Abstract
Autophagy has burgeoned rapidly as a field of study because of its evolutionary conservation, the diversity of intracellular cargoes degraded and recycled by this machinery the mechanisms involved, as well as its physiological relevance to human health and disease. This self-eating process was initially viewed as a non-selective mechanism used by eukaryotic cells to degrade and recycle macromolecules in response to stress; we now know that various cellular constituents, as well as pathogens, can also undergo selective autophagy. In contrast to non-selective autophagy, selective autophagy pathways rely on a plethora of selective autophagy receptors (SARs) that recognize and direct intracellular protein aggregates, organelles and pathogens for specific degradation. Although SARs themselves are not highly conserved, their modes of action and the signalling cascades that activate and regulate them are. Recent yeast studies have provided novel mechanistic insights into selective autophagy pathways, revealing principles of how various cargoes can be marked and targeted for selective degradation.
Autophagy involves the lysosomal degradation (or vacuolar degradation in yeast and plants) of intracellular macromolecular components (FIG. 1). Although it was initially studied as a cellular response to a particular type of stress, namely starvation, it is now apparent that autophagy is in fact a critical regulator of cellular homeostasis with intricate links to cell metabolism, growth control, the balance between cell survival and cell death, as well as ageing1. Therefore, it is not surprising that autophagy has a central role in human health and disease (reviewed in REF. 2). Autophagy is involved in cell death and tumour suppression3, neurodegeneration4, ageing5, inflammation6, immunity7 and genome stability6. We also now know that apart from starvation, autophagy is induced by many other perturbations, including hypoxia and metabolic, osmotic and oxidative stresses8–10.
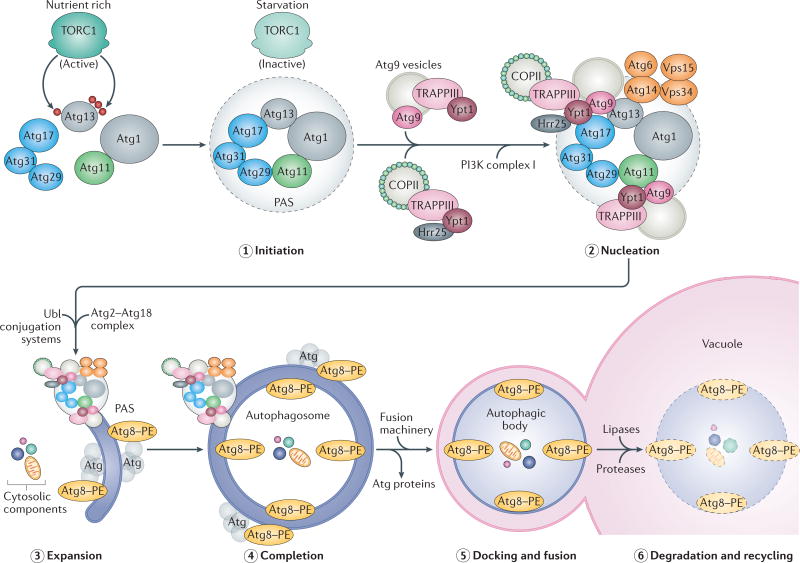
Figure 1. Steps in autophagy.
Autophagy is inhibited under nutrient-rich conditions via the hyperphosphorylation of autophagy-related 13 (Atg13) by target of rapamycin complex 1 (TORC1) kinase; this process prevents a tight interaction between Atg1 kinase and Atg17 (REF. 41). Starvation or rapamycin treatment activates autophagy by inhibiting TORC1, leading to the hypophosphorylation of Atg13, which can then interact with Atg1 and Atg17. The first two steps, initiation (step 1) and nucleation (step 2), involve the recruitment of cytosolic components of the core autophagic machinery to the phagophore assembly site (PAS) in yeast (omegasomes in mammals). In yeast, the non-selective autophagy-specific PAS is organized partly by the scaffold components Atg11 and Atg17, with Atg17 itself being part of a tripartite Atg17–Atg29–Atg31 subcomplex42,53. Scaffold components then recruit additional proteins, including transport protein particle III (TRAPPIII) and Ypt1 (a Rab1 family GTPase), which bring coat protein complex II (COPII) and Atg9 vesicles, to initiate the expansion (step 3) of a double-membrane phagophore. This expansion also involves the activity of phosphoinositide 3-kinase (PI3K) complex I (consisting of Atg6, Atg14, vacuolar protein sorting 34 (Vps34) and Vps15), which generates the phosphatidylinositol 3-phosphate required to recruit other factors involved in phagophore elongation, such as the Atg2–Atg18 complex as well as the ubiquitin-like (Ubl) conjugation systems, Atg8–phosphatidylethanolamine (PE) (Atg8–PE) and Atg5–Atg12–Atg16 (depicted as grey Atg molecules in contact with Atg8–PE); see also FIG. 3 for details on PAS assembly and isolation membrane formation. As a result of this membrane expansion, cargo destined for autophagy is surrounded and engulfed into a double-membrane vesicle called the autophagosome (step 4)11,46–48,115 Autophagosomes are then transported to lysosomes (or vacuoles in yeast and plants). Docking and fusion (step 5) of the outer autophagosomal membrane with that of the lysosome (vacuole) releases the autophagic body into the lysosomal (vacuolar) lumen, where hydrolases degrade and recycle (step 6) the macromolecular components for cellular use.
The autophagic machinery is encoded by autophagy-related (ATG) genes and comprises approximately 19 core Atg proteins that orchestrate the different steps of autophagy (TABLE 1; for a review see REF. 11). In yeast, this machinery can be divided into five multifunctional modules: the Atg8– phosphatidylethanolamine (PE) and the Atg5–Atg12 conjugation systems (Atg3, Atg4, Atg5, Atg7, Atg8, Atg10, Atg12 and Atg16); the Atg1 kinase complex (Atg1, Atg13, Atg17, Atg29 and Atg31); the class III phosphoinositide 3-kinase (PI3K) complex I (Atg6, Atg14, Atg38, vacuolar protein sorting 15 (Vps15) and Vps34); the Atg2–Atg18 complex; and vesicles containing the integral membrane protein Atg9 (REFS 11,12). These core autophagy proteins are often conserved in eukaryotes (TABLE 1), with the exception of red algae13. The autophagic machinery is sequentially engaged, and the process of autophagy can be subdivided into distinct steps. Autophagy starts by establishing a phagophore assembly site (PAS in yeast; omegasomes in mammals), followed by membrane expansion to form a double-membrane phagophore that surrounds and engulfs cargo destined for autophagy. This leads to the formation of a double-membrane vesicle known as the autophagosome, which is then transported to and fuses with the vacuole (yeast and plants; lysosome in mammals) for cargo degradation and recycling (FIG. 1).
Table 1.
Autophagy-related proteins and complexes conserved from yeast to mammals
| Autophagy-related protein (alias) | Function of yeast protein | |
|---|---|---|
| Yeast | Mammals | |
| Signalling | ||
| TORC1 complex | mTORC1 complex | Negative regulator of autophagy |
| Slt2 | ERK5 | MAPK required for pexophagy and mitophagy |
| Hog1 | p38 | MAPK required for mitophagy |
| Pbs2 | MKK4 |
|
| Hrr25 | CK1δ (CSNK1D) | CK1 involved in pexophagy and the Cvt pathway, as well as in non-selective autophagy |
| CK2 | CK2 | Required for mitophagy |
| Factors required for selective autophagy (excluding receptors) | ||
| Atg21 | WIPI1, WIPI2 |
|
| Atg37 (Pichia pastoris) | ACBD5 | Acyl-CoA-binding protein required for pexophagy in Pichia pastoris |
| Pex3 (Pichia pastoris) | PEX3 | Peroxisomal membrane protein involved in peroxisome biogenesis and pexophagy in Pichia pastoris and Hansenula polymorpha |
| Dnm1 | DRP1 | Dynamin-related GTPase required for pexophagy and mitophagy |
| Vps1 | Dynamin | Dynamin-related GTPase required for pexophagy and mitophagy |
| Vesicle formation and completion: PI3K complexes | ||
| Vps30–Atg6 | BECN1 | Subunit of PI3K complexes I (Vps34, Vps15, Vps30/Atg6 and Atg14) and II (Vps34, Vps15, Vps30/Atg6 and Vps38) |
| Atg14 | ATG14 |
|
| Vps34 | PIK3C3 (VPS34) | PI3K catalytic subunit |
| Vps38 | UVRAG |
|
| Vps15 | PIK3R4 (VPS15, p150) | Protein kinase required for Vps34 function |
| Vesicle formation and completion: conjugation systems | ||
| Atg5 | ATG5 | Conjugated to Atg12 |
| Atg7 | ATG7 | E1-like enzyme for both Atg12 and Atg8 |
| Atg8 | LC3A, LC3B, LC3B2, LC3C, GABARAP, GATE16 |
|
| Atg10 | ATG10 | E2-like enzyme that conjugates Atg12 to Atg5 |
| Atg12 | ATG12 | Ubiquitin-like modifier that forms the Atg12–Atg5 conjugate |
| Atg16 | ATG16L1, ATG16L2 | Component of the Atg12–Atg5 complex |
| Atg3 | ATG3 | E2 conjugating enzyme that generates Atg8–PE |
| Atg4 | ATG4A, ATG4B, ATG4C, ATG4D | Cysteine protease involved in Atg8 lipidation and Atg8–PE delipidation |
| Vesicle formation and completion: the Atg1 kinase complex | ||
| Atg1 | ULK1, ULK2 |
|
| Atg13 | ATG13 | Phosphoprotein activated by dephosphorylation under starvation conditions |
| Atg17 | FIP200 (RB1CC1) | Scaffold protein for autophagy and part of the Atg17–Atg29–Atg31 complex |
| Vesicle formation and completion: Atg9 vesicles | ||
| Atg9 | ATG9 |
|
| Vesicle formation and completion: COPII vesicle | ||
| Ypt1 | RAB1 | Rab-family GTPase that recruits Hrr25 |
| TRAPIII complex | TRAPIII complex | Regulates autophagy |
| Vesicle formation and completion: the Atg2–Atg18 complex | ||
| Atg2 | ATG2A, ATG2B |
|
| Atg18 | WIPI1, WIPI2 |
|
ACBD, acyl-CoA binding domain; Atg, autophagy-related; BECN, beclin; CK, casein kinase; Cvt, cytoplasm-to-vacuole targeting; Dnm1, dynamin 1; DRP, dynamin-related protein; ERK5, extracellular-signal related kinase; FIP200, FAK-family kinase interacting protein of 200 kDa; GABARAP, γ-aminobutyric acid-associated protein; HOG, high osmolarity glycerol; LC3, microtubule-associated protein 1A/1B–light chain 3; Pbs, polymyxin B sensitivity; PE, phosphatidylethanolamine; Pex, peroxin; PI3K, phosphatidylinositol 3-kinase class III; PIK3C3, PtdIns3P, phosphatidylinositol-3-phosphate; TORC, target of rapamycin complex; TRAPP, transport protein particle; ULK, Unc51-like kinase; UVRAG, UV-radiation resistance associated gene; Vps, vacuolar protein sorting; WIPI, WD repeat domain, phosphoinositide-interacting protein.
Although autophagy was initially viewed as a non-selective process of self-consumption, it is now well established that a remarkable plethora of cargoes can be degraded with high selectivity (TABLE 2 and references therein). Moreover, these selective autophagy pathways have been linked to various human disease states and in plant host–pathogen interactions14–17. Selective autophagy pathways operate both in normal vegetative conditions (non-induced conditions) and in response to different stimuli (induced conditions) and contribute to intracellular homeostasis. An example of non-induced autophagy is a process known as the cytoplasm-to-vacuole targeting (Cvt) pathway; in this pathway, vegetatively growing yeast cells produce certain proteins, such as vacuolar aminopeptidase 1 (Ape1), aspartyl aminopeptidase 4 (Ape4) and α-mannosidase 1 (Ams1), that are transported to the vacuole where they mature and can serve enzymatic functions18. Conversely, the turnover of superfluous organelles happens in response to environmental stimuli19. Selective autophagy also degrades intracellular protein aggregates, pathogens and damaged organelles20,21. Similar to non-selective autophagy, selective autophagy is also activated by various external stimuli, including stresses such as oxidative, osmotic, hypoxic or starvation conditions8–10.
Table 2.
Types of selective autophagy
| Selective autophagy type | Selective cargo | Organism in which described |
Cargo receptor(s) | Refs |
|---|---|---|---|---|
| Aggrephagy | Protein aggregates | Yeast and mammals | p62*, OPTN*, NBR1*, Cue5*, TOLLIP* | 20,25,118,119 |
| Chaperone-mediated autophagy | Cytosolic proteins with KFERQ-like motifs | Mammals | LAMP2A | 120 |
| Chlorophagy | Chloroplasts | Plants | Unknown | 121 |
| Chromatin autophagy | Chromatin | Mammals | Unknown | 122 |
| Ciliophagy | Cilia | Mammals | HDAC6 | 123 |
| Cytoplasm-to-vacuole targeting | Ape1, Ape4 and Ams1 | Yeast | Atg19, Atg34 | 18 |
| DNA-mediated xenophagy | Mycobacterium tuberculosis | Mammals | p62*, NDP52* | 124 |
| Endoplasmic reticulum-phagy | Peripheral and nuclear endoplasmic reticulum | Yeast and mammals | Atg40, FAM134B | 33,105,125 |
| Ferritinophagy | Ferritin | Zebrafish and mammals | NCOA4 | 126,127 |
| Glycogen autophagy | Glycogen | Mammals | STBD1 | 128–130 |
| Granulophagy | Stress granules and P bodies | Yeast and mammals | Unknown | 131 |
| Lipophagy | Lipid droplets | Yeast and mammals | Unknown | 132,133 |
| Lysophagy | Damaged lysosomes | Mammals | Galectin 8 | 134,135 |
| Midbody ring disposal | Midbody protein CEP55 | Mammals | p62*, NBR1* | 136,137 |
| Mitophagy | Mitochondria | Yeast, plants and mammals | Atg32, NIX, OPTN*, NDP52*, TAX1BP1*, BNIP3, FUNDC1, BCL2L13 | 97,104, 138–142 |
| Myelinophagy | Myelin | Mammals | Unknown | 143 |
| Nucleophagy | Fragments of nucleus | Fungi and mammals | Atg39 | 33,144,145 |
| Pexophagy | Damaged or superfluous peroxisomes | Yeast, plants and mammals | Atg30, Atg36, PEX5*, NBR1*, p62* | 17,26,27,108 |
| Plant p62-like and Nbr1-like | Unknown | Plants | Joka2* | 146 |
| Plant tryptophan-rich sensory protein turnover | Tryptophan-rich sensory protein | Plant (Arabidopsis thaliana) | Unknown | 147 |
| Plastid-to-vacuole pathway | Plastids | Plants | ATI1 | 148 |
| Proteaphagy | Inactive proteasomes | Plants | RPN10* | 149 |
| Ribophagy | Ribosomal proteins | Yeast | Unknown | 150,151 |
| Virophagy | HIV p24 and viral proteins | Mammals | TRIM5α, SMURF1 | 152,153 |
| Xenophagy | Bacterial and viral pathogens | Mammals | Galectin 8, p62*, OPTN*, NDP52*, TAX1BP1*, TECPR1 | 21,154,155 |
| Zymophagy | Pancreatic zymogens | Mammals | p62* | 156 |
Ams1, α-mannosidase 1; Ape1, vacuolar aminopeptidase 1; Ape4, aspartyl aminopeptidase 4; Atg, autophagy-related; ATI1, Atg8-interacting protein; BCL2L13, BCL-2-like 13; BNIP, BCL-2/adenovirus E1B 19 kDa interacting protein 3; CEP55, centrosomal protein of 55 kDa; CUE, coupling of ubiquitin conjugation to ER degradation; FUNDC, FUN-domain containing protein; HDAC6, histone deacetylase 6; LAMP2A, lysosome-associated membrane glycoprotein 2A; NBR, next to BRCA gene 1; NCOA4, NDP, nuclear dot protein; NIX, NIP3-like protein X; OPTN, optineurin; PEX, peroxin; RPN, regulatory particle non-ATPase; SMURF, SMAD ubiquitylation regulatory factor; STBD1, starch-binding domain-containing protein 1; TAX1BP, Tax1-binding protein; TECPR1, tectonin β-propeller repeat containing protein; TOLLIP, Toll-interacting protein; TRIM, tripartite motif.
Ubiquitin-dependent selective autophagy receptors. Data adapted from REF. 22.
Most selective autophagy pathways use a common mechanism, including the ‘core autophagy machinery’ toolbox (TABLE 1), superimposed on which is a set of selectivity factors (TABLE 3). Most important among these selectivity factors are selective autophagy receptors (SARs) (FIG. 2), which mark each specific cargo for selective degradation and initiate the autophagic process. The SARs engage cargo and the core autophagy machinery at the PAS, and activate a particular selective pathway to the exclusion of other selective and non-selective autophagy processes.
Table 3.
Summary of selective autophagy pathways in yeast with known SARs
| Selective autophagy type |
Cargoes | SARs | Requirement for phosphorylation of SARs and interaction between SARs and Atg11 |
SAR kinase(s) |
Atg17– Atg29–Atg31 requirement during selective autophagy |
Additional selectivity factors |
|---|---|---|---|---|---|---|
| Non-induced selective autophagy | ||||||
| Cytoplasm-to-vacuole targeting | prApe1, Ape4, Ams1 | Atg19 (REF. 23) (Sc) | Atg19*–Atg11 (REF. 67) | Hrr25 (REF. 67), Atg1 (REF. 67) | Dispensable | Atg20 (REFS 60,157), Atg21 (REFS 158,159), Atg24 (REF. 157), Vac8 (REF. 160), VFT complex161, actin162,163, Arp2–Arp3 complex164 |
| Induced selective autophagy | ||||||
| Ape1 transport | prApe1 | Atg19 (REF. 23) (Sc) | Atg19 (REF. 23) | N.D. | Required53 | N.D. |
| Ams1 transport | Ams1 | Atg34 (REF. 24) (Sc) | Atg34*–Atg11 (REFS 67,74) | Hrr25 (REFS 67,74) | N.D. | N.D. |
| Pexophagy | Peroxisomes | Atg30 (REF. 26) (Pp) | Atg30*–Atg11 (REF. 26) | N.D. | Required Atg17-Atg28‡ (REF. 36) | Atg21 (REF. 165), Atg24 (REF. 166), Atg26 (REF. 167), Atg35 (REF. 168), Atg37 (REF. 29), Pex3 (REF. 28), Vac8 (REF. 169) |
| Atg36 (REF. 27) (Sc) | Atg36*–Atg11 (REFS 27,59) | Hrr25 (REF. 75) | Required35 | Atg20 (REF. 157), Atg24 (REF. 157), actin162, Arp2–Arp3 complex164, Dnm1 (REF. 90), Vps1 (REF. 90), Fis1 (REF. 90) | ||
| Mitophagy | Mitochondria | Atg32 (REFS 59,71) (Pp) | Atg32*–Atg11 (REF. 72) | N.D. | N.D. | N.D. |
| Atg32 (REFS 30,31) (Sc) | Atg32*–Atg11 (REF. 72) | CK2 (REF. 72) | Required37 | Atg20 (REF. 37), Atg21 (REF. 37), Atg24 (REF. 37), Atg33 (REF. 37), Dnm1 (REF. 91), Fis1 (REF. 91) | ||
| Nucleophagy | Fragments of nucleus | Atg39 (REF. 33) (Sc) | Atg39-Atg11 (REF. 33) | N.D. | Required33 | Actin§ (REF. 170) |
| Endoplasmic reticulum-phagy | Endoplasmic reticulum | Atg40 (REF. 33) (Sc) | Atg40-Atg11 (REF. 33) | N.D. | Required33 | Actin§ (REF. 170) |
| Aggrephagy | Aggregates | Cue5 (REF. 20) (Sc) | Cue5 (REF. 20) | N.D. | N.D. | Rsp5 (REF. 20), Ubc4 and Ubc5 (REF. 20) |
Ams1, α-mannosidase 1; Ape1, vacuolar aminopeptidase 1; Ape4, aspartyl aminopeptidase 4; Atg, autophagy-related; prApe1, precursor of Ape1; Arp, actin-related protein; CK2, casein kinase 2; Cue5, coupling of ubiquitin conjugation to endoplasmic reticulum (ER) degradation 5; Dnm1, dynamin 1; Fis1, mitochondria fission 1; N.D. not determined; Pex, peroxin; Pp, Pichia pastoris; Sc, Saccharomyces cerevisiae; Ubc, ubiquitin carrier protein; Vac, vacuole-related; VFT, Vps fifty three; Vps, vacuolar protein sorting.
SARs known to be phosphorylated.
Atg28 is the potential Pichia pastoris homologue of the Atg29 and Atg31 complex.
Actin is required to engulf ER structure observed by electron microscopy
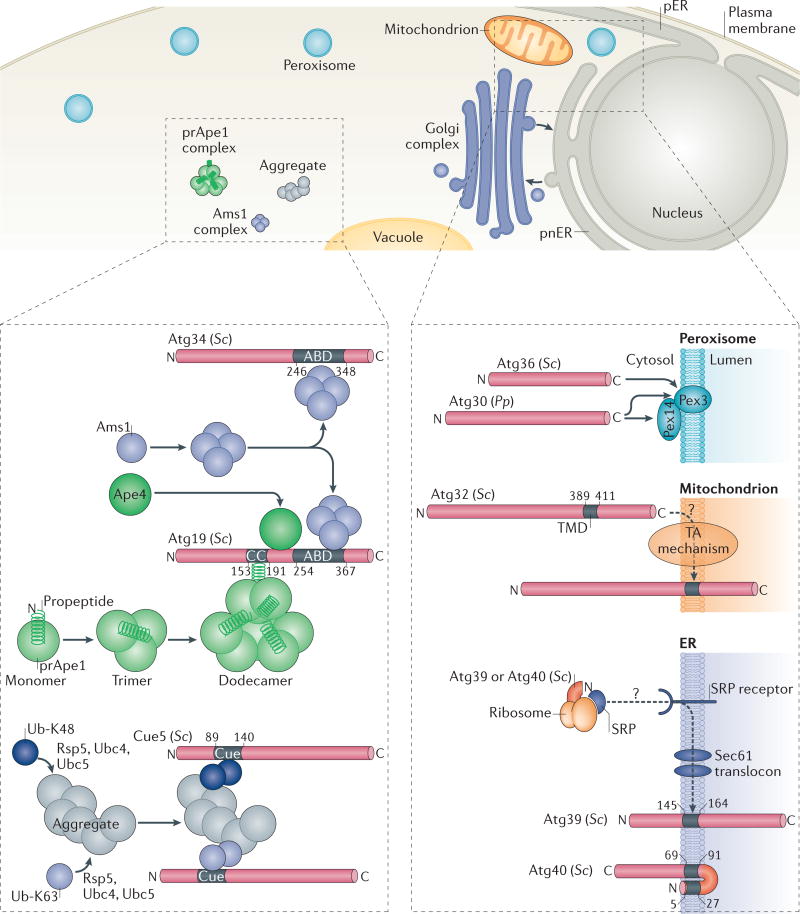
Figure 2. Selective autophagy pathways and cargo recognition by selective autophagy receptors.
The upper panel of the figure shows the specific cargoes, such as oligomeric α-mannosidase 1 (Ams1) or precursor of vacuolar aminopeptidase 1 (prApe1), protein aggregates or organelles (peroxisomes, mitochondria, perinuclear ER (pnER) or peripheral ER (pER), or fragments of the nucleus), that are subject to selective autophagy. The lower left panel depicts soluble selective autophagy receptors (SARs). The prApe1 dodecamer is bound by the coiled-coil (CC) domain of autophagy-related 19 (Atg19)58. Ams1 oligomerizes and associates with Atg19 through the Ams1-binding domain (ABD)116. prApe1, Ams1 and Atg19 assemble into a large complex called the cytoplasm-to-vacuole targeting (Cvt) complex. Atg34, an Atg19 paralogue, is also a receptor for Ams1 (REF. 24), but not for Ape1 or aspartyl aminopeptidase 4 (Ape4)117. Ape4 also binds Atg19. Coupling of ubiquitin conjugation to endoplasmic reticulum (ER) degradation 5 (Cue5) binds aggregates through direct interaction of its Cue domain with lysine 63 (K63)- and lysine 48(K48)-linked ubiquitin (Ub) chains that are covalently attached to cargoes25 by the E3 ubiquitin ligase Rsp5 and the E2 ubiquitin-conjugating enzyme Ubc4 or Ubc5. The lower right panel depicts membrane-associated SARs. Pexophagy receptors of Saccharomyces cerevisiae (Sc) Atg36 and Pichia pastoris (Pp) Atg30 recognize peroxisomal membrane proteins (Pex14 and/or Pex3). The mitophagy receptor Atg32 is embedded in the mitochondrial outer membrane via a single α-helical transmembrane domain (TMD) and probably (indicated by a question mark in the figure) the action of the tail-anchored (TA) mechanism, which refers to the protein machinery that inserts proteins possessing a carboxy-terminal TMD into the membrane such that, topologically, the amino terminus of the protein is cytosolic and the C terminus of the protein is lumenal. The ER-phagy receptors Atg39 and Atg40 have one TMD and two TMDs, respectively, and might insert into the ER membrane co-translationally via the signal recognition particle (SRP), the SRP receptor and the Sec61 translocon.
Because of the complexity and scope of the rapidly expanding modes of selective autophagy (TABLE 2), we focus here on mechanistic insights obtained using yeast models. The amenability of yeast to genetic as well as biochemical manipulations and their ease of imaging have enabled the study of the morphological steps, the molecular machinery and the mechanisms of autophagy in great detail. Importantly, because of the evolutionary conservation of the core autophagy machinery (TABLE 1), many insights gained from studies of selective autophagy in yeast are proving remarkably applicable to mammals. In this Review, we outline the principles governing the selectivity of autophagy, emphasizing the roles played by SARs. We also describe the common features of SARs, their roles and the signalling mechanisms involved in cargo recognition among eukaryotes.
Receptors for selective autophagy
Selective autophagy depends on the recognition of the specific cargo to be degraded. In most cases, this recognition occurs through the binding of specific autophagy receptors, SARs, which allow selective engagement of the autophagy machinery. In yeast, SARs can be divided into two groups: soluble receptors (Atg19, Atg34 and coupling of ubiquitin conjugation to endoplasmic reticulum (ER) degradation 5 (Cue5)) and membrane-associated receptors (Atg30/Atg36, Atg32, Atg39 and Atg40)22 (FIG. 2).
Soluble receptors
Soluble receptors in yeast are involved in the Cvt pathway and in the degradation of misfolded or aggregated proteins (FIG. 2). One of these SARs, Atg19, binds the precursor of Ape1 (prApe1), the primary Cvt cargo23, as well as Ams1 and Ape4. The prApe1, Ams1 and Atg19 proteins assemble into a large complex called the Cvt complex, which is then targeted to and processed in the vacuole. Atg34 functions as an additional receptor protein for Ams1, but not prApe1, and only under starvation conditions24.
A new class of soluble SARs belongs to the conserved CUET protein family25. Cue5 in yeast and Toll-interacting protein (TOLLIP) in mammals are required for the autophagic degradation of ubiquitylated proteins and polyQ proteins. The 50 amino acid long CUE domain of Cue5, which is structurally related to the ubiquitin-binding UBA domain, binds to both monoubiquitylated and polyubiquitylated cargo. Several soluble yeast proteins that aggregate (for example, Abp1, Cpr1, Ent2, Fpr1, Gvp36, Pil1, Rpl14B, Rpl26B, Rpp2B, Seg1, Tma19, Tsa1 and Ygr130c) are degraded by the Cue5-dependent selective autophagy pathway.
Membrane-associated receptors
The first SAR identified for organelles was Atg30 of Pichia pastoris (Atg36 in Saccharomyces cerevisiae), which is the receptor for selective autophagy of peroxisomes (pexophagy)26,27 (FIG. 2; TABLE 2). Many yeast species contain one or the other receptor, but not both. These two SARs do not share amino acid sequence homology, but they fulfil the same function. In silico analysis of their amino acid sequences does not reveal any characteristics that indicate they would associate with membranes or be imported into organelles. Instead, the pexophagy receptors bind directly, in vivo and in vitro, to Pex3, a peroxisomal membrane protein (PMP)26–28, an essential component for peroxisome biogenesis. Atg30 also associates with other PMPs such as Pex14 and Atg37, which are required for peroxisomal matrix protein import and pexophagy, respectively26,29.
Mitochondrial degradation (mitophagy) is mediated by Atg32 (REFS 30,31) (FIG. 2). Despite low overall sequence homology, Atg32 is conserved in most yeast species. Atg32 contains a transmembrane domain (TMD) and is anchored in the mitochondrial outer membrane, with its amino and carboxyl termini exposed to the cytosol and mitochondrial intermembrane space, respectively32.
Atg39 and Atg40 are two specific receptors for ER degradation (ER-phagy)33 (FIG. 2). Atg39 and Atg40 localize at the perinuclear ER and the peripheral ER, respectively.
SARs and the autophagic machinery
The various SARs recognize and mark the cargoes for degradation. However, as outlined above, to target these cargoes to the vacuole, PAS assembly initiation is required. During selective autophagy PAS assembly is mediated by the interactions of activated cargo-bound SARs with the core proteins of the autophagic machinery, which we describe in this section.
Scaffold and Atg8 proteins in autophagy
As outlined above, autophagy involves a sequential recruitment of many proteins that cooperate in the formation of the autophagosome (FIG. 1). The functions of the core autophagy machinery components have been extensively reviewed elsewhere34; however, because they directly interact with SARs and are therefore important mediators of selectivity, we will briefly describe here the functions of three of these core proteins: Atg11 and Atg17, which function as scaffolds, and Atg8 (FIG. 3).
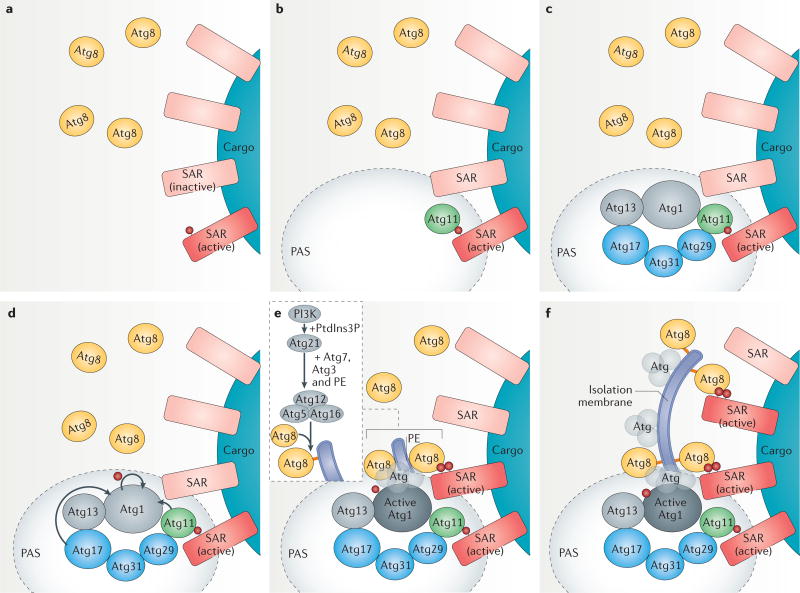
Figure 3. Phagophore assembly site and isolation membrane formation in selective autophagy activation.
a | Phagophore assembly site (PAS) formation starts by activation of selective autophagy receptors (SARs) bound to cargoes. The activation mechanism involves phosphorylation by casein kinases (phosphorylation is indicated by a red ball). b | An activated SAR binds the scaffold protein autophagy-related 11 (Atg11) to initiate PAS formation. c | Atg11 binds the SAR–cargo complex, recruits the Atg17 scaffold complex (composed of Atg17, Atg31 and Atg29) via Atg29, as well as the Atg1 kinase complex (composed of Atg1 and Atg13). d | Atg11 bound to the cargo–SAR complex and the Atg17 complex then activates the Atg1 kinase, which autophosphorylates itself as well as other Atg proteins. e | Activated Atg1 kinase recruits other core autophagy proteins, resulting in the recruitment of Atg8, which is then conjugated to phosphatidylethanolamine (PE) to begin phagophore expansion from the PAS. A second ubiquitin-like conjugate, Atg12–Atg5, forms a complex with Atg16, and is necessary for the recruitment of Atg8 to the PAS and its conjugation to PE (it acts as the E3 ubiquitin ligase). The Atg12–Atg5–Atg16 complex itself is recruited to the PAS by the phosphatidylinositol-3-phosphate (PtdIns3P)-binding protein Atg21, and its localization relies on PtdIns3P synthesis at the PAS by the PI3K complex I (see also FIG. 1)53. Notably, Atg21 is required mostly for selective autophagy pathways and not for non-selective autophagy. Atg8–PE also interacts with neighbouring SARs activated by phosphorylation. f | Isolation membrane expansion then continues around the cargo, engaging other activated SARs.
Atg11 and Atg17 are the autophagic scaffold proteins in yeast and are partially responsible for organizing the PAS (FIG. 3c). The non-inducible Cvt pathway requires Atg11 exclusively, but many selective, induced autophagy pathways, such as mitophagy, nucleophagy, pexophagy and ER-phagy, typically require both Atg11 and the Atg17 complex for efficient organelle degradation33,35–37 (TABLE 3). Many of these pathways, similar to non-selective autophagy, are induced by nitrogen starvation, and this might explain the involvement of Atg17, which is required during starvation. However, selective autophagy often has additional requirements (for example, a change in the carbon source for pexophagy and mitophagy), and requires SARs and their phosphorylation as well as the presence of auxiliary factors (in some cases) (TABLE 3).
The scaffold proteins have several roles. First, they interact with and recruit other core autophagy machinery components, such as the Atg1 complex38 (FIG. 3c – f); during starvation, typically both Atg11 and Atg17 are involved. During nutrient deprivation, the target of rapamycin complex 1 (TORC1), which is a protein kinase, is inactivated, resulting in hypophosphorylation of Atg13 (REF. 39) (FIG. 1). The hypophosphorylated Atg13–Atg1 complex is then bound by the Atg17–Atg29–Atg31 complex and recruited to form the PAS38,40,41. Interestingly, Atg11 can recruit Atg17 to the PAS in the absence of Atg1 and Atg13 through interactions with the Atg29–Atg31 complex42. However, in the presence of Atg1 and Atg13, Atg17 can form the PAS without Atg11, indicating two cooperative mechanisms for PAS formation. Scaffold proteins also promote the activation of the Atg1 kinase43, which is required for isolation membrane elongation and phagophore formation44 (FIG. 3d,e). Active Atg1 phosphorylates itself as well as the autophagy-related integral membrane protein Atg9, which recruits the Atg2–Atg18 complex to initiate phagophore membrane elongation45. Finally, these scaffolds also recruit to the PAS other regulators, such as transport protein particle III (TRAPPIII) and the guanine-nucleotide exchange factor (GEF) for the Ypt1 (a Rab1-family GTPase)18,46–49 (FIG. 1). It is suggested that during nitrogen starvation, normal traffic of coat protein complex II (COPII) vesicles from the ER to the Golgi is inhibited and these vesicles are diverted to the PAS to play some unknown function in autophagy50. TRAPPIII might contribute to the tethering of these COPII-containing, as well as Atg9-containing, vesicles for isolation membrane expansion. By contrast, Ypt1 recruits additional molecules of Atg1 to the PAS. Ypt1 also recruits Hrr25 (a casein kinase 1δ (CK1δ; also known as CSNK1D) homologue) to the PAS and activates its kinase49; the function of Hrr25 in non-selective autophagy is currently unknown. During fed conditions, in which the Cvt pathway is active, Atg11 is the main scaffold required for Atg1 recruitment and activation. During Cvt, Atg11 also recruits Atg9-containing membranes46,51 by interacting with both Ypt1 and directly with Atg9 to promote PAS formation (which, in contrast to nutrient-deprived conditions, occurs independently of COPII vesicles)46,47,52.
Atg8 is a ubiquitin-like protein that functions as a conjugate with the phospholipid PE (Atg8–PE) during autophagy. Atg8–PE localizes to the PAS, isolation membrane and autophagosome, contributes to Atg1 recruitment and is required for autophagosome membrane formation for all types of autophagy34,53 (FIG. 3). The precise function of Atg8–PE during the autophagy process is not yet clear, but this complex is involved in the growth and maturation of autophagosomal structures and it influences autophagosome size54. In addition to its role in autophagosome biogenesis, Atg8–PE appears to be a central factor in mediating cargo selectivity through direct interactions with SARs55 (FIG. 3e,f) (see also below). Interestingly, the functions of Atg8–PE in autophagosome biogenesis and cargo selection depend on different domains of Atg8 and can be separated by mutations in the ATG8 sequence56. Interactions between Atg8–PE and SARs have also been shown to play a part during autophagy termination. In this context, the interaction between Atg8–PE and the SAR leads to the disassembly of the Atg12–Atg5– Atg16–Atg8–PE complex in a reaction that is completed by Atg4-dependent deconjugation of Atg8–PE57.
Interactions of SARs with scaffold proteins
With the exception of Cue5, all known yeast SARs upon activation (for details see below) bind the scaffold protein Atg11 REFS 24,26,27,30,31,33,58). The Atg11-binding regions (A11BRs) in SARs are close (separ ated by 0–62 amino acids) to the Atg8-family interacting motifs (AIMs) (Supplementary information S1 (figure)), precluding simultaneous binding of both Atg8 and Atg11 to the receptor59. As a result, SARs bind either Atg8 or Atg11. Notably, during pexophagy, the same receptor molecules must interact with both Atg8 and Atg11. These interactions, however, occur sequentially, particularly in SARs in which the binding sites for Atg8 and Atg11 are overlapping, or in close proximity, so as to preclude simultaneous binding of both proteins to the SAR59.
The A11BRs in SARs consist of two hydrophobic residues followed by a serine residue and are surrounded by a series of serine or threonine residues and/or acidic amino acids (Supplementary information S1 (figure)). The most frequent Atg11-binding motif signature found in the membrane-associated receptors (Atg30, Atg32, Atg36 and Atg39) is I/VLS (Supplementary information S1 (figure)). Atg19 and Atg34, the two Cvt receptors, bind to Atg11 through DDSSIISTS and DESSIMSTP, respectively. These two sequences do not contain the strict signature motifs of the membrane- associated receptors, although they have in common two hydrophobic residues followed by a serine residue. Atg11 contains four coiled-coil (CC) domains and the last CC interacts directly with SARs60,61. Interestingly, this CC domain is conserved in the C-terminal domain of the mammalian protein FAK family kinase-interacting protein of 200 kDa (FIP200; also known as RB1CC1) and is listed in the protein family (Pfam) database as the Atg11 domain.
Similar to the role of Atg17–Atg29–Atg31 in non-selective autophagy, SAR-bound Atg11 recruits other autophagy proteins, such as the Atg1 kinase complex and Atg9, to the PAS. This process leads to Atg1 activation, which then drives the expansion of the isolation membrane43 (FIG. 3d – f). Atg1 kinase activation (including its autophosphorylation) is normally repressed in nutrient-rich conditions by TORC1 kinase signalling to block non-selective autophagy39,62. However, in nutrient-rich conditions, the cargo–SAR–Atg11 complex activates Atg1 kinase, bypassing the inhibition by TORC1 (REF. 43).
Interactions with Atg8
Atg8 is involved in autophagosome formation in all autophagy-related pathways, but it also tethers the cargo-bound SARs to the isolation membrane during selective autophagy. All known yeast SARs bind to Atg8 through one or more AIMs24,25,27,31,33,59 (Supplementary information S1 (figure)). Most AIMs have a short conserved motif (W/F/Y)xx(L/I/V) surrounded by at least one (often more) proximal acidic residue55. The first and the fourth hydrophobic residues in the AIM bind the hydrophobic pocket of Atg8, and the acidic residue (or residues) upstream of the AIM of SARs contributes a negative charge (or charges) to reinforce the interaction63. In several cases, the acidic microenvironment is further regulated by the phosphorylation of serine or threonine residues of the SARs in, or adjacent to, the AIM33,59 (see below).
Why exactly SARs need to interact with Atg8 is not completely understood. One possibility is to further promote selective autophagic cargo sequestration through tight tethering of the cargo decorated by SARs to the isolation membrane23,64,65. Although Atg8 is essential for all autophagy pathways34, surprisingly, mutations in the AIMs of SARs that bind to both Atg8 and Atg11 only partially impair selective autophagy31,33,59,64.
A possible explanation for this partial defect could come from the finding of multiple AIMs in the Cvt receptor Atg19 (REF. 64) (Supplementary information S1 (figure)). Atg19 contains, in addition to its canonical AIM1, two cryptic upstream AIMs (AIM2 and AIM3). The prApe1 complex is transported both selectively to the vacuole by the Cvt pathway and following initiation of the non-selective autophagy pathway during starvation conditions; in both cases, the transport of the prApe1 complex requires Atg19. It was assumed that AIM1 of Atg19 was required for prApe1 transport by the Cvt pathway but not by non-selective autophagy55,66. However, mutation of AIM1, combined with mutations of one or more of the other AIMs, fully blocks prApe1 transport to the vacuole irrespective of nutrient condition, indicating that the AIMs in Atg19 have some direct or indirect role in cargo selectivity. It has been postulated that one AIM is sufficient for the selectivity of the prApe1 complex, but additional AIMs allow exclusion of non-selective cargo by the Cvt pathway. Thus, it is possible that SARs may contain multiple AIMs such that mutation of a single AIM only abolishes the exclusion of non-selective cargo but not the transport of the selective cargo.
SAR activation through phosphorylation
The mere presence of SARs on the organelle is insufficient to induce selective autophagy; SARs must be activated and this activation is often achieved through phosphorylation (FIGS 3,4). For instance, during organelle biogenesis, SARs such as Atg36 (or Atg30) and Atg32 are present in an inactive hypophosphorylated form in or on the membranes of peroxisomes and mitochondria, respectively. However, a change in media to a carbon source (without nitrogen), which limits metabolism in these organelles, causes SAR activation through their hyperphosphorylation26,27,59, resulting in organelle turnover. In this section we review the role of phosphorylation in the regulation of SARs and we outline the mechanisms governing these phosphorylation events.
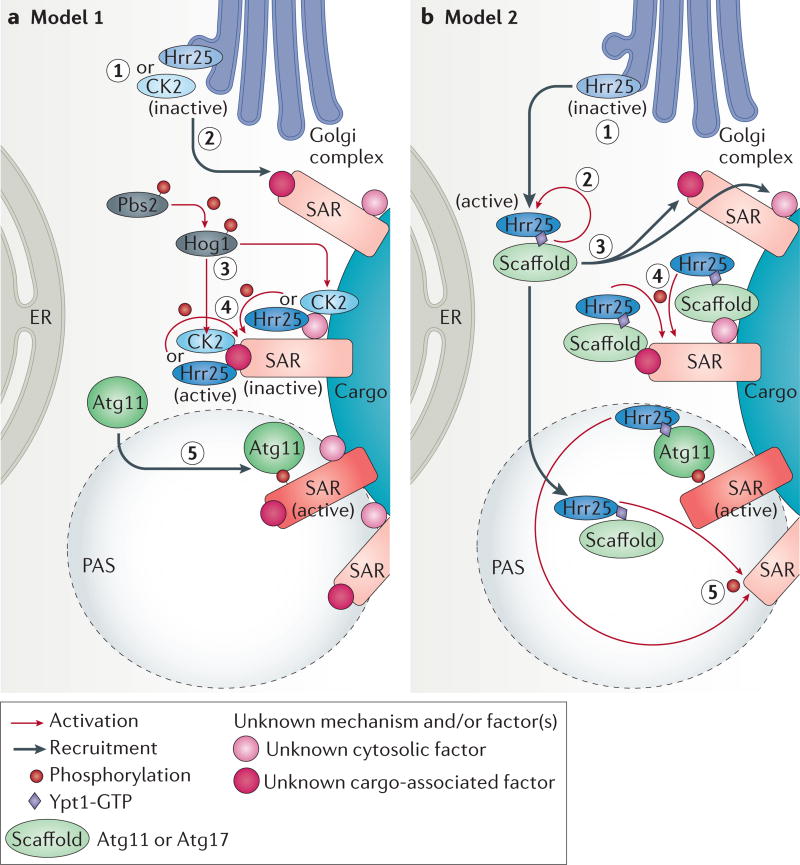
Figure 4. Hypothetical models for activation of selective autophagy receptors by casein kinases.
Yeast selective autophagy receptors (SARs) are generally phosphorylated and casein kinases (Hrr25 or casein kinase 2 (CK2)) have been shown to play a role in the phosphorylation of SARs involved in the mitophagy, pexophagy and Cvt pathways. The SARs for pexophagy as well as the cytoplasm-to-vacuole targeting (Cvt) cargo, precursor of vacuolar aminopeptidase 1 (prApe1), are phosphorylated by the CK1δ homologue, Hrr25, whereas the mitophagy SAR is phosphorylated by CK2. At least two hypothetical models could explain the phosphorylation and activation mechanism of SARs. However, the order and subcellular location of these steps are not currently known. In Model 1 (part a), inactive Hrr25 and CK2 (step 1) are recruited to inactive SARs (step 2) by an unknown factor (or factors) (shown as pink-shaded circles) and by unknown mechanisms and activated close to the SARs (step 3). Activation of CK2 might depend on the MAPKs of the high osmolarity glycerol (HOG) pathway, Hog1 and Pbs2 (REFS 61,72). Activated CK2 and Hrr25 then phosphorylate and activate SARs (step 4), resulting in the recruitment of autophagy-related 11 (Atg11) (step 5). In model 2 (part b), the inactive Hrr25 (step 1) is activated in the cytosol (step 2) and then recruited to the SAR or its vicinity via unknown factors (step 3), as well as to the phagophore assembly site (PAS), in a manner dependent on an activated Ypt1 (Ypt1-GTP) and the scaffold protein Atg17 (REF. 49) or possibly also Atg11 (as Atg11 is known to recruit Ypt1 to the PAS46). Consequently, before PAS formation, the first SAR (shown at the top) will be phosphorylated by the active Hrr25 localized proximal to the SAR by Ypt1-GTP, the scaffold protein and unknown factors associated either with the SAR itself or the cargo (step 4). The direct interaction of the phosphorylated SAR (active) and the scaffold protein Atg11 will initiate PAS formation. Finally, PAS-localized Hrr25 will further propagate the phosphorylating signal and activates other SARs (step 5).
Importance of phosphorylation
Atg19 and Atg30 are phosphorylated at residues upstream of the AIM, as well as in the A11BR59,67. Acidic residues proximal to the AIM increase their affinity for Atg8, and a phosphorylated residue mimics an acidic residue56. Functional studies of the phosphosites near the AIMs of Atg30, Atg32 and Atg36 confirmed the requirement of these phosphosites for these Atg proteins to interact with Atg8 (REF. 59). Similar to the effects of mutations in AIMs (see above), phosphosite mutations upstream of the AIM affect selective autophagy only weakly in vivo. By contrast, phosphorylation in the A11BR, which is conserved in most receptors and is essential for the interaction of SARs with Atg11 (REFS 26,33,61), is strongly required for the respective selective autophagy pathways. Notably, in the case of pexophagy, mutations in both of the phosphosites in the pexophagy receptors — thereby simultaneously affecting SAR-Atg8 and SAR-Atg11 interactions — are necessary to mimic the phenotypes of the deletion of pexophagy SARs (Atg30 or Atg36). These results indicate that interactions with Atg8 and Atg11 both have important and non-redundant roles during pexophagy59.
Signals inducing selective autophagy and SAR phosphorylation
Because organelles often cooperate in multiple metabolic pathways, proliferate under the same conditions and share the division machinery, it should not be surprising that common signals might induce their degradation. The signals and inducers triggering the phosphorylation of SARs are unknown; however, several conditions have been used to study induced selective autophagy pathways. The most common condition is the switching of glucose-rich and nitrogen-rich growth media to glucose without nitrogen. In addition, pexophagy is triggered by replacing the carbon source, such as from media that induces peroxisome proliferation (for example, methanol for P. pastoris) to glucose media without nitrogen. Such nutrient changes results in the degradation of peroxisomes in a manner that requires SAR phosphorylation26,68. Pexophagy and mitophagy can also be induced without changing the growth medium, either by continuous growth into stationary phase or by affecting organelle integrity27,30,43,69. In both cases, SARs, and probably their phosphorylation, are necessary. Mitophagy can also be induced by replacing the carbon source from a non-fermentable source, such as lactate or glycerol, to glucose medium but without nitrogen. Interestingly, mitophagy can be induced by shifting yeast cells from a glucose-rich medium to a glucose-minimal medium without nitrogen; this type of induced mitophagy also depends on SAR phosphorylation59,61,70. Finally, the SAR is not phosphorylated and mitophagy is not induced if only TORC1 is inactivated by rapamycin in glucose-rich media71.
Kinases involved in phosphoregulation
The kinases involved in the direct phosphorylation of SARs are known in S. cerevisiae (FIG. 4, TABLE 3). The mitophagy receptor Atg32 is phosphorylated by CK2 (REF. 72), a vital and highly conserved serine and/or threonine kinase that has a role in diverse cellular processes such as transcription, regulation and cell cycle regulation73. CK2-dependent phosphorylation of Atg32 stabilizes the Atg32–Atg11 interaction, which then leads to PAS assembly, subsequent autophagosome formation and ultimately mitophagy72. In vitro, CK2 phosphorylates two serine residues in the A11BR; one of these serine residues is essential for the in vivo Atg32–Atg11 interaction. CK2 is not important for non-selective autophagy, pexophagy or the Cvt pathway, suggesting that CK2 has a specific role in mitophagy.
Interestingly, the kinase responsible for phosphorylating the Cvt, Ams1 and pexophagy receptors is Hrr25 and not CK2, despite the similar A11BRs in these receptors67,74,75. Hrr25 is a homologue of CK1δ, which functions in ribosomal subunit biogenesis, chromosome segregation, DNA repair and, most importantly, in vesicular trafficking, where it contributes to the directional delivery of ER-derived vesicles to the Golgi76. In vitro experiments have indicated that Hrr25 phosphorylates a residue in the A11BR of Atg36 that is required for the Atg36–Atg11 interaction75 (Supplementary information S1 (figure)). In agreement with the in vitro results, knockdown of Hrr25 diminishes Atg36 phosphorylation and the Atg11–Atg36 interaction. Atg19 and Atg34 are both phosphorylated in their A11BRs by Hrr25 (REF. 67). Similar to the other SARs, these phosphosites are required for the proper interaction of the receptor and the scaffold protein Atg11. In conclusion, most, if not all, selective autophagy pathways are regulated by a uniform mechanism, which is the enhancement of the interaction (or interactions) of receptor–autophagic protein (or proteins) by receptor phosphorylation. It is also interesting that Hrr25 is involved in the regulation of three distinct selective autophagy pathways67,75,77. It is important to note that these different SARs can be phosphorylated by Hrr25 either under nutrient-rich or nitrogen-starvation conditions, indicating once again that the activation mechanism for SARs cannot rely exclusively on TORC1 signalling.
The evidence for the Hrr25-mediated phosphorylation of receptors during selective autophagy is clear67,75,77. However, the involvement of Hrr25 exclusively in selective autophagy has been investigated by only one study75 so far, and the conclusions from this study have been challenged by recent work indicating that Hrr25 is also involved in non-selective autophagy49. As mentioned earlier, the GTPase Ypt1 is involved in both non-selective and selective autophagy and is recruited by Atg17 and Atg11 in the respective pathways. During non-selective autophagy, Ypt1 activates and recruits Hrr25 to the PAS (FIG. 1). The possibility that Atg11–Ypt1 is also involved in Hrr25 activation and recruitment to the PAS during selective autophagy has not been determined, and we do not know whether this interaction is involved in the phosphorylation of SARs. Despite this uncertainty, the recruitment of Hrr25 to the PAS by scaffold proteins is an attractive mechanism to explain the phosphorylation of SARs in selective autophagy.
Interestingly, even though mitophagy and pexophagy receptors are phosphorylated by different casein kinases, these receptors in S. cerevisiae are interchangeable. Atg32 targeted to the peroxisomes facilitates pexophagy and Atg36 targeted to the mitochondria activates mitophagy27,32, suggesting that each SAR may also be phosphorylated by the other kinase. We speculate that SAR phosphorylation might occur at the PAS because Hrr25 localizes there (FIG. 4), and this localization might also explain why these receptors are interchangeable.
Two models could explain the phosphorylation of SARs (FIG. 4). One scenario is that upon induction of selective autophagy the casein kinases (Hrr25 and CK2) are first recruited to the cargo surface and the SARs by unknown factors or mechanisms. These kinases could be locally activated (potentially by kinases of the high osmolarity glycerol (HOG) pathway — Hog1 and Pbs2 (REFS 61,72)) — resulting in the phosphorylation and activation of the receptors. Once phosphorylated, SARs could then recruit Atg11 (FIG. 4a). Another plausible scenario, at least for the phosphorylation of SARs by Hrr25, is that upon induction of selective autophagy, Hrr25 is activated in the cytoplasm and recruited to the SAR in a manner that is dependent on the complex between Ypt1 and scaffold proteins, which would be recruited by some unknown factors or mechanism. Following this initial phosphorylation of the SARs, Hrr25 could continue to be recruited to the PAS via Atg11 and/or Atg17 scaffolds, and phosphorylate additional receptors neighbouring the already activated SAR, thereby propagating receptor activation (FIG. 4b). In Arabidopsis thaliana, the Atg11 homologue interacts with Atg8 (REF. 78), and Atg8 could be the unknown factor that links the SARs to the autophagy machinery. In addition, Atg37 and Pex3 (see below) are potential candidates for the unknown factors bridging the pexophagy SAR and Hrr25.
Further regulation of selectivity
As a consequence of the nature of cargoes, selective autophagy must be tightly regulated and needs to respond to multiple stimuli. It is no surprise therefore that the degree of selective cargo degradation is regulated on multiple levels and by several mechanisms. Signalling pathways, organelle fission as well as transcriptional regulation of SARs have been implicated in controlling selective autophagy. These processes and events that modulate selective degradation during autophagy are described below.
Signalling pathways
Mitophagy and pexophagy, but not non-selective autophagy, are regulated by the MAPK pathways79,80, which play a key part in responding appropriately to external stimuli or environmental conditions81. Mitophagy and pexophagy require the complete cell wall integrity (CWI) signal transduction pathway, which starts from the CWI sensors and ends with the MAPK Slt2. The CWI pathway is normally responsible for maintaining cell wall homeostasis and consequently is activated by cell wall stress; however, this pathway is also activated by nitrogen starvation or rapamycin treatment (a TORC1 signalling inhibition condition)82. The exact function of this pathway during selective autophagy is unclear, but Slt2 activity is needed for the formation of the specific PAS80. The phenotype of the slt2Δ mutant cells resembles that of the A11BR phosphomutant of Atg32 (REF. 61). Thus, it is reasonable to propose that Slt2 kinase is involved in the phosphorylation of SARs.
In addition, mitophagy requires another MAPK signalling pathway, namely the HOG pathway, which is essential for yeast survival in high osmolarity environments. Nitrogen starvation activates at least one component, the MAPK Hog1 (REF. 80). Two distinct roles have been proposed for Hog1. One study proposed that Hog1 and the MAPK kinase Pbs2 are required for mitophagy after PAS assembly, suggesting no direct role of MAPK in SAR phosphorylation. In a second study, an Atg32 phosphorylation defect was observed in the hog1Δ and pbs2Δ mutants of the HOG pathway. However, Hog1 was unable to phosphorylate Atg32 in vitro and it is not known whether the Atg32 phosphosites affected by Δhog1 and Δpbs2 are in the A11BR61,72. If the second finding is true, HOG kinases are most probably involved indirectly in SAR phosphorylation and could potentially activate CK2 (FIG. 4a).
Pexophagy induction in P. pastoris is also regulated by two Atg30-interacting proteins: the acyl-CoA binding protein Atg37 and the PMP Pex3 (REFS 28,29,83) (TABLE 3). Atg37 binds Atg30 and palmitoyl-CoA. Atg37 is required for proper Atg30 phosphorylation and is therefore needed for the Atg30–Atg11 interaction. Moreover, in vitro experiments have indicated that palmitoyl-CoA and Atg30 compete for the same binding region in Atg37. Pex3, as described earlier, recruits Atg30 to the peroxisomes and is also needed for Atg30 phosphorylation and its interaction with Atg11 (REF. 28). So, what is the role of palmitoyl-CoA in pexophagy? One possible role is that the presence of sufficient local concentrations of palmitoyl- CoA, generated locally by peroxisomal fatty-acid oxidation, might prevent the activation of pexophagy by preventing Atg37 from interacting with Atg30. Once peroxisomal β-oxidation declines, the palmitoyl- CoA concentration is reduced. In result, the inhibition of the Atg37-Atg30 interaction is alleviated, thereby allowing Atg30 phosphorylation and consequently pexophagy. This model, however, remains to be tested.
Organelle fission is required for selective autophagy
Organelles frequently divide (through fission), and most (except peroxisomes) also fuse together. Peroxisomes are subject to fission by Pex11 (which is a PMP) as well as two GTPases, dynamin 1 (Dnm1) and Vps1 (which are dynamin-like proteins)84–86, and this fission probably facilitates selective organelle degradation. The recruitment of Dnm1 to the peroxisomes requires both mitochondria fission 1 (Fis1) and mitochondrial division protein 1 (Mdv1), which together form a protein complex required for the recruitment of Dnm1. Remarkably, this molecular complex (Fis1–Mdv1–Dnm1) is also necessary for mitochondrial fission87 and, recently, this fission machinery was reported to be necessary for mitophagy and pexophagy88–90. During mitophagy, Dnm1, which fragments mitochondria, additionally relies on Atg11 for its recruitment to mitochondria91. Autophagosomes may have a size limit because overexpression of the Cvt pathway cargo prApe1 causes larger complexes to form that cannot be engulfed92, suggesting that fission may facilitate autophagy of large organelle cargoes by decreasing their size. Notably however, the fission machinery seems to regulate selective autophagy also independently of organelle size. As an example, peroxisomes are dispersed in the cytosol as individual compartments, and when induced by oleate treatment they have an average diameter of ~150 nm, which is much smaller than the largest autophagosome (~900 nm in diameter). Nevertheless, S. cerevisiae pexophagy also requires the fission machinery90. Similarly, the methylotrophic yeast Hansenula polymorpha uses the fission machinery indirectly, as explained below, to degrade peroxisomes and to remove large intra-peroxisomal protein aggregates by pexophagy88. The protein aggregate is first separated from the mother peroxisome by Dnm1- and Pex11-dependent asymmetric fission and degraded in an Atg1- and Atg11-dependent manner. During pexophagy, although both Dnm1 and Vps1 interact with Atg11 and the receptor Atg36 (REF. 90), the dynamin-like proteins also interact with Atg36 that has mutations in both its AIM and A11BR. This finding suggests that the interactions of dynamin-like proteins with Atg36 are direct but independent of pexophagy activation.
Transcriptional regulation of receptors
Atg19 is expressed in nutrient-rich media to mediate the biosynthetic Cvt pathway. Notably, nitrogen starvation substantially increases the amount of the Cvt pathway cargo prApe1, and this increase is associated with a parallel, several- fold increase in Atg19 levels23. This result indicates that the modulation of SAR expression is an important regulatory mechanism during selective autophagy23.
The pexophagy receptor Atg30 is associated with peroxisomes during their biogenesis, long before pexophagy induction26. Similarly, in S. cerevisiae, Atg32 and Atg36 localize to the mitochondria and peroxisomes, respectively, during organelle biogenesis. As discussed above, the mere presence of SARs on the organelle is insufficient to induce selective autophagy, and SARs must be activated. Nevertheless, also the SAR levels appear to be important for the fate of the organelle because their overexpression induces their respective selective-autophagy pathways26,27,30.
By contrast, some SARs, such as Atg32 in P. pastoris71 (which is responsible for mitochondrial degradation), as well as Atg39 and Atg40 in S. cerevisiae33 (which mediate ER-phagy), are not expressed in vegetative conditions. Their expression is only induced when cells encounter nitrogen starvation or are treated with rapamycin, which indicates a regulation of the expression of these receptors by TORC1 signalling93. In support of this finding, it has been revealed that Atg32 expression in P. pastoris and S. cerevisiae is inhibited by TORC1 and a histone deacetylase complex comprising Sin3 and Rpd3 (REF. 71). Atg32 levels increase dramatically when TORC1 is inhibited or when the Rpd3 or Sin3 proteins are absent, suggesting that these proteins suppress ATG32 gene transcription. Interestingly, Atg8 expression is also regulated by this histone deacetylase complex, and Atg8 levels determine the size, but not the number, of autophagosomes, thus influencing autophagic activity94. Controlling the levels of SARs and their interacting partner Atg8 by the same pathway may be the mechanism that maintains the correct ratio of SAR with respect to Atg8 during the sequestration of large and numerous cargoes.
Termination of selective autophagy
The termination of signalling for selective autophagy is poorly understood but is probably achieved at multiple levels, including destruction of the cargo, together with the SAR–Atg8 complex and/or attenuation of the signalling pathway that activates selective autophagy26,30,31. Attenuation of the signalling pathway that activates selective autophagy can be achieved through the transient inactivation of the signalling enzymes, such as kinases, or through the activation of enzymes, such as phosphatases, deubiquitylation enzymes or C-terminal ubiquitin hydrolases that reverse the chemical protein modifications involved in autophagy.
Functional conservation of SARs
Notably, approximately 90% of the core autophagy genes are conserved across eukaryotes95 (TABLE 1). By contrast, genes encoding SARs on average exhibit lower conservation in ancient taxa, with the majority having evolved in Eumetazoan evolution (estimated 650 million years ago). The exceptions are next to BRCA1 gene 1 (NBR1) and FUN14 domain-containing protein 1 (FUNDC1) (TABLE 2), which exist in metazoans and older taxa. However, despite the low primary sequence conservation of SARs, even between closely related yeasts, recent reports have indicated that proteins exhibiting functional equivalence to yeast SARs do exist in higher eukaryotes and that the principles of their activity during autophagy are conserved96,97. In this section, we provide an overview of these common principles governing selective cargo recognition.
Similar to yeast, higher eukaryotes have ubiquitin-dependent and ubiquitin-independent SARs22 (TABLE 2). Interestingly, in higher eukaryotes, common tags, such as ubiquitylation, are used much more prevalently and this prevalence of tagging may offer a simpler (more efficient) solution to marking the cargo for degradation because it allows the use of common adaptors for multiple cargoes. Thus, the selective autophagy pathways that require ubiquitin-dependent SARs can use the same receptors (which include p62 (also known as sequestosome 1), NBR1, NDP52 (also known as CALCOCO2), optineurin (OPTN), Tax1-binding protein 1 (TAX1BP1) and TOLLIP in mammals). In some cases, a single mammalian ubiquitin-dependent SAR recognizes the ubiquitylated cargo destined for degradation; frequently, more than one SAR participates in cargo recognition. For example, two ubiquitin-dependent SARs (NBR1 and p62) are involved in pexophagy, three in mitophagy (OPTN, NDP52, TAX1BP1) and four (p62, NDP52, OPTN, TAX1BP1) during elimination of bacteria and viruses (xenophagy). Additionally, some processes such as xenophagy and lysophagy may also use ubiquitin-independent receptors (such as galectin 8), and the same is true for mitophagy, which, as recently shown, uses the BCL-2-like protein 13 (BCL2L13) as a ubiquitin-independent receptor97. Understandably, despite common mechanisms of cargo tagging, different SARs are still required for efficient selective autophagy in mammals because the cargoes can differ and organelles are not fixed entities but dynamic structures interacting with each other and remodelling themselves in response to various stimuli.
The theme of phosphoregulation of receptors by kinases also extends to mammalian systems. During pexophagy, p62 recognizes peroxisomes through its interaction with monoubiquitylated PEX5 (REFS 17,98,99) but, as discussed above, p62 also recognizes other selective cargoes such as ubiquitylated aggregates, bacteria and zymogens, allowing a common tag on multiple cargoes to be recognized by the same receptor22,100. p62 is phosphorylated by several kinases, including Unc51-like kinase 1 (ULK1; the mammalian homologue of Atg1), CK2 and TANK-binding kinase 1 (TBK1), and, in each case, this activation allows p62 to bind to its relevant cargoes100. In yeast, the key regulatory step in selective autophagy appears not to be cargo binding perse, but rather SAR phosphorylation in response to appropriate stimuli to engage the autophagic machinery. By contrast, mammals exploit phosphorylation of the SAR for binding both to the cargo and the autophagy machinery, reflecting the greater complexity of phosphoregulation of selectivity101–104. Unfortunately not enough is known at this point about the role of phosphorylation as a regulatory step for the ubiquitin-dependent CUET pathway in yeast25 to make meaningful comparisons with the ubiquitin-dependent pathways in higher eukaryotes.
As is true for yeast, in mammals, most of the ubiquitin-independent SARs associated with organelles contain TMDs that allow them to associate with their cargoes using the intrinsic organelle import machinery. The closest mammalian examples that mimic a yeast ubiquitin-independent SAR is the mitophagy receptor BCL2L13 (REF. 97) and the ER-phagy receptor FAM134B33,105. BCL2L13, similar to yeast Atg32, localizes to the outer mitochondrial membrane, contains a TMD at its C-terminal region and its N terminus is exposed to the cytosol. BCL2L13 is imported into the outer mitochondrial membrane via its C-terminal tail anchor. Remarkably, despite the absence of sequence homology, BCL2L13 compensates for the function of Atg32 in yeast. FAM134B, similar to Atg40, localizes at the peripheral ER and contains two TMDs with characteristics of a reticulon-like domain.
The mode of recruitment of Atg8 and its mammalian homologues, microtubule-associated protein 1A/1B-light chain 3 (LC3; also known as MAP1LC3) or γ-aminobutyric acid receptor-associated protein (GABARAP), to the growing autophagosome is also conserved. As previously discussed, in yeast, the SARs have one or more AIMs. Similarly, the mammalian receptors have one or more LC3-interacting regions (LIRs). For example, the ubiquitin-dependent SARs (NBR1 and p62) and the ubiquitin- independent SARs (BCL2L13 and FAM134B) directly bind the Atg8-like protein (or proteins) LC3 and/or GABARAP97,105–107. NBR1 has two LIRs, LIR1 and LIR2, which have major and minor roles in the binding process, respectively. BCL2L13 and p62 have a single LIR55,107. Interestingly, and similar to organelle SARs and Atg8 interactions in yeast, NBR1-LC3 binding is only partially required for pexophagy108. Finally, in both yeast and mammals, the AIM and the LIR are activated via phosphorylation of serine or threonine residues within or adjacent to these domains59,101.
The interaction of the cargo–SAR complex with scaffold proteins is not well established in higher eukaryotes. There is some homology between the yeast autophagy scaffolds Atg11 and Atg17, and two metazoan protein families represented in humans by FIP200 and huntingtin109–112. Similar to yeast Atg11, which binds SARs and also activates the Atg1 kinase43, these scaffolds interact on the one hand with autophagy receptors and on the other hand with the metazoan counterpart of the Atg1 kinase ULK1 (REFS 109–111,113).
Conclusions and perspective
Many intracellular and extracellular components are selectively degraded by autophagy. Selective autophagy relies on selectivity factors and the core autophagy machinery to degrade its cargoes. The main selectivity factors are the SARs, which exist as two types in mammals and yeast: the ubiquitin-independent and ubiquitin-dependent SARs. The principal regulatory mechanism activating selective autophagy pathways is the phosphorylation of the SARs, leading to the engagement of the core autophagy machinery and/or recognition of the ubiquitylated cargo.
Despite the rapid and impressive progress in unravelling selective autophagy mechanisms, many details of this process are unknown. To start, most, if not all, yeast SARs are phosphoproteins and most are phosphorylated at the A11BR by casein kinases (CK2 for Atg32 and Hrr25 for Atg19, Atg34 and Atg36), but the kinase (or kinases) for some SARs, such as Atg30, as well as the kinase (or kinases) for AIM phosphorylation remain unknown. In addition, the phosphorylation status of Atg39 and Atg40 has not been determined. The A11BR of different receptors, such as Atg32 and Atg36, are relatively well conserved, as is the signal (nitrogen starvation condition) that triggers mitophagy and pexophagy, but, surprisingly, these SARs are phosphorylated by different casein kinases. The mechanisms responsible for this selective regulation, as well as the signalling cascades that activate the casein kinases, are currently elusive. Furthermore, the involvement of MAPK pathways in SAR activation, although inferred, remains a mystery. Although SAR phosphorylation is the primary activation mechanism for selective autophagy, it also needs to be considered that it might not be the only one. For example, despite some controversy in this area, the mitophagy receptor Atg32 seems to be activated by an additional mechanism involving the proteolytic maturation of its C-terminal region by the protease Yme1 (REF. 114). Finally, it needs to be determined whether yeast, as in mammals, have more than one SAR for each organelle, with each responding perhaps to a different stimulus. Together, resolving these questions will shed important new light onto how selectivity and precision during autophagy can be achieved.
Supplementary Material
Acknowledgments
This work was supported by a US National Institutes of Health grant (DK41737) and by the Chancellor’s Associates Endowed Chair of the University of California San Diego, USA, to S.S. The authors regret not being able to cite all the primary literature relating to this topic owing to limitations on the number of citations permitted.
Glossary
- Ubiquitin-like (Ubl) conjugation systems
Proteins in these systems behave like ubiquitin and are conjugated to other proteins (or lipids) using E1, E2 and E3 enzymes, similar to ubiquitin.
- PolyQ proteins
Proteins with a long stretch of glutamine (Q) residues that form aggregates mimicking the mutant huntingtin protein of Huntington disease.
- GTPase
A GTP hydrolysing enzyme that coverts GTP to GDP.
- Coat protein complex II
(COPII). A type of vesicle coat protein present on vesicles that transport cargoes from the rough endoplasmic reticulum to the Golgi apparatus.
- Tail-anchored (TA) mechanism
A mechanism of insertion of proteins into organelle membranes that operates posttranslationally and occurs through a carboxy-terminal transmembrane domain of the protein; the exact mechanism of insertion (apart from endoplasmic reticulum membranes) remains elusive.
- Sec61 translocon
An evolutionarily conserved protein complex that forms a channel in the membrane of the endoplasmic reticulum and mediates protein translocation across the membrane as well as membrane insertion of proteins.
- Fission
Membrane fission is the process by which a continuous cellular membrane divides into two distinct membranes.
- Dynamin
A type of GTPase involved in membrane fission events.
- Oleate
A monounsaturated omega-9 fatty acid used as a carbon source to induce peroxisomes biogenesis in yeast because its oxidation requires peroxisomal enzymes.
- Histone deacetylase
An enzyme reversing acetylation of lysine residues of histones, thereby playing a crucial role in chromatin remodelling and in the regulation of gene transcription.
- Zymogens
Inactive enzymes that are activated by proteolytic processing.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Ryter SW, Cloonan SM, Choi AM. Autophagy: a critical regulator of cellular metabolism and homeostasis. Mol. Cells. 2013;36:7–16. doi: 10.1007/s10059-013-0140-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N. Engl. J. Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 3.Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–2906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- 4.Frake RA, Ricketts T, Menzies FM, Rubinsztein DC. Autophagy and neurodegeneration. J. Clin. Invest. 2015;125:65–74. doi: 10.1172/JCI73944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 6.Netea-Maier RT, Plantinga TS, Van De Veerdonk FL, Smit JW, Netea MG. Modulation of inflammation by autophagy: consequences for human disease. Autophagy. 2015;12:1–16. doi: 10.1080/15548627.2015.1071759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomes LC, Dikic I. Autophagy in antimicrobial immunity. Mol. Cell. 2014;54:224–233. doi: 10.1016/j.molcel.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 8.Nowikovsky K, Devenish RJ, Froschauer E, Schweyen RJ. Determination of yeast mitochondrial KHE activity, osmotic swelling and mitophagy. Methods Enzymol. 2009;457:305–317. doi: 10.1016/S0076-6879(09)05017-4. [DOI] [PubMed] [Google Scholar]
- 9.Walter KM, et al. Hif-2α promotes degradation of mammalian peroxisomes by selective autophagy. Cell. Metab. 2014;20:882–897. doi: 10.1016/j.cmet.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Li S, Liu Y, Ma C. Redox regulated peroxisome homeostasis. Redox Biol. 2015;4:104–108. doi: 10.1016/j.redox.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohsumi Y. Historical landmarks of autophagy research. Cell Res. 2014;24:9–23. doi: 10.1038/cr.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki SW, et al. Atg13 HORMA domain recruits Atg9 vesicles during autophagosome formation. Proc. Natl Acad. Sci. USA. 2015;112:3350–3355. doi: 10.1073/pnas.1421092112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shemi A, Ben-Dor S, Vardi A. Elucidating the composition and conservation of the autophagy pathway in photosynthetic eukaryotes. Autophagy. 2015;11:701–715. doi: 10.1080/15548627.2015.1034407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kubo Y. Function of peroxisomes in plant–pathogen interactions. Subcell. Biochem. 2013;69:329–345. doi: 10.1007/978-94-007-6889-5_18. [DOI] [PubMed] [Google Scholar]
- 15.Mizumura K, Choi AM, Ryter SW. Emerging role of selective autophagy in human diseases. Front. Pharmacol. 2014;5:244. doi: 10.3389/fphar.2014.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Vaart A, Mari M, Reggiori F. A picky eater: exploring the mechanisms of selective autophagy in human pathologies. Traffic. 2008;9:281–289. doi: 10.1111/j.1600-0854.2007.00674.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, et al. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat. Cell Biol. 2015;17:1259–1269. doi: 10.1038/ncb3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010;584:1359–1366. doi: 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuttle DL, Lewin AS, Dunn WA., Jr Selective autophagy of peroxisomes in methylotrophic yeasts. Eur. J. Cell Biol. 1993;60:283–290. [PubMed] [Google Scholar]
- 20.Lu K, Psakhye I, Jentsch S. A new class of ubiquitin-Atg8 receptors involved in selective autophagy and polyQ protein clearance. Autophagy. 2014;10:2381–2382. doi: 10.4161/15548627.2014.981919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steele S, Brunton J, Kawula T. The role of autophagy in intracellular pathogen nutrient acquisition. Front. Cell. Infect. Microbiol. 2015;5:51. doi: 10.3389/fcimb.2015.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khaminets A, Behl C, Dikic I. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 2016;26:6–16. doi: 10.1016/j.tcb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 23.Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol. Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki K, Kondo C, Morimoto M, Ohsumi Y. Selective transport of alpha-mannosidase by autophagic pathways: identification of a novel receptor, Atg34p. J. Biol. Chem. 2010;285:30019–30025. doi: 10.1074/jbc.M110.143511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu K, Psakhye I, Jentsch S. Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell. 2014;158:549–563. doi: 10.1016/j.cell.2014.05.048. Extends the concept derived from mammalian models of ubiquitin as a tag for the selective autophagy of protein aggregates to a new and conserved class of CUET (CUE-domain targeting proteins, Cue5 in yeast and TOLLIP in mammals) proteins. Cue5 recognizes ubiquitylated proteins and collaborates with the ubiquitin ligase Rsp5 in this new pathway. [DOI] [PubMed] [Google Scholar]
- 26.Farre JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. Cell. 2008;14:365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Motley AM, Nuttall JM, Hettema EH. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 2012;31:2852–2868. doi: 10.1038/emboj.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burnett SF, Farre JC, Nazarko TY, Subramani S. Peroxisomal Pex3 activates selective autophagy of peroxisomes via interaction with the pexophagy receptor Atg30. J. Biol. Chem. 2015;290:8623–8631. doi: 10.1074/jbc.M114.619338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nazarko TY, et al. Peroxisomal Atg37 binds Atg30 or palmitoyl-CoA to regulate phagophore formation during pexophagy. J. Cell Biol. 2014;204:541–557. doi: 10.1083/jcb.201307050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 32.Kondo-Okamoto N, et al. Autophagy-related protein 32 acts as autophagic degron and directly initiates mitophagy. J. Biol. Chem. 2012;287:10631–10638. doi: 10.1074/jbc.M111.299917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mochida K, et al. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015;522:359–362. doi: 10.1038/nature14506. [DOI] [PubMed] [Google Scholar]
- 34.Jin M, Klionsky DJ. In: Autophagy and Cancer. Wang H-G, editor. Springer New York; 2013. pp. 25–45. [Google Scholar]
- 35.Cheong H, et al. Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nazarko TY, Farre JC, Subramani S. Peroxisome size provides insights into the function of autophagy-related proteins. Mol. Biol. Cell. 2009;20:3828–3839. doi: 10.1091/mbc.E09-03-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanki T, et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol. Biol. Cell. 2009;20:4730–4738. doi: 10.1091/mbc.E09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kabeya Y, et al. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol. Biol. Cell. 2005;16:2544–2553. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamada Y, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabeya Y, et al. Characterization of the Atg17- Atg29-Atg31 complex specifically required for starvation-induced autophagy in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2009;389:612–615. doi: 10.1016/j.bbrc.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 41.Fujioka Y, et al. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat. Struct. Mol. Biol. 2014;21:513–521. doi: 10.1038/nsmb.2822. [DOI] [PubMed] [Google Scholar]
- 42.Mao K, et al. Atg29 phosphorylation regulates coordination of the Atg17-Atg31-Atg29 complex with the Atg11 scaffold during autophagy initiation. Proc. Natl Acad. Sci. USA. 2013;110:E2875–E2884. doi: 10.1073/pnas.1300064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamber RA, Shoemaker CJ, Denic V. Receptor-bound targets of selective autophagy use a scaffold protein to activate the Atg1 kinase. Mol. Cell. 2015;59:372–381. doi: 10.1016/j.molcel.2015.06.009. Reports the dynamic role that the Atg11 scaffold has in the Cvt and pexophagy pathways. Atg11 activates the Atg1 kinase when it interacts with the SAR-cargo complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheong H, Nair U, Geng J, Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 2008;19:668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papinski D, et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol. Cell. 2014;53:471–483. doi: 10.1016/j.molcel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lipatova Z, et al. Regulation of selective autophagy onset by a Ypt/Rab GTPase module. Proc. Natl Acad. Sci. USA. 2012;109:6981–6986. doi: 10.1073/pnas.1121299109. Shows that the Ypt1/Rab GTPase also regulates autophagy by interacting with the scaffold protein Atg11 to regulate PAS assembly and subsequent autophagosome formation during selective autophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kakuta S, et al. Atg9 vesicles recruit vesicle-tethering proteins Trs85 and Ypt1 to the autophagosome formation site. J. Biol. Chem. 2012;287:44261–44269. doi: 10.1074/jbc.M112.411454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J, et al. Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proc. Natl Acad. Sci. USA. 2013;110:9800–9805. doi: 10.1073/pnas.1302337110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J, et al. Ypt1/Rab1 regulates Hrr25/CK1delta kinase activity in ER-Golgi traffic and macroautophagy. J. Cell Biol. 2015;210:273–285. doi: 10.1083/jcb.201408075. Shows that the Rab GTPase Ypt1/Rab1 recruits Hrr25/CK1δ to COPII vesicles and to the PAS during non-selective autophagy. Hrr25 is activated by Ypt1 and is required for non-selective autophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan D, et al. The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc. Natl Acad. Sci. USA. 2013;110:19432–19437. doi: 10.1073/pnas.1316356110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishihara N, et al. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Backues SK, et al. Atg23 and Atg27 act at the early stages of Atg9 trafficking in S. cerevisiae. Traffic. 2015;16:172–190. doi: 10.1111/tra.12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 54.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Noda NN, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 56.Ho KH, Chang HE, Huang WP. Mutation at the cargo-receptor binding site of Atg8 also affects its general autophagy regulation function. Autophagy. 2009;5:461–471. doi: 10.4161/auto.5.4.7696. [DOI] [PubMed] [Google Scholar]
- 57.Kaufmann A, Beier V, Franquelim HG, Wollert T. Molecular mechanism of autophagic membrane-scaffold assembly and disassembly. Cell. 2014;156:469–481. doi: 10.1016/j.cell.2013.12.022. Provides a detailed account of how autophagy activation and termination occur. [DOI] [PubMed] [Google Scholar]
- 58.Shintani T, Huang WP, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Farre JC, Burkenroad A, Burnett SF, Subramani S. Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Rep. 2013;14:441–449. doi: 10.1038/embor.2013.40. Lays the foundation for the mechanism by which selective autophagy receptors act by bridging selective cargo and the core autophagy machinery in a phosphoregulated fashion in yeast and mammals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol. Biol. Cell. 2005;16:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aoki Y, et al. Phosphorylation of Serine 114 on Atg32 mediates mitophagy. Mol. Biol. Cell. 2011;22:3206–3217. doi: 10.1091/mbc.E11-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. The To r and PKA signaling pathways independently target the Atg1/ Atg13 protein kinase complex to control autophagy. Proc. Natl Acad. Sci. USA. 2009;106:17049–17054. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010;584:1379–1385. doi: 10.1016/j.febslet.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 64.Sawa-Makarska J, et al. Cargo binding to Atg19 unmasks additional Atg8 binding sites to mediate membrane-cargo apposition during selective autophagy. Nat. Cell Biol. 2014;16:425–433. doi: 10.1038/ncb2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sawa-Makarska J, Martens S. Excluding the unwanted during autophagy. Cell Cycle. 2014;13:2313–2314. doi: 10.4161/cc.29826. [DOI] [PubMed] [Google Scholar]
- 66.Kim J, et al. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pfaffenwimmer T, et al. Hrr25 kinase promotes selective autophagy by phosphorylating the cargo receptor Atg19. EMBO Rep. 2014;15:862–870. doi: 10.15252/embr.201438932. Shows that a casein kinase, Hrr25, is required for Atg19 phosphorylation in the Cvt pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tuttle DL, Dunn WA., Jr Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J. Cell Sci. 1995;108:25–35. doi: 10.1242/jcs.108.1.25. [DOI] [PubMed] [Google Scholar]
- 69.Eiyama A, Kondo-Okamoto N, Okamoto K. Mitochondrial degradation during starvation is selective and temporally distinct from bulk autophagy in yeast. FEBS Lett. 2013;587:1787–1792. doi: 10.1016/j.febslet.2013.04.030. [DOI] [PubMed] [Google Scholar]
- 70.Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J. Biol. Chem. 2008;283:32386–32393. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aihara M, et al. To r and the Sin3-Rpd3 complex regulate expression of the mitophagy receptor protein Atg32 in yeast. J. Cell Sci. 2014;127:3184–3196. doi: 10.1242/jcs.153254. [DOI] [PubMed] [Google Scholar]
- 72.Kanki T, et al. Casein kinase 2 is essential for mitophagy. EMBO Rep. 2013;14:788–794. doi: 10.1038/embor.2013.114. Provides evidence that CK2 phosphorylates the mitophagy receptor Atg32 and regulates its interaction with Atg11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Canton DA, Litchfield DW. The shape of things to come: an emerging role for protein kinase CK2 in the regulation of cell morphology and the cytoskeleton. Cell Signal. 2006;18:267–275. doi: 10.1016/j.cellsig.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 74.Mochida K, Ohsumi Y, Nakatogawa H. Hrr25 phosphorylates the autophagic receptor Atg34 to promote vacuolar transport of alpha-mannosidase under nitrogen starvation conditions. FEBS Lett. 2014;588:3862–3869. doi: 10.1016/j.febslet.2014.09.032. [DOI] [PubMed] [Google Scholar]
- 75.Tanaka C, et al. Hrr25 triggers selective autophagy-related pathways by phosphorylating receptor proteins. J. Cell Biol. 2014;207:91–105. doi: 10.1083/jcb.201402128. Demonstrates that two SARs, Atg19 and Atg36, are phosphorylated by Hrr25. Hrr25-mediated phosphorylation enhances the interactions of these receptors with the scaffold protein Atg11, which is required for most selective autophagy pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lord C, et al. Sequential interactions with Sec23 control the direction of vesicle traffic. Nature. 2011;473:181–186. doi: 10.1038/nature09969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nakatogawa H. Hrr25: an emerging major player in selective autophagy regulation in Saccharomyces cerevisiae. Autophagy. 2015;11:432–433. doi: 10.1080/15548627.2015.1017195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li F, Vierstra RD. Arabidopsis ATG11, a scaffold that links the ATG1-ATG13 kinase complex to general autophagy and selective mitophagy. Autophagy. 2014;10:1466–1467. doi: 10.4161/auto.29320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Manjithaya R, Jain S, Farre JC, Subramani S. A yeast MAPK cascade regulates pexophagy but not other autophagy pathways. J. Cell Biol. 2010;189:303–310. doi: 10.1083/jcb.200909154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mao K, Klionsky DJ. MAPKs regulate mitophagy in Saccharomyces cerevisiae. Autophagy. 2011;7:1564–1565. doi: 10.4161/auto.7.12.17971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen RE, Thorner J. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2007;1773:1311–1340. doi: 10.1016/j.bbamcr.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Torres J, Di Como CJ, Herrero E, De La Torre-Ruiz MA. Regulation of the cell integrity pathway by rapamycin-sensitive TOR function in budding yeast. J. Biol. Chem. 2002;277:43495–43504. doi: 10.1074/jbc.M205408200. [DOI] [PubMed] [Google Scholar]
- 83.Nazarko TY. Atg37 regulates the assembly of the pexophagic receptor protein complex. Autophagy. 2014;10:1348–1349. doi: 10.4161/auto.29073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li X, Gould SJ. The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J. Biol. Chem. 2003;278:17012–17020. doi: 10.1074/jbc.M212031200. [DOI] [PubMed] [Google Scholar]
- 85.Kuravi K, et al. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J. Cell Sci. 2006;119:3994–4001. doi: 10.1242/jcs.03166. [DOI] [PubMed] [Google Scholar]
- 86.Motley AM, Ward GP, Hettema EH. Dnm1p–dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J. Cell Sci. 2008;121:1633–1640. doi: 10.1242/jcs.026344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schrader M. Shared components of mitochondrial and peroxisomal division. Biochim. Biophys. Acta. 2006;1763:531–541. doi: 10.1016/j.bbamcr.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 88.Manivannan S, de Boer R, Veenhuis M, van der Klei IJ. Lumenal peroxisomal protein aggregates are removed by concerted fission and autophagy events. Autophagy. 2013;9:1044–1056. doi: 10.4161/auto.24543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mao K, Klionsky DJ. Participation of mitochondrial fission during mitophagy. Cell Cycle. 2013;12:3131–3132. doi: 10.4161/cc.26352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mao K, Liu X, Feng Y, Klionsky DJ. The progression of peroxisomal degradation through autophagy requires peroxisomal division. Autophagy. 2014;10:652–661. doi: 10.4161/auto.27852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mao K, Wang K, Liu X, Klionsky DJ. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev. Cell. 2013;26:9–18. doi: 10.1016/j.devcel.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J. Cell Sci. 2013;126:2534–2544. doi: 10.1242/jcs.122960. [DOI] [PubMed] [Google Scholar]
- 93.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 94.Backues SK, Lynch-Day MA, Klionsky DJ. The Ume6-Sin3-Rpd3 complex regulates ATG8 transcription to control autophagosome size. Autophagy. 2012;8:1835–1836. doi: 10.4161/auto.21845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Till A, et al. Evolutionary trends and functional anatomy of the human expanded autophagy network. Autophagy. 2015;11:1652–1667. doi: 10.1080/15548627.2015.1059558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat. Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 97.Murakawa T, et al. Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat. Commun. 2015;6:7527. doi: 10.1038/ncomms8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nordgren M, Wang B, Apanasets O, Fransen M. Peroxisome degradation in mammals: mechanisms of action, recent advances, and perspectives. Front. Physiol. 2013;4:145. doi: 10.3389/fphys.2013.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Subramani S. A mammalian pexophagy target. Nat. Cell Biol. 2015;17:1371–1373. doi: 10.1038/ncb3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Katsuragi Y, Ichimura Y, Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015;282:4672–4678. doi: 10.1111/febs.13540. [DOI] [PubMed] [Google Scholar]
- 101.Wild P, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. Describes the role of TBK1-mediated phosphorylation of OPTN. This modification enhances the affinity of OPTN for LC3, showing the phosphoregulation of the LIR in OPTN. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Richter B, et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc. Natl Acad. Sci. USA. 2016;113:4039–4044. doi: 10.1073/pnas.1523926113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/ NDP52 recruitment and TBK1 activation to promote mitophagy. Mol. Cell. 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lazarou M, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Khaminets A, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522:354–358. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- 106.Waters S, Marchbank K, Solomon E, Whitehouse C, Gautel M. Interactions with LC3 and polyubiquitin chains link NBR1 to autophagic protein turnover. FEBS Lett. 2009;583:1846–1852. doi: 10.1016/j.febslet.2009.04.049. [DOI] [PubMed] [Google Scholar]
- 107.Kirkin V, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 108.Deosaran E, et al. NBR1 acts as an autophagy receptor for peroxisomes. J. Cell Sci. 2013;126:939–952. doi: 10.1242/jcs.114819. [DOI] [PubMed] [Google Scholar]
- 109.Lin L, et al. The scaffold protein EPG-7 links cargo- receptor complexes with the autophagic assembly machinery. J. Cell Biol. 2013;201:113–129. doi: 10.1083/jcb.201209098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nagy P, et al. Atg17/FIP200 localizes to perilysosomal Ref(2)P aggregates and promotes autophagy by activation of Atg1 in Drosophila. Autophagy. 2014;10:453–467. doi: 10.4161/auto.27442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ochaba J, et al. Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proc. Natl Acad. Sci. USA. 2014;111:16889–16894. doi: 10.1073/pnas.1420103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hara T, Mizushima N. Role of ULK-FIP200 complex in mammalian autophagy: FIP200, a counterpart of yeast Atg17? Autophagy. 2009;5:85–87. doi: 10.4161/auto.5.1.7180. [DOI] [PubMed] [Google Scholar]
- 113.Rui YN, et al. Huntingtin functions as a scaffold for selective macroautophagy. Nat. Cell Biol. 2015;17:262–275. doi: 10.1038/ncb3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang K, Jin M, Liu X, Klionsky DJ. Proteolytic processing of Atg32 by the mitochondrial i-AAA protease Yme1 regulates mitophagy. Autophagy. 2013;9:1828–1836. doi: 10.4161/auto.26281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Axe EL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Watanabe Y, et al. Selective transport of α-mannosidase by autophagic pathways: structural basis for cargo recognition by Atg19 and Atg34. J. Biol. Chem. 2010;285:30026–30033. doi: 10.1074/jbc.M110.143545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yuga M, Gomi K, Klionsky DJ, Shintani T. Aspartyl aminopeptidase is imported from the cytoplasm to the vacuole by selective autophagy in Saccharomyces cerevisiae. J. Biol. Chem. 2011;286:13704–13713. doi: 10.1074/jbc.M110.173906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hyttinen JM, et al. Clearance of misfolded and aggregated proteins by aggrephagy and implications for aggregation diseases. Ageing Res. Rev. 2014;18:16–28. doi: 10.1016/j.arr.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 119.Svenning S, Johansen T. Selective autophagy. Essays Biochem. 2013;55:79–92. doi: 10.1042/bse0550079. [DOI] [PubMed] [Google Scholar]
- 120.Orenstein SJ, Cuervo AM. Chaperone-mediated autophagy: molecular mechanisms and physiological relevance. Semin. Cell Dev. Biol. 2010;21:719–726. doi: 10.1016/j.semcdb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Seay M, Hayward AP, Tsao J, Dinesh-Kumar SP. Something old, something new: plant innate immunity and autophagy. Curr. Top. Microbiol. Immunol. 2009;335:287–306. doi: 10.1007/978-3-642-00302-8_14. [DOI] [PubMed] [Google Scholar]
- 122.Changou CA, et al. Arginine starvation-associated atypical cellular death involves mitochondrial dysfunction, nuclear DNA leakage, and chromatin autophagy. Proc. Natl Acad. Sci. USA. 2014;111:14147–14152. doi: 10.1073/pnas.1404171111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cloonan SM, Lam HC, Ryter SW, Choi AM. “Ciliophagy”: the consumption of cilia components by autophagy. Autophagy. 2014;10:532–534. doi: 10.4161/auto.27641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lipatova Z, Segev N. A role for macro-ER-phagy in ER quality control. PLoS Genet. 2015;11:e1005390. doi: 10.1371/journal.pgen.1005390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mancias JD, et al. Ferritinophagy via NCOA4 is required for erythropoiesis and is regulated by iron dependent HERC2-mediated proteolysis. eLife. 2015;4:e10308. doi: 10.7554/eLife.10308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105–109. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jiang S, Wells CD, Roach PJ. Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem. Biophys. Res. Commun. 2011;413:420–425. doi: 10.1016/j.bbrc.2011.08.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy. Microsc. Res. Tech. 2004;64:10–20. doi: 10.1002/jemt.20046. [DOI] [PubMed] [Google Scholar]
- 130.Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy in glucose homeostasis. Pathol. Res. Pract. 2006;202:631–638. doi: 10.1016/j.prp.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 131.Buchan JR, Kolaitis RM, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu K, Czaja MJ. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 2013;20:3–11. doi: 10.1038/cdd.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.van Zutphen T, et al. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2014;25:290–301. doi: 10.1091/mbc.E13-08-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Maejima I, et al. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 2013;32:2336–2347. doi: 10.1038/emboj.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hung YH, Chen LM, Yang JY, Yang WY. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat. Commun. 2013;4:2111. doi: 10.1038/ncomms3111. [DOI] [PubMed] [Google Scholar]
- 136.Kuo TC, et al. Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nat. Cell Biol. 2011;13:1214–1223. doi: 10.1038/ncb2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pohl C, Jentsch S. Midbody ring disposal by autophagy is a post-abscission event of cytokinesis. Nat. Cell Biol. 2009;11:65–70. doi: 10.1038/ncb1813. [DOI] [PubMed] [Google Scholar]
- 138.Kanki T, Furukawa K, Yamashita S. Mitophagy in yeast: molecular mechanisms and physiological role. Biochim. Biophys. Acta. 2015;1853:2756–2765. doi: 10.1016/j.bbamcr.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 139.Liu L, Sakakibara K, Chen Q, Okamoto K. Receptor-mediated mitophagy in yeast and mammalian systems. Cell Res. 2014;24:787–795. doi: 10.1038/cr.2014.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Muller M, Lu K, Reichert AS. Mitophagy and mitochondrial dynamics in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2015;1853:2766–2774. doi: 10.1016/j.bbamcr.2015.02.024. [DOI] [PubMed] [Google Scholar]
- 141.Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc. Natl Acad. Sci. USA. 2014;111:E4439–E4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gomez-Sanchez JA, et al. Schwann cell autophagy, myelinophagy, initiates myelin clearance from injured nerves. J. Cell Biol. 2015;210:153–168. doi: 10.1083/jcb.201503019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Krick R, et al. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol. Biol. Cell. 2008;19:4492–4505. doi: 10.1091/mbc.E08-04-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mijaljica D, Devenish RJ. Nucleophagy at a glance. J. Cell Sci. 2013;126:4325–4330. doi: 10.1242/jcs.133090. [DOI] [PubMed] [Google Scholar]
- 146.Zientara-Rytter K, et al. Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy. 2011;7:1145–1158. doi: 10.4161/auto.7.10.16617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Vanhee C, Zapotoczny G, Masquelier D, Ghislain M, Batoko H. The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell. 2011;23:785–805. doi: 10.1105/tpc.110.081570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Michaeli S, Honig A, Levanony H, Peled-Zehavi H, Peled-Zehavi Galili G. Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell. 2014;26:4084–4101. doi: 10.1105/tpc.114.129999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Marshall RS, Li F, Gemperline DC, Book AJ, Vierstra RD. Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/Ubiquitin receptor RPN10 in Arabidopsis. Mol. Cell. 2015;58:1053–1066. doi: 10.1016/j.molcel.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 151.Ossareh-Nazari B, et al. Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Rep. 2010;11:548–554. doi: 10.1038/embor.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mandell MA, et al. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev. Cell. 2014;30:394–409. doi: 10.1016/j.devcel.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Orvedahl A, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Castrejon-Jimenez NS, Leyva-Paredes K, Hernandez-Gonzalez JC, Luna-Herrera J, Garcia-Perez BE. The role of autophagy in bacterial infections. Biosci. Trends. 2015;9:149–159. doi: 10.5582/bst.2015.01035. [DOI] [PubMed] [Google Scholar]
- 155.Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Vaccaro MI. Zymophagy: selective autophagy of secretory granules. Int. J. Cell Biol. 2012;2012:396705. doi: 10.1155/2012/396705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nice DC, Sato TK, Stromhaug PE, Emr SD, Klionsky DJ. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J. Biol. Chem. 2002;277:30198–30207. doi: 10.1074/jbc.M204736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Juris L, et al. PI3P binding by Atg21 organises Atg8 lipidation. EMBO J. 2015;34:955–973. doi: 10.15252/embj.201488957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Stromhaug PE, Reggiori F, Guan J, Wang CW, Klionsky DJ. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol. Biol. Cell. 2004;15:3553–3566. doi: 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Scott SV, et al. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J. Biol. Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 161.Reggiori F, Wang CW, Stromhaug PE, Shintani T, Klionsky DJ. Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion. J. Biol. Chem. 2003;278:5009–5020. doi: 10.1074/jbc.M210436200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Reggiori F, Monastyrska I, Shintani T, Klionsky DJ. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16:5843–5856. doi: 10.1091/mbc.E05-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.He C, et al. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J. Cell Biol. 2006;175:925–935. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Monastyrska I, et al. Arp2 links autophagic machinery with the actin cytoskeleton. Mol. Biol. Cell. 2008;19:1962–1975. doi: 10.1091/mbc.E07-09-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tamura N, Oku M, Sakai Y. Atg21 regulates pexophagy via its PI(3)P-binding activity in Pichia pastoris. FEMS Yeast Res. 2014;14:435–444. doi: 10.1111/1567-1364.12132. [DOI] [PubMed] [Google Scholar]
- 166.Ano Y, et al. A sorting nexin PpAtg24 regulates vacuolar membrane dynamics during pexophagy via binding to phosphatidylinositol-3-phosphate. Mol. Biol. Cell. 2005;16:446–457. doi: 10.1091/mbc.E04-09-0842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Oku M, et al. Peroxisome degradation requires catalytically active sterol glucosyltransferase with a GRAM domain. EMBO J. 2003;22:3231–3241. doi: 10.1093/emboj/cdg331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Nazarko VY, et al. Atg35, a micropexophagy-specific protein that regulates micropexophagic apparatus formation in Pichia pastoris. Autophagy. 2011;7:375–385. doi: 10.4161/auto.7.4.14369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Fry MR, Thomson JM, Tomasini AJ, Dunn WA., Jr Early and late molecular events of glucose-induced pexophagy in Pichia pastoris require Vac8. Autophagy. 2006;2:280–288. doi: 10.4161/auto.3164. [DOI] [PubMed] [Google Scholar]
- 170.Hamasaki M, Noda T, Baba M, Ohsumi Y. Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic. 2005;6:56–65. doi: 10.1111/j.1600-0854.2004.00245.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.