Abstract
The X-linked Trap1a gene encodes the tumor rejection antigen P1A, which is expressed in fetal tissues and multiple lineages of tumor cells. The function of this gene remains unknown. Using chimeric mice with wild-type (WT) and Trap1a−/y bone marrow, we show that Trap1a−/y donor cells are capable of generating most lineages of hematopoietic cells, with the notable exception of T cells. Deletion of Trap1a selectively arrests T-cell development at double-negative stage 1 (DN1, with a CD4−CD8−CD25−CD44+ phenotype). Because Trap1a is expressed in Lin−Sca-1+c-Kit+ and common lymphoid progenitors but not in immature thymocytes (DN1-DN4), Trap1a mutations affect the differentiation potential of progenitor cells without directly acting on T cells. Despite a similarity in the blockade of DN1 to DN2 transition, the Trap1a−/y DN1 cells have normal expression of c-Kit, in contrast to what was reported in the Notch1−/− DN1. Complementary DNA profiling of Trap1a−/y and WT embryonic stem cells shows that Trap1a does not regulate the Notch pathway. Our data reveal that Trap1a is an X-linked regulator that affects the differentiation potential of progenitor cells into T cells through a Notch-independent mechanism and identify an important function for the Trap1a gene.
Keywords: Notch, tumor antigens, T cell development
Introduction
Thymopoiesis consists of both the orderly differentiation and rapid expansion of T-cell progenitors arriving from peripheral blood.1 The early differentiation of immature thymocytes can be marked by the expression of CD44 and CD25 among the CD3−CD4−CD8− thymocytes.2 Recent studies suggest that a subset of blood Lin−Sca1+c-Kit+ cells (LSK) are likely the precursors of thymocytes. In the thymus, the LSK are part of the double-negative stage 1 (DN1) (CD44+CD25−) subset.1 Notch/RBP-J signaling is responsible for commitment of progenitors to T-cell lineage (early T-cell progenitor) and the transitions of early T-cell progenitorsfrom DN1 to DN2 (CD44+CD25+) and from DN3 (CD44−CD25+) to DN4 (CD44−CD25−).3 Notch/RBP-J signaling also controls the massive expansion of immature thymocytes.4
The X-linked Trap1a gene encodes the first known tumor antigen (P1A) recognized by cytotoxic T cells in mice5 and is overexpressed in multiple lineages of tumor cells.6 P1A is known to be overexpressed in the testis, placenta7, 8 and embryonic stem (ES) cells (this study). We reported that the transgenic overexpression of Trap1a is sufficient to cause thymic lymphoma in an immune-deficient host.9 However, the physiological function of P1A remains largely unknown due to the lack of genetic models. To address this issue, we produced ES cells with a floxed Trap1a locus. Because these ES cells were not germline-transmitted, we analyzed the contribution of these mutant ES cells to various tissues in chimeras. Using both blastocyst and bone marrow (BM) chimeras, we observed an unexpected cell-autonomous function of P1A in early thymocyte differentiation, specifically in the DN1 to DN2 transition. Our data reveal an X-linked regulator of the DN1 to DN2 transition in thymocyte development.
Materials and methods
Generation of Trap1a −/y mutant ES cells
A mouse Trap1a genomic clone was obtained by screening a 129/SvJ BAC library. A neomycin/TK cassette flanked by two LoxP sites was cloned between Exon 1 and Exon 2, introducing a new BamHI site for genotyping purposes. A third LoxP site was located before Exon 1. This construct was electroporated into murine male X:Y R1 ES cells,10 and homologous recombination was confirmed by Southern blot analysis after BamHI digestion. Positive ES cell clones were then electroporated with a Cre vector to eliminate the neomycin/TK cassette and Trap1a Exon 1 (Figure 1a). ES cells were co-cultured with mouse embryonic fibroblast feeder cells in Dulbecco’s modified Eagle’s medium containing 15% fetal calf serum, 0.1 mM β-mercaptoethanol, 103 u/ml leukemia inhibitory factor and 4 mM glutamine as previously described.11
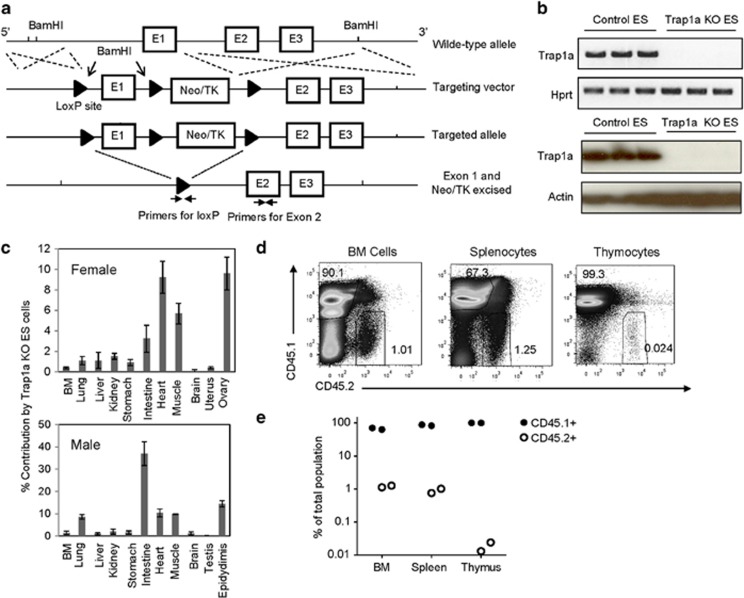
Figure 1.
Contribution of Trap1a−/y ES cells to mouse tissues through blastocyst chimera analysis. (a) Diagram of constructs and mutant alleles. (b) Confirmation of Trap1a inactivation in ES cell clones by RT-PCR and western blot analysis. The Trap1afl/y ES cells were electroporated with a vector expressing Cre recombinase. Three clones in control and Cre-transduced groups were compared for RNA and protein expression. (c) Contribution of Trap1a−/y ES cells to various tissues and organs. Chimera mice with greater than 50% contribution based on coat color were analyzed for contribution of Trap1a−/y cells based on quantitative PCR. (d) Flow cytometric quantitation of Trap1a−/y (CD45.2+) hematopoietic cells in the BM, spleen and thymus. Data from the female chimera mouse are presented; similar results were obtained in the male chimera. (e) % Contribution of CD45.2+ cells in the male and female chimera mice. BM, bone marrow; ES cells, embryonic stem cells; RT-PCR, reverse transcriptase-polymerase chain reaction.
Contribution of mutant ES cells to different organs
Trap1a−/y ES cells (CD45.2) were microinjected into B6.SJL-PtprcaPep3b/BoyJ blastocysts (CD45.1) by the Transgenic Animal Model Core at the University of Michigan. The resulting pups were assessed initially for chimerism based on coat coloration. Genomic DNA was isolated from tissues of chimeric mice using a DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, USA). ES cell contribution was initially tested by quantitative PCR. The contribution of Trap1a−/y ES cells to various tissues of chimeric mice was determined by using primers specific for the LoxP site. The amount of Trap1a Exon 2 was used as an internal control. The contribution ratio of Trap1a−/y ES cells in different organs was calculated as 2 to the power of the Ct value of the specific LoxP site subtracted from the corresponding Ct value of the P1A Exon 2. This value was normalized against the value from Trap1a−/y ES cells, which is defined as 100%. The following primers were used for detecting the LoxP site: forward, 5′-CCCTAATGTATGCTATACGAAGTTAT-3′ reverse, 5′-GGGGTCCTGTAAAGGAAAGG-3′ The following primers were used for Trap1a Exon 2: forward, 5′-TCCACGACCCTAATTTCC-3′ reverse, 5′-GCATGCCTAAGGTGAGAAGC-3′.
Reverse transcription (RT)-PCR
Total RNA was isolated from Trap1a+/y and Trap1a−/y ES cells using a RNeasy Mini Kit (Qiagen). Complementary DNA was synthesized using a SuperScript first-strand synthesis system kit (Invitrogen). For quantitative PCR, the expression of Trap1a was amplified by the forward primer 5′-AGAGATGAGCGTGGAAATGG-3′ and the reverse primer 5′-CAGGAAATTAGGGTCGTGGA-3′. The Hprt gene was used as an internal control, and the primers were 5′-CAGGCCAGACTTTGTTGGAT-3′ (forward) and 5′-GCGCTCATCTTAGGCTTTGT-3′ (reverse). For conventional PCR, primers specific for Trap1a (forward: 5′-ATCTAGGGTGGCTGGTCTTC-3′ reverse: 5′-CCATTTCCTCTTCCTCCTCC-3′) were used to validate its expression in the mouse LSK population.
Western blot analysis
Samples were lysed in protein lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% NP-40), and protease inhibitor cocktails (Sigma, St Louis, MO, USA) including 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, Aprotinin, Bestatin, E-64, Leupeptin and Pepstatin A were added. Cell lysates were prepared and 50 μg of protein per lane was loaded and separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to polyvinylidene fluoride membranes, and incubated with corresponding antibodies, including rabbit anti-P1A antibody9 (1:500 dilution), anti-actin mouse antibody (Sigma, clone AC-15, 1:5000 dilution). Anti-rabbit or anti-mouse IgG horseradish peroxidase-linked antibody at 1:3500 dilution (GE Healthcare, Little Chalfont, UK) was used as a secondary antibody. Antibodies were detected with a chemiluminescence reaction using an enhanced chemiluminescence kit (Amersham Biosciences, Little Chalfont, UK) and visualized with exposure to film.
BM transplantation
Because Trap1a−/y ES only contributed ~1% to bone marrow cells in the chimeric mice, some experiments used anti-fluorescein isothiocyanate microbeads (Miltenyi Biotec, San Diego, CA, USA) to partially delete fluorescein isothiocyanate-labeled CD45.1+ cells and enrich the CD45.2+Trap1a−y cell population. After CD45.2 enrichment, or without enrichment, a total of 5-7 × 106 chimeric BM cells were injected intravenously into lethally irradiated (129/Sv × C57BL/6) F1 mice (CD45.1+ CD45.2+) by a 137Cs γ-ray source delivering a total of 1100 rads. All animal experiments were conducted in accordance with accepted standards of animal care and approved by the Institutional Animal Care and Use Committee of the University of Michigan and the Children’s National Medical Center Research Institute.
Flow cytometric analysis and cell sorting
Peripheral blood was collected from the orbital sinus of mice. BM cells were collected from the tibia and femur of hind legs. Spleen and thymus were homogenized and passed through a cell strainer to generate a single-cell suspension. Cells were stained in phosphate-buffered saline plus 2% fetal bovine serum for 20 min on ice with fluorochrome-conjugated monoclonal antibodies (mAbs) at 1:200 or 1:100 dilution. The following mAbs from BD Biosciences (San Diego, CA, USA), eBioscience (San Diego, CA, USA) or BioLegend (San Diego, CA, USA) were used: anti-CD45.1 (A20), anti-CD45.2 (104), anti-CD3 (145-2C11), anti-CD4 (GK1.5), anti-CD8 (53-6.7), anti-CD25 (PC61.5), anti-CD44 (IM7), anti-CD11b (M1/70), anti-B220 (RA3-6B2), anti-TER-119 (TER-119), anti-Gr-1 (RB6-8C5), anti-Sca-1 (D7) and anti-c-Kit (2B8). After surface staining, cells were subsequently analyzed with a BD LSR II flow cytometer. The data were analyzed using FlowJo software (Tree Star, San Diego, CA, USA). Cell sorting was performed using a FACSAria (BD Biosciences).
cDNA microarray analysis
Gene expression profiling analysis of Trap1a+/y and the Trap1a−/y ES cells was carried out using an Affymetrix GeneChip Mouse Genome 430 2.0 array. The RNA samples were sent to the Affymetrix (Cleveland, OH, USA) and Microarray Core Facility at the University of Michigan for full service sample preparation. Probe intensities were measured and then processed with Affymetrix GeneChip operating software into image analysis (CEL) files. The Affymetrix CEL files were normalized using the Robust Multi-array Averaging method12 and affy.13 Then, we used limma14 to identify differentially expressed genes. Functional analysis (gene ontology enrichment) of differentially expressed genes was assessed using the DAVID database (http://david.abcc.ncifcrf.gov/).
In silico data analysis
The RNA-seq raw data of thymocytes (double negative (DN) cells, double positive cells and natural regulatory T cells) were accessed through GEO Series accession number GSE48138.15 Alignment of sequencing reads to the reference genome (version mm10) was performed using TopHat.16 Then, the gene expression level was measured by RPKM and calculated by DEGseq.17 The expression values of hematopoietic stem cells, LSK and common lymphoid progenitors (CLP) were extracted from the expression result file through GEO Series accession numbers GSE5089618 and GSE50739.19
Statistics
Data (shown as bar graphs) are presented as the mean±s.e.m. (Figure 1c) or mean±s.d. (Figure 5e). Two-tailed Student t-tests were used for comparison between the two experimental groups. Solid lines and dotted lines in the scatter plots represent mean values for the indicated groups. Statistical significance was determined as *P<0.05, **P<0.01 and ***P<0.001.
Results
Chimera analysis revealed differential contributions of Trap1a-deficient ES cells to different organs
To understand the function of Trap1a, we generated ES cell lines containing a Trap1a triple LoxP gene-targeting vector that included a selectable neomycin/TK cassette and LoxP-flanked Trap1a Exon 1 in the mouse X chromosome (Figure 1a). ES clones devoid of P1A protein were obtained by electroporation with a Cre recombinase expression vector (Figure 1b). Unfortunately, the ES cells did not give germline transmission even before Trap1a deletion despite repeated attempts, perhaps because the neomycin/TK cassette may have subtly affected Trap1a expression in the hemizygous male-derived ES cells.
As an alternative approach, we removed the neomycin/TK cassette and the Trap1a Exon 1 by electroporation with a Cre recombinase expression vector into ES cells. Cre excised the intervening DNA segment and resulted in a single remaining LoxP site (Figure 1a). Several single Trap1a-null clones were selected and cultured for further analysis. As shown in Figure 1b by RT-PCR and western blotting, we successfully knocked out Trap1a expression in ES cells after removing Trap1a Exon 1. This deletion did not affect ES cell colony morphology or growth rate (Supplementary Figure S1). Moreover, the mutant ES cells formed teratomas of comparable sizes to those derived from WT ES cells. Moreover, histological analyses demonstrated that the mutant ES cells were capable of forming mesoderm, endoderm, ectoderm and germ cells (Supplementary Figure S2).
To determine whether Trap1a−/y ES cells could contribute to different organs, we injected them (derived from 129 ES cell line R1, CD45.2) into C57BL/6 (CD45.1) WT blastocysts. We characterized two chimeras with greater than 50% coat color contribution from Trap1a−/y ES cells. Through qPCR, the contribution ratio of Trap1a−/y ES cells in different organs was calculated as 2 to the power of the Ct value of the specific LoxP site subtracted from the corresponding Ct value of P1A Exon 2 (internal control). This value was normalized against the value from Trap1a−/y ES cells, which is defined as 100%. The organs and tissues that we analyzed included the lung, liver, kidney, stomach, intestine, heart, muscle, brain, uterus, ovary, testis, epididymis and BM. The primer positions are depicted in the upper panel of Figure 1c. As shown in the lower panel of Figure 1c, the Trap1a−/y ES cells contributed to various internal organs, although the contribution ratios differed among organs. Consistent with its abundant expression in the testis,20 Trap1a-deficient ES cells failed to contribute to testis tissues.
Using the CD45 congenic markers, we quantified the relative number of Trap1a−/y ES cell-derived cells in major hematopoietic tissues, including the BM, spleen and thymus. As shown in Figures 1d and e, the number of CD45.2 cells in the spleen and BM was ~1%. Surprisingly, the contribution to thymocytes was further reduced by 40-fold, to 0.024%. These data suggest that Trap1a may affect thymocyte development.
Cell-intrinsic function of Trap1a in thymocyte development
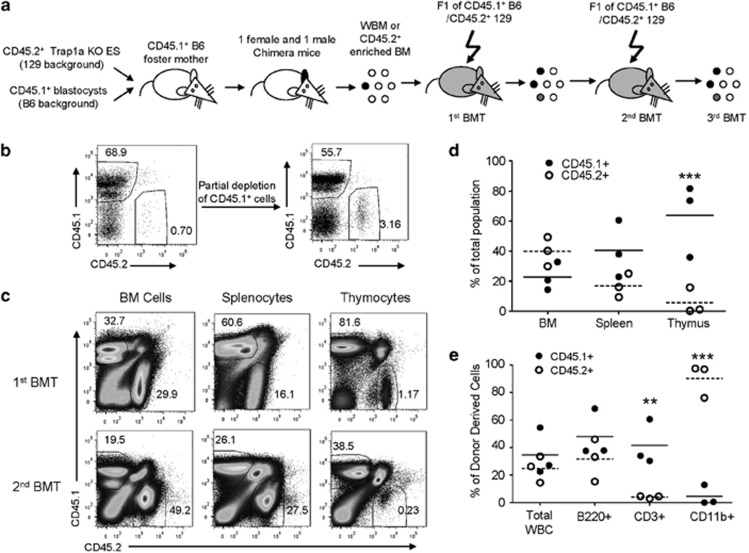
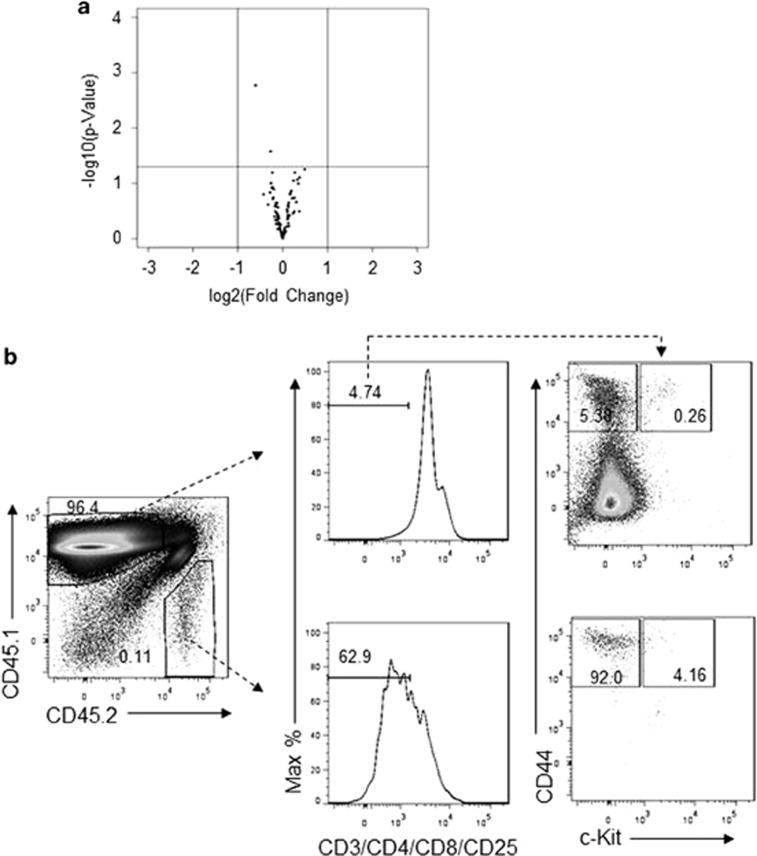
To determine whether the Trap1a gene function in hematopoiesis is cell-intrinsic, we transplanted BM cells from mouse chimeras (either CD45.1+ or CD45.2+) into lethally irradiated (129/Sv × C57BL/6) F1 mice (CD45.1+CD45.2+) (Figure 2a). In this setting, the WT cells were CD45.1+CD45.2−, the Trap1a−/y cells were CD45.1−CD45.2+, whereas the residual recipient leukocytes were CD45.1+CD45.2+ (Figure 2a). Because the number of CD45.2+ BM cells in the female chimera was exceedingly low, we partially enriched CD45.1−CD45.2+cells by using negative selection (Figure 2b). This partial enrichment is sufficient because the proportion of stem/progenitor cells is higher among the mutant hematopoietic cells (data not shown). For comparison, male BM cells were directly used without enrichment. A similar trend was found with or without enrichment. We present data mainly from the female chimera because the higher proportion of mutant-derived cells in the female made the quantitation more reliable. As shown in Figure 2c, with the partial enrichment of CD45.1− CD45.2+ donor cells, the proportions of mutant CD45.1− CD45.2+ cells were comparable to WT CD45.1+CD45.2− donor cells in the BM and spleen. However, the thymus was largely depleted of CD45.1−CD45.2+ cells (Figure 2c). The defects were even more striking in repeated transplantation (Figures 2c and d). The marked and selective reduction of thymocytes from the CD45.1−CD45.2+ donor, as summarized in Figure 2d, demonstrated a critical cell-intrinsic function of Trap1a in T-cell development. Consistent with a defective T-cell development, the proportion of T cells in the peripheral blood was also reduced, whereas that of myeloid cells was increased (Figure 2e).
Figure 2.
Cell-autonomous and long-lasting effect of Trap1a mutation on thymocyte production. (a) Diagram of experimental design. (b) Evidence of partial enrichment of mutant bone marrow cells by negative selection with anti-CD45.1 beads. (c) Impact of Trap1a mutation on thymocyte production in serial transplantation. Data shown are representative profiles of first and second transplantation. (d) Selective reduction of thymocyte production after Trap1a deletion. Data shown are % of WT and Trap1a−/y leukocytes at 4 or 11 weeks after second transplantation. (e) Trap1a deletion causes selective reduction of T cells and increase of myeloid cells in the peripheral blood at 11 weeks after second transplantation. **P<0.01; ***P<0.001. WT, wild type.
A critical role for Trap1a in DN1 to DN2 transition
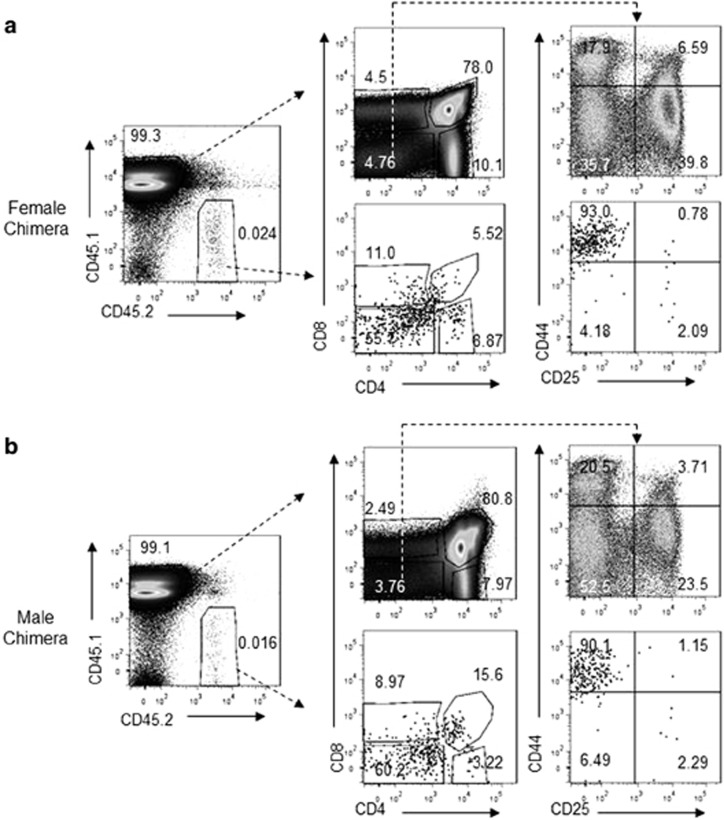
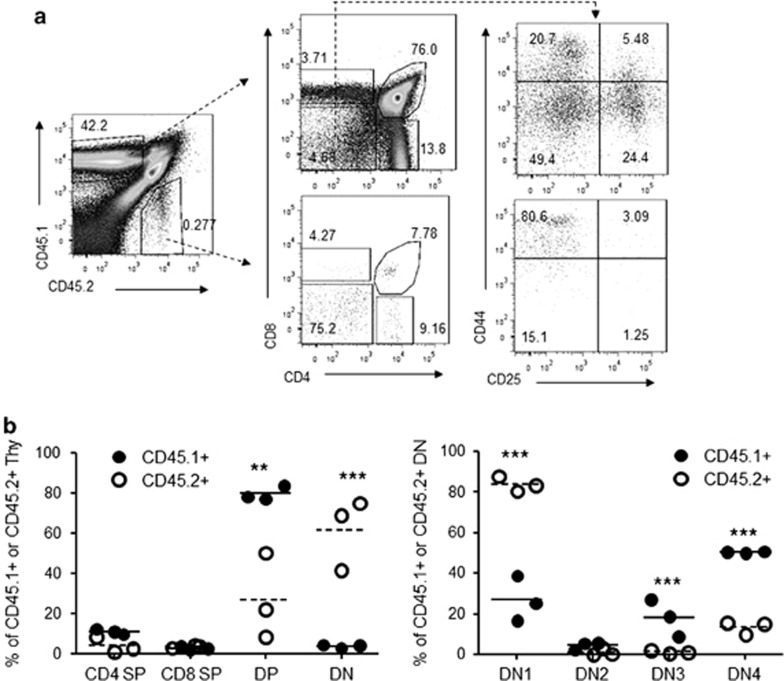
To understand which developmental stage is regulated by Trap1a, we further analyzed the thymocyte composition of the blastocyst mouse chimeras based on the expression of CD4, CD8, CD44 and CD25. As shown in Figure 3, although the CD4−CD8− thymocytes were a minority among the WT cells (CD45.1+), they represented the overwhelming majority of the CD45.2 thymocytes. After the CD4−CD8− thymocytes were further subdivided into DN1-DN4 based on the distribution of CD44 and CD25 markers, it was clear that essentially all DN had a phenotype of CD44hi CD25− in the CD45.2 thymocytes. The selective defects in the blastocyst chimera were substantiated in multiple rounds of BM transplantation (Figures 4a and b). Therefore, Trap1a deletion caused cell-intrinsic defects in DN1-DN2 transition.
Figure 3.
Selective blockade of DN1-DN2 transition by Trap1a deletion as revealed by flow cytometry. Data shown are profiles of thymocytes from the female (a) and male (b) blastocyst chimera mice.
Figure 4.
Cell-intrinsic Trap1a mutation is responsible for blockade of DN1-DN2 transition. (a) Representative profile of bone marrow chimera mouse at 11 weeks after the second round of transplantation. (b) Accumulation of DN1 cells in Trap1a−/y thymocytes. The thymocytes were harvested respectively at 11 or 12 weeks after the second round of transplantation or 7 months after the first round of transplantation.
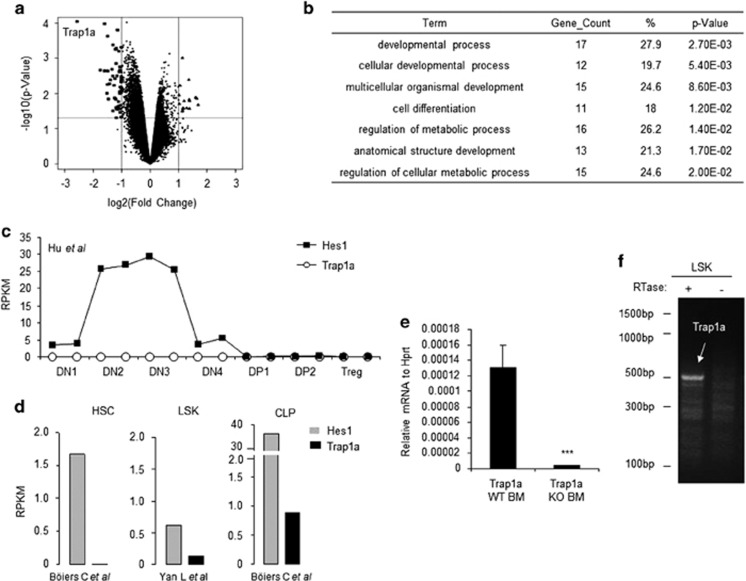
Because there were too few Trap1a−/y thymocytes for expression profiling, we profiled the Trap1a+/y and the Trap1a−/y ES cells and compared the Notch signaling signature. Using twofold changes and a P value of 0.05 as the cutoff, we observed 51 downregulated genes and 17 upregulated genes after Trap1a deletion (Figure 5a and Supplementary Dataset 1). As expected, the most downregulated gene was Trap1a. Functional enrichment analysis suggested that these genes affect cellular metabolism and development (Figure 5b).
Figure 5.
Function of Trap1a gene based on global expression analysis. (a) Identification of genes that are affected by Trap1a deletion in ES cells. The X-axis shows the ratios of expression levels between WT and Trap1a−/y ES cells, while the Y-axis shows –log10 P values. The large symbols indicate genes that are either upregulated (triangles) or downregulated (squares) by Trap1a deletion. (b) Functional enrichment analysis suggests that Trap1a may be involved in cellular development and metabolic pathways. (c–e) Trap1a is expressed in LSK and CLP but not in thymocytes and hematopoietic stem cells. (c, d) In silico analyses of RNAseq data from previous publications. (e) qPCR analysis of bone, marrow cells sorted from bone marrow chimera mice. (f) RT-PCR verification of Trap1a expression in the sorted LSK from CD45.1 (WT) and CD45.2 (mutant) LSK. CLP, common lymphoid progenitor; LSK, Lin−/SCA+/Kit+; qPCR, quantitative polymerase chain reaction; RT-PCR, reverse transcriptase-polymerase chain reaction.
Theoretically, Trap1a may act directly either to promote the DN1-DN2 transition or to determine the potential of T-cell progenitors. The former predicts the expression of Trap1a in the thymocytes, whereas the latter does not have such a requirement. We carried out in silico analyses for Trap1a expression. As shown in Figure 5c, RNAseq data15 showed that Trap1a was not expressed in thymocytes, whereas databases generated by others18, 19 revealed low levels of expression in BM LSK stem/progenitor cells and CLP cells (Figure 5d). Because thymocyte progenitors were identified as part of the LSK population,21 we used PCR to validate Trap1a expression in LSK cells. As shown in Figure 5e, Trap1a was detected in the WT but not the CD45.2+ mutant BM cells by qPCR. Further investigation by conventional PCR confirmed the expression of Trap1a in sorted LSK cells from the BM of WT mice (Figure 5f). Therefore, the expression pattern suggested that Trap1a affects the potential of T cell progenitors rather than directly acting on DN1 to promote the DN1 to DN2 transition.
The nearly complete inhibition of the DN1 to DN2 transition is reminiscent of the phenotype of Notch1−/− thymocytes.4 To determine whether the Notch signaling pathway was affected, we compared the expression of genes involved in Notch pathways in Trap1a+/y and Trap1a−/y ES cells. As shown in Figure 6a and Supplementary Dataset 2, none of the genes in the Notch pathway were affected by twofold or more. Because Notch1 deletion eliminated c-Kit expression in DN1 cells,21 we compared the expression of c-Kit in DN1 thymocytes. As shown in Figure 6b, the distribution of c-Kit expression in DN1 was unaffected by Trap1a deletion. These data suggest that Trap1a deletion does not affect thymocyte development through inhibition of Notch signaling.
Figure 6.
Trap1a mutation does not affect Notch signaling. (a) None of the 43 Notch signaling genes are affected by more than twofold. The X-axis shows the ratios of expression levels between WT and Trap1a−/y ES cells, while the Y-axis shows –log10 P values. (b) Distribution of c-Kit expression among the DN1 thymocytes from WT and Trap1a−/y stem cells. Data shown are representative profiles of thymocytes based on CD3/CD4/CD8/CD25/CD44 and c-Kit markers and have been reproduced in three independent experiments. ES cells, embryonic stem cells; WT, wild type.
Discussion
Taken together, our data revealed Trap1a as a new regulator of the DN1-DN2 transition. This checkpoint is distinct from Notch1 signaling at three levels. First, phenotypically, Notch1-deficient DN1 thymocytes were blocked prior to the expression of c-Kit, the earliest marker of T-cell progenitors, whereas the Trap1a-deficient thymocytes had normal expression of c-Kit. Second, although Notch signaling is active at the DN1-DN4 based on the expression of its target genes, Trap1a is expressed only in progenitor cells before they reach the thymus. Third, gene signature analysis of ES cells suggests that Trap1a deletion does not affect the expression of genes involved in Notch signaling. It is intriguing to note that because Trap1a expression cannot be found in developing thymocytes the potential for T-cell fate can be determined before T-cell progenitors reach the thymus.
P1A was the first tumor antigen to be identified. However, no physiological function has been attributed to P1A. By showing a critical role for P1A in T-cell development, our study filled in this major gap. Furthermore, based on the lack of contribution of the Trap1a−/y ES cells to several tissues, particularly the uterus, testis and brain, it is likely that P1A plays a fundamental role in the development of vital organs. Additional studies will be needed to determine the molecular mechanism by which P1A controls development of multiple cell types.
Acknowledgments
This work is supported by grants from the National Institutes of Health (AI64350, CA171972, CA58033 and AG036690).
Footnotes
Supplementary Information for this article can be found on the Cellular & Molecular Immunology website (http://www.nature.com/cmi)
The authors declare no conflict of interest.
Supplementary Material
References
- Schwarz BA, Bhandoola A. Circulating hematopoietic progenitors with T lineage potential. Nat Immunol 2004; 5: 953–960. [DOI] [PubMed] [Google Scholar]
- Ceredig R, Rolink T. A positive look at double-negative thymocytes. Nat Rev Immunol 2002; 2: 888–897. [DOI] [PubMed] [Google Scholar]
- Schmitt TM, Ciofani M, Petrie HT, Zuniga-Pflucker JC. Maintenance of T cell specification and differentiation requires recurrent notch receptor-ligand interactions. J Exp Med 2004; 200: 469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allman D, Karnell FG, Punt JA, Bakkour S, Xu L, Myung P et al. Separation of Notch1 promoted lineage commitment and expansion/transformation in developing T cells. J Exp Med 2001; 194: 99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Eynde B, Lethe B, Van Pel A, De Plaen E, Boon T. The gene coding for a major tumor rejection antigen of tumor P815 is identical to the normal gene of syngeneic DBA/2 mice. J Exp Med 1991; 173: 1373–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramarathinam L, Sarma S, Maric M, Zhao M, Yang G, Chen L et al. Multiple lineages of tumors express a common tumor antigen, P1A, but they are not cross-protected. J Immunol 1995; 155: 5323–5329. [PubMed] [Google Scholar]
- Boon T, Cerottini JC, Van den Eynde B, van der Bruggen P, Van Pel A. Tumor antigens recognized by T lymphocytes. Annu Rev Immunol 1994; 12: 337–365. [DOI] [PubMed] [Google Scholar]
- Brandle D, Bilsborough J, Rulicke T, Uyttenhove C, Boon T, Van den Eynde BJ. The shared tumor-specific antigen encoded by mouse gene P1A is a target not only for cytolytic T lymphocytes but also for tumor rejection. Eur J Immunol 1998; 28: 4010–4019. [DOI] [PubMed] [Google Scholar]
- Li CS, Chen C, Zheng P, Liu Y. Transgenic expression of P1A induced thymic tumor: a role for onco-fetal antigens in tumorigenesis. PLoS One 2010; 5: e13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA 1993; 90: 8424–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ED, Saunders TL. Gene Targeting in Embryonic Stem Cells. Springer-Verlag: Berlin. 2001. [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003; 4: 249–264. [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004; 20: 307–315. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA et al. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol 2013; 14: 1190–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013; 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Feng Z, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010; 26: 136–138. [DOI] [PubMed] [Google Scholar]
- Boiers C, Carrelha J, Lutteropp M, Luc S, Green JC, Azzoni E et al. Lymphomyeloid contribution of an immune-restricted progenitor emerging prior to definitive hematopoietic stem cells. Cell Stem Cell 2013; 13: 535–548. [DOI] [PubMed] [Google Scholar]
- Yan L, Womack B, Wotton D, Guo Y, Shyr Y, Dave U et al. Tgif1 regulates quiescence and self-renewal of hematopoietic stem cells. Mol Cell Biol 2013; 33: 4824–4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyttenhove C, Godfraind C, Lethe B, Amar-Costesec A, Renauld JC, Gajewski TF et al. The expression of mouse gene P1A in testis does not prevent safe induction of cytolytic T cells against a P1A-encoded tumor antigen. Int J Cancer 1997; 70: 349–356. [DOI] [PubMed] [Google Scholar]
- Tan JB, Visan I, Yuan JS, Guidos CJ. Requirement for Notch1 signals at sequential early stages of intrathymic T cell development. Nat Immunol 2005; 6: 671–679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.