization and serial analysis of gene expression analysis of breast tumor progression. Cancer Res. 2006;66:4065–78. doi: 10.1158/0008-5472.CAN-05-4083. [DOI] [PubMed] [Google Scholar]
- 6.Nagai MA, Medeiros AC, Brentani MM, et al. Five distinct deleted regions on chromosome 17 defining different subsets of human primary breast tumors. Oncology. 1995;52:448–53. doi: 10.1159/000227509. [DOI] [PubMed] [Google Scholar]
- 7.Shackney SE, Singh SG, Yakulis R, et al. Aneuploidy in breast cancer: a fluorescence in situ hybridization study. Cytometry. 1995;22:282–91. doi: 10.1002/cyto.990220404. [DOI] [PubMed] [Google Scholar]
- 8.Chin SF, Wang Y, Thorne NP, et al. Using array-comparative genomic hybridization to define molecular portraits of primary breast cancers. Oncogene. 2007;26:1959–70. doi: 10.1038/sj.onc.1209985. [DOI] [PubMed] [Google Scholar]
- 9.Fridlyand J, Snijders AM, Ylstra B, et al. Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer. 2006;6:96. doi: 10.1186/1471-2407-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Botti C, Pescatore B, Mottolese M, et al. Incidence of chromosomes 1 and 17 aneusomy in breast cancer and adjacent tissue: an interphase cytogenetic study. J Am Coll Surg. 2000;190:530–39. doi: 10.1016/s1072-7515(00)00252-0. [DOI] [PubMed] [Google Scholar]
- 11.Bergamaschi A, Kim YH, Wang P, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45:1033–40. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- 12.Persons DL, Robinson RA, Hsu PH, et al. Chromosome-specific aneusomy in carcinoma of the breast. Clin Cancer Res. 1996;2:883–88. [PubMed] [Google Scholar]
- 13.Sato T, Tanigami A, Yamakawa K, et al. Allelotype of breast cancer: cumulative allele losses promote tumor progression in primary breast cancer. Cancer Res. 1990;50:7184–89. [PubMed] [Google Scholar]
- 14.Varley JM, Brammar WJ, Lane DP, Swallow JE, Dolan C, Walker RA. Loss of chromosome 17p13 sequences and mutation of p53 in human breast carcinomas. Oncogene. 1991;6:413–21. [PubMed] [Google Scholar]
- 15.Sigurdsson S, Bodvarsdottir SK, Anamthawat-Jonsson K, et al. p53 abnormality and chromosomal instability in the same breast tumor cells. Cancer Genet Cytogenet. 2000;121:150–55. doi: 10.1016/s0165-4608(00)00260-0. [DOI] [PubMed] [Google Scholar]
- 16.Jarvinen TA, Liu ET. HER-2/neu and topoisomerase IIalpha in breast cancer. Breast Cancer Res Treat. 2003;78:299–311. doi: 10.1023/a:1023077507295. [DOI] [PubMed] [Google Scholar]
- 17.Jarvinen TA, Liu ET. Topoisomerase IIalpha gene (TOP2A) amplification and deletion in cancer—more common than anticipated. Cytopathology. 2003;14:309–13. doi: 10.1046/j.0956-5507.2003.00105.x. [DOI] [PubMed] [Google Scholar]
- 18.Jarvinen TA, Tanner M, Barlund M, Borg A, Isola J. Characterization of topoisomerase II alpha gene amplification and deletion in breast cancer. Genes Chromosomes Cancer. 1999;26:142–50. [PubMed] [Google Scholar]
- 19.Beser AR, Tuzlali S, Guzey D, Dolek Guler S, Hacihanefioglu S, Dalay N. HER-2, TOP2A and chromosome 17 alterations in breast cancer. Pathol Oncol Res. 2007;13:180–85. doi: 10.1007/BF02893497. [DOI] [PubMed] [Google Scholar]
- 20.Bofin AM, Ytterhus B, Hagmar BM. TOP2A and HER-2 gene amplification in fine needle aspirates from breast carcinomas. Cytopathology. 2003;14:314–19. doi: 10.1046/j.0956-5507.2003.00088.x. [DOI] [PubMed] [Google Scholar]
- 21.Slamon D, Eiermann W, Robert N, et al. San Antonio Breast Cancer Seminar. San Antonio, TX, USA: Dec 14–17, 2006. BCIRG 006: 2nd interim analysis phase III randomized trial comparing doxorubicin and cyclophosphamide followed by docetaxel (ACT) with doxorubicin and cyclophosphamide followed by docetaxel and trastuzumab (ACTH) with docetaxel, carboplatin and trastuzumab (TCH) in ERBB2neu positive early breast cancer patients. [Google Scholar]
- 22.Arriola E, Rodriguez-Pinilla SM, Lambros MB, et al. Topoisomerase II alpha amplification may predict benefit from adjuvant anthracyclines in ERBB2 positive early breast cancer. Breast Cancer Res Treat. 2007;106:181–89. doi: 10.1007/s10549-006-9492-5. [DOI] [PubMed] [Google Scholar]
- 23.Perez E, Romond E, Suman V, et al. Updated results of the combined analysis of NCCTG N9831 and NSABP B-31 adjuvant chemotherapy with/without trastuzumab in patients with ERBB2-positive breast cancer. J Clin Oncol. 2007;25(suppl):512. [Google Scholar]
- 24.Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable ERBB2-positive breast cancer. N Engl J Med. 2005;353:1673–84. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 25.Hofmann M, Stoss O, Gaiser T, et al. Central ERBB2 IHC and FISH analysis in a trastuzumab (Herceptin(R)) Phase II monotherapy study: assessment of test sensitivity and impact of chromosome 17 polysomy. J Clin Pathol. 2007;1:89–94. doi: 10.1136/jcp.2006.043562. [DOI] [PubMed] [Google Scholar]
- 26.Kaufman PA, Broadwater G, Lezon-Geyda K, et al. CALGB 150002: correlation of ERBB2 and chromosome 17 (ch17) copy number with trastuzumab (T) efficacy in CALGB 9840, paclitaxel (P) with or without T in ERBB2+ and ERBB2− metastatic breast cancer (MBC) J Clin Oncol. 2007;25:1009. [Google Scholar]
- 27.Risio M, Casorzo L, Redana S, Montemurro F. ERBB2 gene-amplified breast cancers with monosomy of chromosome 17 are poorly responsive to trastuzumab-based treatment. Oncol Rep. 2005;13:305–09. [PubMed] [Google Scholar]
- 28.Reinholz M, Jenkins RB, Hillman D, et al. The clinical significance of polysomy 17 in the ERBB2+ N9831 intergroup adjuvant trastuzumab trial. Breast Cancer Res Treat. 2007;106(supp 1):11. [Google Scholar]
- 29.Herrington CS, Leek RD, McGee JO. Correlation of numerical chromosome 11 and 17 imbalance with metastasis of primary breast cancer to lymph nodes. J Pathol. 1995;176:353–59. doi: 10.1002/path.1711760406. [DOI] [PubMed] [Google Scholar]
- 30.Ichikawa D, Hashimoto N, Hoshima M, et al. Analysis of numerical aberrations in specific chromosomes by fluorescent in situ hybridization as a diagnostic tool in breast cancer. Cancer. 1996;77:2064–69. doi: 10.1002/(SICI)1097-0142(19960515)77:10<2064::AID-CNCR15>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 31.McManus DT, Patterson AH, Maxwell P, et al. Interphase cytogenetics of chromosomes 11 and 17 in fine needle aspirates of breast cancer. Hum Pathol. 1999;30:137–44. doi: 10.1016/s0046-8177(99)90267-8. [DOI] [PubMed] [Google Scholar]
- 32.Tsukamoto F, Miyoshi Y, Egawa C, et al. Clinicopathologic analysis of breast carcinoma with chromosomal aneusomy detected by fluorescence in situ hybridization. Cancer. 2001;93:165–70. doi: 10.1002/cncr.9024. [DOI] [PubMed] [Google Scholar]
- 33.Fehm T, Morrison L, Saboorian H, Hynan L, Tucker T, Uhr J. Patterns of aneusomy for three chromosomes in individual cells from breast cancer tumors. Breast Cancer Res Treat. 2002;75:227–39. doi: 10.1023/a:1019901010758. [DOI] [PubMed] [Google Scholar]
- 34.Nakopoulou L, Giannopoulou I, Trafalis D, Gakiopoulou H, Keramopoulos A, Davaris P. Evaluation of numeric alterations of chromosomes 1 and 17 by in situ hybridization in invasive breast carcinoma with clinicopathologic parameters. Appl Immunohistochem Mol Morphol. 2002;10:20–28. doi: 10.1097/00129039-200203000-00004. [DOI] [PubMed] [Google Scholar]
- 35.Wang S, Hossein Saboorian M, Frenkel EP, et al. Aneusomy 17 in breast cancer: its role in HER-2/neu protein expression and implication for clinical assessment of HER-2/neu status. Mod Pathol. 2002;15:137–45. doi: 10.1038/modpathol.3880505. [DOI] [PubMed] [Google Scholar]
- 36.Watters AD, Going JJ, Cooke TG, Bartlett JM. Chromosome 17 aneusomy is associated with poor prognostic factors in invasive breast carcinoma. Breast Cancer Res Treat. 2003;77:109–14. doi: 10.1023/a:1021399923825. [DOI] [PubMed] [Google Scholar]
- 37.Ma Y, Lespagnard L, Durbecq V, et al. Polysomy 17 in HER-2/neu status elaboration in breast cancer: effect on daily practice. Clin Cancer Res. 2005;11:4393–99. doi: 10.1158/1078-0432.CCR-04-2256. [DOI] [PubMed] [Google Scholar]
- 38.Merola R, Mottolese M, Orlandi G, et al. Analysis of aneusomy level and HER-2 gene copy number and their effect on amplification rate in breast cancer specimens read as 2+ in immunohistochemical analysis. Eur J Cancer. 2006;42:1501–06. doi: 10.1016/j.ejca.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 39.Takehisa M, Sasa M, Bando Y, et al. Chromosomal aneusomy (chr 1, 11, 17) detected by fluorescence in situ hybridization may be a prognostic factor in breast cancer. Anticancer Res. 2007;27:1073–78. [PubMed] [Google Scholar]
- 40.Hyun CL, Lee HE, Kim KS, et al. The effect of chromosome 17 polysomy on HER-2/neu status in breast cancer. J Clin Pathol. 2008;61:317–21. doi: 10.1136/jcp.2007.050336. [DOI] [PubMed] [Google Scholar]
- 41.Mezzelani A, Alasio L, Bartoli C, et al. c-erbB2/neu gene and chromosome 17 analysis in breast cancer by FISH on archival cytological fine-needle aspirates. Br J Cancer. 1999;80:519–25. doi: 10.1038/sj.bjc.6690387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farabegoli F, Ceccarelli C, Santini D, et al. c-erbB-2 over-expression in amplified and non-amplified breast carcinoma samples. Int J Cancer. 1999;84:273–77. doi: 10.1002/(sici)1097-0215(19990621)84:3<273::aid-ijc13>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 43.Jimenez RE, Wallis T, Tabasczka P, Visscher DW. Determination of Her-2/Neu status in breast carcinoma: comparative analysis of immunohistochemistry and fluorescent in situ hybridization. Mod Pathol. 2000;13:37–45. doi: 10.1038/modpathol.3880007. [DOI] [PubMed] [Google Scholar]
- 44.Bose S, Mohammed M, Shintaku P, Rao PN. Her-2/neu gene amplification in low to moderately expressing breast cancers: possible role of chromosome 17/Her-2/neu polysomy. Breast J. 2001;7:337–44. doi: 10.1046/j.1524-4741.2001.21018.x. [DOI] [PubMed] [Google Scholar]
- 45.Tubbs RR, Pettay JD, Roche PC, Stoler MH, Jenkins RB, Grogan TM. Discrepancies in clinical laboratory testing of eligibility for trastuzumab therapy: apparent immunohistochemical false-positives do not get the message. J Clin Oncol. 2001;19:2714–21. doi: 10.1200/JCO.2001.19.10.2714. [DOI] [PubMed] [Google Scholar]
- 46.Varshney D, Zhou YY, Geller SA, Alsabeh R. Determination of HER-2 status and chromosome 17 polysomy in breast carcinomas comparing HercepTest and PathVysion FISH assay. Am J Clin Pathol. 2004;121:70–77. doi: 10.1309/FUQH-92B0-3902-5LHG. [DOI] [PubMed] [Google Scholar]
- 47.Salido M, Tusquets I, Corominas JM, et al. Polysomy of chromosome 17 in breast cancer tumors showing an overexpression of ERBB2: a study of 175 cases using fluorescence in situ hybridization and immunohistochemistry. Breast Cancer Res. 2005;7:267–73. doi: 10.1186/bcr996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Downs-Kelly E, Yoder BJ, Stoler M, et al. The influence of polysomy 17 on ERBB2 gene and protein expression in adenocarcinoma of the breast: a fluorescent in situ hybridization, immunohistochemical, and isotopic mRNA in situ hybridization study. Am J Surg Pathol. 2005;29:1221–27. doi: 10.1097/01.pas.0000165528.78945.95. [DOI] [PubMed] [Google Scholar]
- 49.Torrisi R, Rotmensz N, Bagnardi V, et al. ERBB2 status in early breast cancer: relevance of cell staining patterns, gene amplification and polysomy 17. Eur J Cancer. 2007;43:2339–44. doi: 10.1016/j.ejca.2007.07.033. [DOI] [PubMed] [Google Scholar]
- 50.Vanden Bempt I, Van Loo P, Drijkoningen M, et al. Polysomy 17 in breast cancer: clinicopathologic significance and impact on HER-2 testing. J Clin Oncol. 2008;26:4869–74. doi: 10.1200/JCO.2007.13.4296. [DOI] [PubMed] [Google Scholar]
- 51.Visscher D, Jimenez RE, Grayson M, Mendelin J, Wallis T. Histopathologic analysis of chromosome aneuploidy in ductal carcinoma in situ. Hum Pathol. 2000;31:201–07. doi: 10.1016/s0046-8177(00)80220-8. [DOI] [PubMed] [Google Scholar]
- 52.McManus DT, Patterson AH, Maxwell P, Humphreys MW, Anderson NH. Fluorescence in situ hybridisation detection of erbB2 amplification in breast cancer fine needle aspirates. Mol Pathol. 1999;52:75–77. doi: 10.1136/mp.52.2.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmitt FC, Soares R, Leitao D. Detection of numerical chromosome 17 abnormalities in fine-needle aspirates of breast cancer using a novel in situ hybridization signal amplification method. Diagn Cytopathol. 1998;19:141–46. doi: 10.1002/(sici)1097-0339(199808)19:2<141::aid-dc16>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 54.Murphy DS, McHardy P, Coutts J, et al. Interphase cytogenetic analysis of erbB2 and topoII alpha co-amplification in invasive breast cancer and polysomy of chromosome 17 in ductal carcinoma in situ. Int J Cancer. 1995;64:18–26. doi: 10.1002/ijc.2910640106. [DOI] [PubMed] [Google Scholar]
- 55.Marinho AF, Botelho M, Schmitt FC. Evaluation of numerical abnormalities of chromosomes 1 and 17 in proliferative epithelial breast lesions using fluorescence in situ hybridization. Pathol Res Pract. 2000;196:227–33. doi: 10.1016/S0344-0338(00)80071-0. [DOI] [PubMed] [Google Scholar]
- 56.Micale MA, Visscher DW, Gulino SE, Wolman SR. Chromosomal aneuploidy in proliferative breast disease. Hum Pathol. 1994;25:29–35. doi: 10.1016/0046-8177(94)90167-8. [DOI] [PubMed] [Google Scholar]
- 57.Visscher DW, Wallis TL, Crissman JD. Evaluation of chromosome aneuploidy in tissue sections of preinvasive breast carcinomas using interphase cytogenetics. Cancer. 1996;77:315–20. doi: 10.1002/(SICI)1097-0142(19960115)77:2<315::AID-CNCR14>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 58.Mendelin J, Grayson M, Wallis T, Visscher DW. Analysis of chromosome aneuploidy in breast carcinoma progression by using fluorescence in situ hybridization. Lab Invest. 1999;79:387–93. [PubMed] [Google Scholar]
- 59.Sneige N, Liu B, Yin G, Gong Y, Arun BK. Correlation of cytologic findings and chromosomal instability detected by fluorescence in situ hybridization in breast fine-needle aspiration specimens from women at high risk for breast cancer. Mod Pathol. 2006;19:622–29. doi: 10.1038/modpathol.3800571. [DOI] [PubMed] [Google Scholar]
- 60.Murphy DS, Hoare SF, Going JJ, et al. Characterization of extensive genetic alterations in ductal carcinoma in situ by fluorescence in situ hybridization and molecular analysis. J Natl Cancer Inst. 1995;87:1694–704. doi: 10.1093/jnci/87.22.1694. [DOI] [PubMed] [Google Scholar]
- 61.Lal P, Salazar PA, Ladanyi M, Chen B. Impact of polysomy 17 on HER-2/neu immunohistochemistry in breast carcinomas without HER-2/neu gene amplification. J Mol Diagn. 2003;5:155–59. doi: 10.1016/S1525-1578(10)60467-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCormick SR, Lillemoe TJ, Beneke J, Schrauth J, Reinartz J. ERBB2 assessment by immunohistochemical analysis and fluorescence in situ hybridization: comparison of HercepTest and PathVysion commercial assays. Am J Clin Pathol. 2002;117:935–43. doi: 10.1309/3643-F955-7Q6B-EWWL. [DOI] [PubMed] [Google Scholar]
- 63.Lamy PJ, Nanni I, Fina F, et al. Reliability and discriminant validity of ERBB2 gene quantification and chromosome 17 aneusomy analysis by real-time PCR in primary breast cancer. Int J Biol Markers. 2006;21:20–29. doi: 10.1177/172460080602100104. [DOI] [PubMed] [Google Scholar]
- 64.Dal Lago L, Durbecq V, Desmedt C, et al. Correction for chromosome-17 is critical for the determination of true Her-2/neu gene amplification status in breast cancer. Mol Cancer Ther. 2006;5:2572–79. doi: 10.1158/1535-7163.MCT-06-0129. [DOI] [PubMed] [Google Scholar]
- 65.Corzo C, Bellosillo B, Corominas JM, et al. Does polysomy of chromosome 17 have a role in ERBB2 and topoisomerase IIalpha expression? Gene, mRNA and protein expression: a comprehensive analysis. Tumour Biol. 2007;28:221–28. doi: 10.1159/000107583. [DOI] [PubMed] [Google Scholar]
- 66.Vanden Bempt I, Vanhentenrijk V, Drijkoningen M, Wlodarska I, Vandenberghe P, De Wolf-Peeters C. Real-time reverse transcription-PCR and fluorescence in-situ hybridization are complementary to understand the mechanisms involved in HER-2/neu overexpression in human breast carcinomas. Histopathology. 2005;46:431–41. doi: 10.1111/j.1365-2559.2005.02112.x. [DOI] [PubMed] [Google Scholar]
- 67.Zhao J, Wu R, Au A, Marquez A, Yu Y, Shi Z. Determination of ERBB2 gene amplification by chromogenic in situ hybridization (CISH) in archival breast carcinoma. Mod Pathol. 2002;15:657–65. doi: 10.1038/modpathol.3880582. [DOI] [PubMed] [Google Scholar]
- 68.Vanden Bempt I, Drijkoningen M, Van Loo P, et al. HER-2 gene amplification but not chromosome 17 polysomy defines a distinct breast cancer phenotype. Breast Disease. 2006;25:45. [Google Scholar]
- 69.Adeyinka A, Mertens F, Idvall I, Bondeson L, Pandis N. Multiple polysomies in breast carcinomas: preferential gain of chromosomes 1, 5, 6, 7, 12, 16, 17, 18, and 19. Cancer Genet Cytogenet. 1999;111:144–48. doi: 10.1016/s0165-4608(98)00233-7. [DOI] [PubMed] [Google Scholar]
- 70.Chen T, Dhingra K, Sahin A, Sneige N, Hortobagyi G, Aldaz CM. Technical approach for the study of the genetic evolution of breast cancer from paraffin-embedded tissue sections. Breast Cancer Res Treat. 1996;39:177–85. doi: 10.1007/BF01806184. [DOI] [PubMed] [Google Scholar]
- 71.Vanden Bempt I, Drijkoningen M, De Wolf-Peeters C. The complexity of genotypic alterations underlying ERBB2-positive breast cancer: an explanation for its clinical heterogeneity. Curr Opin Oncol. 2007;19:552–57. doi: 10.1097/CCO.0b013e3282f0ad8e. [DOI] [PubMed] [Google Scholar]
- 72.Di Leo A, Gomez HL, Aziz Z, et al. Phase III, double-blind, randomized study comparing lapatinib plus paclitaxel with placebo plus paclitaxel as first-line treatment for metastatic breast cancer. J Clin Oncol. 2008;26:5542–52. doi: 10.1200/JCO.2008.16.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Livingston RB, Downey L, Di Leo A, et al. Evaluation of chromosome 17 (Chr-17) polysomy in ERBB2 FISH-negative metastatic breast cancer (MBC) patients enrolled in a randomized phase III study of paclitaxel and lapatinib. J Clin Oncol. 2008;26(suppl) abstr 1006. [Google Scholar]
- 74.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 75.Walker RA, Bartlett J, Dowsett M, et al. ERBB2 testing in the UK—further update to recommendations. J Clin Pathol. 2008;61:818–24. doi: 10.1136/jcp.2007.054866. [DOI] [PubMed] [Google Scholar]
- 76.Rosenberg CL. Polysomy 17 and HER-2 Amplification: true, true, and unrelated. J Clin Oncol. 2008;26:4856–58. doi: 10.1200/JCO.2008.17.2684. [DOI] [PubMed] [Google Scholar]