Abstract
Purpose
Gemtuzumab ozogamicin (GO), a CD33-targeted immunoconjugate, is a re-emerging therapy for acute myeloid leukemia (AML). CD33 single nucleotide polymorphism rs12459419 C>T in the splice enhancer region regulates the expression of an alternatively spliced CD33 isoform lacking exon2 (D2-CD33), thus eliminating the CD33 IgV domain, which is the antibody-binding site for GO, as well as diagnostic immunophenotypic panels. We aimed to determine the impact of the genotype of this splicing polymorphism in patients with AML treated with GO-containing chemotherapy.
Patients and Methods
CD33 splicing single nucleotide polymorphism was evaluated in newly diagnosed patients with AML randomly assigned to receive standard five-course chemotherapy alone (No-GO arm, n = 408) or chemotherapy with the addition of two doses of GO once during induction and once during intensification (GO arm, n = 408) as per the Children’s Oncology Group AAML0531 trial.
Results
The rs12459419 genotype was CC in 415 patients (51%), CT in 316 patients (39%), and TT in 85 patients (10%), with a minor allele frequency of 30%. The T allele was significantly associated with higher levels of D2-CD33 transcript (P < 1.0E−6) and with lower diagnostic leukemic cell surface CD33 intensity (P < 1.0E−6). Patients with the CC genotype had significantly lower relapse risk in the GO arm than in the No-GO arm (26% v 49%; P < .001). However, in patients with the CT or TT genotype, exposure to GO did not influence relapse risk (39% v 40%; P = .85). Disease-free survival was higher in patients with the CC genotype in the GO arm than in the No-GO arm (65% v 46%, respectively; P = .004), but this benefit of GO addition was not seen in patients with the CT or TT genotype.
Conclusion
Our results suggest that patients with the CC genotype for rs12459419 have a substantial response to GO, making this a potential biomarker for the selection of patients with a likelihood of significant response to GO.
INTRODUCTION
Acute myeloid leukemia (AML) is a heterogeneous disease that is associated with poor outcome. Although intensive chemotherapy and hematopoietic stem-cell transplantation (SCT) remain the mainstay of current AML therapy, targeted therapies such as monoclonal antibodies and small-molecule inhibitors have emerged as promising novel approaches. Gemtuzumab ozogamicin (GO), a humanized anti-CD33 antibody linked to the toxin calicheamicin, targets CD33 antigen present on the majority of AML blasts.1,2 GO received an accelerated approval in 2000 by the US Food and Drug Administration for the treatment of relapsed AML in patients older than 60 years of age,3 but it was withdrawn because of a lack of benefit and a high early mortality in the SWOG-S0106 study.4 However, four large randomized studies conducted in Europe demonstrated congruently that GO lowered the relapse risk (RR) and prolonged survival in favorable-risk and, to a lesser degree, intermediate-risk, AML.4-11 In pediatric AML, GO was investigated in the Children’s Oncology Group (COG) trials AAML03P1 and AAML0531.12,13 Results from these trials suggested that GO has the potential to improve outcomes in pediatric AML and may have been withdrawn prematurely from the market.13-15
We reported previously in a small pilot study in the St Jude AML02 trial that the CD33-coding single nucleotide polymorphism (SNP) rs12459419 was associated with response.16 A follow-up study showed significant association of this SNP with CD33 cell surface expression in leukemic blasts.17 rs12459419 (C > T; Ala14Val) in exon2 is present within 4 base pairs of the intron/exon junction and resides within the exonic splicing enhancer (ESE) binding site for SRSF2.18 It has been related to the skipping of exon2, resulting in the shorter CD33 isoform (D2-CD33), which lacks the IgV domain.19-21 The exon2-encoding IgV domain is an epitope for the hP67.6-CD33 antibody, which is used for diagnostic immunophenotyping, and for the antibody that is conjugated to calicheamicin in GO. As a result, loss of this domain can not only interfere with the detection of total CD33, but also affect the binding, internalization, and clinical efficacy of GO (Fig 1A).
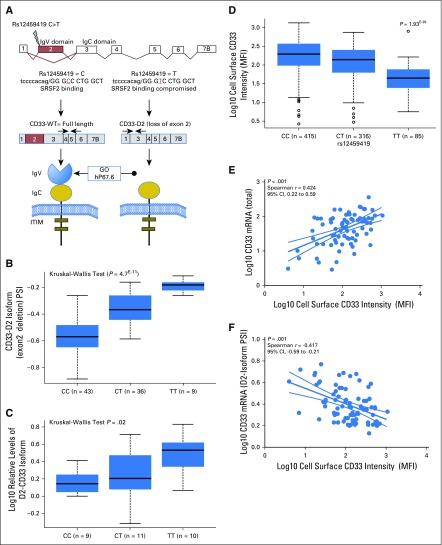
Fig 1.
CD33 exon2 single nucleotide polymorphism (SNP) rs12459419 influences the alternative splicing of CD33. (A) CD33 gene and location and potential functional consequences of rs12459419 SNP. The presence of rs12459419 in exon2 affects the exonic enhancer binding site for SRFS2, thereby resulting in the loss of exon2 (shown in red) in the T allele. The loss of exon2 results in a shorter CD33 isoform lacking the IgV domain, which is recognized by gemtuzumab ozogamicin (GO) and currently, used antibodies. (B) Transcriptome-sequencing data from AAML0531 patients (n = 88) shown as percent spliced isoform (PSI) with loss of exon2 for different rs12459419 SNP genotypes. (C) Quantitative real-time polymerase chain reaction (Data Supplement) using isoform-specific primers shown in (A) in an independent set of patient specimens (n = 30) obtained at diagnosis. Log10 relative levels of the D2-CD33 isoform are shown for the rs12459419 genotypes (patient numbers in parentheses). Box plots show medians as a line between the boxes representing the first and third quartiles; the whiskers represent the range after excluding the outliers. The outliers are defined as data points that fall outside the first and third quartiles by > 1.5 times the interquartile range. Circles outside the whiskers represent outliers. Cell membrane was created using clipart from clker.com. (D) Association of rs12459419 with CD33 expression determined as mean fluorescence intensity (MFI) in diagnostic leukemic blasts from patients treated in AAML0531 (n = 816). CD33 levels were determined in the diagnostic leukemic blasts by multiparameter flow cytometry. Log10 CD33 MFI is shown for different rs12459419 genotypes (patient numbers in parentheses). Correlation plots show the association of CD33 MFI with levels of (E) the CD33 total mRNA levels and (F) D2-CD33 isoform. Plots were created using the GraphPad Prism software. ITIM, immunoreceptor tyrosine inhibitory motif; WT, wild-type.
In this study, we evaluated rs12459419 for an association with CD33 cell surface expression in leukemic blasts and clinical outcome in patients treated with or without the addition of GO to standard chemotherapy in the AAML0531 trial.
PATIENTS AND METHODS
Patients and Treatment
Details of the AAML0531 study design, treatment regimen, and clinical outcome have been reported previously.13 Overall, patients with de novo AML (0 to 29 years of age) were randomly assigned to either standard five-course chemotherapy (No-GO arm) or to the same chemotherapy with the addition of two doses of 3 mg/m2 GO (GO arm). Risk groups were defined as follows: low risk (LR), which included t(8;21), inv(16), or t(16;16) presence of CEBP-α and NPM mutations; high risk, which included monosomy 7, monosomy 5/5q deletion, FLT3-ITD, or persistent disease at the end of induction 1; and intermediate or standard risk, which included all other patients.13,22 This study included 816 patients with specimens and CD33 cell surface expression data available. The institutional review boards of all participating institutions approved the clinical protocol, and the COG Myeloid Disease Biology Committee approved this research.
Genotyping CD33 SNPs
CD33-coding SNP rs12459419-Ala14Val and linked promoter SNP rs3865444 were genotyped using the Sequenome (San Diego, CA) platform at the Biomedical Genomics Center, University of Minnesota. Both SNPs had a call rate of > 0.98 and were in Hardy-Weinberg equilibrium (P > .05).
CD33 Expression Levels on Leukemic Blasts
CD33 expression levels were determined by measuring the mean fluorescence intensity (MFI) of myeloid blasts obtained at diagnosis by flow cytometry, as described previously.22-24
mRNA Levels of CD33 and Its Isoforms
CD33 wild-type and alternatively spliced transcript levels were extracted from RNA sequencing (RNA-seq) data obtained from diagnostic leukemia blasts from 88 patients. In addition, CD33 wild-type and splice variants lacking exon2 were quantitated in 30 samples by quantitative real-time polymerase chain reaction (qRT-PCR), using isoform-specific primers (Data Supplement).
Statistical Analyses
The Kruskal-Wallis exact test was used to compare CD33 cell surface levels or CD33 transcript levels across genotype groups. Clinical outcome data for patients treated in AAML0531 were analyzed as of September 30, 2014. The Kaplan-Meier method was used to estimate overall survival (OS), event-free survival (EFS), and disease-free survival (DFS; Data Supplement).25 For patients in complete remission (CR), RR was defined as the time from the end of induction 1 to relapse or death, where deaths without a relapse were considered competing events. All the time-to-event end points are reported at 5 years; for DFS, it was 5 years from the end of induction 1. The significance of predictor variables was tested with the log-rank statistic for OS, EFS, and DFS and with Gray’s statistic for RR. Cox proportional hazards models were used to estimate the hazard ratio (HR) for OS and EFS.26 The χ2 test was used to test the significance of observed differences in proportions. The Mann-Whitney or Wilcoxon signed-rank test was used to compare differences in medians. A P value < .05 was considered statistically significant (Data Supplement).
RESULTS
Genotype Frequency of rs12459419
Of the 816 patients genotyped for rs12459419, 415 (51%) had the CC genotype, 316 (39%) had the CT genotype, and 85 (10%) had the TT genotype, with a corresponding minor allele frequency of 30%.
Comparison of the demographics and disease characteristics across the different genotype cohorts showed no difference in genotype frequency by sex, treatment arm, disease characteristics, or receipt of SCT during treatment (Data Supplement). However, frequency of the SNP was significantly higher in white patients (0.31) than in black patients (0.13; P < .001). There was no difference in the clinical outcome measures between patients included in the study and those who were part of the parent clinical trial but were not included in this study (Data Supplement).
rs12459419 C>T at the ESE Binding Site Is Associated With Expression of Exon2-Skipped Transcript Lacking GO Binding Site
Splicing SNP rs12459419 is located within 4 base pairs of the exonic junction of exon2, a known binding domain for splicing factor SRSF2. The C>T change results in an Ala14Val change and alters the splicing enhancer binding site, thereby altering CD33-exon2 splicing (Fig 1A). We used RNA-seq data from 88 patients treated in AAML0531 to determine whether the minor allele (T) is associated with alternative splicing of CD33 transcript in AML. The presence of rs12459419 variant T allele was significantly associated with increased levels of an alternatively spliced D2-CD33 isoform lacking exon2 (P < 1.0E−6; Fig 1B). qRT-PCR of RNA from leukemic blasts obtained at diagnosis in a representative set of 30 patients in AAML0531 (CC = 9, CT = 11, and TT = 10) using isoform-specific primers (Fig 1A; Data Supplement) revealed a significant association of the T allele with increased levels of the D2-CD33 isoform (Fig 1C; P = .02). There was no association between total CD33 transcript levels and rs12459419 in samples from this cohort (P > .05) in either the RNA-seq or the qRT-PCR analysis.
Association of rs12459419 With CD33 Cell Surface Intensity in Leukemic Blasts
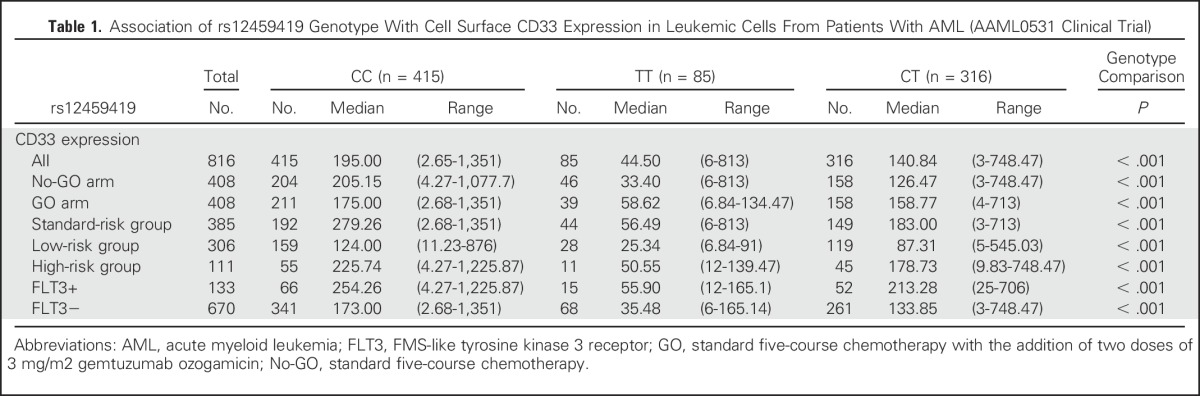
Given that commonly used antibodies in diagnostic immunophenotypic panels target the IgV domain (coded by exon2), we reasoned that those with minor allele (D2 isoform) would not be amenable to detection by available antibodies. rs12459419 genotype data and CD33 cell surface expression of the blast population were available for 816 patients (No-GO arm, n = 408; GO arm, n = 408). The MFI was determined using the commonly used P67.6 antibody that targets the IgV domain of CD33. Our results demonstrated that the presence of rs12459419 T allele was highly associated with lower cell surface CD33 MFI (P < 1.0E−6; Table 1; Fig 1D) and that genotype associations were balanced within each arm, risk group, and FLT3-ITD status (Table 1; Data Supplement).
Table 1.
Association of rs12459419 Genotype With Cell Surface CD33 Expression in Leukemic Cells From Patients With AML (AAML0531 Clinical Trial)
Correlation of RNA-seq data with CD33 cell surface expression levels by MFI showed significant positive correlation with total CD33 mRNA expression (Spearman r = 0.424; P < .001; Fig 1E) and negative correlation of the D2-CD33 isoform with CD33 MFI (Spearman r = –0.417; P = .001; Fig 1F), further supporting our speculation that the lower cell surface expression of CD33 might be, in part, a result of the lack of detection of the expressed D2-CD33 isoform by current diagnostic immunophenotypic panels.
rs12459419 Genotype Defines the Clinical Response to GO
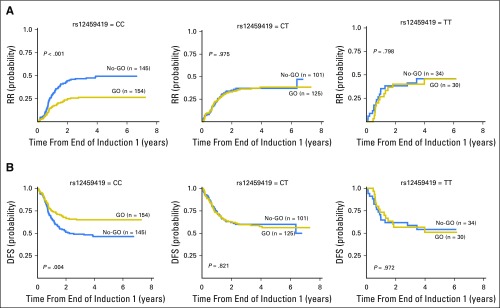
Given that the rs12459419 variant influences the ESE binding site and is associated with the expression of the variant CD33 lacking the GO binding domain, we hypothesized that the response to GO would be limited to the rs12459419 genotype encoding the full-length protein. We correlated the rs12459419 SNP genotype with clinical end points OS, EFS, DFS, RR, and treatment-related mortality (TRM) in 816 patients in the GO versus the No-GO arms (No-GO arm, n = 408; GO arm, n = 408). Comparisons between patients included in the analyses with eligible AAML0531 patients who consented to banking (n = 816) versus those who were not included in the analyses (n = 163) showed no statistically significant differences in the outcome (Data Supplement). Patients with the rs12459419 homozygous CC had an RR of 26% ± 7% in the GO arm, whereas those in the No-GO arm had an RR of 49% ± 9% (HR, 0.468; P < .001). The corresponding DFS rates for patients with the CC genotype in the GO and No-GO arms were 65% ± 7% and 46% ± 9%, respectively (HR, 0.597; P = .004; Table 2; Fig 2). In contrast, GO exposure provided no clinical benefit in RR in patients with the heterozygous CT or the homozygous TT genotype (CT: 38% ± 9% v 37% ± 10% [GO v No-GO]; P = .975; TT: 46% ± 20% v 46% ± 20% [GO v No-GO]; P = .798) or DFS (CT: 56% ± 9% v 60% ± 10% [GO v No-GO]; P = .821; TT: 51% ± 20% v 54% ± 18% [GO v No-GO]; P = .972; Fig 2). Given the similar impact of GO on the CT and TT genotypes, these two cohorts were combined in all additional analyses (Data Supplement). We also performed tests of interaction, and our results show a significant interaction between SNP genotype and treatment of DFS (P = .008) and RR (.031).
Table 2.
Association of CD33 SNP rs12459419 With Clinical Outcome by Treatment Arm in Patients Treated in the COG AAML0531 Clinical Trial
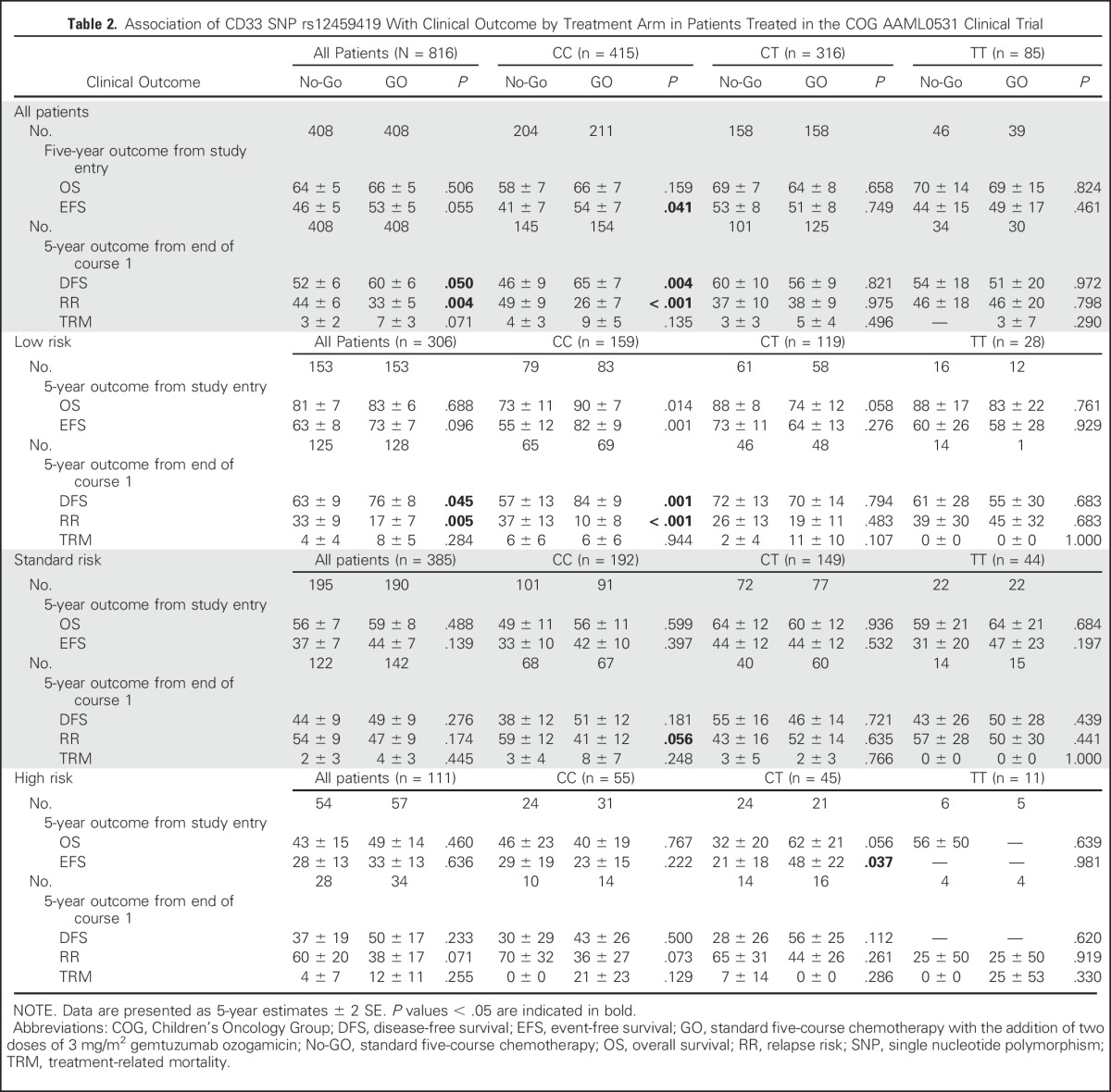
Fig 2.
Association of rs12459419 genotypes with clinical response by treatment arm in patients treated in AAML0531. (A) Differences in relapse risk (RR) and (B) disease-free survival (DFS) from the end of course 1 in patients with the CC, CT, or TT genotype in the standard five-course chemotherapy with the addition of two doses of 3 mg/m2 gemtuzumab ozogamicin (GO) versus the standard five-course chemotherapy alone (No-GO) arm.
Clinical Outcome for Patients With rs12459419 Genotypes by Risk Group and CD33 Expression
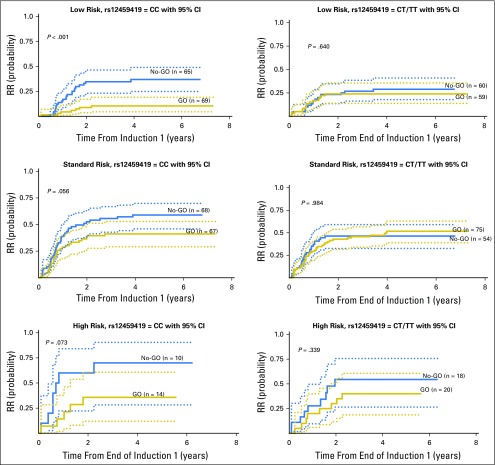
Previous studies have reported that the clinical impact of GO is limited to those with favorable cytogenetics or to those with high CD33 expression.12,13 Thus, we evaluated the impact of the CD33 genotype in the efficacy of GO in different risk groups, as well as in high versus low CD33 expression cohorts. Patients in the LR group, as defined by the presence of t(8;21) or Inv(16) translocations or NPM1 or CEBPa mutations, with the CC genotype treated with GO had an RR from remission of 10% ± 8% versus 37% ± 13% for No-GO patients (P < .001). In contrast, those with LR disease with the CT or TT genotype did not benefit from GO (Table 2; Fig 3). Standard-risk patients with the CC genotype had an RR of 41% ± 12% versus 59% ± 12% (P = .056); high-risk patients had an RR of 36% ± 27% versus 70% ± 32% (P = .073) for the GO and No-GO arms, respectively. As with the LR patients, addition of GO in patients with the CT or TT genotype showed no benefit (Table 2; Fig 3). These data provide the rationale that GO is efficacious in patients with the CC genotype (expression of antibody-binding domain), with a significant effect in patients in the LR group and a moderate impact in those in the high- and standard-risk groups.
Fig 3.
Association of rs12459419 genotypes with relapse risk (RR) for patients treated in AAML0531 in the low-, standard-, and high-risk groups in the standard five-course chemotherapy with the addition of two doses of 3 mg/m2 gemtuzumab ozogamicin (GO) versus the standard five-course chemotherapy alone (No-GO) arm.
On the basis of our recently published study on the impact of CD33 expression and the efficacy of GO, we evaluated the association of rs12459419 genotypes in patients with low and high CD33 cell surface expression quartile 1 (n = 206) and quartiles 2 to 4 (n = 610), respectively, as defined by Pollard et al.22 As expected, patients with the CC genotype (n = 353 [58%]) were more common than those with the CT or TT genotype (n = 260 [42%]) in the higher CD33 expression cohort (quartiles 2 to 4). Among patients in quartile 1, the CT or TT genotype (n = 144 [70%]) was more common than the CC genotype (n = 62 [30%]). The Data Supplement provides the genotype distribution of patients in the whole patient cohort, as well as within each risk group for quartile 1 and quartiles 2 to 4. In patients with high CD33 expression, those with the CC genotype in the GO arm showed significant improvement in RR compared with those in the No-GO arm (P = .001), but this effect was not observed in patients with the CT or TT genotype (P = .125). As expected, there were fewer patients with the CC genotype (n = 44 [28%]) than with the CT or TT genotype (n = 110 [72%]) in the lower CD33 expression (quartile 1) cohort. However, even within this small subset, patients with the CC genotype in the GO arm showed a similar magnitude and a trend toward improvement in RR compared with those in the No-GO arm (P = .055), but this trend was not seen in patients with the CT or TT genotype (P = .125; Data Supplement), thus further emphasizing the CD33 genotype as a significant biomarker, irrespective of CD33 cell surface expression. We further evaluated the impact of this SNP in response to GO within the high CD33 expression cohort on the basis of risk status. Despite a limited sample size for LR and standard-risk patients with high CD33 expression, LR patients with the CC genotype who received GO had a significantly lower RR compared with those in the No-GO arm (P = .003). In standard-risk patients with the CC genotype, there was a trend in a consistent direction toward GO response (P = .09; Data Supplement).
A multivariate Cox regression analysis that included CD33 genotype, risk status, and CD33 expression demonstrated that the CD33 CC genotype was an independent predictor of response to GO (HR, 0.45 and P < .001 for RR; HR, 0.57 and P = .003 for DFS). In this model, LR status remained significantly associated with response to GO, whereas CD33 expression was no longer associated with response to GO (Data Supplement).
All the results were consistent for the rs3865444 promoter SNP that occurs in linkage disequilibrium (LD) with rs12459419 (Data Supplement).
DISCUSSION
The addition of GO to standard chemotherapy has shown promising results in AML, with variable outcomes in different trials. Results from recent meta-analyses in adults with AML show that GO, when added to standard induction chemotherapy, improves survival in patients with favorable-risk and intermediate-risk cytogenetics.10,11 In pediatric AML, COG AAML0531 demonstrated that patient outcomes are modestly improved when GO is added to standard chemotherapy.13 This study further demonstrated wide interpatient variation (approximately two log-fold) in CD33 expression in leukemic cells and indicated that the impact of GO was limited to those with higher CD33 expression.27-29 These results were validated in the French ALFA trial30 and are consistent with in vitro studies relating GO-induced cytotoxicity to an abundance of cell surface CD33.31
CD33 SNP rs12459419 has been associated with the expression of an alternatively spliced variant of CD33 lacking the GO binding site (IgV domain).19 Results from this study show that this SNP defines a genotypic-driven prediction of response to GO. In addition to defining a clinical response to GO, and given that all available diagnostic antibodies also target the CD33 IgV domain, this SNP defines the ability to reliably identify CD33 expression level.
We reported previously on the coding and regulatory polymorphisms in CD3316,17 and showed an association between these SNPs and cell surface CD33 expression as well as clinical outcome. In the context of the recent COG randomized trial, we were able to extend these findings and highlight the significance of SNP rs12459419 in defining response to GO. This SNP is unique by virtue of its location in the ESE region of exon2, and it regulates the splicing of CD33 exon2. Using transcriptome-sequencing data from AML diagnostic leukemic cells, we demonstrated that the presence of the T allele leads to a higher expression of a shorter, alternatively spliced CD33 transcript that lacks exon2 (the D2-CD33 isoform), thus lacking the GO-binding IgV domain of CD33. We further demonstrated a strong association between the rs12459419 genotype and CD33 cell surface expression (detection) on leukemic blasts.
Consistent with the fact that GO recognizes the IgV domain, our studies identified rs12459419 as a significant determinant of an eventual benefit from GO when added to conventional chemotherapy. Patients with the CC genotype had an almost 50% reduction in RR and experienced superior DFS with the addition of GO, compared with those receiving standard chemotherapy alone. Conversely, patients with at least one T allele (which increases the production the CD33 isoform lacking the IgV domain) did not experience any improvement in outcome when GO was administered. This was true for patients in all risk categories. As an example, the OS from study entry for patients with the CC genotype in the LR group was 90% in the GO arm versus 73% for patients with the CC genotype in the No-GO arm (Table 2).
This finding has multiple significant clinical as well as biologic implications. Because most available CD33 antibodies used for the detection or generation of CD33-targeted antibody-drug conjugates (ADCs) are directed against the IgV domain of the CD33 protein, the truncated CD33 variant lacking the IgV domain cannot be detected by hP67.7 (used in the generation of GO and the most common antibody in diagnostic panels). As a result, one would predict that patients with the CC genotype would respond to GO, whereas those with the CT genotype would have no response to GO. It is of interest that the heterozygous CT genotype, which would be expected to have both the full length as well as the short isoform, shows no response to GO. This is a presence of biologically important observation because it would indicate that the short CD33 isoform interferes with the cytotoxicity of GO. This might be a result of an interference with internalization of the ligated CD33 or of altered trafficking of the GO-CD33 conjugated complex.
CD33 rs12459419 and linked promoter SNP rs3865444 have been associated previously to late-onset Alzheimer’s disease in genome-wide association analyses32 and to reduced CD33 expression levels in the hematopoietic-derived microglial cells.33,34 Although in-depth mechanistic studies of CD33 SNPs are warranted, our results show that the CD33 rs12459419 genotype allows for the identification of patients expressing the CD33 isoform lacking the antibody-binding site for GO. In a recent report, we characterized CD33 splice variants and demonstrated that this variant is unable to serve as a target of GO.35 Perhaps more importantly, it holds promise as a marker for selecting patients who are likely to benefit from the addition of GO to chemotherapy (CC genotype) regardless of the clinical risk or the CD33 cell surface expression levels. The fact that patients heterozygous for rs12459419 (CT) do not experience any benefit from the addition of GO indicates that the prediction of outcome by SNP is more powerful than the prediction by CD33 cell surface expression and suggests that CD33 expression alone may not fully inform on the GO responsiveness of the leukemic cell subsets most relevant to disease progression or relapse.
Knowledge of the CD33 genotype and prediction of response to GO provides opportunities to use patient genotypes for selecting CD33-targeted therapies. Given that only one half of patients are expected to have a response to GO, we propose that all prior GO-containing studies be re-evaluated for response on the basis of the patients’ CD33 genotype. Furthermore, the fact that patients with the full-length CD33 respond to GO argues for the potential efficacy of a CD33-ADC using an antibody targeting the IgC domain, which is shared by all CD33 isoforms, thus creating the next generation of CD33 ADCs with potential benefit for all patients with AML. Given the significance of this observation and its impact in therapeutic decision making regarding which patients should receive GO, external validation of these findings is required. These findings will likely be relevant to other CD33-targeted therapies such as SGN-CD33A and lintuzumab.
ACKNOWLEDGMENT
We thank the COG AML reference laboratory for providing specimens for the study. We also thank Sommer Castro and Rhonda Ries for providing specimens and help with RNA-seq analysis.
Footnotes
Supported by the National Institutes of Health under Award Nos. U10CA180899, U10CA180886, U10CA98413, and U10CA098543, as well as by NCI-R21CA155524 (J.K.L and R.B.W) and R01CA133881 (R.A).
Clinical trial information: NCT00372593.
AUTHOR CONTRIBUTIONS
Conception and design: Jatinder K. Lamba, Soheil Meshinchi
Collection and assembly of data: All authors
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
CD33 Splicing Polymorphism Determines Gemtuzumab Ozogamicin Response in De Novo Acute Myeloid Leukemia: Report From Randomized Phase III Children’s Oncology Group Trial AAML0531
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Jatinder K. Lamba
Patents, Royalties, Other Intellectual Property: Patent application No. 62/319284, entitled CD33-Targeted Cancer Therapy
Lata Chauhan
No relationship to disclose
Miyoung Shin
No relationship to disclose
Michael R. Loken
Employment: HematoLogics
Leadership: HematoLogics
Stock or Other Ownership: HematoLogics
Jessica A. Pollard
Research Funding: Bayer HealthCare Pharmaceuticals
Yi-Cheng Wang
No relationship to disclose
Rhonda E. Ries
No relationship to disclose
Richard Aplenc
Honoraria: Sigma-Tau Pharmaceuticals
Expert Testimony: Dana Wiggins
Travel, Accommodations, Expenses: Sigma-Tau Pharmaceuticals
Betsy A. Hirsch
No relationship to disclose
Susana C. Raimondi
No relationship to disclose
Roland B. Walter
Stock or Other Ownership: Amphivena Therapeutics
Consulting or Advisory Role: Amphivena Therapeutics, Covagen, Emergent Biosolutions, Seattle Genetics, Agios, Tolero Pharmaceuticals, Race Oncology, BiolineRx
Research Funding: Amgen, Amphivena Therapeutics, Abbvie, Stemline Therapeutics, Celator, Arog, Pharmacyclics, ADC Therapeutics, Seattle Genetics, Covagen AG
Irwin D. Bernstein
Patents, Royalties, Other Intellectual Property: Royalty from CD33 antibody used for leukemia diagnosis from Becton Dickinson; royalty for intellectual property (patent) on Notch-induced cord blood stem cell expansion
Alan S. Gamis
Consulting or Advisory Role: Pfizer, Novartis
Todd A. Alonzo
No relationship to disclose
Soheil Meshinchi
No relationship to disclose
REFERENCES
- 1.Walter RB, Appelbaum FR, Estey EH, et al. : Acute myeloid leukemia stem cells and CD33-targeted immunotherapy. Blood 119:6198-6208, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krupka C, Kufer P, Kischel R, et al. : CD33 target validation and sustained depletion of AML blasts in long-term cultures by the bispecific T-cell-engaging antibody AMG 330. Blood 123:356-365, 2014 [DOI] [PubMed] [Google Scholar]
- 3.Bross PF, Beitz J, Chen G, et al. : Approval summary: Gemtuzumab ozogamicin in relapsed acute myeloid leukemia. Clin Cancer Res 7:1490-1496, 2001 [PubMed] [Google Scholar]
- 4.Petersdorf SH, Kopecky KJ, Slovak M, et al. : A phase 3 study of gemtuzumab ozogamicin during induction and postconsolidation therapy in younger patients with acute myeloid leukemia. Blood 121:4854-4860, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burnett AK, Hills RK, Hunter AE, et al. : The addition of gemtuzumab ozogamicin to low-dose Ara-C improves remission rate but does not significantly prolong survival in older patients with acute myeloid leukaemia: Results from the LRF AML14 and NCRI AML16 pick-a-winner comparison. Leukemia 27:75-81, 2013 [DOI] [PubMed] [Google Scholar]
- 6.Burnett AK, Hills RK, Milligan D, et al. : Identification of patients with acute myeloblastic leukemia who benefit from the addition of gemtuzumab ozogamicin: Results of the MRC AML15 trial. J Clin Oncol 29:369-377, 2011 [DOI] [PubMed] [Google Scholar]
- 7.Burnett AK, Russell NH, Hills RK, et al. : Addition of gemtuzumab ozogamicin to induction chemotherapy improves survival in older patients with acute myeloid leukemia. J Clin Oncol 30:3924-3931, 2012 [DOI] [PubMed] [Google Scholar]
- 8.Castaigne S, Pautas C, Terré C, et al. : Effect of gemtuzumab ozogamicin on survival of adult patients with de-novo acute myeloid leukaemia (ALFA-0701): A randomised, open-label, phase 3 study. Lancet 379:1508-1516, 2012 [DOI] [PubMed] [Google Scholar]
- 9.Thol F, Schlenk RF: Gemtuzumab ozogamicin in acute myeloid leukemia revisited. Expert Opin Biol Ther 14:1185-1195, 2014 [DOI] [PubMed] [Google Scholar]
- 10.Hills RK, Castaigne S, Appelbaum FR, et al. : Addition of gemtuzumab ozogamicin to induction chemotherapy in adult patients with acute myeloid leukaemia: A meta-analysis of individual patient data from randomised controlled trials. Lancet Oncol 15:986-996, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loke J, Khan JN, Wilson JS, et al. : Mylotarg has potent anti-leukaemic effect: A systematic review and meta-analysis of anti-CD33 antibody treatment in acute myeloid leukaemia. Ann Hematol 94:361-373, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper TM, Franklin J, Gerbing RB, et al. : AAML03P1, a pilot study of the safety of gemtuzumab ozogamicin in combination with chemotherapy for newly diagnosed childhood acute myeloid leukemia: A report from the Children’s Oncology Group. Cancer 118:761-769, 2012 [DOI] [PubMed] [Google Scholar]
- 13.Gamis AS, Alonzo TA, Meshinchi S, et al. : Gemtuzumab ozogamicin in children and adolescents with de novo acute myeloid leukemia improves event-free survival by reducing relapse risk: Results from the randomized phase III Children’s Oncology Group trial AAML0531. J Clin Oncol 32:3021-3032, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarke WT, Marks PW: Gemtuzumab ozogamicin: Is there room for salvage? Blood 116:2618-2619, 2010 [DOI] [PubMed] [Google Scholar]
- 15.Ravandi F, Estey EH, Appelbaum FR, et al. : Gemtuzumab ozogamicin: Time to resurrect? J Clin Oncol 30:3921-3923, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamba JK, Pounds S, Cao X, et al. : Coding polymorphisms in CD33 and response to gemtuzumab ozogamicin in pediatric patients with AML: A pilot study. Leukemia 23:402-404, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mortland L, Alonzo TA, Walter RB, et al. : Clinical significance of CD33 nonsynonymous single-nucleotide polymorphisms in pediatric patients with acute myeloid leukemia treated with gemtuzumab-ozogamicin-containing chemotherapy. Clin Cancer Res 19:1620-1627, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. ESE Finder: http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home.
- 19.Malik M, Chiles J, III, Xi HS, et al. : Genetics of CD33 in Alzheimer’s disease and acute myeloid leukemia. Hum Mol Genet 24:3557-3570, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malik M, Simpson JF, Parikh I, et al. : CD33 Alzheimer’s risk-altering polymorphism, CD33 expression, and exon 2 splicing. J Neurosci 33:13320-13325, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raj T, Ryan KJ, Replogle JM, et al. : CD33: Increased inclusion of exon 2 implicates the Ig V-set domain in Alzheimer’s disease susceptibility. Hum Mol Genet 23:2729-2736, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollard JA, Loken M, Gerbing RB, et al. : CD33 expression and its association with gemtuzumab ozogamicin response: Results from the randomized phase III Children’s Oncology Group trial AAML0531. J Clin Oncol 34:747-755, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pollard JA, Alonzo TA, Loken M, et al. : Correlation of CD33 expression level with disease characteristics and response to gemtuzumab ozogamicin containing chemotherapy in childhood AML. Blood 119:3705-3711, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. doi: 10.1016/j.leukres.2007.10.002. Wells DA, Loken MR: Flow cytometric mean fluorescence intensity: The biophysics behind the number. Leuk Res 32:845-846, 2008. [DOI] [PubMed] [Google Scholar]
- 25. Kaplan EL, Meier P: Nonparametric estimation from incomplete observations. J Am Stat Assoc 53:457-481, 1958. [Google Scholar]
- 26. Cox DR: Regression models and life tables. J R Stat Soc Series B 34:187-220, 1972. [Google Scholar]
- 27.Jager E, van der Velden VH, te Marvelde JG, et al. : Targeted drug delivery by gemtuzumab ozogamicin: Mechanism-based mathematical model for treatment strategy improvement and therapy individualization. PLoS One 6:e24265, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jedema I, Barge RM, van der Velden VH, et al. : Internalization and cell cycle-dependent killing of leukemic cells by gemtuzumab ozogamicin: Rationale for efficacy in CD33-negative malignancies with endocytic capacity. Leukemia 18:316-325, 2004 [DOI] [PubMed] [Google Scholar]
- 29.Walter RB, Gooley TA, van der Velden VH, et al. : CD33 expression and P-glycoprotein-mediated drug efflux inversely correlate and predict clinical outcome in patients with acute myeloid leukemia treated with gemtuzumab ozogamicin monotherapy. Blood 109:4168-4170, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olombel G, Guerin E, Guy J, et al. : The level of blast CD33 expression positively impacts the effect of gemtuzumab ozogamicin in patients with acute myeloid leukemia. Blood 127:2157-2160, 2016 [DOI] [PubMed] [Google Scholar]
- 31.Walter RB, Raden BW, Kamikura DM, et al. : Influence of CD33 expression levels and ITIM-dependent internalization on gemtuzumab ozogamicin-induced cytotoxicity. Blood 105:1295-1302, 2005 [DOI] [PubMed] [Google Scholar]
- 32.Naj AC, Jun G, Beecham GW, et al. : Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet 43:436-441, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bradshaw EM, Chibnik LB, Keenan BT, et al. : CD33 Alzheimer’s disease locus: Altered monocyte function and amyloid biology. Nat Neurosci 16:848-850, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griciuc A, Serrano-Pozo A, Parrado AR, et al. : Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron 78:631-643, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laszlo GS, Harrington KH, Gudgeon CJ, et al. : Expression and functional characterization of CD33 transcript variants in human acute myeloid leukemia. Oncotarget 7:43281-43294, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]