Abstract
Background:
Panton-Valentine leukocidin (PVL) is a gamma-toxin produced by Staphylococcus aureus encoded by genes lukS/lukF-PV with several single-nucleotide polymorphisms. A mutation at nucleotide position 527 results in substitution of histidine (H) to arginine (R) at amino acid 176. The groups defined based on the amino acid change, the “R isoform” group and the “H isoform” group. The purpose of this study was to determine the frequency of PVL gene isoforms in S. aureus strains isolated from patients at Al-Zahra Hospital Isfahan and molecular characterization of PVL-positive methicillin-resistant S. aureus (MRSA) strains including the detection of mecA gene and staphylococcal chromosomal cassette mec (SCCmec) typing.
Materials and Methods:
In this study, 130 isolates of S. aureus were collected from Al-Zahra Hospital. The PVL gene identified using polymerase chain reaction (PCR); PCR products were sequenced to identify the type of isoform. The molecular characterization of isolates of PVL-positive MRSA including detection of mecA gene by PCR and also SCCmec typing was performed by multiplex PCR.
Results:
Out of 130 isolates, 23% were positive for the presence of PVL genes. The PVL positive isolates were comprised 37% (11/30) of methicillin-resistant isolates and 63% (19/30) of methicillin-susceptible S. aureus (MSSA) isolates. The results showed that 17 isolated carrying isoform H and 13 isolated carrying the R isoform.
Conclusion:
The PVL gene was predominantly found in MSSA isolates. There was no relation between PVL isoforms and the presence of mecA and SCCmec types.
Keywords: Isoform Panton-Valentine leukocidin, mecA gene, Panton-Valentine leukocidin, staphylococcal chromosomal cassette mec typing, Staphylococcus aureus
Introduction
Staphylococcus aureus is able to produce a vast range of virulence factors including toxins.[1] Panton-Valentine leukocidin (PVL) was, for the first time, described by Panton and Valentine from strain S. aureus V8, which was isolated from a patient with chronic furunculosis.[2] PVL is a gamma-toxin produced by S. aureus,[1] which consists of two subunits F and S. Toxin is encoded by lukS/lukF-PV genes.[3] This toxin targets the outer membrane of polymorphonuclear cells, monocytes, and macrophages. This toxin increases the cell membrane permeability that leads to the degradation and necrosis of leukocytes.[4,5] “PVL positive” S. aureus increases virulence and are responsible for intense infections such as furuncles, cutaneous abscesses, and intensive necrotic skin infections.[1,6] Although PVL has been forcefully associated with community acquired methicillin-resistant S. aureus (CA-MRSA), lukS/lukF-PV genes can be carried as well by methicillin-susceptible S. aureus (MSSA) isolates.[7,8] CA-MRSA usually carries staphylococcal chromosomal cassette mec (SCCmec) type IV or V, and frequently express the PVL genes.[4,9] lukF and lukS genes have 12 main single nucleotide polymorphisms, the majority of which are synonymous. A nonsynonymous mutation at position 527, leads to an (A → G) His to Arg substitution at amino acid 176 (H and R isoforms).[10,11,12] The R isoform was strongly related to isolates from the USA and exclusively the USA300 MRSA clone. The H isoform was related to isolates outside the USA and both MSSA and MRSA.[11,13] Molecular modeling suggests these isoforms may affect pore formation and leukotoxicity.[8,11,12] Moreover, molecular models suggest that the R isoform can be increased resulting in pore formation and greater leukotoxicity[14] which is also dependent on the presence of mecA.[4] Therefore, evaluating and determining PVL gene isoforms distribution of the isolates of S. aureus is an important issue. This study attempts to examine the prevalence of PVL gene and determine its isoforms in clinical samples, the molecular characterization of PVL-positive MRSA isolates including identification of the mecA gene and SCCmec typing.
Materials and Methods
In an 8-month study from September 2013 to April 2014, 130 clinical isolates of S. aureus were collected from Al-Zahra Hospital in Isfahan. These isolates were taken from wound, blood, urine, abscess, trachea, throat, cerebrospinal fluid, and catheter samples. S. aureus isolates were identified by biochemical tests including gram-staining, catalase, coagulase, DNase test, and mannitol fermentation.[15]
Genomic DNA extraction
Genomic DNAs of S. aureus isolates were extracted using phenol-chloroform method.[16]
Polymerase chain reaction tests for detection of Panton-Valentine leukocidin gene
To identify the PVL genes, we designed the forward and reverse PVL genes primers using computer software Gene-Runner (Hastings Software Inc; http://www.genelink.com/tools/gl-downloads.asp). Sequences of primers are listed in Table 1. Polymerase chain reaction (PCR) technology was used to characterize all S. aureus isolates on the basis of the presence or absence of the PVL gene determinants. S. aureus strain NCTC13300 was used as positive control for detecting of PVL gene. DNA was amplified on an Eppendorf thermocycler with the final volume of 25 μl containing 2.5 μl of × 10 buffer, 0.75 μl of MgCl2 (25 mM) and 0.5 μl of dNTP mix, 10 pMol of each primers, 0.25 U of Taq polymerase, 2 μl of template DNA, and 17 μl of distilled water were taken.
Table 1.
Primers used in this study

PCR was performed with the following thermal setting: Initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 55.8°C for 30 s, and extension at 72°C for 1-min with final extension at 72°C for 10 min.
The PCR products were loaded on 1.5% agarose gel and were analyzed by gel electrophoresis.
PCR Identification of mecA for all isolates and Multiplex PCR for SCCmec typing of isolates PVL-positive MRSA were performed as previously described.[17,18,19] The primers used for the PCR are shown in Table 1.
Nucleotide sequencing
The PCR products of lukS-PV-positive isolates (n = 30) were sequenced, and R/H isoform for all isolates was defined. All sequences were aligned with the NCBI reference sequence (S. aureus lukS-PV-AB 006796).
Statistical analysis
The data were analyzed using SPSS version 18.0 (SPSS Inc., Chicago, Illinois, USA). Chi-square test and Fisher's exact test were used for data analysis (P < 0.05 was considered as significant).
Results
The 23% of S. aureus isolates were positive for PVL and most the PVL-positive isolates related to S. aureus isolated from wounds (37%). PVL positive isolates comprised 37% (11/30) of methicillin-resistant isolates and 63% (30/19) of MSSA isolates [Table 2]. Thirteen sequences have a G at position 527, which results in an arginine (R) at aa 176, and 17 sequences have an A at 527, resulting in a histidine (H) at aa 176. These will be referred as the “isoform R” and the “isoform H,” respectively [Table 3].
Table 2.
Association of Panton-Valentine leukocidin-positive isolates with types of staphylococcal infection
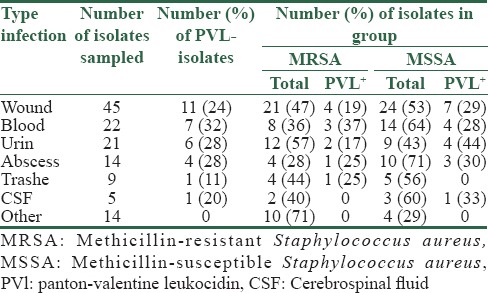
Table 3.
The relationship between Panton-Valentine leukocidin isoforms and mecA gene
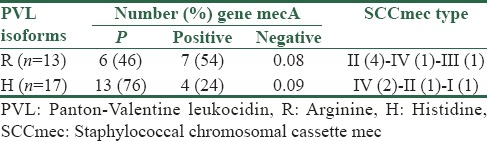
Polymerase chain reaction for mecA gene detection and typing staphylococcal chromosomal cassette mec
Of 130 S. aureus isolates, the authors found 61 isolates (45%) were MRSA and positive for the presence of genes mecA. SCCmec typing was used for all MRSA PVL + isolates. The five (45.4%) MRSA strains were SCCmec type II, 3 (27.2%) were type IV, 1 (9%) were type III, and 1 (9%) were type I. The MRSA strains 1 (9%) could not be typed [Table 3].
Discussion
PVL-positive S. aureus has received significant attention in the recent years.[13] In this study, the prevalence of PVL gene in S. aureus strains were 23%, and the majority were associated with skin and soft tissue infections. In studies from Iran, the prevalence of 14.3% and 18% of this gene has been reported among staphylococcal strains,[20] which is smaller than the PVL percentage reported by authors. In a study conducted by Nickerson et al. in Thailand, it was shown that PVL gene positive S. aureus isolates were 49%, and all of them were MSSA.[21] The frequency of PVL positive isolates was found 27% in Chini et al. study.[22] In the USA from 1055 examined isolates, 36% carried PVL gene which was higher than the percentage reported by the authors.[13] The overall difference in results of these studies could be due to differences in geographic location and type of samples.
Many prior studies have mainly concentrate on community-associated MRSA; one of the key results of our study is the high rate of PVL positive in MSSA isolates. Nearly, 60% of total PVL-positive S. aureus isolates in England in the past 5 years found to be susceptible to methicillin.[10] A high PVL positive MSSA spread (70%) was reported from France from surgically drained abscesses.[23]
O'Hara et al. in a study conducted in the USA (2008) detected the nucleotide variation at the lukSF-PV sequence. Their results showed the lukS-PV sequence was more variable than the lukF-PV sequence. In their study, 111 isolates were found to carry the H isoform of PVL, whereas 63 isolates harbored the R isoform.[4] This study results were similar to their study. In a study by Tong et al. in northern Australia (2010), the frequency of the isoforms carrying the PVL genes in S. aureus strains was examined. PVL positive isolates comprised 54% of CA-MRSA isolates and 40% of MSSA isolates. There were 113 H isoform–and 110 R isoform–harboring isolates.[14] We found distribution to be similar to the mentioned study, however, PVL positive isolates comprised 37% of MRSA and 63% of MSSA isolates. In a study by Brown et al., among the 86 strains of S. aureus typed, 72 R isoforms and 14 H isoforms were identified.[13] Results obtained by O'Hara et al. found R isoform to be strongly associated with the presence of mecA, however, Tong et al. found no relation between PVL isoforms and the presence of mecA.[4,11] In our study, Chi-square test and Fisher exact tests indicated that there was no correlation between the isoforms of PVL and mecA gene (P > 0.05) [Table 3].
O'Hara et al. suggested that H isoform is older than the R isoform.[4] This hypothesis is supported by wider geographical distribution, distributed among several clonal complexes, and increase in the diversity of H isoform sequence rather than R isoform. Older isoform is more related to MSSA strains, whereas newer isoform is related to CA-MRSA strains. This hypothesis indicates that PVL-positive MSSA strains act as a reservoir for PVL genes and then have been incorporated in CA-MRSA lineage.[4]
CA-MRSA generally carries SCCmec type IV or V and often produces PVL.[9] SCCmec typing of PVL-positive MRSA isolates in the samples showed that SCCmec type II and type IV are significantly prevalent. Chi-square test showed that there was no correlation between PVL isoforms and SCCmec types (P > 0.05). Keeping in mind that types IV and II are, respectively, the origins of community- and nosocomial-related infections, so hospital infection control measures should primarily focus on preventing cross-contamination.
Conclusion
Since the infections caused by PVL-positive S. aureus strains have high virulence[10] and regional differences in the prevalence of PVL gene and its isoforms may affect the clinical spectrum of staphylococcal infections, so knowledge about the prevalence of strains containing PVL gene and isoform distribution can be helpful in estimating their pathogenicity and implementing better treatment policies.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Cupane L, Pugacova N, Berzina D, Cauce V, Gardovska D, Miklaševics E. Patients with Panton-Valentine leukocidin positive Staphylococcus aureus infections run an increased risk of longer hospitalisation. Int J Mol Epidemiol Genet. 2012;3:48–55. [PMC free article] [PubMed] [Google Scholar]
- 2.Colin DA, Mazurier I, Sire S, Finck-Barbançon V. Interaction of the two components of leukocidin from Staphylococcus aureus with human polymorphonuclear leukocyte membranes: Sequential binding and subsequent activation. Infect Immun. 1994;62:3184–8. doi: 10.1128/iai.62.8.3184-3188.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mariem BJ, Ito T, Zhang M, Jin J, Li S, Ilhem BB, et al. Molecular characterization of methicillin-resistant Panton-Valentine leukocidin positive Staphylococcus aureus clones disseminating in Tunisian hospitals and in the community. BMC Microbiol. 2013;13:2. doi: 10.1186/1471-2180-13-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Hara FP, Guex N, Word JM, Miller LA, Becker JA, Walsh SL, et al. A geographic variant of the Staphylococcus aureus Panton-Valentine leukocidin toxin and the origin of community-associated methicillin-resistant S. aureus USA300. J Infect Dis. 2008;197:187–94. doi: 10.1086/524684. [DOI] [PubMed] [Google Scholar]
- 5.Khosravi AD, Hoveizavi H, Farshadzadeh Z. The prevalence of genes encoding leukocidins in Staphylococcus aureus strains resistant and sensitive to methicillin isolated from burn patients in Taleghani hospital, Ahvaz, Iran. Burns. 2012;38:247–51. doi: 10.1016/j.burns.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–32. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 7.Vandenesch F, Naimi T, Enright MC, Lina G, Nimmo GR, Heffernan H, et al. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: Worldwide emergence. Emerg Infect Dis. 2003;9:978–84. doi: 10.3201/eid0908.030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Besseyre des Horts T, Dumitrescu O, Badiou C, Thomas D, Benito Y, Etienne J, et al. A histidine-to-arginine substitution in Panton-Valentine leukocidin from USA300 community-acquired methicillin-resistant Staphylococcus aureus does not impair its leukotoxicity. Infect Immun. 2010;78:260–4. doi: 10.1128/IAI.00843-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khokhlova O, Tomita Y, Hung WC, Takano T, Iwao Y, Higuchi W, et al. Elderly infection in the community due to ST5/SCCmecII methicillin-resistant Staphylococcus aureus (the New York/Japan clone) in Japan: Panton-Valentine leukocidin-negative necrotizing pneumonia. J Microbiol Immunol Infect. 2015;48:335–9. doi: 10.1016/j.jmii.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Otokunefor K, Sloan T, Kearns AM, James R. Molecular characterization and Panton-Valentine leucocidin typing of community-acquired methicillin-sensitive Staphylococcus aureus clinical isolates. J Clin Microbiol. 2012;50:3069–72. doi: 10.1128/JCM.00602-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong SY, Lilliebridge RA, Holt DC, Coombs GW, Currie BJ, Giffard PM. Rapid detection of H and R Panton-Valentine leukocidin isoforms in Staphylococcus aureus by high-resolution melting analysis. Diagn Microbiol Infect Dis. 2010;67:399–401. doi: 10.1016/j.diagmicrobio.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 12.Dumitrescu O, Tristan A, Meugnier H, Bes M, Gouy M, Etienne J, et al. Polymorphism of the Staphylococcus aureus Panton-Valentine leukocidin genes and its possible link with the fitness of community-associated methicillin-resistant S. aureus. J Infect Dis. 2008;198:792–4. doi: 10.1086/590914. [DOI] [PubMed] [Google Scholar]
- 13.Brown ML, O'Hara FP, Close NM, Mera RM, Miller LA, Suaya JA, et al. Prevalence and sequence variation of Panton-Valentine leukocidin in methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains in the United States. J Clin Microbiol. 2012;50:86–90. doi: 10.1128/JCM.05564-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tong SY, Lilliebridge RA, Bishop EJ, Cheng AC, Holt DC, McDonald MI, et al. Clinical correlates of Panton-Valentine leukocidin (PVL), PVL isoforms, and clonal complex in the Staphylococcus aureus population of Northern Australia. J Infect Dis. 2010;202:760–9. doi: 10.1086/655396. [DOI] [PubMed] [Google Scholar]
- 15.Forbes BA, Sahm DF, Welssfeld AS. Bailey and Scott's Diagnostic Microbiology. 11th ed. St. Louis: Mosby Inc; 2002. pp. 285–92. [Google Scholar]
- 16.Darabi N, Habibollahi H, Shahbabian K. Molecular Epidemiology of Staphylococcus aureus Isolated from patients and personnel in Army hospital. Ann Mil Health Sci Res. 2010;8:193–199. [Google Scholar]
- 17.Omar NY, Ali HA, Harfoush RA, El Khayat EH. Molecular typing of methicillin resistant Staphylococcus aureus clinical isolates on the basis of protein A and coagulase gene polymorphisms. Int J Microbiol. 2014 doi: 10.1155/2014/650328. 650328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Azimian A, Havaei SA, Fazeli H, Naderi M, Ghazvini K, Samiee SM, Soleimani M, Peerayeh SN. Genetic characterization of a vancomycin-resistant Staphylococcus aureus isolate from the respiratory tract of a patient in a university hospital in northeastern Iran. J Clin Microbiol. 2012;50:3581–5. doi: 10.1128/JCM.01727-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molla-abbaszadeh H, Mobayen H, Mirzaei H. Identification of Panton Valentine Leukocidin (pvl) Genes in Staphylococcus aureus isolated from In- patients of Emam Reza and Shohada Hospitals of Tabriz by Real-Time PCR. Iran J Med Microbiol. 2013;6:72–80. [Google Scholar]
- 20.Ohadian Moghadam S, Havaei SA, Pourmand MR. Prevalence of methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin gene in cutaneous infections in the city of Isfahan. J Med Bacteriol. 2012;1:9–16. [Google Scholar]
- 21.Nickerson EK, Wuthiekanun V, Wongsuvan G, Limmathurosakul D, Srisamang P, Mahavanakul W, et al. Factors predicting and reducing mortality in patients with invasive Staphylococcus aureus disease in a developing country. PLoS One. 2009;4:e6512. doi: 10.1371/journal.pone.0006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chini V, Petinaki E, Foka A, Paratiras S, Dimitracopoulos G, Spiliopoulou I. Spread of Staphylococcus aureus clinical isolates carrying Panton-Valentine leukocidin genes during a 3-year period in Greece. Clin Microbiol Infect. 2006;12:29–34. doi: 10.1111/j.1469-0691.2005.01295.x. [DOI] [PubMed] [Google Scholar]
- 23.del Giudice P, Blanc V, de Rougemont A, Bes M, Lina G, Hubiche T, et al. Primary skin abscesses are mainly caused by Panton-Valentine leukocidin-positive Staphylococcus aureus strains. Dermatology. 2009;219:299–302. doi: 10.1159/000232391. [DOI] [PubMed] [Google Scholar]


