Abstract
High levels of RNA polymerase III gene transcription are achieved by facilitated recycling of the polymerase on transcription factor IIIB (TFIIIB)-DNA complexes that are stable through multiple rounds of initiation. TFIIIB-DNA complexes in yeast comprise the TATA-binding protein (TBP), the TFIIB-related factor TFIIIB70, and TFIIIB90. The high stability of the TFIIIB-DNA complex is conferred by TFIIIB90 binding to TFIIIB70-TBP-DNA complexes. This stability is thought to result from compound bends introduced in the DNA by TBP and TFIIIB90 and by protein–protein interactions that obstruct DNA dissociation. Here we present biochemical evidence that the high stability of TFIIIB-DNA complexes results from kinetic trapping of the DNA. Thermodynamic analysis shows that the free energies of formation of TFIIIB70-TBP-DNA (ΔG° = −12.10 ± 0.12 kcal/mol) and TFIIIB-DNA (ΔG° = −11.90 ± 0.14 kcal/mol) complexes are equivalent whereas a kinetic analysis shows that the half-lives of these complexes (46 ± 3 min and 95 ± 6 min, respectively) differ significantly. The differential stability of these isoenergetic complexes demonstrates that TFIIIB90 binding energy is used to drive conformational changes and increase the barrier to complex dissociation.
RNA polymerase (pol) III transcribes a variety of nontranslated RNA genes encoding transfer RNAs, 5S ribosomal RNA, U6 snRNA, and other small cellular RNAs (1). In Saccharomyces cerevisiae, transcription of these genes is directed by the initiation factor TFIIIB, which is assembled upstream of the start site by other factors (TFIIIA and/or TFIIIC), bound to downstream promoter elements. Yeast TFIIIB is a heterotrimeric complex comprising the TATA-binding protein (TBP), a TFIIB-related component, TFIIIB70 (Brf1), and a SANT domain protein, TFIIIB90 (B′′). Structural and functional homologs of these proteins have been identified in human cells and confer TFIIIB activity (termed TFIIIB-α in ref. 2) for the transcription of tRNA and related pol III genes having internal promoter elements (2–4). Additionally, human cells contain a second TFIIIB activity (TFIIIB-β) that is used by pol III genes whose promoter elements are located upstream of the start site (e.g., U6 snRNA and 7SK RNA). TFIIIB-β differs from TFIIIB-α in that it contains a different TFIIB-related component (termed BRFU or hTFIIIB50) and associated proteins (2, 3). Further complexity among TFIIIB complexes is suggested by the identification of three splice variants of human Brf1 (4). One of these variants, Brf2, appears to be active in the transcription U6 snRNA.
Pol III genes are among the most actively transcribed genes in eukaryotic cells. High rates of pol III gene transcription are achieved through the facilitated recycling of pol molecules (5–8) on TFIIIB complexes that remain bound to the DNA for multiple rounds of initiation (9, 10). The stability of TFIIIB-DNA complexes is therefore a key property that enables rapid reinitiation by eliminating rate-limiting steps in transcription complex assembly. Yeast TFIIIB-DNA complexes are considered to be exceptionally stable based on their ability to resist dissociation by heparin and molar salt concentrations that strip TFIIIC and TFIIIA from the DNA (9). Interestingly, this resistance to dissociation does not depend on the DNA sequence, which is highly variable in the region bound by TFIIIB, but reflects a unique property of the nucleoprotein complex.
The structure of TFIIIB-DNA has been probed extensively by site-specific DNA-protein photocrosslinking, hydroxyl radical and DNase I footprinting, missing-nucleoside interference, native gel analysis of complex assembly and DNA bending, studies with mutant proteins, and other methods (refs. 11–16 and references therein). These studies provide a conceptual view of the TFIIIB-DNA complex as a proteinaceous cage (terminology coined by E. P. Geiduschek at Asilomar, 1998) that makes close contacts with both DNA strands over a distance of ≈40 bp (11, 14, 15). Within the complex, the path of the DNA is distorted by compound bends induced by TBP at the TATA box (14, 17, 18) and by TFIIIB90 in the region between the TATA box and the transcription start site (15). Heparin resistance is an intrinsic property of TFIIIB-DNA complexes (19) that is attained, in the stepwise assembly of a pol III transcription complex, after the binding of TFIIIB90 to TBP-TFIIIB70-TFIIIC-DNA complexes (13). Thus, DNA bending by TFIIIB90 and TFIIIB90-dependent protein–protein interactions are thought to play a critical role in TFIIIB-DNA complex stabilization (15).
The ability to assemble TBP-DNA, TBP-TFIIIB70-DNA (B′-DNA), and TFIIIB-DNA on TATA box-containing templates, independent of TFIIIC, has been exploited to study the structure, function, and biochemical properties of these complexes. Using this approach, we have previously found that TFIIIB70 makes a positive cooperative contribution to the assembly of B′-DNA, increasing the affinity of TBP for the TATA box up to 17-fold (20, 21). In the present study, we have examined the energetic and kinetic contribution of TFIIIB90 to the assembly of the TFIIIB-DNA complex. Our biochemical data indicate that the high stability of the TFIIIB-DNA complex is achieved by kinetic trapping of the DNA rather than by an increase in the free energy of the complex relative to B′-DNA.
Materials and Methods
Protein Preparations.
Recombinant S. cerevisiae TBP was prepared, quantified, and stored as described (22, 23). Recombinant TFIIIB70 was prepared under denaturing conditions and purified on Ni2+-NTA agarose (Qiagen, Chatsworth, CA) following the manufacturer's recommendations. Further purification was achieved by chromatography on a POROS HS column. Most of the TFIIIB70 remained bound to this resin at 2.0 M NaCl and was eluted with 6 M guanidine hydrochloride. The nonionic detergent Brij-58 was added to a final concentration of 0.01%, and the protein was renatured by dialysis as described (20) except that EDTA was omitted and ZnCl2 was increased to 20 μM. TFIIIB70 concentration and purity was determined as described (20).
Recombinant TFIIIB90 with a C-terminal histidine tag (pET21d clone provided by G. Kassavetis, University of California, San Diego) was expressed in Escherichia coli BL21(DE3) containing the ArgU expression plasmid pUBS520 (20). The protein was extracted under native conditions and purified on Ni2+-NTA agarose (Qiagen) following the manufacturer's recommendations. After elution with buffer D500 (20 mM Hepes⋅KOH, pH 7.8/500 mM NaCl/7 mM MgCl2/10 mM β-mercaptoethanol/0.01% Tween-20/10% glycerol) containing 250 mM imidizole, the protein (approximately 20% pure) was dialyzed into buffer containing 20 mM Hepes⋅KOH (pH 7.8), 250 mM KCl, 1 mM EDTA, 1 mM DTT, 0.02% Tween-20, and 10% glycerol, loaded onto a column of POROS HS 50 and eluted with a 0.25–1.0 M KCl gradient (24). TFIIIB90 eluted at about 500 mM KCl and was more than 70% pure. The concentration of TFIIIB90 was determined by absorbance at 280 nm by using ɛ = 26,920 M−1⋅cm−1 (calculated by using gcg software).
Preparation of DNA Probe.
Labeling and purification of a probe derived from the S. cerevisiae SNR6 gene (plasmid pM4,21) was performed following published protocols (25). The pM4 DNA was end-labeled after HindIII digestion and then cleaved with BsrDI. This process generates a fragment of 204 bp in which the SNR6 TATA element is located 134 bp from the labeled 3′ end.
Quantitative Equilibrium and Kinetic DNase I Footprint Experiments.
Equilibrium DNase I footprint titration experiments were conducted as described (20, 22, 25). In all experiments, the concentration of 32P-labeled DNA (1–10 pM) was significantly lower than the Kd of the TBP-DNA interaction. This allows the assumption to be made that [protein]total ≈ [protein]free in the analysis of equilibrium binding data. The 32P-labeled DNA and proteins were allowed to equilibrate at 30°C for at least 45 min in 100-μl volumes of assay buffer [25 mM Bis⋅Tris/100 mM KCl/5 mM MgCl2/1 mM CaCl2/1 mM DTT/1 μg/ml poly(dG⋅dC)/0.01% Brij-58 at pH 7.0]. Independent experiments showed that under these conditions, TBP-, B′-, and TFIIIB-DNA complex assembly had reached equilibrium and that the TFIIIB-DNA complexes were resistant to dissociation by heparin (200 μg/ml) as reported (19). The reaction mixtures were exposed to DNase I for 2 min and then quenched by addition of 35 μl of 50 mM EDTA followed by 800 μl of precipitation solution (26).
In kinetic experiments designed to determine the stability of the protein-DNA complexes, sufficient protein was added to saturate the 32P-labeled DNA, and the reactions were equilibrated as described above. The dissociation reaction was initiated by the addition of poly[dA-dT]/[dA-dT] oligonucleotide competitor (211 μM/bp) in excess of the concentration of TBP. At various times after the addition of the competitor, samples were exposed to DNase I for 30 s, and the reaction was quenched as described above.
The DNase I reaction products were separated on denaturing 8%
polyacrylamide gels and imaged by exposure to a phosphor storage plate
by using a PhosphorImager (Molecular Dynamics). Quantitation of the
digital images was conducted by using imagequant (Molecular
Dynamics) software as described (25, 27). Briefly, the intensities of
the bands corresponding to the TATA box were corrected for background,
standardized for the total amount of DNA loaded in each lane, and
matched with the corresponding TBP concentration or time after the
addition of competitor. A plot of band intensity versus TBP
concentration or time yields a transition curve of the fractional
protection, pi, of the TATA box for the reaction. Nonlinear
least-squares analysis of the data were performed by using Microcal
ORIGIN 5.0 or nonlin software.
The fractional saturation
(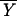 TATA) of the TATA box in
equilibrium and kinetic experiments was obtained by fitting the data
against:
TATA) of the TATA box in
equilibrium and kinetic experiments was obtained by fitting the data
against:
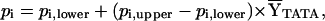 |
1 |
where
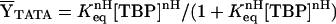 |
2 |
or
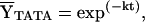 |
3 |
and where pi,lower and pi,upper are the lower and upper end points for the equilibrium or kinetic transition curves, respectively, and nH is the Hill coefficient. Fitting of equilibrium binding data to the Hill binding polynomial (Eq. 2) yields the equilibrium association constant, Keq. Fitting of kinetic data to the single-exponential dissociation function (Eq. 3) yields the dissociation rate constant, k. The Gibbs free energy of binding ΔG° is related to the equilibrium association constant, Keq, by ΔG° = −RT ln Keq. Eq. 2 reduces to the single-site binding expression when nH = 1.0. The reported thermodynamic and kinetic parameters were determined by global (i.e., simultaneous) analysis of multiple scaled data sets to Eqs. 2 and 3, respectively.
Results
TFIIIB90 Alone Does Not Contribute to the TBP-DNA Binding Reaction.
As a starting point for equilibrium binding studies of the TFIIIB-DNA complex, a series of quantitative DNase I footprint titrations were performed to investigate possible intrinsic DNA binding by TFIIIB90 and to assess the effect, if any, of TFIIIB90 on the interaction of TBP with DNA. The latter experiment was prompted by studies showing that TFIIIB90 binds to TBP-DNA complexes (24) and photocrosslinks weakly to DNA in the presence of TBP (12). Titrations of TFIIIB90 by itself on the yeast SNR6 promoter (yU6) resulted in no detectable DNase I protections anywhere on the DNA fragment at concentrations of the factor up to 0.25 μM (data not shown). Thus, under the experimental conditions used in this work, TFIIIB90 exhibits no intrinsic DNA binding activity. Based on these data, a simulated single-site binding curve (Eq. 2 with nH = 1) yielded a lower limit for the intrinsic DNA binding affinity of TFIIIB90 of ≈50 μM. We next performed DNase I footprint titrations of TBP in the presence or absence of TFIIIB90 (500 nM). This concentration of TFIIIB90 is 2-fold greater than that reported to quantitatively convert TBP-DNA into TFIIIB90-TBP-DNA complexes (24). No extension of the TBP footprint by TFIIIB90 was observed (data not shown). Moreover, the binding isotherms derived from these experiments demonstrated that TFIIIB90 did not contribute to the free energy of formation of TBP-DNA complexes. Similarly, titrations of TFIIIB90 up to 250 nM performed at a constant limiting concentration of TBP (10 nM, which is sufficient to bind 5–10% of the yU6 DNA) showed no qualitative or quantitative change in the DNase I footprint. Thus, either the binding of TFIIIB90 to TBP-DNA does not occur under our reaction conditions or the interaction is noncooperative with respect to the TBP-DNA binding reaction.
Energetics of TFIIIB-DNA Complex Assembly.
To examine the energetics of TFIIIB-DNA complex assembly, we extended the approach used previously to quantify the cooperative assembly of B′-DNA complexes on the SNR6 TATA box (21). The methodology and experimental design for this study involved the performance of DNase I footprint titrations of TBP (i) in the absence of additional factors, (ii) as a function of TFIIIB70 concentration, and (iii) as a function of TFIIIB70 concentration at a series of TFIIIB90 concentrations. In each of these experiments, protein-DNA complex assembly was quantified by measuring saturation of the TATA box by TBP. The resulting equilibrium binding isotherms were individually fit to Eqs. 1 and 2 and then replicate experiments of each type were analyzed globally (i.e., simultaneously) to find the best overall fit. All of the TBP binding isotherms adequately described the single-site binding polynomial (Table 1, nH = 1 within experimental error), consistent with a single molecule of TBP binding to the TATA box. The quality and reproducibility of the data are demonstrated by the high confidence limits of the free energies determined from curve fitting (Table 1, ΔG°).
Table 1.
Summary of thermodynamic and kinetic data
| TBP-DNA | B′-DNA | TFIIIB-DNA | |
|---|---|---|---|
| ΔG°Total* | −10.50 ± 0.10 | −12.10 ± 0.12 | −11.90 ± 0.14 |
| nH† | 1.12 ± 0.20 | 1.30 ± 0.30 | 1.04 ± 0.20 |
| kd‡ | 80 (± 5) × 10−5 | 25 (± 1.4) × 10−5 | 12 (± 0.8) × 10−5 |
| t1/2 | 15.0 ± 1 min | 46.0 ± 3 min | 95.0 ± 6 min |
Total Gibbs free energy of association for TBP binding to the TATA element on yU6 DNA was determined for each complex from a global analysis of multiple data sets by using the Hill binding polynomial (see Fig. 1 and Materials and Methods). Gibbs free energy is measured in kcal/mol.
Hill coefficient.
Kinetic dissociation constants (kd) were determined from a global analysis of three or more kinetic progress curves for each complex by using a single exponential dissociation function. Dissociation constants are measured in sec−1. The half-life, t1/2 = ln2/kd. The errors associated with all of the parameters were determined during curve fitting with nonlin (thermodynamics) or Microcal Origin 5.0 (kinetics) software.
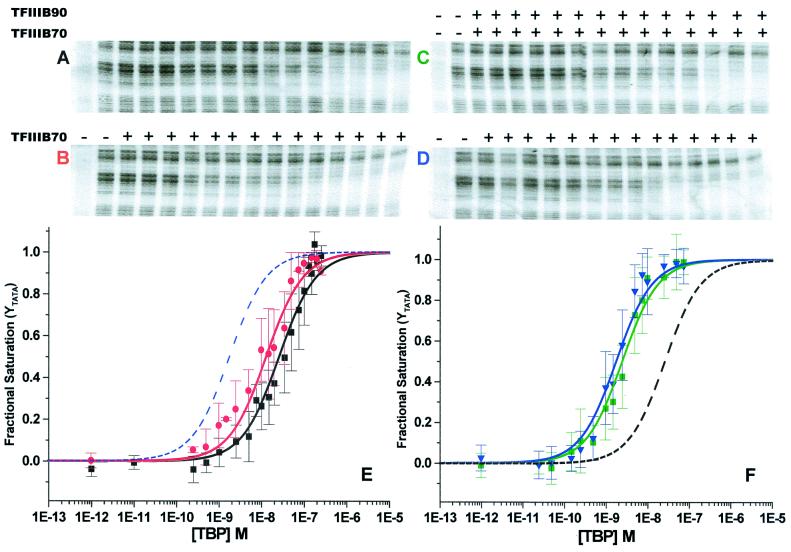
A representative set of DNase I footprints involving TBP titrations on the yU6 TATA probe are shown in Fig. 1 A–D. As reported previously, TBP binding to the yU6 TATA box results in a clear and specific DNase I protection pattern (Fig. 1A). The equilibrium binding isotherm resulting from a global analysis of TATA box protection in TBP alone titrations (Fig. 1E, black isotherm) yields a ΔG° of −10.50 ± 0.10 kcal/mol (Table 1), in good agreement with previous studies performed under comparable experimental conditions (21). When a constant amount of TFIIIB70 is added to the TBP titrations (Fig. 1D), TATA box protection commences at lower TBP concentrations. This dependence of the TBP-DNA interaction on TFIIIB70 is clearly seen in the global binding isotherm (Fig. 1F, blue isotherm) as a shift to lower TBP concentrations relative to titrations of TBP alone (Fig. 1F, dashed line). The concentration of TFIIIB70 (100 nM) in this experiment had previously been determined to be saturating for TBP-TFIIIB70 binding cooperativity (21), a result confirmed in the present experiments. The free energy of complex formation for B′-DNA (Table 1, ΔG° = −12.10 ± 0.12 kcal/mol, which represents an increase in TBP affinity of ≈16-fold) includes the cooperative contribution of TFIIIB70 to the TBP-DNA binding reaction and is comparable to previous studies of this complex (21).
Figure 1.
DNase I footprints of TBP titrations on the yU6 TATA element in the presence or absence of TFIIIB70 and/or TFIIIB90 and global binding isotherms. (A) DNase I footprint of a TBP titration on the yU6 DNA. The leftmost lane is a no-DNase I control. Adjacent lanes show reactions with increasing concentrations of TBP corresponding to 0.0, 0.5, 1.0, 2.5, 5, 10, 15, 20, 35, 50, 75, 100, 150, 200, and 250 nM. (B) The TBP titration in A was performed in the presence (+) of subsaturating TFIIIB70 (5 nM). (C) DNase I footprint of a TBP titration on the yU6 DNA in the presence (+) of subsaturating TFIIIB70 (5 nM) and saturating TFIIIB90 (100 nM, see Fig. 2). The leftmost lane is a no-DNase I control. Adjacent lanes show reactions with increasing concentrations of TBP corresponding to 0, 0.025, 0.05, 0.15, 0.25, 0.5, 1.0, 1.5, 2.5, 5.0, 7.5, 10, 25, 50, and 75 nM. (D) The TBP titration in C was performed in the presence (+) of saturating TFIIIB70 (100 nM). (E) Single-site binding isotherms determined from a global analysis of multiple TBP titrations performed under the conditions of A (no additional factors, black isotherm) and B (subsaturating TFIIIB70, 5 nM, red isotherm). For reference, the dotted blue line shows the position of the blue isotherm from F. (F) Single-site binding isotherms determined from a global analysis of multiple TBP titrations performed under the conditions of C (subsaturating TFIIIB70, 5 nM and saturating TFIIIB90, 100 nM, green isotherm) and D (saturating TFIIIB70, 100 nM, blue isotherm). For reference, the dotted black line shows the position of the black isotherm from E. The error bars show the SD of the data at each TBP concentration.
TBP titrations performed at TFIIIB70 concentrations that are subsaturating for TBP-TFIIIB70 cooperativity (<70 nM, ref. 21) display a reduced association free energy for B′-DNA complexes. For example, at 5 nM TFIIIB70, the concentration of TBP needed to saturate the yU6 TATA box is reduced only slightly (Fig. 1B). Under these conditions, the global binding isotherm (Fig. 1E, red isotherm) yields a ΔG° of −10.90 ± 0.12 kcal/mol, representing ≈25% of the maximal TFIIIB70-specific cooperativity (compare Fig. 1 A and B and Fig. 1E, red and black isotherms). Addition of 100 nM TFIIIB90 to these reactions (Fig. 1C) increases the observed ΔG° of binding in individual experiments and in the global analysis to a value identical, within experimental error, to that obtained for B′-DNA complexes (Table 1; compare Fig. 1 C and D and Fig. 1F, green and blue isotherms). These data demonstrate that TFIIIB90 contributes cooperative binding energy in the assembly of TFIIIB-DNA complexes.
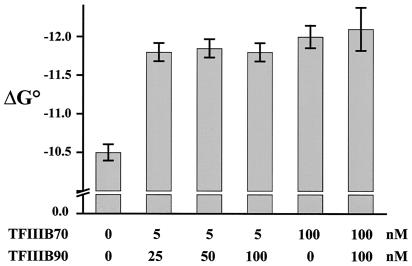
A comparison of DNase I footprint titrations of TBP performed at saturating levels of TFIIIB70 (100 nM) in the presence or absence of TFIIIB90 (100 nM) revealed no qualitative difference in the protection patterns and no quantitative effect on the free energy of complex assembly (Fig. 2 and data not shown). This result, together with the data from Fig. 1, indicates that the energetic contribution of TFIIIB90 to the TBP-DNA binding reaction is apparent only at subsaturating levels of TFIIIB70 and that 100 nM TFIIIB90 represents a saturating concentration for the assembly of TFIIIB-DNA complexes under these conditions. Indeed, titrations of TBP performed at a subsaturating concentration of TFIIIB70 (5 nM) in the presence of 25, 50, or 100 nM TFIIIB90 yielded free energies of TFIIIB-DNA formation that were identical within experimental error (Fig. 2 and Table 1). Thus, the cooperative assembly of TFIIIB-DNA is complete with as little as 5 nM TFIIIB70 and 25 nM TFIIIB90.
Figure 2.
B′-DNA and TFIIIB-DNA complexes have equivalent free energies of formation. Equilibrium binding data are shown for multiple independent TBP titrations performed under the specified conditions. Three different concentrations of TFIIIB90 are compared for their ability to increase the cooperative binding of TBP to the yU6 TATA element in the presence of a subsaturating concentration of TFIIIB70 (5 nM, as in Fig. 1C). The free energies of formation of TBP-DNA, B′-DNA, and TFIIIB-DNA (performed at a saturating concentration of TFIIIB70, 100 nM) were determined from global binding isotherms (Fig. 1 A and D and data not shown). The error bars represent the confidence limits obtained from the global analysis.
Stability of TBP-, B′-, and TFIIIB-DNA Complexes.
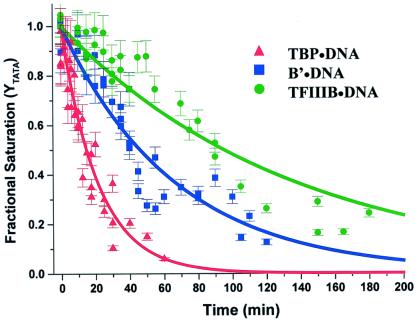
One of the characteristic physical properties of TFIIIB-DNA complexes that distinguishes them from other protein-DNA complexes required for pol III-dependent transcription is their resistance to dissociation by molar salt concentrations and heparin (9, 28). TFIIIB-DNA complexes treated with heparin (200 μg/ml) for 3–5 min resist dissociation whereas B′-TFIIIC-DNA complexes are quantitatively disrupted (9, 13, 19). Heparin resistance is therefore commonly used as an indicator of TFIIIB-DNA complex assembly and was demonstrated for this complex in the present work (data not shown). However, quantitative studies comparing the stability of TBP-DNA, B′-DNA, and TFIIIB-DNA complexes have not been reported to date. We measured the stability of these complexes assembled on the SNR6 promoter by DNase I footprinting after the addition of an excess of duplex poly[dA-dT] competitor (to trap free TBP). Replicate experiments for each complex were performed and analyzed globally by nonlinear least-squares fitting to a single-exponential dissociation function (see Materials and Methods, Eq. 3).
The global kinetic progress curve for the dissociation of TBP-DNA (Fig. 3) yields a dissociation rate constant of 80 ± 5 × 10−5 sec−1, which equates to a complex half-life of 15 ± 1 min (Table 1), in good agreement with studies performed under the same ionic conditions (29). In the determination of dissociation rates for B′- and TFIIIB-DNA, it is important that the assembly conditions yield a specific complex rather than mixtures of TBP-DNA and higher-order forms. Accordingly, B′-DNA, complexes were assembled at a subsaturating concentration of TBP (20 nM) in the presence of excess TFIIIB70 (100 nM) and TFIIIB complexes were assembled at subsaturating levels of TBP (20 nM) and TFIIIB70 (20 nM) in the presence of excess TFIIIB90 (100 nM). Saturation of the TATA box under these conditions depends on the factor added in excess (Fig. 1 A, C, and D, ref. 21, and data not shown). In addition, each B′-DNA and TFIIIB-DNA dissociation experiment was performed with controls for TBP-DNA complex saturation. The global kinetic progress curves for the B′-DNA and TFIIIB-DNA complexes yield half-lives that are three and six times longer, respectively, than that for TBP-DNA (Fig. 3 and Table 1). As expected, the data show that TFIIIB-DNA is significantly more stable than the other two complexes.
Figure 3.
Kinetics of TBP dissociation from TBP-DNA, B′-DNA, and TFIIIB-DNA complexes. Proteins and DNA were allowed to assemble for 45 min at 30°C before addition of the duplex poly[dA⋅dT] (211 μM/bp) competitor. At various times, samples were removed for DNase I digestion as described in Materials and Methods. The dissociation rate constant, kd, for each complex was obtained by global analysis of the data from three or more experiments using a single exponential dissociation function (see Materials and Methods, Eq. 3). The error bars show the SD for each time point from multiple experiments.
Discussion
In previous studies (21) and in experiments reproduced here (Fig. 1D, blue isotherm and Table 1), we found that TFIIIB70 makes a substantial (1.6–1.7 kcal/mol) cooperative contribution to the assembly of B′-DNA complexes on the SNR6 TATA box. We have now extended these studies to the assembly of TFIIIB on the same promoter. By measuring TATA box protection as a function of TBP concentration we found that both TFIIIB70 and TFIIIB90 contribute cooperative binding energy to this reaction. This cooperative binding of TBP to DNA is distinct from but energetically related to the cooperative binding of TFIIIB90 to preassembled TBP-DNA complexes in the presence of TFIIIB70 (24). At subunit concentrations typical for yeast cell extracts and partially purified fractions (i.e., less than 100 nM, ref. 30), TFIIIB does not form a stable complex in the absence of DNA (31). Nonetheless, pairwise solution interactions have been reported between TBP and the other TFIIIB subunits (31, 32), and complex assembly experiments have identified interaction sites/domains involving all three components (11, 12, 33). Thus, it is likely that protein–protein interactions between all of the components of TFIIIB contribute to cooperativity in the TBP-DNA binding reaction by bringing TFIIIB70 and TFIIIB90 into close proximity to the DNA.
Given the well-documented qualitative differences in the stability of B′-DNA and TFIIIB-DNA complexes, we were surprised to find that these complexes have equivalent free energies of formation (Table 1). One interpretation of this result is that TFIIIB90 contributes to TFIIIB-DNA complex assembly only through protein–protein interactions and thus does not appreciably affect the energetics of DNA binding. However, this explanation is not compatible with a large body of work on the assembly of TFIIIB-DNA complexes (refs. 11, 12, and 14–16 and references therein). A subset of observations that demonstrate this point and that are relevant to the interpretation of our thermodynamic data are discussed below. (i) Studies of complex assembly using DNA probes of different length have shown that stable B′-DNA complexes require 1 bp upstream and 15 bp downstream of the TATA box whereas TFIIIB-DNA complexes require an additional 14 bp of upstream DNA (11). Consistent with this result, hydroxyl radical footprinting has shown that the binding of TFIIIB90 to B′-DNA extends protection upstream of the TATA box by 7 bp on the nontranscribed strand and by 2 bp on the transcribed strand. Downstream extension of the footprint on both strands also is observed (11). These data indicate direct interactions of TFIIIB90 with the DNA, indirect effects of TFIIIB90 on TFIIIB70-DNA interactions, and/or a combination of these possibilities. Regardless of which of these apply, the close proximity of TFIIIB90 and/or TFIIIB70 to the deoxyribose backbone in the region of extended protection is expected to contribute additional free energy to the formation of the TFIIIB-DNA complex relative to B′-DNA. (ii) Extensive photocrosslinking studies have positioned TFIIIB90 and TFIIIB70 predominantly on opposite sides of the TBP-DNA complex. TFIIIB90 is photocrosslinked with high efficiency relative to TFIIIB70 at sites upstream of the TATA box (e.g., −38/−37 and −35/−34 on the SUP4 gene, refs. 16 and 34) and is also accessible to photoprobes positioned in the central core of the TFIIIB-DNA complex, underneath and adjacent to TBP (14). Importantly, molecular modeling of the photoprobes used for much of this work shows that the most distal location of the photoactive site is at the edge of the major groove (14). Together with the use of highly reactive phenyldiazirine crosslinkers (DB probes), which do not exhibit a preference for amino acid side chains, it has been possible to identify the proteins making close contacts with the major groove at different positions along the DNA. With regard to TFIIIB90, DB probes positioned within the TATA-like sequence of the SUP4 gene indicate interactions at −27 and −23 on the transcribed strand and at −30 on the nontranscribed strand. Neither TBP nor TFIIIB70 are crosslinked at −30 and only minor crosslinking is seen to TBP at −23 and to TFIIIB70 at −27 relative to TFIIIB90 at these sites (14). These TFIIIB90-DNA interactions are thought to play a critical role in modifying the interaction of TBP with the DNA in TFIIIB versus B′-DNA complexes and are therefore energetically linked to the TBP-DNA binding reaction. In light of the preceding observations, our thermodynamic data (Table 1) indicate that the formation of B′- and TFIIIB-DNA complexes is isoenergetic despite the input of binding energy from TFIIIB90.
The isoenergetic assembly of B′- and TFIIIB-DNA complexes raises the question of how the energy derived from TFIIIB90 binding is expended. One obvious use for this energy is to drive TFIIIB90-dependent bending of the DNA (15). In addition, it is possible that some of the input binding energy is offset at least partially by a loss of interactions between TBP and DNA. This loss is indicated by the elimination of TBP photocrosslinking at −33/−32 and at other sites on the nontranscribed strand of the SUP4 gene upon TFIIIB90 binding to the B′-DNA complex (14). A third way of using TFIIIB90 binding energy, which is not mutually exclusive from the other two, involves increasing the energy barrier for complex dissociation as required by the higher stability of the TFIIIB-DNA complex (Fig. 3 and Table 1). Because the equivalent free energies of the B′- and TFIIIB-DNA complexes preclude thermodynamic stabilization of the DNA, the higher energy barrier for TFIIIB-DNA dissociation must be achieved by TFIIIB90-dependent structural changes that kinetically trap the DNA. Kinetic trapping of DNA in the TFIIIB complex is implicit in the hypothesis advanced by Grove et al. (15) that TFIIIB90-dependent complex stabilization and DNA bending are interrelated and together with the ensemble of protein–protein interactions in the complex, sterically obstruct DNA dissociation. The thermodynamic and kinetic data described here provide direct biochemical support for this mechanism.
Currently, it is not known how TFIIIB-DNA complexes are disrupted. However, the dissociation rate constant for TFIIIB-DNA (Table 1) yields a complex half-life that is equivalent to the doubling time of wild-type yeast cells in rich medium (90–100 min). Thus, the lifetime of the TFIIIB-DNA complex in vitro exceeds the length of the yeast cell cycle by a significant margin. The kinetic data are therefore consistent with the idea that TFIIIB, once assembled onto DNA within the yeast nucleus, is likely to remain associated until displaced, presumably by a passing DNA replication fork (35). The high stability of TFIIIB-DNA complexes in the cell is supported by in vivo footprinting studies in asynchronously growing cell populations and stationary phase cells (36). These experiments show protection of the region upstream of the transcription start site of pol III genes that corresponds to the region protected by TFIIIB in vitro. The apparent stability of the TFIIIB-DNA complex in vivo underscores the importance of TFIIIB recruitment to the promoters of pol III genes and not to other sites in the genome where silencing of adjacent pol II genes may occur (37).
The 2-fold difference in stability between B′-DNA and TFIIIB-DNA determined by using duplex poly[dA-dT] as the competitor (Table 1) is smaller than we had anticipated given the dramatic difference in the heparin resistance of these complexes (described above). An explanation for these apparently disparate observations is suggested by the differential stability of B′-DNA in the presence of heparin versus an excess of duplex poly[dA-dT]. Using DNA as the competitor, B′-DNA complexes dissociate with a half-life of 46 min, comparable to the analogous pol II ternary complex (TFIIB-TBP-DNA), determined in a similar manner (38). However, in the presence of heparin, B′-TFIIIC-DNA complexes dissociate with a half-life of less than 2 min (13). This finding indicates that heparin is not acting in a passive manner to bind free protein but rather is an active participant in the dissociation of B′-TFIIIC-DNA and presumably B′-DNA complexes. Accordingly, we suggest that the facilitated dissociation of B′-DNA by heparin contributes to the marked difference in the stability of pol III transcription complexes upon challenge with this polyanion.
In previous work we found that the sequence of the TATA element as well as TATA flanking sequences affect the cooperative binding of TBP and TFIIIB70 (20, 21). TATA flanking sequences also have been shown to affect the binding of TBP alone (21, 39) and the cooperative binding of TBP and TFIIB (38). Whereas TFIIB stabilizes TBP against dissociation on all promoters tested, the ability of TFIIB to influence the rate of association of TBP with the DNA depends on specific sequences and combinations of the TATA element and the flanking DNA (38). The origin of these sequence-dependent association rate effects is presently unknown. However, the sequential two-intermediate model for TBP binding proposed by Parkhurst and coworkers (29) provides a framework for interpreting these effects. In this scheme, intermediate 1 (I1) is bent similar to the final complex and is present at a significant mole fraction during the binding reaction. However, the I1 intermediate is not the thermodynamically most stable form (29). As suggested by Parkhurst et al. (29), TFIIB binding to the I1 intermediate would provide kinetic control of the association reaction. In addition, if different TATA and flanking sequences were to modulate the abundance of this species, association rate effects of TFIIB would be modulated similarly. The structural and functional similarities between TFIIB and TFIIIB70 together with the unusual kinetic mechanism for TFIIIB-DNA complex stabilization suggest that it will be interesting to determine how the cooperative binding of TFIIIB70 and TFIIIB90 is partitioned in the assembly of B′- and TFIIIB-DNA complexes.
Acknowledgments
We thank Liz Jamison for providing the TBP for these studies. Thanks also to Robyn Moir and John Blanchard for stimulating discussions and comments on the manuscript. This work was supported by National Institutes of Health Grants GM42728 and GM39929 and Predoctoral Training Grant GM07491.
Abbreviations
- pol
polymerase
- B′-DNA
TFIIIB70-TBP-DNA
- TBP
TATA binding protein
- TF
transcription factor
- yU6
yeast SNR6
References
- 1.Paule M R, White R J. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teichmann M, Wang Z, Roeder R G. Proc Natl Acad Sci USA. 2000;97:14200–14205. doi: 10.1073/pnas.97.26.14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schramm L, Pendergrast P S, Sun Y, Hernandez N. Genes Dev. 2000;14:2650–2663. doi: 10.1101/gad.836400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCulloch V, Hardin P, Peng W, Ruppert J M, Lobo-Ruppert S M. EMBO J. 2000;19:4134–4143. doi: 10.1093/emboj/19.15.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dieci G, Sentenac A. Cell. 1996;84:245–252. doi: 10.1016/s0092-8674(00)80979-4. [DOI] [PubMed] [Google Scholar]
- 6.Maraia R J, Kenan D J, Keene J D. Mol Cell Biol. 1994;14:2147–2158. doi: 10.1128/mcb.14.3.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maraia R J. Proc Natl Acad Sci USA. 1996;93:3383–3387. doi: 10.1073/pnas.93.8.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Roeder R G. Mol Cell. 1998;1:749–757. doi: 10.1016/s1097-2765(00)80074-x. [DOI] [PubMed] [Google Scholar]
- 9.Kassavetis G A, Braun B R, Nguyen L H, Geiduschek E P. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 10.Goodier J L, Fan H, Maraia R J. Mol Cell Biol. 1997;17:5823–5832. doi: 10.1128/mcb.17.10.5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colbert T, Lee S, Schimmack G, Hahn S. Mol Cell Biol. 1998;18:1682–1691. doi: 10.1128/mcb.18.3.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kassavetis G A, Kumar A, Ramirez E, Geiduschek E P. Mol Cell Biol. 1998;18:5587–5599. doi: 10.1128/mcb.18.9.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar A, Kassavetis G A, Geiduschek E P, Hambalko M, Brent C J. Mol Cell Biol. 1997;17:1868–1880. doi: 10.1128/mcb.17.4.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Persinger J, Sengupta S M, Bartholomew B. Mol Cell Biol. 1999;19:5218–5234. doi: 10.1128/mcb.19.7.5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grove A, Kassavetis G A, Johnson T E, Geiduschek E P. J Mol Biol. 1999;285:1429–1440. doi: 10.1006/jmbi.1998.2347. [DOI] [PubMed] [Google Scholar]
- 16.Shah S M, Kumar A, Geiduschek E P, Kassavetis G A. J Biol Chem. 1999;274:28736–28744. doi: 10.1074/jbc.274.40.28736. [DOI] [PubMed] [Google Scholar]
- 17.Kim J L, Nikolov D B, Burley S K. Nature (London) 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 18.Kim Y, Geiger J H, Hahn S, Sigler P B. Nature (London) 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 19.Joazeiro C A, Kassavetis G A, Geiduschek E P. Mol Cell Biol. 1994;14:2798–2808. doi: 10.1128/mcb.14.4.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Librizzi M D, Moir R D, Brenowitz M, Willis I M. J Biol Chem. 1996;271:32695–32701. doi: 10.1074/jbc.271.51.32695. [DOI] [PubMed] [Google Scholar]
- 21.Librizzi M D, Brenowitz M, Willis I M. J Biol Chem. 1998;273:4563–4568. doi: 10.1074/jbc.273.8.4563. [DOI] [PubMed] [Google Scholar]
- 22.Petri V, Hsieh M, Brenowitz M. Biochemistry. 1995;34:9977–9984. doi: 10.1021/bi00031a020. [DOI] [PubMed] [Google Scholar]
- 23.Parkhurst K M, Brenowitz M, Parkhurst L J. Biochemistry. 1996;35:7459–7465. doi: 10.1021/bi9530301. [DOI] [PubMed] [Google Scholar]
- 24.Roberts S, Miller S J, Lane W S, Lee S, Hahn S. J Biol Chem. 1996;271:14903–14909. doi: 10.1074/jbc.271.25.14903. [DOI] [PubMed] [Google Scholar]
- 25.Brenowitz M, Senear D, Jamison E, Dalma-Weiszhausz D. In: Footprinting of Nucleic Acid-Protein Complexes. Revzin A, editor. San Diego: Academic; 1993. pp. 1–43. [Google Scholar]
- 26.Gaillard C, Strauss F. Nucleic Acids Res. 1990;18:378. doi: 10.1093/nar/18.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brenowitz M, Senear D F, Shea M A, Ackers G K. Methods Enzymol. 1986;130:132–181. doi: 10.1016/0076-6879(86)30011-9. [DOI] [PubMed] [Google Scholar]
- 28.Kassavetis G A, Nguyen S T, Kobayashi R, Kumar A, Geiduschek E P, Pisano M. Proc Natl Acad Sci USA. 1995;92:9786–9790. doi: 10.1073/pnas.92.21.9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parkhurst K M, Richards R M, Brenowitz M, Parkhurst L J. J Mol Biol. 1999;289:1327–1341. doi: 10.1006/jmbi.1999.2835. [DOI] [PubMed] [Google Scholar]
- 30.Sethy-Coraci I, Moir R D, Lopez-de-Leon A, Willis I M. Nucleic Acids Res. 1998;26:2344–2352. doi: 10.1093/nar/26.10.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huet J, Conesa C, Manaud N, Chaussivert N, Sentenac A. Nucleic Acids Res. 1994;22:2282–2288. doi: 10.1093/nar/22.12.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khoo B, Brophy B, Jackson S P. Genes Dev. 1994;8:2879–2890. doi: 10.1101/gad.8.23.2879. [DOI] [PubMed] [Google Scholar]
- 33.Shen Y, Kassavetis G A, Bryant G O, Berk A J. Mol Cell Biol. 1998;18:1692–1700. doi: 10.1128/mcb.18.3.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartholomew B, Kassavetis G A, Geiduschek E P. Mol Cell Biol. 1991;11:5181–5189. doi: 10.1128/mcb.11.10.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deshpande A M, Newlon C S. Science. 1996;272:1030–1033. doi: 10.1126/science.272.5264.1030. [DOI] [PubMed] [Google Scholar]
- 36.Huibregtse J M, Engelke D R. Mol Cell Biol. 1989;9:3244–3252. doi: 10.1128/mcb.9.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hull M W, Erickson J, Johnston M, Engelke D R. Mol Cell Biol. 1994;14:1266–1277. doi: 10.1128/mcb.14.2.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolner B S, Gralla J D. J Biol Chem. 2001;276:6260–6266. doi: 10.1074/jbc.M008273200. [DOI] [PubMed] [Google Scholar]
- 39.Bareket-Samish A, Cohen I, Haran T E. J Mol Biol. 2000;299:965–977. doi: 10.1006/jmbi.2000.3797. [DOI] [PubMed] [Google Scholar]