Abstract
DNA damage outcomes depend upon the efficiency and fidelity of DNA damage responses (DDRs) for different cells and damage. As such, DDRs represent tightly regulated prototypical systems for linking nanoscale biomolecular structure and assembly to the biology of genomic regulation and cell signaling. However, the dynamic and multifunctional nature of DDR assemblies can render elusive the correlation between the structures of DDR factors and specific biological disruptions to the DDR when these structures are altered. In this chapter, we discuss concepts and strategies for combining structural, biophysical, and imaging techniques to investigate DDR recognition and regulation, and thus bridge sequence-level structural biochemistry to quantitative biological outcomes visualized in cells. We focus on representative DDR responses from PARP/PARG/AIF damage signaling in DNA single-strand break repair and nonhomologous end joining complexes in double-strand break repair. Methods with exemplary experimental results are considered with a focus on strategies for probing flexibility, conformational changes, and assembly processes that shape a predictive understanding of DDR mechanisms in a cellular context. Integration of structural and imaging measurements promises to provide foundational knowledge to rationally control and optimize DNA damage outcomes for synthetic lethality and for immune activation with resulting insights for biology and cancer interventions.
1. INTRODUCTION
DNA holds the cellular information essential for life, but is constantly being damaged. To maintain genetic homeostasis, cells must continually repair DNA: a process that relies on the redundant information in the DNA double helix and efficient DNA damage signaling and repair (DR) processes known collectively as the DNA damage response (DDR). Inadequacies in the DDR lead to the accumulation of oncogenic mutations and genomic instability that is a hallmark of cancer (Hanahan & Weinberg, 2011). Proliferating cells require efficient transcription and replication, and damage can block both transcription and replication polymerases, requiring either repair or the means to bypass lesions. Cell cycle checkpoints ensure DNA is correctly copied and repaired before cell division (Zhou & Elledge, 2000). Where DNA damage is beyond repair, apoptosis is triggered. Thus, DR is at the heart of replication and transcription and forms a vital signaling interface linking damaged DNA to cell cycle progression and cell death.
Collectively, DDRs are essential for genomic integrity and therefore cell fate. Due to their pleiotropic potential, evolutionary pressures result in the tight control of DDR initiation and activation. Mutant DDR proteins that deregulate DDR can fuel genetic alterations sufficient for aggressive cancer phenotypes (Loeb, Loeb, & Anderson, 2003; Lord & Ashworth, 2012). Furthermore, as evident in the contributions in this volume, DNA repair complexes have emerged in discoveries of many aspects of cancer—as tumor suppressors, intrinsic barriers against cancer initiation, resistance factors in cancer therapy, markers for cancer prognosis and immune-therapy efficacy, and, due to intrinsic oncogenic replication and repair stresses, as therapeutic targets (Pearl, Schierz, Ward, Al-Lazikani, & Pearl, 2015). Indeed, many cancer treatments cause complex DNA damages that are repaired by different DNA repair pathways. Accordingly, how these pathways interconnect and interface with replication and transcription within the chromatin environment are key information for nanoscale biological understanding and for the design of advanced cancer therapeutics. Yet, these nanoscale interconnections are among the most mysterious aspects of DR. In fact, gaining a predictive mechanistic understanding of the behavior of interconnected DR systems is one of the great scientific challenges that we face in the next decades for biology and for medicine.
At the nanoscale of DNA repair and signaling complexes, different types of forces (electrostatic, chemical, and mechanical) operate on similar scales. Macromolecules can thus achieve their often extreme efficiencies by creating metastable (dynamic) assemblies that can interconvert these different types of forces. As such, DDR assemblies can capitalize upon evolving architectures and compositions to drive repair progression and interface with cellular signaling. These functionally critical, yet often transient, assembly architectures can be difficult to capture by conventional structural methodologies, such as macromolecular X-ray crystallography (MX), which optimally report upon static molecular arrangements. Even with newer free electron X-ray laser (FEL) methods employing diffract-and-destroy approaches (Kern et al., 2013), we remain a factor of ~1000 from being able to solve single-molecule structures of these dynamic complexes (see http://bl831.als.lbl.gov/~jamesh/fastBragg/xfel/). Moreover, function can also be encoded in macromolecular flexibility, as seen in replication and repair complexes (e.g., Hegde et al., 2013; Perry, Hitomi, & Tainer, 2009; Querol-Audi et al., 2012). As such, there may not be a single correct structure of many DDR macromolecules and complexes, but rather ensembles of relevant conformations, whose population-based properties drive DDR.
Attempts to dissect DDR assembly and function through cellular approaches also present challenges. DDR complexes are often multifunctional, participating in more than one repair or signaling pathway, interfacing with different DNA/chromatin substrates, and responding to variable homeostatic states of the cell (such as in the cell cycle), as seen for the Mre11–Rad50–Nbs1 complex (Limbo et al., 2012; Roset et al., 2014; Schlacher et al., 2011a; Shibata et al., 2014; Williams, Lees-Miller, & Tainer, 2010; Williams, Moncalian, et al., 2008). Thus, defining unambiguous, functional DR phenotypes through conventional knockout/ knockdown experiments may fail to provide a clear interpretation of how specific DR factors contribute to repair. Using nanoscale structure to define dissection-of-function mutants or develop high specificity chemical inhibitors for DR factors is a powerful way to collect data on mechanism. Such separation-of-function approaches are likely critical to defining functional contributions in specific repair and signaling events.
To pursue mechanisms of the DDR on a nanoscale, we propose that it is necessary to create dynamic models of DDR structural assemblies and link these models to quantitative biological outcomes in the cellular environment. Accurate characterization of DDR assembly architecture and dynamics requires an integrative approach: solution measurements by small-angle X-ray scattering (SAXS), validated by high-resolution methods such as MX (Putnam, Hammel, Hura, & Tainer, 2007), biophysical techniques, and computational methodologies. High-resolution visualization of the structures acting in a pathway can provide the first step toward understanding mechanisms of coordination and regulation (Parikh, Mol, Hosfield, & Tainer, 1999; Parikh, Mol, & Tainer, 1997). However, by analogy to the structure of a running horse, one needs accurate structural information integrated with conformation, i.e., snapshots of the leg positions plus the knowledge that the legs are flexible and moving and the timescales of their motion. SAXS allows comprehensive measurements of flexibility and conformations at sequence-level resolution when combined with MX or other high-resolution methods, such as solution NMR as noted elsewhere in this volume (Rambo & Tainer, 2010; Rambo & Tainer, 2013). Single-cell imaging allows accurate and quantitative assessment of dynamic DDR models by monitoring changes in local concentrations and interactions, assembly stoichiometry, and timescales of assembly—all quantitative information, which can be lost in averaged measurements such as coimmunoprecipitation (IP) or qualitative pull-down methods that identify complexes but lose local information. With these points in mind, we focus here on representative systems demonstrating integrated approaches to structural biochemistry and/or imaging from our recently published work and from systems with which we are familiar. We apologize to those whose work in similar areas is not highlighted. Rather than provide detailed protocols for measurements, we point to examples and publications that can guide these methods. The goal is to provide pathways to combine measurements from structures and imaging rather than to correlate increasingly large data sets.
2. METABOLIC SIGNALING: NAD AND DDR BY PARP, PARG, AND AIF
One of the prototypic networks connecting DNA damage to signaling and the choice of repair or cell death involves the coenzyme nicotinamide adenine dinucleotide (NAD), in which adenine and nicotinamide nucleotides are joined via their phosphate groups. The fundamental functions of NAD metabolism span energy homeostasis, signal transduction, genomic stability, and cell death or survival. In all cells, NAD occurs in oxidized (NAD+) and reduced (NADH) forms. NADH produced in the cytoplasm carries electrons into the mitochondrion to act in the electron transport chain, which pumps protons across a membrane and generates ATP through oxidative phosphorylation. Besides being a coenzyme in redox reactions, NAD acts as a precursor of the second messenger cyclic ADP-ribose, as sirtuin cofactors to remove protein acetyl groups, and as an ADP-ribose donor in ADP-ribosylation reactions for the DDR.
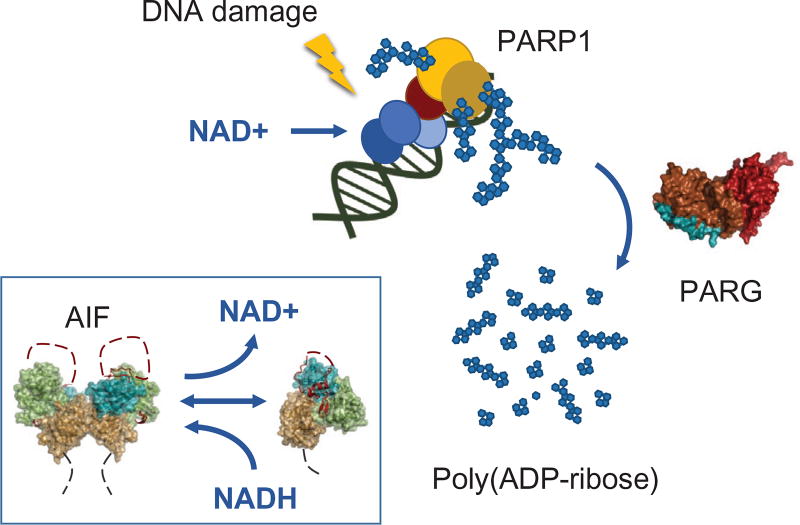
For the DDR, poly(ADP-ribosyl)ation (PAR) in the nucleus is catalyzed by poly(ADP-ribose) polymerase (PARP) (Burkle, 2005). DDR–PARPs couple their damaged DNA interactions to PAR production, which acts in DDR coordination (Fig. 1). The PAR glycohydrolase (PARG) whose structure uncovers a tyrosine clasp mechanism to allow it to act as an endo- and exo-glycohydrolase (Kim et al., 2012), rapidly hydrolyzes the O-glycosidic linkage of ADP-ribose polymers, releasing free ADP-ribose. Mitochondrial apoptosis-inducing factor (AIF) is a downstream effecter of DDR signaling in response to PARP1 hyper-activation. Excessive PARP1 activity and poly(ADP-ribose) synthesis stimulates nuclear accumulation of AIF, chromatin degradation, and cell death (parthanatos) (Fatokun, Dawson, & Dawson, 2014; Yu et al., 2002). AIF’s participation in parthanatos is hypothesized to result from modulation of AIF allostery via cellular depletion of NAD+ during PARP1 hyperactivation (Alano, Ying, & Swanson, 2004; Brosey et al., 2016). Thus, the collective actions of PARP, PARG, and AIF link the DDR to metabolism, aging, transcription, cell death, inflammation, and stress resistance through NAD. Measurements of functional flexibility, conformational changes, and allostery along with cellular localizations provide insights into the mechanisms controlling the regulated activities and functions of PARP, PARG, and AIF.
Fig. 1.
NAD as a metabolic cofactor, damage sensor, and signal in the interrelated PARP, PARG, and AIF allosteric responses that control cell responses to DNA damage. PARP1 acts in the cellular response to ssDNA breaks, where it links damage binding to acute production of poly(ADP-ribose) (PAR) that recruits multiple repair factors to the damage site. PARP1 activation by DNA damage entails unfolding of an autoinhibitory domain to enable proper positioning of NAD+ that leads to a burst in PAR production (PARP1 [based upon PDB 4DQY]—ART domain, gold; HD domain, yellow; WGR domain, red; zinc fingers 1–3, blue. PARG—macrodomain fold, red; mitochondrial targeting sequence, cyan). AIF is an allosteric sensor controlling mitochondrial metabolism and programmed cell death based upon NAD levels that control AIF monomer to dimer assembly by local unfolding (FAD domain, green; NADH domain, gold; C-terminal domain, teal; C-loops, red-dotted lines; mitochondrial innermembrane tethers, dotted gray lines).
2.1 Measuring Allostery in PARP and PARG
Like many cellular processes, the DDR utilizes reversible protein posttranslational modifications (PTMs) to increase protein functional diversity and enable switching between “on” and “off” states (Walsh, 2006). Most PTMs alter binding and regulate enzyme–substrate complexes or protein–protein interactions, and this is the case for the transient PTM of proteins with poly (ADP-ribose). The (PAR) polymerase PARP1 is activated by binding to DNA breaks, which primarily causes PTM of PARP1 plus other DDR proteins, e.g., histones, chromatin remodeling enzymes, and DNA repair factors (Schreiber, Dantzer, Ame, & de Murcia, 2006). The DDR–PARPs (PARP1, PARP2, and PARP3) are associated with multiple DNA repair pathways, including base excision repair, homologous recombination, and nonhomologous end joining (NHEJ) (Beck, Robert, Reina-San-Martin, Schreiber, & Dantzer, 2014; De Vos, Schreiber, & Dantzer, 2012). In the DDR, PARP binds to damaged DNA leading to a dramatic increase in PAR production to facilitate repair. To control this process and predict outcomes of mutations, it is critical to define the mechanism for this regulated activation, which has been uncovered by integrating X-ray structural and solution measurements. The PAR production burst response to genotoxic stress recruits repair proteins to DNA damage sites and alters chromatin architecture to expedite repair. PARP1 synthesizes long branched PAR chains that change protein size and charge. These changes create binding sites for PAR-specific binding proteins that mediate downstream responses to this PTM (Zaja, Mikoc, Barkauskaite, & Ahel, 2012).
2.1.1 Measuring Structural Interactions for Beads-on-a-String’ Assembly
PARP1 is exemplary of the class of DDR proteins that contain multiple domains joined by a flexible “beads-on-a-string” assembly by unstructured linkers that become more ordered upon binding to damage and partners. PARP1 has six domains from N- to C-terminus: zinc finger 1 (Zn1; PDB: 3ODA), zinc finger 2 (Zn2; PDB: 3ODE), zinc finger 3 (Zn3; PDB: 2JVN), BRCA-1 C-terminus fold (BRCT; PDB: 2COK), Tryptophan–Glycine–Arginine domain (WGR; PDB: 2CR9), and catalytic domain (CAT; PDB: 1A26). The CAT consists of an alpha-helical subdomain (HD) and ADP-ribosyltransferase fold (ART).
There are multiple useful means to predict and measure this type of beads-on-a-string assembly and more generally highly flexible and unstructured regions joining domains. Flexible regions can be predicted from sequence to some extent (e.g., IUPred, PONDR, and PrDOS) as seen for the NBS1 phosphoprotein binding subunit of the DSBR MRE11– RAD50–NDS1 complex, where sequence prediction was validated by X-ray scattering measures of flexibility, and domain rotation of the folded regions was evidently transferred by the flexible regions to the Mre11– Rad50 core (Williams et al., 2009). A simple and objective experimental way to assess such flexible regions is to examine proteolytic cleavage. High susceptibility to proteolysis is characteristic of unstructured regions in proteins, since flexibility increases protease access to the polypeptide main chain. Indeed, this feature was used to make a high-throughput assay to examine BRCA1 mutants without requiring purification, where it showed that many mutants resulted in poorly folded protein (Williams et al., 2003). In fact, this proteolysis method has been successfully used on several DDR proteins, such as PARP2, to uncover disordered regions (Riccio, Cingolani, & Pascal, 2016).
The crystal structure of human PARP1 in complex with DNA (PDB: 4DQY) reveals that PARP1 domains collapse into a network of DNA–protein and protein–protein interactions upon binding damaged DNA. This structural collapse allosterically bridges DNA damage detection domains and the CAT domain to effect damage-dependent catalytic activation (Langelier, Planck, Roy, & Pascal, 2012; Steffen, McCauley, & Pascal, 2016). This represents a common theme among DDR proteins: advancement to a new global architecture upon rearrangement of DNA-binding domains on damaged DNA. Also revealed in the PARP1 crystal structure is the allosteric rearrangement and destabilization of the HD in the catalytic region of PARP1. These mechanistic observations of PARP1 allostery arising from the crystal structure were more fully investigated using complementary solution techniques to probe PARP1 dynamics. Application of hydrogen/deuterium exchange-mass spectrometry to PARP1 in the absence and presence of DNA substrate captured allosteric unfolding of the HD upon DNA binding and established its role as an autoinhibitory domain, whose destabilization enables PARP1 to bind NAD+ and become catalytically active (Dawicki-McKenna et al., 2015). In a separate study, DNA-induced collapse of PARP1 was monitored and tested using full-length PARP1 engineered to contain FRET labels on its opposing termini. With this system, the authors were able to observe interdomain collapse as a rise in FRET signal and test residues involved in allosteric communication between domains (Steffen et al., 2016).
Another means to assess structural detail and flexible domain interactions is to combine X-ray crystal structures with SAXS, as done for the poly(ADP-ribose) polymerase enzyme Tankyrase-1 (TNKS), which regulates multiple cellular processes and interacts with other proteins via five ankyrin repeat clusters (ARCs) (Eisemann et al., 2016). SAXS provides a means to comprehensively measure conformation and assembly in solution under near physiological conditions (Hura, Budworth, et al., 2013; Putnam et al., 2007; Rambo & Tainer, 2013). The crystal structure of ARC1–3 combined with SAXS uncovered the solution conformations of the ankyrin repeat. Specific structural restraints and points of flexibility dictate how multiple ARCs collectively function to interact with target proteins. These rigid and flexible ankyrin repeat elements form an adaptable binding platform. The functional cooperation between specific ARC combinations explains how target proteins may be positioned for modification with poly(ADP-ribose).
2.1.2 Implicated Allosteric Interactions in the PARG Crystal Structure
In vivo, the PARG endo- and exo-glycohydrolase rapidly reverses PAR modifications by hydrolyzing the O-glycosidic linkage of ADP-ribose polymers, releasing free ADP-ribose to maintain DRR homeostasis. The terminal products of the PARG reaction are ADP-ribose, and a mono(ADP-ribose)ylated protein that is a substrate for mono(ADP-ribose) glycohydrolases (Sharifi et al., 2013). PARP1 synthesis and PARG turnover of PAR are required for DDR (Hakme, Wong, Dantzer, & Schreiber, 2008), and both enzymes are being evaluated as targets for cancer therapy. As the only mammalian enzyme that efficiently catalyzes PAR turnover, PARG is an essential determinant of cell fate in response to genotoxic stress and inflammatory conditions.
The PARG crystal structure shows that its signature motif (GGG-X6–8-GEE) extends from the glycine-rich loop and precisely orients the catalytic E752 toward the scissile O-glycosidic bond of the ribose moiety. The mammalian Tyr clasp, which is absent in the bacterial enzyme, positions Y791 at its apex to coordinate with the O5′ of the diphosphate of ADP-HPD and edge-stack with its adenine ring (Kim et al., 2012). The exposed substrate-binding channel of mammalian PARG that is capped by the Tyr clasp, may enable both endo- and exo-glycosidic hydrolysis of PAR chains to release oligo(ADP-ribose) and mono(ADP-ribose), both of which may activate signaling pathways. PARG appears to be allosterically regulated by an extended loop containing the mitochondrial targeting sequence (MTS) that wraps around the catalytic domain and stabilizes the tyrosine clasp (Tyr clasp). The MTS buttresses the Tyr clasp, orienting Y791 toward the active site cleft, explaining why the MTS is required for PARG activity.
How PARP and PARPs act as hubs in many signaling pathways and how they are regulated by allostery is a subject of ongoing investigations. The abundance of PARP enzymes in the cell and the efficiency of their reaction converting NAD+ into chains of PAR rapidly depletes the cell of NAD+ and ultimately of its ability to produce ATP. Changes in NAD+ levels upon DNA damage provide a direct link to metabolism and possible programmed cell death by allosterically controlling the assembly of AIF.
2.2 AIF: A Case Study for Allosteric Switching
Mitochondrial AIF is a key downstream effecter of DNA damage signaling in response to PARP1 hyperactivation during single-strand break repair (Moubarak et al., 2007; Yu et al., 2006). Excessive PARP1 activity and poly(ADP-ribose) synthesis stimulates nuclear accumulation of AIF, chromatin degradation, and cell death (parthanatos) (Fatokun et al., 2014). AIF undergoes allosteric monomer–dimer switching in response to NADH binding and charge-transfer complex (CTC) formation with its FAD cofactor (Fig. 2A). Thus, it has been hypothesized that AIF’s participation in parthanatos may arise through modulation of AIF allostery via cellular depletion of NAD+ during PARP1 hyperactivation, stimulating interest in defining the allosteric mechanism.
Fig. 2.
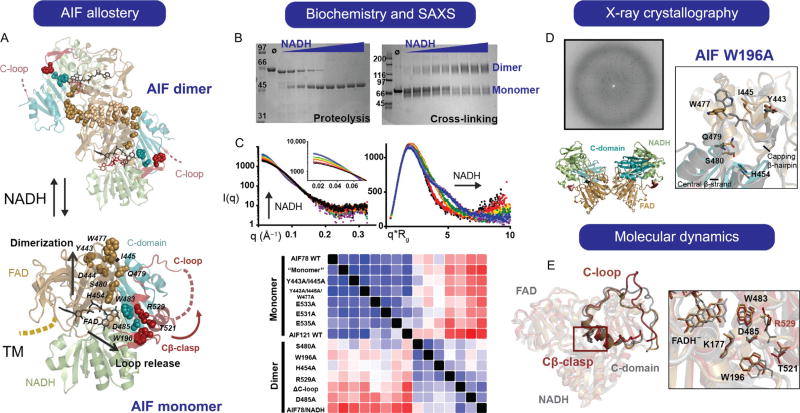
AIF as a Case Study for Allosteric Switching. AIF is allosterically modulated by binding of NADH, dimerizing, and releasing its 50-residue C-loop from the protein core when ligand is present (AIF Allostery) (A). Allosteric mutants of AIF were assessed for C-loop release and ability to dimerize by limited proteolysis and cross-linking assays, respectively (B), and were structurally validated as monomeric or dimeric using SAXS (C). Crystallization of the allosteric AIF W196A mutant allowed elucidation of the molecular pathway linking AIF’s NADH-binding site (residue H454) to the dimer interface (Y443, I445, W477) (D), while molecular dynamics simulations of NADH-bound AIF supported correlation between AIF’s active site and Cβ-clasp in promoting C-loop release (E). MD snapshots from the AIF-CTC trajectory demonstrate mobility in the C-loop (25ns (gray), 225ns (gold), 500ns (red)), while the zoomed inset shows reorientation of R529 within the Cβ-clasp.
The AIF system exemplifies the necessity of bringing multiple approaches to bear upon investigation of allosteric pathways. While high-resolution crystal and NMR structures provide valuable information on three-dimensional biomolecular architecture, their ability to capture dynamic intramolecular pathways that drive conformational transition can be limited. Monomeric AIF and dimeric AIF/NADH CTC complexes have been crystallized at high resolution (Ferreira et al., 2014; Sevrioukova, 2009). Deriving AIF’s allosteric trajectory using these static monomer/ dimer snapshots, however—from NADH binding to dimerization and release of AIF’s 50-residue C-loop insert—has been elusive, motivating combined experimental and computational methods. Sets of structures plus computation can provide specific mechanisms for steps that may be missing, as seen for mechanisms of DNA base repair (Ivanov, Tainer, & McCammon, 2007). Using the crystal structures as a starting point, we applied biochemical, structural, and computational methods to identify molecular pathways linking AIF’s active site to allosteric endpoints on the protein surface at the dimerization interface and the Cβ-clasp. Later, we discuss our strategy for interrogating AIF allostery and selecting complementary approaches to characterize AIF’s allosteric endpoints: dimerization (biochemistry, structural analysis) and C-loop release (computational simulation).
2.2.1 Identifying Allosteric Triggers Through Rationale Mutagenesis
Our first task was to identify AIF mutants exhibiting hindered allosteric responses upon NADH binding or permissive allostery in the absence of NADH ligand. These mutants were then to be analyzed structurally to uncover specific molecular pathways driving allosteric transitions. We first targeted structural elements known to be endpoints of AIF allostery—the dimerization interface and the C-loop insert—and neutralized them by alanine point mutation (dimerization) or deletion (C-loop). Mutants were subsequently screened for impaired or enhanced allostery using simple biochemical assays: for dimerization, cross-linking by BS3 (bis(sulfosuccinimidyl)suberate); for C-loop release, limited proteolysis with Proteinase K. Examples of these assays are presented in Fig. 2B for wild-type AIF, where increasing NADH shifts AIF to a dimeric molecular weight and stimulates proteolytic digestion, reflecting release of the C-loop. These assays can be assembled in less than an hour for multiple reactions, allowing for rapid assessment of mutant allostery.
Biochemical analysis of the AIF C-loop deletion mutant revealed permissive dimerization in the absence of NADH ligand, indicating activation of an intrinsic allosteric pathway. Creation and analysis of a second, focused set of alanine point mutations within and around the C-loop pinpointed the Cβ-clasp region of the C-loop as the source of pathway activation, both for C-loop release and dimerization. To ensure that mutagenic activation of allostery invoked the same structural pathways stimulated by NADH ligand binding, it was critical to screen mutants by SAXS to verify their structural similarity to wild-type monomeric and dimeric AIF, which yield highly distinct scattering profiles (Fig. 2C). High-throughput SAXS acquisition in 96-well plate format facilitates the screening of multiple mutants and NADH concentrations in one experiment, and SAXS similarity matrix analysis software, available from the SIBYLS beamline website (http://sibyls.als.lbl.gov), allows for rapid comparison between mutant and wild-type AIF. (Details of sample preparation for SAXS, data acquisition, and analysis can be found in Classen et al., 2013; Dyer et al., 2014).
2.2.2 Tracing NADH-Driven Dimerization Pathways Through Structural Analysis
Having identified allosterically activated mutants, we proceeded to crystallize the AIF W196A mutant, which disrupts AIF’s Cβ-clasp, releases the C-loop, and is permissive for dimerization without NADH ligand. Altered crystal packing should be considered when analyzing conformations by crystallography. Fortunately, the allostery mutant crystallized in a space group similar to the wild-type protein and also formed the dimer interface observed for the wild-type AIF complex (Fig. 2D). Density for the released C-loop was absent due to its disordered conformation. To identify motifs and residues relevant for allosteric activation, we overlayed the mutant structure with coordinates from monomeric and dimeric AIF and systematically compared backbone and side-chain orientations among the three structures. The W196A allostery mutant exhibited a subset of the total structural differences observed between monomeric and dimeric AIF, specifically displacement and rotation of the central β-strand linking active site residue H454 to the dimerization surface. Subsequent tracking of residue reorientation from the active site to the dimerization surface identified three hydrophobic residues (Y443,I445, and W477) exposed to the solvent upon allosteric activation and forming an outer edge to the dimerization interface (Fig. 2D), a change reminiscent of the conformational switch for PARP1 HD activation described earlier. We verified this putative allosteric pathway by preparing and testing mutants of active-site residue H454, its hydrogen-binding partner S480 from the central β-strand, and surface hydrophobic residues Y443,I445, and W477. Application of our biochemical and SAXS assays confirmed that we were able to manipulate AIF dimerization in a predictable manner with these mutations, thus validating this molecular pathway.
2.2.3 Understanding Initiation of C-Loop Displacement With Computational Methods
Computational approaches provide a means to incorporate missing structural elements into a unified model. Whereas crystallographic analysis of an allostery mutant illuminated molecular pathways associated with AIF dimerization, the absence of C-loop density in the AIF W196A structure made it difficult to identify molecular pathways associated with C-loop release. To gain insight into NADH-stimulated C-loop release, we utilized all-atom molecular dynamics simulations of an AIF/FADH− /NAD+ CTC to capture structural states just prior to release of the C-loop. Inspection of these trajectories (~500-ns each) consistently revealed destabilization of the Cβ-clasp with loss of hydrogen bond contacts and reorientation of the R529 side chain. Visual inspection of the trajectories was complemented by mutual information analysis of torsion angle correlations between residues of the active site and the remainder of AIF (Bowman & Geissler, 2012; Singh & Bowman, 2017). This analysis also pointed toward the Cβ-clasp region and residue R529 as highly correlated to the AIF–CTC active site. The initial AIF mutant screens had identified Cβ-clasp residues as critical for maintaining clasp integrity, and these computational data were able to implicate these residues as participating in a second molecular pathway from the active site to the Cβ-clasp to facilitate C-loop release (Fig. 2E).
2.2.4 Looking Toward Allostery in Complex Protein Machinery: Switches and Ensembles
Full elucidation of AIF allostery required integration of biochemical, structural, and computational information to derive molecular pathways promoting dimerization and C-loop release in response to NADH binding. Simple biochemical assays and SAXS analysis allowed efficient determination and categorization of AIF allostery mutants. Crystallographic analysis of the most informative of these mutants (AIF W196A) allowed identification and testing of specific residues contributing to dimerization, while computational simulation of the AIF–CTC revealed residues significant for coupling NADH binding to C-loop release.
AIF’s NADH-driven exchange encompasses two well-defined structural states with discrete trigger points for allostery originating within AIF’s NADH active site. Allostery among the universe of DNA signaling and repair proteins, however, is often more complex, encompassing ensembles of structural states (particularly for modular protein architectures with disordered segments) and multiple biomolecular actors dynamically remodeling protein structure during DNA repair (Lafrance-Vanasse, Williams, & Tainer, 2015; Perry, Cotner-Gohara, Ellenberger, & Tainer, 2010; Sugitani & Chazin, 2015). Nevertheless, our strategy and toolbox for investigating AIF allostery provides a pathway for considering these more complex ensemble phenomena. For example, integration of SAXS and computational analysis has provided insight into complex formation between ubiquitinated PCNA and translesion polymerases (Tsutakawa et al., 2011), while application of NMR, SAXS, and molecular dynamics simulations has illuminated the dynamic architecture and binding states of the multidomain ssDNA-binding protein Replication Protein A (Brosey et al., 2009, 2015, 2013).
More challenging still is extending investigations of architectural transitions from single protein factors to the multifactor complexes acting in DNA double-strand break (DSB) damage signaling and repair, which we turn to next for NHEJ.
3. DNA DOUBLE-STRAND BREAK RESPONSES
DNA double-strand breaks (DSBs) are the most cytotoxic form of DNA damage. DSBs have major impacts on the cell biology of transcription, replication and repair, and interface with metabolic responses, such as those outlined earlier. DSBs are generated by ionizing radiation (IR), including radiation-based cancer therapy and many DNA damaging drugs, including topoisomerase poisons and DNA cross-linking agents widely used in chemotherapy (Ciccia & Elledge, 2010). Yet, DSBs are also produced endogenously due to collapsed replication forks, reactive oxygen species and are a critical component of essential cellular processes such as V(D)J recombination and class switch recombination in the immune system. Understanding how DSBs are detected and repaired has important implications for enhancing existing cancer therapies, developing new more precise approaches to target tumor cells, and understanding essential cellular processes and factors that predispose to cancer.
The detection and accurate and timely repair of DSBs require a tightly coordinated DDR: proteins that detect the lesion, convert nonligatable end groups to ligatable ends and seal the break. In addition to this, DSBs activate a signaling network to arrest cell cycle progression and regulate other cellular processes in response to the DNA lesion. This sophisticated response is orchestrated by a network of protein–protein, protein–DNA, and possible protein–RNA, interactions that are regulated by PTM, primarily phosphorylation and ubiquitination, as well as SUMO and Nedd-ylation.
To illustrate the methods that have proven useful to investigate NHEJ, we describe here exemplary biochemical, cell biology, biophysical and structural approaches, highlighting the advantages and limitations of current technologies, and opportunities for new understanding of the DDR.
3.1 NHEJ Repair: A Dynamic Assembly Process
DSBs occur when two single-strand breaks occur a short distance apart on opposite strands of the double helix. In the case of IR, DNA termini frequently contain nonligatable ends groups such as 3′-phosphates, 3′-phosphoglycolates, and 5′-hydoxyls that must be converted to 3′-hydroxyl groups and 5′-phosphate groups prior to ligation. These two reactions are both accomplished by polynucleotide kinase phosphatase (PNKP). Combined structural, mutational, and computational results uncovered flexibly linked domains and a bipartite DNA-binding surface that together explain the striking specificity that allows PNKP to restore 5′-phosphate and 3′-hydroxyl termini at DNA damage sites (Bernstein et al., 2009). Similarly, topoisomerase poisons, such as camptothecin, leave blocked DNA ends that must be removed prior to ligation, which can be accomplished by the DSBR nuclease MRE11 (Lee et al., 2012), as well as by the tyrosyl DNA phosphodiesterase-2 (Tdp2), which hydrolyzes the 5′-phosphotyrosine bond formed by many topoisomerases and is the target of several classes of anticancer drugs (Shi et al., 2012). Structures of Tdp2 bound to DNA reveal a deep and narrow basic groove to selectively bind the 5′ end of single-stranded DNA in a stretched conformation and allow an acidic peptide stretch that suggests a mechanism for autoregulation. Geometric restrictions, such as seen in Tdp2 are a powerful and frequently observed means to enforce specificity.
The two main pathways for repair of DSBs in human cells are NHEJ and homologous recombination repair (HRR), which is considered elsewhere in this volume. One of the nanomachines initiating HRR vs NHEJ DSBR is the Mre11–Rad50–Nbs1 complex, which forms a flexing scaffold for the repair of DSBs by HRR (Hopfner et al., 2002; Williams, Williams, & Tainer, 2007). As with NHEJ, the MRN complex activates and is regulated by conformational changes and a kinase, in this case ATM (Deshpande et al., 2014; Rahal et al., 2010). NHEJ is active in nondividing cells and in G1, S, and G2. In contrast, HRR is active only in S and G2 phases of the cell cycle. Yet, understanding the interplay between NHEJ and HRR remains an area of intense study.
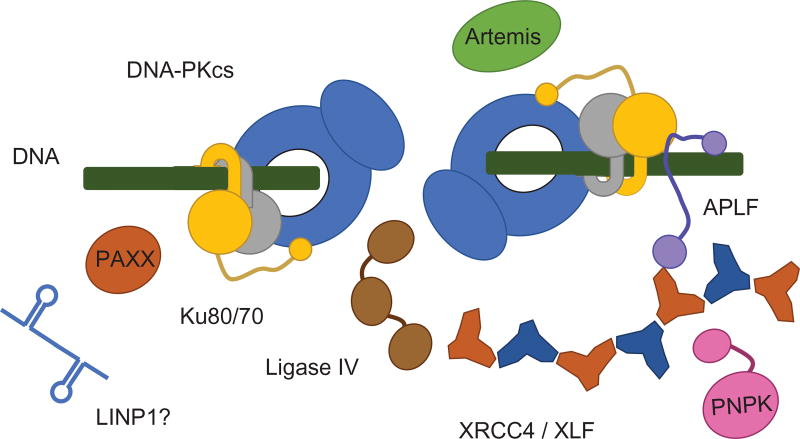
NHEJ is initiated by binding of the Ku70/80 heterodimer to DSB ends. Ku then recruits other components of the NHEJ pathway, including DNA-dependent protein kinase catalytic subunit (DNA-PKcs), XRCC4-like Factor (XLF), Paralog of XRCC4 and XLF (PAXX), and the XRCC4–DNA ligase IV complex to the break (Fig. 3). DNA end processing proteins such as PNKP and the nuclease Artemis are recruited through interaction with XRCC4 and DNA-PKcs, respectively, while the intrinsically disordered Aprataxin and PNKP-like factor (APLF) acts as a molecular thread, interacting with Ku80, XRCC4, and chromatin, to stabilize the NHEJ complex (Radhakrishnan, Jette, & Lees-Miller, 2014; Wang & Lees-Miller, 2013). Critical to NHEJ function is protein phosphorylation by both DNA-PKcs and the related ATM, which phosphorylate each other and other components of the DDR to regulate NHEJ and other cellular processes. Recently, noncoding RNAs have also been shown to regulate DSB repair (Lees-Miller, Beattie, & Tainer, 2016; Zhang et al., 2016), adding an additional level of largely unexplored regulation to the system.
Fig. 3.
NHEJ as a dynamic assembly around Ku dimers at DNA ends. The architecture of the NHEJ core complex based on combined crystallographic and SAXS structures uncovers mechanisms for the synergy of DNA-PKcs/Ku/XRCC4/XLF/LigaseIV interactions in ligating DSBs. Unstructured linkers join DNA-PKcs and Ku dimers in the architectural control of DNA ends by XRCC4–XLF filaments and flexible scaffolding by APLF, PAXX, and possibly RNA (LINP1). The resulting dynamic assembly positions and protects ends while allowing access to Artemis and PNKP for end processing and Ligase IV for end joining (DNA-PKcs, blue; Ku70, gray; Ku80, yellow; XRCC4, orange; XLF, dark blue, PAXX, dark orange; Artemis, green; PNKP, pink; APLF, purple; Ligase IV, brown).
3.2 Combined Structural and Biophysical Measurements on NHEJ Complexes
The crystal structure of the Ku70/80 DNA-binding core revealed a basket structure with a base composed of extensive interactions between Ku70 and Ku80 subunits and a preformed ring. The ring is composed of both Ku70 and Ku80 polypeptides and optimally sized to accommodate a single strand of dsDNA, providing a structural explanation for Ku’s DNA end binding properties. The DNA–protein interface is rich in basic amino acids to provide a surface for electrostatic interactions with the DNA in a sequence-independent manner (Walker, Corpina, & Goldberg, 2001). Missing from the Ku structure were the flexible C-terminal domains on Ku70 (the SAP domain) and Ku80 (the Ku80 C-terminal domain) that contains a critical DNA-PKcs interaction site (Falck, Coates, & Jackson, 2005; Gell & Jackson, 1999). The structures of these C-terminal domains were solved separately by NMR (Harris et al., 2004; Zhang et al., 2004, 2001).
Using SAXS, we were able to analyze the intact Ku70/80 heterodimer, showing that the Ku80 CTR forms a highly flexible extension from the Ku70/80 core (Hammel, Yu, Mahaney, et al., 2010). Moreover, SAXS revealed that the Ku80 CTR remains in an extended conformation in the DNA-PKcs–Ku-DNA complex, allowing us to propose a novel model for the NHEJ complex in which Ku binds at the DNA end, but DNA-PKcs is flexibly associated with the NHEJ complex via the Ku80 CTR (Hammel, Yu, et al., 2016).
Similarly, for XRCC4 and XLF, X-ray crystallography showed that the core domains of both proteins form homodimers with a structured head domain and coiled coil stem (Ahnesorg, Smith, & Jackson, 2006; Andres, Modesti, Tsai, Chu, & Junop, 2007; Junop et al., 2000; Li et al., 2008). Yet, the structure of the full-length proteins remained enigmatic because of the flexibility of the unstructured C-terminal tails. SAXS revealed that XRCC4/XLF dimers interact via their head domains to form multimers in solution (Hammel, Yu, Fang, Lees-Miller, & Tainer, 2010), a fact that was confirmed by crystallography of truncated proteins (Hammel et al., 2011). Moreover, HDX-MS provided information on the interactions of the head domains and disordered tails of XRCC4 and XLF with each other and with DNA (Hammel et al., 2011). X-ray crystallography of XRCC4/ XLF dimers also confirmed the presence of multimers, revealing that XRCC4 and XLF interact via their head groups to form long alpha helical filaments (Andres et al., 2012; Hammel et al., 2011; Wu et al., 2011). The ability of XRCC4 and XLF to form long helical bundles in vitro was confirmed using electron microscopy (Ropars et al., 2011), and XRCC4 and XLF have been shown to coat chromatin on either side of the break in cells using high-resolution microscopy (Reid et al., 2015), hinting at their formation in vivo. Biochemical studies suggest that these filaments play a role in bridging DNA ends (Andres et al., 2012), providing a sliding sleeve across the DNA (Brouwer et al., 2016). XRCC4–XLF filaments may play a structural or scaffolding role in bridging and noncovalently tethering DSB ends prior to ligation (Mahaney, Hammel, Meek, Tainer, & Lees-Miller, 2013), but roles in protecting DSB ends from nuclease-mediated resection to regulate DSB pathway choice are also possible. Further, genetic studies will be useful to enhance our understanding of the function of these filaments in vivo.
DNA ligase IV interacts with the coiled coil region of XRCC4 through its tandem BRCT domains (Sibanda et al., 2001; Wu et al., 2009), tethering the C-terminus of DNA ligase IV to the XRCC4 scaffold. SAXS shows that the N-terminal catalytic domain of DNA ligase IV is flexibly linked to the XRCC4 scaffold (Hammel, Yu, et al., 2016). By integrating crystallography, SAXS, cryo-EM, and biochemistry, we have proposed a model in which Ku70/80 bind to the ends of the break with DNA-PKcs flexibly attached via the extended Ku80CTR (Hammel, Yu, et al., 2016). XRCC4 and XLF then form a scaffold around the break to which critical processing enzymes such as PNKP can be recruited via phosphorylation-dependent FHA domain interactions with XRCC4 (Bernstein et al., 2009, 2005). In contrast, the largely intrinsically disordered protein APLF can bind phosphorylated XRCC4 via its N-terminal FHA domain and Ku70/80 via its mid domain, effectively threading through the NHEJ complex to provide additional stability (Hammel, Yu, et al., 2016). APLF can also interact with histones (Mehrotra et al., 2011) and poly(ADP-ribose) (Ahel et al., 2008; Eustermann et al., 2010), suggesting possible mechanisms to tethering the NHEJ complex to chromatin.
3.3 DNA-PK Architecture From Cryo-EM and X-ray Crystallography
DNA-PKcs presents considerable challenges for structural biology due to its large size: 4128 amino acids and over 460kDa. The first studies using cryo-EM revealed a globular protein with dimensions of approximately 130 by 180Å with a clear head domain, a large central channel suitable for accommodating dsDNA, and a base (Chiu, Cary, Chen, Peterson, & Stewart, 1998; Williams, Lee, Shi, Chen, & Stewart, 2008). At the low resolution available at the time, it was not clear whether the kinase domain of DNA-PKcs was located in the head or the base of the structure (Brewerton, Dore, Drake, Leuther, & Blundell, 2004). The 6.6Å crystal structure of DNA-PKcs in complex with the C-terminal region of Ku80 provided the first insights into this dilemma. Although the resolution of this structure did not allow mapping of amino acids side chains and was incomplete, it did allow the kinase FAT and FAT-C domains to be mapped to the head region of DNA-PKcs (Sibanda, Chirgadze, & Blundell, 2010). Further resolution of this structure to 4Å revealed the presence of the Ku80 binding sites at the base of the molecule and the location of autophosphorylation sites critical for regulation of DNA-PKcs function (Sibanda, Chirgadze, Ascher, & Blundell, 2017). The detailed story of the DNA-PK structure determination and analysis is considered elsewhere in this volume, so we focus here on the implications for the control of NHEJ by phosphorylation.
3.4 Measuring Allosteric Regulation in DNA-PK Interactions
DNA-PKcs is highly phosphorylated in vitro (Douglas et al., 2002) and in vivo (Chan et al., 2002; Chen et al., 2005; Douglas et al., 2007; Meek, Douglas, Cui, Ding, & Lees-Miller, 2007). Two major clusters of phosphorylation sites have been identified: the ABCDE/Threonine 2609 cluster (Ding et al., 2003) and the 2056 or PQR cluster (Chen et al., 2005; Cui et al., 2005). Disruption of the ABCDE sites leads to reduced dissociation of DNA-PKcs from Ku-DNA in vitro (Hammel, Yu, Mahaney, et al., 2010; Jette & Lees-Miller, 2015) and in vivo (Uematsu et al., 2007), suggesting that DNA-PKcs autophosphorylation is a mechanism to regulate access of DNA-PKcs with DNA ends. Indeed, failure of DNA-PKcs to autophosphorylate leads to radiation sensitivity, reduced NHEJ (Ding et al., 2003), and delayed HR (Shibata et al., 2011) in cells and loss of hematopoetic stem cells and lethality in mice (Zhang et al., 2011).
The serine 2056 autophosphorylation site was located at the base of DNA-PKcs close to the Ku binding site in the most recent MX structure (Sibanda et al., 2017), suggesting it could contribute to autophosphorylation-induced dissociation of DNA-PKcs from DNA-bound Ku (Chan & Lees-Miller, 1996; Hammel, Yu, Mahaney, et al., 2010; Merkle et al., 2002). The other major and functionally important DNA-PKcs phosphorylation sites, the ABCDE or threonine 2609 cluster, were positioned midway between the kinase domain and the Ku binding site suitable to dynamically switch between protruding into solution and collapse toward the central cavity in a phosphorylation dependent manner. Other approaches, such as HDX-MS, may be particularly useful in probing solvent exposed regions or regions of flexibility. Indeed, in preliminary studies, the region containing the ABCDE/T2609 phosphorylation cluster was highly accessible to deuterium, consistent with the presence of an exposed region of the protein (Sheff, Hepburn, Yu, Lees-Miller, & Schriemer, 2017).
Given the flexibility and extended nature of the DNA-PKcs–Ku-DNA complex (Hammel, Yu, Mahaney, et al., 2010; Hammel, Yu, et al., 2016), crystallography of the trimeric complex would be challenging, but SAXS-based structures have provided insight (Hammel, Yu, Mahaney, et al., 2010; Hammel, Yu, et al., 2016), and the availability of the higher resolution DNA-PKcs structure will facilitate more detailed analysis. As described, MS methods also have potential to investigate these assemblies, particularly if combined with crosslinking (Lossl, van de Waterbeemd, & Heck, 2016; Sinz, 2017). Furthermore, advances in cryo-EM make this a highly promising tool to study large protein–DNA complexes such as the NHEJ complexes in vitro. Analyzing these complexes in vivo presents major challenges; however, super-resolution microscopy has already provided evidence for XRCC4/XLF filaments (Reid et al., 2015) and Ku binding to DSBs (Britton, Coates, & Jackson, 2013) in vivo. Thus, we can look forward to combined measurements providing an integrated model for NHEJ for biology and medicine. To test models and mechanisms for complex DDR such as NHEJ, it will be critical to examine kinetics in cells, and Section 4 considers these methods for going forward.
4. TAKING STRUCTURAL MECHANISMS INTO CELLS BY ADVANCED IMAGING
As illustrated earlier, dynamic multiprotein large complexes play critical roles in the DDR. Although the molecular mechanisms of many DDR complexes have been interrogated diligently in vitro, less has been done to validate these learned functions in cells, in the context of the entire repair process, largely due to technological obstacles. PTMs play a big role in the timing and the dynamic nature of DDR complex formation and dissociations, which are not fully understood. Therefore, investigation of DDR in cells will need to incorporate all essential proteins and the relevant modifications.
Advancement in fluorescence imaging methods offer an opportunity to investigate DDR by measuring protein localizations, interactions, concentrations, diffusion characteristics, and enzymatic rates. For example, fluorescence recovery after photobleaching has been used to compare and contrast XPC and ERCC1 binding kinetics that correlates with known molecular functions. XPC displays a faster diffusion rate suggesting its scanning function for DNA damage, while the endonuclease ERCC1 exhibits a much longer residency on individual damage sites during processing (Houtsmuller et al., 1999; Luijsterburg et al., 2010).
4.1 Testing Mechanisms by Measuring Kinetics by Fluorescence Imaging
Methods that have not been widely used for DDR that may prove useful for monitoring the dynamics of DDR complexes are fluorescence correlation spectroscopy (FCS) and fluorescence cross correlation spectroscopy (FCCS), where multiple molecules can be monitored in live cells for the diffusion alone or in a complex (Bacia, Kim, & Schwille, 2006; Dean & Palmer, 2014; Tiwari & Kinjo, 2015). FCCS detects local spontaneous fluctuations in fluorescence intensity due to molecular diffusion, where a small number of molecules at a given time contribute to the measured signal. Improved photostable fluorescence dyes, stable light sources, and ultrasensitive detectors make FCCS promising for robust single molecule movement and dynamics in living cells. The intensity fluctuations are autocorrelated over time giving accurate behavior of protein complexes as the cellular conditions are manipulated.
The power of FCS and FCCS are demonstrated when they are combined with the confocal microscope, which allows the detection of nanomolar range fluorescence molecules in femtoliter volumes at a microsecond time scale with high signal-to-noise ratio. Both FCS and FCCS have been used to measure local protein concentrations, diffusion coefficients, and reaction rates, as well as protein–protein, protein–DNA, protein– RNA and protein–drug interactions, dynamics and translocation processes (Huang et al., 2015; Laurence et al., 2008).
There are two important biophysical parameters measured by FCCS: the average number of fluorescent molecules in the detection volume and the translational diffusion time of the molecules through the open volume. The diffusional mobility reflects molecular size, shape, and the apparent viscosity of the medium. When observing protein–protein interactions in living cells, this technique could also be used to report on the dynamic changes in molecular complexes. As this technique is not size limiting, any changes in the molecular complex can be correlated with the diffusion coefficient of the labeled molecule to estimate the size of the DR complex during damage repair. We expect that FCCS will be valuable in monitoring the movements of DNA repair machines during DNA-repair in microsecond to second timescales in living cells.
Emerging single-molecule super-resolution microscopy methods now also permit direct visualization of individual proteins and DNA repair events in vivo with the potential to link cellular heterogeneity to mechanistic observations at the molecular level. Phosphorylation of histone H2AX in chromatin flanking DNA damage by ATM and ATR checkpoint kinase establishes a recruitment platform for checkpoint and repair proteins that merits examination by combined structures and imaging. Initial tests of super-resolution imaging of PCNA and gamma H2AX revealed intriguing new insight into the damage foci itself, which shows an apparent structure of ~500nm rather than a dot inside the nucleus (Fig. 4A and B). These images hold promise for integration with structural results. For example, genetic, biochemical, SAXS, and X-ray crystallographic results on Schizosaccharomyces pombe Brc1, a 6-BRCT-domain protein that is structurally related to mammalian PTIP, show how Brc1 binds gamma H2A to form nuclear foci (Williams, Williams, et al., 2010). Spontaneous Brc1 foci are associated with ribosomal DNA repeat regions, which are prone to fork stalling and genomic instability. DNA damage-induced Brc1 foci colocalize with DSB response factors. The structurally implied DNA-mimicking Brc1 surface and chromatin-specific response to replication-associated DNA damage can now be assessed in cells. Such protein mimicry of DNA may be a general feature of DDR regulation (Putnam & Tainer, 2005).
Fig. 4.
DNA repair machines at super-resolution. (A) STED 3D image of Rad50 and Mre11 colocalization in Hela cells. 7.5Gy of X-ray irradiation was delivered to cells growing on coverslips using Radsource RS-2000, and cells were allowed to recover for 8h before fixation. Endogenous Rad50 and Mre11 were detected by immunofluorescence with complementary mouse Atto488 (Mre11) and rabbit Atto549 (Rad50) secondary antibodies. Dotted circles represent approximate nuclear membrane boundaries. Insets show zoomed rectangular boxes. Images were captured at 100 × with oil immersion using a Leica TCS SP8 STED 3× microscope capable of continuous-wave stimulated emission depletion (cwSTED) and gated STED imaging, equipped with a 405 nm diode laser and a tuneable super-continuum White Light Laser (WLL, 470–670 nm) for excitation, as well as a 592 nm continuous-wave depletion laser. (B) Super-resolution image of γH2AX foci in Hela cells. Cells were X-ray irradiated, fixed as above and stained with rabbit antiphospho serine 139 specific H2AX antibody followed by antirabbit Cy3B. Image was captured using a Vutara352 microscope. (C) STED imaging of PCNA using the experimental conditions in (A) with anti-PCNA rabbit primary and rabbit Atto594 secondary antibodies.
Similarly, STED nanoscopy of RAD50–MRE11 also reveals specific localization and apparent structure, as well as sequestering of Mre11 within the nucleus rather than a diffused distribution (Fig. 4C). Super-resolution imaging of flexible scaffolds such as MRE11–RAD50 provides a means to test implied structural mechanisms and their functional impacts (Lafrance-Vanasse et al., 2015; Williams, Lees-Miller, et al., 2010). We know that Mre11 complex-dependent DDR signaling, replication fork processing, end processing or tethering, homeostasis, and tumorigenesis dependent on its localization, architecture, and conformations (Deshpande et al., 2014; Roset et al., 2014; Schlacher et al., 2011a; Shibata et al., 2014). Similarly, for NHEJ the DSBR process may proceed as a canonical linear repair pathway with sequential steps or as the dynamic assembly of a macromolecular machine. These two distinct mechanisms should be testable by imaging in cells, i.e., is NHEJ blocked by mutating a given step or is there an altered machine assembled with some break repair capacity? Thus, imaging of separation-of-function mutations and inhibitors will open the door to joining structural mechanisms to kinetics and outcomes in cells. More generally, such imaging results promise to illuminate the connections between structural mechanisms and biological outcomes that lie hidden in the nanoscale between proteins and cells.
4.2 Fluorescence Lifetime Imaging Microscopy With FRET
The integration of imaging with structures relies on the robust ability to make quantitative measurements free from factors producing noise and error. These considerations make fluorescence lifetime imaging microscopy (FLIM) an important, underused technique. FLIM is a direct quantitative assay independent of fluorescence intensity, optical path of the microscope, photobleaching, and fluorescence detection efficiency (Chen, Lloyd, Chang, Sud, & Mycek, 2013). Environmental factors such as pH, ion and oxygen concentration, as well as resonance energy transfer due to proximity to energy acceptor are the determining factors for fluorescence lifetime (Boreham, Brodwolf, Walker, Haag, & Alexiev, 2017; Levitt, Matthews, Ameer-Beg, & Suhling, 2009; Suhling, French, & Phillips, 2005). As fluorescence lifetime is defined by the average time, a molecule remains in the excited state before returning to its ground state. The contrast of the image is provided by differences in the excited state decay rates, and since the intrinsic property of the fluorescent molecule is measured, intensity-based artifacts of fluorescence experiments are not an issue. Importantly, FLIM can be effectively used for FRET, which only occurs when the donor and acceptor fluorophores are within close proximity (typically <10 nm), and the emission spectrum of the donor and the absorption spectrum of the acceptor overlap. FLIM with FRET provides the means to monitor both interactions and conformations, as seen for FRET on PARP1 allosteric conformations discussed earlier (Steffen et al., 2016). However due to its versatility, FLIM has also been used for imaging viscosity, temperature, pH, refractive index, and ion and oxygen concentrations (Suhling et al., 2015).
4.2.1 Application of FLIM in DDR Pathways: (PARP to NADH)
The use of FLIM in DDR pathways is tremendously underrepresented. An environment sensor, FLIM could be particularly useful for monitoring the cellular concentration of NAD+, which is utilized by poly(ADP-ribose) polymerases (PARPs) that undergo auto-ADP-ribosylation on DNA damage sites resulting in the recruitment of DDR proteins (Satoh & Lindahl, 1992; Schreiber et al., 2006). FLIM has been used to monitor accurately cellular NAD+/NADH level (Blacker et al., 2014; Skala et al., 2007) and could provide valuable insight to the level of NAD+ in the vicinity of DNA damage foci and indeed the redox state of the entire cell following DNA damage. Moreover, FLIM would also allow quantitative monitoring of allosteric changes to AIF assembly and location during transition from mitochondria to the nucleus during cell death.
Chromatin condensation is associated with transcriptional repression as well as cell death and cell cycle. FLIM is emerging as a valuable method for mapping differentially compacted regions of chromatin along the chromosome and bears enormous potential for structural and diagnostic studies (Estandarte, Botchway, Lynch, Yusuf, & Robinson, 2016; Spagnol & Dahl, 2016). FLIM has also been successfully used to show the presence of G-quadruplex (G4) DNA in the mitochondria of mammalian cells (Huang et al., 2015). We can expect a growing development of FLIM for the quantitative evaluation of the DDR in cells.
4.2.2 FLIM Limitations
FLIM experiments are conducted in fixed samples. Therefore, it is not currently possible to measure interactions or changes in environment in the live cell setting. However, with improved high efficiency detectors, rapid acquisition with single photon sensitivity, and combining FLIM with FCS, FLIM techniques are evolving with the ability to measure lifetime in live cells. Like all other advanced imaging methodologies, the investment in specific hardware and software, as well as technical and operational expertise will be required. However, all commercial microscope manufacturers now offer FLIM hardware and software options for their microscopes, which will allow greater accessibility to the scientific community and spur its use in DDR.
5. WHAT’S AHEAD
Although biology is written at the level of sequences, we suggest that it is read in the context of dynamic molecular shapes and assemblies, particularly for the DDR. We are now able to define multiple regulation aspects that are built into the structures of protein–DNA complexes, as shown for structure-specific nucleases that act in replication and repair (Tsutakawa, Lafrance-Vanasse, & Tainer, 2014). Aided by mass spectrometry methods, we can see DNA–protein interactions that extend beyond the highly ordered regions seen by crystallography (Roberts et al., 2012). Furthermore, we are able to combine sequence and biophysical information to computationally predict non-B-DNA sequences and their role in DDR success and failure based upon the Cancer Genome Atlas (Bacolla, Tainer, Vasquez, & Cooper, 2016). We are starting to structurally define core replication complexes by combined SAXS and crystallography (Wallen et al., 2017). This core replication complex will open the door for examining DDR mechanisms in replication fork protection and processing by proteins such as MRE11, RAD51, and BRCA2 (Schlacher, Wu, & Jasin, 2012; Schlacher et al., 2011b). Within chromatin DNA is protected from damage but also restricted from repair, so the interactions of DDR in chromatin will be of central importance going forward. Based upon current results, we can expect multiple PTMs including ubiquitin and SUMO-ylation to act in switching conformations and interactions to coordinate repair in chromatin (e.g., Groocock et al., 2014; Hodge et al., 2016).
Sets of structures can help identify functional motifs, conformational changes in DNA binding, and protein sculpting of DNA, as seen in the repair of alkylation damage repair that can be a chemotherapy resistance factor (Tubbs, Pegg, & Tainer, 2007). As such, we need to continue to develop and apply integrated structural and imaging methods to define sequence-level resolution models for multiprotein DDR complexes and conformations. This structural and mechanistic knowledge will specify pivot points, switches, and crosstalk for control of DDR pathways and their interactions. It will delineate how DDR pathway choices are made and identify targets for controlling repair pathways. By defining combinatorial chromatin–DDR structures, we will be able to dissect DDR multifunctionality as tested by separation-of-function mutations and structurally informed chemical inhibitors. The initial success of PARP inhibitors in cancer therapy (Bryant et al., 2005; Farmer et al., 2005) underscores the value of such actionable knowledge where interaction with replication plus crosstalk among DR pathways adds important layers of complexity that are not readily or even accurately defined by correlative studies. Separation-of-function mutants and inhibitors are key as the components and complexes can be multifunctional and cannot be tested by genetic knockouts alone, as seen for our specific inhibitors of MRE11 exonuclease and endonuclease activities (Shibata et al., 2014). To integrate structural mechanisms with biological outcomes, it is important to test structural mechanisms by either design of specific inhibitors, as done for MRE11, or by mutations to make predetermined alterations in function rather than mutations that simply break function, which may not prove a specific mechanism. Thus, the structural implied specificity of uracil-DNA glycosylase (Mol et al., 1995) was tested by redesign to instead specifically recognize cytosine and thymine in DNA in cells (Kavli et al., 1996). Similarly, based upon the first full-length Rad51 structure, mutations of an archeal Rad51 ortholog allowed the creation of a designed mutant archeal Rad51 binding interaction with BRCA2 in human cells that verified the biological recognition mechanism (Shin et al., 2003). For validation, SAXS provides an objective means to assess design success and structural similarity (Brunette et al., 2015; Lai et al., 2016, 2014).
We can furthermore expect to see multiple links of metabolism to DDRs, as shown by NAD as well as by systems to remove damaged oxidized and deaminated nucleotide components, as seen for human dUTP pyrophosphatase (Mol, Harris, McIntosh, & Tainer, 1996). Although cross-link repair can be accomplished even by damage reversal proteins such as human O(6)-alkylguanine-DNA alkyltransferase (Fang et al., 2008), we can expect to see important advances in this area given its importance in cancer and the complex repair systems acting to remove not only DNA but also protein cross-links (Stingele et al., 2016). The recognition of bulky lesions by nucleotide excision repair NER machinery, which has strong links to transcription (Sarker et al., 2005), will help target cancer interventions. NER, which can interface with base repair (e.g., Tubbs et al., 2009), requires coordinated helicase and nuclease interactions (Fan et al., 2006, 2008; Fuss & Tainer, 2011). There is much to learn about these NER complexes which are also providing new insights into possible charge transfer through DNA (Mui, Fuss, Ishida, Tainer, & Barton, 2011), as discussed in detail elsewhere in this volume.
There are also largely unexplored levels of interactions beyond the level of specific assemblies. It would be useful to follow DNA or protein conformations in reconstituted multiprotein DR pathways, so methods that allow structural interrogation of complex mixtures are likely to become more important, such as SAXS with gold labeled DNA (Hura, Tsai, et al., 2013). There are also implications of interactions that resemble phase transitions that may change local concentrations in cells in functional important ways, e.g., do unstructured regions allow long-range localizations of repair factors and is controlling these localizations one function of PTMs to these regions? Perhaps, fluorescence methods plus X-ray scattering with X-ray tomography in cells (Hammel, Amlanjyoti, et al., 2016) will address this issue and help with other gaps in knowledge at the transition from nanoscale DDR complexes to mesoscale subcellular structures.
These bottom-up mechanistic approaches to developing a quantitative and predictive knowledge promise medical and biological impact. DNA damage is used to treat cancer due to the high replication stress in cancer cells and often compromised DR processes that are an apparent “Achilles heel” for cancers. Radiation and chemotherapy treatments, the current standard care for many cancers, exploit this by delivering high levels of DNA damage to cancer cells to trigger apoptosis (Cancer Genome Atlas Research Network et al., 2013; Weinstein, 2002). These DNA damaging treatments are inherently toxic, relatively indiscriminant, and unable to cure in many cases. Resistance often arises through partial restoration of DR, accumulation of further genetic instability, and increased metastatic potential. The concept of synthetic lethality promises reduced toxicity to normal cells by directly targeting the DDR rather than brute-force DNA damage to overwhelm repair in cancer cells. The synthetic lethality approach has for example been exploited with the use of PARP1 inhibitors for the treatment of BRCA deficient ovarian cancer (Brown & Baltimore, 2000; Brown, Kaye, & Yap, 2016) and studies in cell lines suggest this concept may also be extended to ATM-deficient tumors (Wang, Jette, Moussienko, Bebb, & Lees-Miller, 2017). DNA damage by IR and other means has added therapeutic potential for tipping the threshold level of unrepaired DNA breaks to activate the immune response. The rare abscopal response, which can mediate regression of metastatic lesions outside the field of radiation to eliminate cancer in patients, depends upon immune system activation by a threshold level of unrepaired DNA breaks (Grass, Krishna, & Kim, 2016). Measurements defining the structural mechanisms for the DDR pathways and their network interactions with replication and transcription may thus provide foundational knowledge to rationally control key steps and optimize damage levels for synthetic lethality and for immune activation—a potential double impact for cancer therapy.
Acknowledgments
This work was supported in part by the National Institutes of Health (NIH) Structural Cell Biology of DNA Repair Machines P01 Grant CA92584 (JAT, SPLM), Canadian Institutes of Health Research operating Grant MOP13639 (SPLM), the Robert A. Welch Distinguished Chair in Chemistry ( J.A.T.), and the Engineered Air Chair in Cancer Research (S.P.L.M.). J.A.T. acknowledges startup funds from the Cancer Prevention and Research Institute of Texas and the University of Texas STARs program. SAXS at the Advanced Light Source SIBYLS beamline is supported in part the United States Department of Energy program Integrated Diffraction Analysis Technologies (IDAT).
References
- Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451(7174):81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124(2):301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- Alano CC, Ying W, Swanson RA. Poly(ADP-ribose) polymerase-1-mediated cell death in astrocytes requires NAD+ depletion and mitochondrial permeability transition. The Journal of Biological Chemistry. 2004;279(18):18895–18902. doi: 10.1074/jbc.M313329200. [DOI] [PubMed] [Google Scholar]
- Andres SN, Modesti M, Tsai CJ, Chu G, Junop MS. Crystal structure of human XLF: A twist in nonhomologous DNA end-joining. Molecular Cell. 2007;28(6):1093–1101. doi: 10.1016/j.molcel.2007.10.024. [DOI] [PubMed] [Google Scholar]
- Andres SN, Vergnes A, Ristic D, Wyman C, Modesti M, Junop M. A human XRCC4-XLF complex bridges DNA. Nucleic Acids Research. 2012;40(4):1868–1878. doi: 10.1093/nar/gks022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacia K, Kim SA, Schwille P. Fluorescence cross-correlation spectroscopy in living cells. Nature Methods. 2006;3(2):83–89. doi: 10.1038/nmeth822. [DOI] [PubMed] [Google Scholar]
- Bacolla A, Tainer JA, Vasquez KM, Cooper DN. Translocation and deletion breakpoints in cancer genomes are associated with potential non-B DNA-forming sequences. Nucleic Acids Research. 2016;44(12):5673–5688. doi: 10.1093/nar/gkw261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck C, Robert I, Reina-San-Martin B, Schreiber V, Dantzer F. Poly(ADP-ribose) polymerases in double-strand break repair: Focus on PARP1, PARP2 and PARP3. Experimental Cell Research. 2014;329(1):18–25. doi: 10.1016/j.yexcr.2014.07.003. [DOI] [PubMed] [Google Scholar]
- Bernstein NK, Hammel M, Mani RS, Weinfeld M, Pelikan M, Tainer JA, et al. Mechanism of DNA substrate recognition by the mammalian DNA repair enzyme, polynucleotide kinase. Nucleic Acids Research. 2009;37(18):6161–6173. doi: 10.1093/nar/gkp597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein NK, Williams RS, Rakovszky ML, Cui D, Green R, Karimi-Busheri F, et al. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Molecular Cell. 2005;17(5):657–670. doi: 10.1016/j.molcel.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Blacker TS, Mann ZF, Gale JE, Ziegler M, Bain AJ, Szabadkai G, et al. Separating NADH and NADPH fluorescence in live cells and tissues using FLIM. Nature Communications. 2014;5:3936. doi: 10.1038/ncomms4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boreham A, Brodwolf R, Walker K, Haag R, Alexiev U. Time-resolved fluorescence spectroscopy and fluorescence lifetime imaging microscopy for characterization of dendritic polymer nanoparticles and applications in nanomedicine. Molecules. 2017;22(1):17. doi: 10.3390/molecules22010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GR, Geissler PL. Equilibrium fluctuations of a single folded protein reveal a multitude of potential cryptic allosteric sites. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(29):11681–11686. doi: 10.1073/pnas.1209309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewerton SC, Dore AS, Drake ACB, Leuther KK, Blundell TL. Structural analysis of DNA-PKcs: Modelling of the repeat units and insights into the detailed molecular architecture. Journal of Structural Biology. 2004;145(3):295–306. doi: 10.1016/j.jsb.2003.11.024. [DOI] [PubMed] [Google Scholar]
- Britton S, Coates J, Jackson SP. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. The Journal of Cell Biology. 2013;202(3):579–595. doi: 10.1083/jcb.201303073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosey CA, Chagot ME, Ehrhardt M, Pretto DI, Weiner BE, Chazin WJ. NMR analysis of the architecture and functional remodeling of a modular multidomain protein, RPA. Journal of the American Chemical Society. 2009;131(18):6346–6347. doi: 10.1021/ja9013634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosey CA, Ho C, Long WZ, Singh S, Burnett K, Hura GL, et al. Defining NADH-driven allostery regulating apoptosis-inducing factor. Structure. 2016;24(12):2067–2079. doi: 10.1016/j.str.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosey CA, Soss SE, Brooks S, Yan C, Ivanov I, Dorai K, et al. Functional dynamics in replication protein A DNA binding and protein recruitment domains. Structure. 2015;23(6):1028–1038. doi: 10.1016/j.str.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosey CA, Yan C, Tsutakawa SE, Heller WT, Rambo RP, Tainer JA, et al. A new structural framework for integrating replication protein A into DNA processing machinery. Nucleic Acids Research. 2013;41(4):2313–2327. doi: 10.1093/nar/gks1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer I, Sitters G, Candelli A, Heerema SJ, Heller I, de Melo AJ, et al. Sliding sleeves of XRCC4-XLF bridge DNA and connect fragments of broken DNA. Nature. 2016;535(7613):566–569. doi: 10.1038/nature18643. [DOI] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes & Development. 2000;14(4):397–402. [PMC free article] [PubMed] [Google Scholar]
- Brown JS, Kaye SB, Yap TA. PARP inhibitors: The race is on. British Journal of Cancer. 2016;114(7):713–715. doi: 10.1038/bjc.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunette TJ, Parmeggiani F, Huang PS, Bhabha G, Ekiert DC, Tsutakawa SE, et al. Exploring the repeat protein universe through computational protein design. Nature. 2015;528(7583):580–584. doi: 10.1038/nature16162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(7035):913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Burkle A. Poly(ADP-ribose)—The most elaborate metabolite of NAD. FEBS Journal. 2005;272(18):4576–4589. doi: 10.1111/j.1742-4658.2005.04864.x. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The cancer genome atlas pan-cancer analysis project. Nature Genetics. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, et al. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes & Development. 2002;16(18):2333–2338. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DW, Lees-Miller SP. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. The Journal of Biological Chemistry. 1996;271(15):8936–8941. doi: 10.1074/jbc.271.15.8936. [DOI] [PubMed] [Google Scholar]
- Chen BP, Chan DW, Kobayashi J, Burma S, Asaithamby A, Morotomi-Yano K, et al. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. The Journal of Biological Chemistry. 2005;280(14715):14709–14715. doi: 10.1074/jbc.M408827200. [DOI] [PubMed] [Google Scholar]
- Chen LC, Lloyd WR, 3rd, Chang CW, Sud D, Mycek MA. Fluorescence lifetime imaging microscopy for quantitative biological imaging. Methods in Cell Biology. 2013;114:457–488. doi: 10.1016/B978-0-12-407761-4.00020-8. [DOI] [PubMed] [Google Scholar]
- Chiu CY, Cary RB, Chen DJ, Peterson SR, Stewart PL. Cryo-EM imaging of the catalytic subunit of the DNA-dependent protein kinase. Journal of Molecular Biology. 1998;284(4):1075–1081. doi: 10.1006/jmbi.1998.2212. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: Making it safe to play with knives. Molecular Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classen S, Hura GL, Holton JM, Rambo RP, Rodic I, McGuire PJ, et al. Implementation and performance of SIBYLS: A dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the advanced light source. Journal of Applied Crystallography. 2013;46:1–13. doi: 10.1107/S0021889812048698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Yu Y, Gupta S, Cho YM, Lees-Miller SP, Meek K. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Molecular and Cellular Biology. 2005;25(24):10842–10852. doi: 10.1128/MCB.25.24.10842-10852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawicki-McKenna JM, Langelier MF, DeNizio JE, Riccio AA, Cao CD, Karch KR, et al. PARP-1 activation requires local unfolding of an autoinhibitory domain. Molecular Cell. 2015;60(5):755–768. doi: 10.1016/j.molcel.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos M, Schreiber V, Dantzer F. The diverse roles and clinical relevance of PARPs in DNA damage repair: Current state of the art. Biochemical Pharmacology. 2012;84(2):137–146. doi: 10.1016/j.bcp.2012.03.018. [DOI] [PubMed] [Google Scholar]
- Dean KM, Palmer AE. Advances in fluorescence labeling strategies for dynamic cellular imaging. Nature Chemical Biology. 2014;10(7):512–523. doi: 10.1038/nchembio.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande RA, Williams GJ, Limbo O, Williams RS, Kuhnlein J, Lee JH, et al. ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling. The EMBO Journal. 2014;33(5):482–500. doi: 10.1002/embj.201386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Reddy YV, Wang W, Woods T, Douglas P, Ramsden DA, et al. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Molecular and Cellular Biology. 2003;23(16):5836–5848. doi: 10.1128/MCB.23.16.5836-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas P, Cui X, Block WD, Yu Y, Gupta S, Ding Q, et al. The DNA-dependent protein kinase catalytic subunit is phosphorylated in vivo on threonine 3950, a highly conserved amino acid in the protein kinase domain. Molecular and Cellular Biology. 2007;27(5):1581–1591. doi: 10.1128/MCB.01962-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas P, Sapkota GP, Morrice N, Yu Y, Goodarzi AA, Merkle D, et al. Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA-dependent protein kinase. The Biochemical Journal. 2002;368(1):243–251. doi: 10.1042/BJ20020973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer KN, Hammel M, Rambo RP, Tsutakawa SE, Rodic I, Classen S, et al. High-throughput SAXS for the characterization of biomolecules in solution: A practical approach. Methods in Molecular Biology. 2014;1091:245–258. doi: 10.1007/978-1-62703-691-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisemann T, McCauley M, Langelier MF, Gupta K, Roy S, Van Duyne GD, et al. Tankyrase-1 ankyrin repeats form an adaptable binding platform for targets of ADP-ribose modification. Structure. 2016;24(10):1679–1692. doi: 10.1016/j.str.2016.07.014. [DOI] [PubMed] [Google Scholar]
- Estandarte AK, Botchway S, Lynch C, Yusuf M, Robinson I. The use of DAPI fluorescence lifetime imaging for investigating chromatin condensation in human chromosomes. Scientific Reports. 2016;6:31417. doi: 10.1038/srep31417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustermann S, Brockmann C, Mehrotra PV, Yang JC, Loakes D, West SC, et al. Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose) Nature Structural & Molecular Biology. 2010;17(2):241–243. doi: 10.1038/nsmb.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434(7033):605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- Fan L, Arvai AS, Cooper PK, Iwai S, Hanaoka F, Tainer JA. Conserved XPB core structure and motifs for DNA unwinding: Implications for pathway selection of transcription or excision repair. Molecular Cell. 2006;22(1):27–37. doi: 10.1016/j.molcel.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, et al. XPD helicase structures and activities: Insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133(5):789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang QM, Noronha AM, Murphy SP, Wilds CJ, Tubbs JL, Tainer JA, et al. Repair of O(6)-G-alkyl-O(6)-G interstrand cross-links by human O(6)-alkylguanine-DNA alkyltransferase. Biochemistry. 2008;47(41):10892–10903. doi: 10.1021/bi8008664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Fatokun AA, Dawson VL, Dawson TM. Parthanatos: Mitochondrial-linked mechanisms and therapeutic opportunities. British Journal of Pharmacology. 2014;171(8):2000–2016. doi: 10.1111/bph.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira P, Villanueva R, Martinez-Julvez M, Herguedas B, Marcuello C, Fernandez-Silva P, et al. Structural insights into the coenzyme mediated monomer-dimer transition of the pro-apoptotic apoptosis inducing factor. Biochemistry. 2014;53(25):4204–4215. doi: 10.1021/bi500343r. [DOI] [PubMed] [Google Scholar]
- Fuss JO, Tainer JA. XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase. DNA Repair. 2011;10(7):697–713. doi: 10.1016/j.dnarep.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gell D, Jackson SP. Mapping of protein-protein interactions within the DNA-dependent protein kinase complex. Nucleic Acids Research. 1999;27(17):3494–3502. doi: 10.1093/nar/27.17.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass GD, Krishna N, Kim S. The immune mechanisms of abscopal effect in radiation therapy. Current Problems in Cancer. 2016;40(1):10–24. doi: 10.1016/j.currproblcancer.2015.10.003. [DOI] [PubMed] [Google Scholar]
- Groocock LM, Nie MH, Prudden J, Moiani D, Wang T, Cheltsov A, et al. RNF4 interacts with both SUMO and nucleosomes to promote the DNA damage response. EMBO Reports. 2014;15(5):601–608. doi: 10.1002/embr.201338369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakme A, Wong HK, Dantzer F, Schreiber V. The expanding field of poly (ADP-ribosyl)ation reactions. ‘Protein modifications: Beyond the usual suspects’ review series. EMBO Reports. 2008;9(11):1094–1100. doi: 10.1038/embor.2008.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M, Amlanjyoti D, Reyes FE, Chen JH, Parpana R, Tang HY, et al. HU multimerization shift controls nucleoid compaction. Science Advances. 2016b;2(7):e1600650. doi: 10.1126/sciadv.1600650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M, Rey M, Yu Y, Mani RS, Classen S, Liu M, et al. XRCC4 protein interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double strand break repair. The Journal of Biological Chemistry. 2011;286(37):32638–32650. doi: 10.1074/jbc.M111.272641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M, Yu Y, Fang S, Lees-Miller SP, Tainer JA. XLF regulates filament architecture of the XRCC4.ligase IV complex. Structure. 2010b;18(11):1431–1442. doi: 10.1016/j.str.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, et al. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. The Journal of Biological Chemistry. 2010a;285(2):1414–1423. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M, Yu Y, Radhakrishnan SK, Chokshi C, Tsai MS, Matsumoto Y, et al. An intrinsically disordered APLF links Ku, DNA-PKcs, and XRCC4-DNA ligase IV in an extended flexible non-homologous end joining complex. The Journal of Biological Chemistry. 2016a;291(53):26987–27006. doi: 10.1074/jbc.M116.751867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Harris R, Esposito D, Sankar A, Maman JD, Hinks JA, Pearl LH, et al. The 3D solution structure of the C-terminal region of Ku86 (Ku86CTR) Journal of Molecular Biology. 2004;335(2):573–582. doi: 10.1016/j.jmb.2003.10.047. [DOI] [PubMed] [Google Scholar]
- Hegde ML, Tsutakawa SE, Hegde PM, Holthauzen LMF, Li J, Oezguen N, et al. The disordered C-terminal domain of human DNA glycosylase NEIL1 contributes to its stability via intramolecular interactions. Journal of Molecular Biology. 2013;425(13):2359–2371. doi: 10.1016/j.jmb.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge CD, Ismail IH, Edwards RA, Hura GL, Xiao AT, Tainer JA, et al. RNF8 E3 ubiquitin ligase stimulates Ubc13 E2 conjugating activity that is essential for DNA double strand break signaling and BRCA1 tumor suppressor recruitment. Journal of Biological Chemistry. 2016;291(18):9396–9410. doi: 10.1074/jbc.M116.715698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BAL, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418(6897):562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- Houtsmuller AB, Rademakers S, Nigg AL, Hoogstraten D, Hoeijmakers JH, Vermeulen W. Action of DNA repair endonuclease ERCC1/XPF in living cells. Science. 1999;284(5416):958–961. doi: 10.1126/science.284.5416.958. [DOI] [PubMed] [Google Scholar]
- Huang WC, Tseng TY, Chen YT, Chang CC, Wang ZF, Wang CL, et al. Direct evidence of mitochondrial G-quadruplex DNA by using fluorescent anticancer agents. Nucleic Acids Research. 2015;43(21):10102–10113. doi: 10.1093/nar/gkv1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hura GL, Budworth H, Dyer KN, Rambo RP, Hammel M, McMurray CT, et al. Comprehensive macromolecular conformations mapped by quantitative SAXS analyses. Nature Methods. 2013a;10(6):453–454. doi: 10.1038/nmeth.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hura GL, Tsai CL, Claridge SA, Mendillo ML, Smith JM, Williams GJ, et al. DNA conformations in mismatch repair probed in solution by X-ray scattering from gold nanocrystals. Proceedings of the National Academy of Sciences of the United States of America. 2013b;110(43):17308–17313. doi: 10.1073/pnas.1308595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I, Tainer JA, McCammon JA. Unraveling the three-metal-ion catalytic mechanism of the DNA repair enzyme endonuclease IV. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(5):1465–1470. doi: 10.1073/pnas.0603468104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jette N, Lees-Miller SP. The DNA-dependent protein kinase: A multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Progress in Biophysics and Molecular Biology. 2015;117(2–3):194–205. doi: 10.1016/j.pbiomolbio.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junop MS, Modesti M, Guarne A, Ghirlando R, Gellert M, Yang W. Crystal structure of the Xrcc4 DNA repair protein and implications for end joining. The EMBO Journal. 2000;19(22):5962–5970. doi: 10.1093/emboj/19.22.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavli B, Slupphaug G, Mol CD, Arvai AS, Petersen SB, Tainer JA, et al. Excision of cytosine and thymine from DNA by mutants of human uracil-DNA glycosylase. The EMBO Journal. 1996;15(13):3442–3447. [PMC free article] [PubMed] [Google Scholar]
- Kern J, Alonso-Mori R, Tran R, Hattne J, Gildea RJ, Echols N, et al. Simultaneous femtosecond X-ray spectroscopy and diffraction of photosystem II at room temperature. Science. 2013;340(6131):491–495. doi: 10.1126/science.1234273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IK, Kiefer JR, Ho CMW, Stegeman RA, Classen S, Tainer JA, et al. Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element. Nature Structural & Molecular Biology. 2012;19(6):653–656. doi: 10.1038/nsmb.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafrance-Vanasse J, Williams GJ, Tainer JA. Envisioning the dynamics and flexibility of Mre11-Rad50-Nbs1 complex to decipher its roles in DNA replication and repair. Progress in Biophysics and Molecular Biology. 2015;117(2–3):182–193. doi: 10.1016/j.pbiomolbio.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai YT, Hura GL, Dyer KN, Tang HYH, Tainer JA, Yeates TO. Designing and defining dynamic protein cage nanoassemblies in solution. Science Advances. 2016;2(12):e1501855. doi: 10.1126/sciadv.1501855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai YT, Reading E, Hura GL, Tsai KL, Laganowsky A, Asturias FJ, et al. Structure of a designed protein cage that self-assembles into a highly porous cube. Nature Chemistry. 2014;6(12):1065–1071. doi: 10.1038/nchem.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Planck JL, Roy S, Pascal JM. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012;336(6082):728–732. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence TA, Kwon Y, Johnson A, Hollars CW, O’Donnell M, Camarero JA, et al. Motion of a DNA sliding clamp observed by single molecule fluorescence spectroscopy. The Journal of Biological Chemistry. 2008;283(34):22895–22906. doi: 10.1074/jbc.M800174200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KC, Padget K, Curtis H, Cowell IG, Moiani D, Sondka Z, et al. MRE11 facilitates the removal of human topoisomerase II complexes from genomic DNA. Biology Open. 2012;1(9):863–873. doi: 10.1242/bio.20121834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees-Miller SP, Beattie TL, Tainer JA. Noncoding RNA joins Ku and DNA-PKcs for DNA-break resistance in breast cancer. Nature Structural & Molecular Biology. 2016;23(6):509–510. doi: 10.1038/nsmb.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt JA, Matthews DR, Ameer-Beg SM, Suhling K. Fluorescence lifetime and polarization-resolved imaging in cell biology. Current Opinion in Biotechnology. 2009;20(1):28–36. doi: 10.1016/j.copbio.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Li Y, Chirgadze DY, Bolanos-Garcia VM, Sibanda BL, Davies OR, Ahnesorg P, et al. Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ. The EMBO Journal. 2008;27(1):290–300. doi: 10.1038/sj.emboj.7601942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O, Moiani D, Kertokalio A, Wyman C, Tainer JA, Russell P. Mre11 ATLD17/18 mutation retains Tel1/ATM activity but blocks DNA double-strand break repair. Nucleic Acids Research. 2012;40(22):11435–11449. doi: 10.1093/nar/gks954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb LA, Loeb KR, Anderson JP. Multiple mutations and cancer. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(3):776–781. doi: 10.1073/pnas.0334858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481(7381):287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- Lossl P, van de Waterbeemd M, Heck AJ. The diverse and expanding role of mass spectrometry in structural and molecular biology. The EMBO Journal. 2016;35(24):2634–2657. doi: 10.15252/embj.201694818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijsterburg MS, Bornstaedt Gv, Gourdin AM, Politi AZ, Mone MJ, Warmerdam DO, et al. Stochastic and reversible assembly of a multiprotein DNA repair complex ensures accurate target site recognition and efficient repair. The Journal of Cell Biology. 2010;189(3):445–463. doi: 10.1083/jcb.200909175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaney BL, Hammel M, Meek K, Tainer JA, Lees-Miller SP. XRCC4 and XLF form long helical protein filaments suitable for DNA end protection and alignment to facilitate DNA double strand break repair. Biochemistry and Cell Biology = Biochimie et Biologie Cellulaire. 2013;91(1):31–41. doi: 10.1139/bcb-2012-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek K, Douglas P, Cui X, Ding Q, Lees-Miller SP. Trans autophosphorylation at DNA-dependent protein kinase’s two major autophosphorylation site clusters facilitates end processing but not end joining. Molecular and Cellular Biology. 2007;27(10):3881–3890. doi: 10.1128/MCB.02366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra PV, Ahel D, Ryan DP, Weston R, Wiechens N, Kraehenbuehl R, et al. DNA repair factor APLF is a histone chaperone. Molecular Cell. 2011;41(1):46–55. doi: 10.1016/j.molcel.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle D, Douglas P, Moorhead GB, Leonenko Z, Yu Y, Cramb D, et al. The DNA-dependent protein kinase interacts with DNA to form a protein-DNA complex that is disrupted by phosphorylation. Biochemistry. 2002;41(42):12706–12714. doi: 10.1021/bi0263558. [DOI] [PubMed] [Google Scholar]
- Mol CD, Arvai AS, Slupphaug G, Kavli B, Alseth I, Krokan HE, et al. Crystal-structure and mutational analysis of human uracil-DNA glycosylase—Structural basis for specificity and catalysis. Cell. 1995;80(6):869–878. doi: 10.1016/0092-8674(95)90290-2. [DOI] [PubMed] [Google Scholar]
- Mol CD, Harris JM, McIntosh EM, Tainer JA. Human dUTP pyrophosphatase: Uracil recognition by a beta hairpin and active sites formed by three separate subunits. Structure. 1996;4(9):1077–1092. doi: 10.1016/s0969-2126(96)00114-1. [DOI] [PubMed] [Google Scholar]
- Moubarak RS, Yuste VJ, Artus C, Bouharrour A, Greer PA, Murcia JMD, et al. Sequential activation of poly(ADP-ribose) polymerase 1, calpains, and bax is essential in apoptosis-inducing factor-mediated programmed necrosis. Molecular and Cellular Biology. 2007;27(13):4844–4862. doi: 10.1128/MCB.02141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mui TP, Fuss JO, Ishida JP, Tainer JA, Barton JK. ATP-stimulated, DNA-mediated redox signaling by XPD, a DNA repair and transcription helicase. Journal of the American Chemical Society. 2011;133(41):16378–16381. doi: 10.1021/ja207222t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh SS, Mol CD, Hosfield DJ, Tainer JA. Envisioning the molecular choreography of DNA base excision repair. Current Opinion in Structural Biology. 1999;9(1):37–47. doi: 10.1016/s0959-440x(99)80006-2. [DOI] [PubMed] [Google Scholar]
- Parikh SS, Mol CD, Tainer JA. Base excision repair enzyme family portrait: Integrating the structure and chemistry of an entire DNA repair pathway. Structure. 1997;5(12):1543–1550. doi: 10.1016/s0969-2126(97)00303-1. [DOI] [PubMed] [Google Scholar]
- Pearl LH, Schierz AC, Ward SE, Al-Lazikani B, Pearl FMG. Therapeutic opportunities within the DNA damage response. Nature Reviews. Cancer. 2015;15(3):166–180. doi: 10.1038/nrc3891. [DOI] [PubMed] [Google Scholar]
- Perry JJ, Cotner-Gohara E, Ellenberger T, Tainer JA. Structural dynamics in DNA damage signaling and repair. Current Opinion in Structural Biology. 2010;20(3):283–294. doi: 10.1016/j.sbi.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JJP, Hitomi K, Tainer JA. Flexibility promotes fidelity. Structure. 2009;17(5):633–634. doi: 10.1016/j.str.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: Defining accurate macromolecular structures, conformations and assemblies in solution. Quarterly Reviews of Biophysics. 2007;40(3):191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- Putnam CD, Tainer JA. Protein mimicry of DNA and pathway regulation. DNA Repair. 2005;4(12):1410–1420. doi: 10.1016/j.dnarep.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Querol-Audi J, Yan CL, Xu XJ, Tsutakawa SE, Tsai MS, Tainer JA, et al. Repair complexes of FEN1 endonuclease, DNA, and Rad9-Hus1-Rad1 are distinguished from their PCNA counterparts by functionally important stability. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(22):8528–8533. doi: 10.1073/pnas.1121116109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan SK, Jette N, Lees-Miller SP. Non-homologous end joining: Emerging themes and unanswered questions. DNA Repair. 2014;17:2–8. doi: 10.1016/j.dnarep.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahal EA, Henricksen LA, Li YL, Williams RS, Tainer JA, Dixon K. ATM regulates Mre11-dependent DNA end-degradation and microhomologymediated end joining. Cell Cycle. 2010;9(14):2866–2877. doi: 10.4161/cc.9.14.12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Bridging the solution divide: Comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Current Opinion in Structural Biology. 2010;20(1):128–137. doi: 10.1016/j.sbi.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Super-resolution in solution X-ray scattering and its applications to structural systems biology. Annual Review of Biophysics. 2013;42:415–441. doi: 10.1146/annurev-biophys-083012-130301. [DOI] [PubMed] [Google Scholar]
- Reid DA, Keegan S, Leo-Macias A, Watanabe G, Strande NT, Chang HH, et al. Organization and dynamics of the nonhomologous end-joining machinery during DNA double-strand break repair. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(20):E2575–E2584. doi: 10.1073/pnas.1420115112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio AA, Cingolani G, Pascal JM. PARP-2 domain requirements for DNA damage-dependent activation and localization to sites of DNA damage. Nucleic Acids Research. 2016;44(4):1691–1702. doi: 10.1093/nar/gkv1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts VA, Pique ME, Hsu S, Li S, Slupphaug G, Rambo RP, et al. Combining H/D exchange mass spectroscopy and computational docking reveals extended DNA-binding surface on uracil-DNA glycosylase. Nucleic Acids Research. 2012;40(13):6070–6081. doi: 10.1093/nar/gks291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropars V, Drevet P, Legrand P, Baconnais S, Amram J, Faure G, et al. Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(31):12663–12668. doi: 10.1073/pnas.1100758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roset R, Inagaki A, Hohl M, Brenet F, Lafrance-Vanasse J, Lange J, et al. The Rad50 hook domain regulates DNA damage signaling and tumorigenesis. Genes & Development. 2014;28(5):451–462. doi: 10.1101/gad.236745.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker AH, Tsutakawa SE, Kostek S, Ng C, Shin DS, Peris M, et al. Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: Insights for transcription-coupled repair and Cockayne syndrome. Molecular Cell. 2005;20(2):187–198. doi: 10.1016/j.molcel.2005.09.022. [DOI] [PubMed] [Google Scholar]
- Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA-repair. Nature. 1992;356(6367):356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11 (vol 145, p 529, 2011) Cell. 2011a;145(6):993. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011b;145(4):529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22(1):106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): Novel functions for an old molecule. Nature Reviews. Molecular Cell Biology. 2006;7(7):517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- Sevrioukova IF. Redox-linked conformational dynamics in apoptosis-inducing factor. Journal of Molecular Biology. 2009;390(5):924–938. doi: 10.1016/j.jmb.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi R, Morra R, Appel CD, Tallis M, Chioza B, Jankevicius G, et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. The EMBO Journal. 2013;32(9):1225–1237. doi: 10.1038/emboj.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff JG, Hepburn M, Yu Y, Lees-Miller SP, Schriemer DC. Nanospray HX-MS configuration for structural interrogation of large protein systems. The Analyst. 2017;142:904–910. doi: 10.1039/c6an02707e. [DOI] [PubMed] [Google Scholar]
- Shi K, Kurahashi K, Gao R, Tsutakawa SE, Tainer JA, Pommier Y, et al. Structural basis for recognition of 5′-phosphotyrosine adducts by Tdp2. Nature Structural & Molecular Biology. 2012;19(12):1372–1377. doi: 10.1038/nsmb.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata A, Conrad S, Birraux J, Geuting V, Barton O, Ismail A, et al. Factors determining DNA double-strand break repair pathway choice in G2 phase. The EMBO Journal. 2011;30(6):1079–1092. doi: 10.1038/emboj.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois MM, et al. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Molecular Cell. 2014;53(1):7–18. doi: 10.1016/j.molcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DS, Pellegrini L, Daniels DS, Yelent B, Craig L, Bates D, et al. Full-length archaeal Rad51 structure and mutants: Mechanisms for RAD51 assembly and control by BRCA2. The EMBO Journal. 2003;22(17):4566–4576. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibanda BL, Chirgadze DY, Ascher DB, Blundell TL. DNA-PKcs structure suggests an allosteric mechanism modulating DNA double-strand break repair. Science. 2017;355(6324):520–524. doi: 10.1126/science.aak9654. [DOI] [PubMed] [Google Scholar]
- Sibanda BL, Chirgadze DY, Blundell TL. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature. 2010;463(7277):118–121. doi: 10.1038/nature08648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibanda BL, Critchlow SE, Begun J, Pei XY, Jackson SP, Blundell TL, et al. Crystal structure of an Xrcc4-DNA ligase IV complex. Nature Structural Biology. 2001;8(12):1015–1019. doi: 10.1038/nsb725. [DOI] [PubMed] [Google Scholar]
- Singh S, Bowman GR. Quantifying allosteric communication via both concerted structural changes and conformational disorder with CARDS. Journal of Chemical Theory and Computation. 2017;13:1509–1517. doi: 10.1021/acs.jctc.6b01181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinz A. Divide and conquer: Cleavable cross-linkers to study protein conformation and protein-protein interactions. Analytical and Bioanalytical Chemistry. 2017;409(1):33–44. doi: 10.1007/s00216-016-9941-x. [DOI] [PubMed] [Google Scholar]
- Skala MC, Riching KM, Gendron-Fitzpatrick A, Eickhoff J, Eliceiri KW, White JG, et al. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(49):19494–19499. doi: 10.1073/pnas.0708425104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnol ST, Dahl KN. Spatially resolved quantification of chromatin condensation through differential local rheology in cell nuclei fluorescence lifetime imaging. PLoS One. 2016;11(1):e0146244. doi: 10.1371/journal.pone.0146244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen JD, McCauley MM, Pascal JM. Fluorescent sensors of PARP-1 structural dynamics and allosteric regulation in response to DNA damage. Nucleic Acids Research. 2016;44(20):9771–9783. doi: 10.1093/nar/gkw710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele J, Bellelli R, Alte F, Hewitt G, Sarek G, Maslen SL, et al. Mechanism and regulation of DNA-protein crosslink repair by the DNA-dependent metalloprotease SPRTN. Molecular Cell. 2016;64(4):688–703. doi: 10.1016/j.molcel.2016.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugitani N, Chazin WJ. Characteristics and concepts of dynamic hub proteins in DNA processing machinery from studies of RPA. Progress in Biophysics and Molecular Biology. 2015;117(2–3):206–211. doi: 10.1016/j.pbiomolbio.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhling K, French PM, Phillips D. Time-resolved fluorescence microscopy. Photochemical & Photobiological Sciences. 2005;4(1):13–22. doi: 10.1039/b412924p. [DOI] [PubMed] [Google Scholar]
- Suhling K, Hirvonen LM, Levitt JLP-HC, Tregidgo C, Marois AL, Rusakov DA, et al. Fluorescence lifetime imaging (FLIM): Basic concepts and some recent developments. Medical Photonics. 2015;27:3–40. [Google Scholar]
- Tiwari M, Kinjo M. Determination of the dissociation constant of the NFkappaB p50/p65 heterodimer in living cells using fluorescence cross-correlation spectroscopy. Methods in Molecular Biology. 2015;1228:173–186. doi: 10.1007/978-1-4939-1680-1_14. [DOI] [PubMed] [Google Scholar]
- Tsutakawa SE, Lafrance-Vanasse J, Tainer JA. The cutting edges in DNA repair, licensing, and fidelity: DNA and RNA repair nucleases sculpt DNA to measure twice, cut once. DNA Repair (Amst) 2014;19:95–107. doi: 10.1016/j.dnarep.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa SE, Van Wynsberghe AW, Freudenthal BD, Weinacht CP, Gakhar L, Washington MT, et al. Solution X-ray scattering combined with computational modeling reveals multiple conformations of covalently bound ubiquitin on PCNA. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(43):17672–17677. doi: 10.1073/pnas.1110480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs JL, Latypov V, Kanugula S, Butt A, Melikishvili M, Kraehenbuehl R, et al. Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature. 2009;459(7248):808–813. doi: 10.1038/nature08076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs JL, Pegg AE, Tainer JA. DNA binding, nucleotide flipping, and the helix-turn-helix motif in base repair by O-6-alkylguanine-DNA alkyltransferase and its implications for cancer chemotherapy. DNA Repair. 2007;6(8):1100–1115. doi: 10.1016/j.dnarep.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, et al. Autophosphorylation of DNA-PKcs regulates its dynamics at DNA double-strand breaks. The Journal of Cell Biology. 2007;177(2):219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412(6847):607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- Wallen JR, Zhang H, Weis C, Cui W, Foster BM, Ho CM, et al. Hybrid methods reveal multiple flexibly linked DNA polymerases within the bacteriophage T7 replisome. Structure. 2017;25(1):157–166. doi: 10.1016/j.str.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C. Posttranslational modification of proteins: Expanding nature’s inventory. Englewood, CO: Roberts and Company Publishers; 2006. [Google Scholar]
- Wang C, Jette N, Moussienko D, Bebb DG, Lees-Miller SP. ATM-deficient colorectal cancer cells are sensitive to the PARP inhibitor olaparib. Translational Oncology. 2017;10(2):190–196. doi: 10.1016/j.tranon.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Lees-Miller SP. Detection and repair of ionizing radiation-induced DNA double strand breaks: New developments in nonhomologous end joining. International Journal of Radiation Oncology, Biology, Physics. 2013;86(3):440–449. doi: 10.1016/j.ijrobp.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB. Cancer. Addiction to oncogenes—The Achilles heal of cancer. Science. 2002;297(5578):63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- Williams RS, Chasman DI, Hau DD, Hui B, Lau AY, Glover JN. Detection of protein folding defects caused by BRCA1-BRCT truncation and missense mutations. The Journal of Biological Chemistry. 2003;278(52):53007–53016. doi: 10.1074/jbc.M310182200. [DOI] [PubMed] [Google Scholar]
- Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, et al. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139(1):87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DR, Lee KJ, Shi J, Chen DJ, Stewart PL. Cryo-EM structure of the DNA-dependent protein kinase catalytic subunit at subnanometer resolution reveals alpha helices and insight into DNA binding. Structure. 2008b;16(3):468–477. doi: 10.1016/j.str.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GJ, Lees-Miller SP, Tainer JA. Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair. 2010a;9(12):1299–1306. doi: 10.1016/j.dnarep.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008a;135(1):97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Williams RS, Dovey CL, Guenther G, Tainer JA, Russell P, et al. The EMBO Journal. 2010b;29(6):1136–1148. doi: 10.1038/emboj.2009.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochemistry and Cell Biology. 2007;85(4):509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- Wu PY, Frit P, Meesala S, Dauvillier S, Modesti M, Andres SN, et al. Structural and functional interaction between the human DNA repair proteins DNA ligase IV and XRCC4. Molecular and Cellular Biology. 2009;29(11):3163–3172. doi: 10.1128/MCB.01895-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Ochi T, Matak-Vinkovic D, Robinson CV, Chirgadze DY, Blundell TL. Non-homologous end-joining partners in a helical dance: Structural studies of XLF-XRCC4 interactions. Biochemical Society Transactions. 2011;39(5)(Suppl. 2):1387–1392. doi: 10.1042/BST0391387. following 1392. [DOI] [PubMed] [Google Scholar]
- Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, et al. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(48):18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, et al. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297(5579):259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- Zaja R, Mikoc A, Barkauskaite E, Ahel I. Molecular insights into poly(ADP-ribose) recognition and processing. Biomolecules. 2012;3(1):1–17. doi: 10.3390/biom3010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, He Q, Hu Z, Feng Y, Fan L, Tang Z, et al. Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nature Structural & Molecular Biology. 2016;23:522–530. doi: 10.1038/nsmb.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hu W, Cano L, Lee TD, Chen DJ, Chen Y. Solution structure of the C-terminal domain of Ku80 suggests important sites for protein-protein interactions. Structure (Camb) 2004;12(3):495–502. doi: 10.1016/j.str.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Zhang S, Yajima H, Huynh H, Zheng J, Callen E, Chen HT, et al. Congenital bone marrow failure in DNA-PKcs mutant mice associated with deficiencies in DNA repair. The Journal of Cell Biology. 2011;193(2):295–305. doi: 10.1083/jcb.201009074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhu L, Lin D, Chen F, Chen DJ, Chen Y. The three-dimensional structure of the C-terminal DNA-binding domain of human Ku70. The Journal of Biological Chemistry. 2001;276(41):38231–38236. doi: 10.1074/jbc.M105238200. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ. The DNA damage response: Putting checkpoints in perspective. Nature. 2000;408(6811):433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]