Abstract
The analysis of intact proteins (top-down strategy) by mass spectrometry has great potential to elucidate proteoform variation, including patterns of post-translational modifications (PTMs), which may not be discernable by analysis of peptides alone (bottom-up approach). To maximize sequence coverage and localization of PTMs, various fragmentation modes have been developed to produce fragment ions from deep within intact proteins. Ultraviolet photodissociation (UVPD) has recently been shown to produce high sequence coverage and PTM retention on a variety of proteins, with increasing evidence of efficacy on a chromatographic time scale. However, utilization of UVPD for high-throughput top-down analysis to date has been limited by bioinformatics. Here, we detected 153 proteins and 489 proteoforms using UVPD and 271 proteins and 982 proteoforms using higher-energy collisional dissociation (HCD) in a comparative analysis of HeLa whole-cell lysate by qualitative top-down proteomics. Of the total detected proteoforms, 286 overlapped between the UVPD and HCD datasets, with 68% of proteoforms having C-scores greater than 40 for UVPD and 63% for HCD. The average sequence coverage (28% ± 20% for UVPD versus 17% ± 8% for HCD, p < 0.0001) found to be higher for UVPD than HCD, and with a trend toward improvement in q-value for the UVPD dataset. This study demonstrates the complementarity of UVPD and HCD for more extensive protein profiling and proteoform characterization.
Keywords: proteomics, protein, top-down, ultraviolet photodissociation, higher-energy collisional dissociation, Orbitrap mass spectrometer, proteoform, HeLa
Introduction
Despite the enormous popularity and tremendous success of bottom-up mass spectrometry strategies,1, 2 top-down approaches aimed at the analysis of intact proteins rather than mixtures of proteolytic peptides are a compelling alternative for large-scale proteomics studies.3, 4 Top-down methods offer the potential to pinpoint all modifications and mutations of a protein without loss of key features which might be overlooked owing to peptides that are not sampled in bottom-up shotgun workflows.5 Performance gains and technological advances in mass spectrometers, separation methods, and bioinformatics have accelerated the arena of top- down mass spectrometry in recent years. Both the number of proteins and proteoforms identified by high-throughput top-down mass spectrometry has increased dramatically6–13 because of the use of proteome fractionation via molecular weight separation7 or through orthogonal improvements in HPLC methods.11, 12 The use of these separations has extended the number of intact proteins and proteoforms detected from the tens or hundreds14–18 to the thousands,6–9, 19 while at the same time increasing both the depth and confidence of assignments of post-translational modifications (PTMs) for a diverse range of proteins. Concurrently with the development of pre-mass spectrometry techniques, the development of new scoring metrics for top-down data (e.g., C-score20) has facilitated a more complete understanding of both the confidence of protein identification and how well PTMs are localized or constrained by fragmentation data. Many high-throughput top-down analyses use higher-energy collisional dissociation (HCD) for fragmentation6, 9, 13, 21–24 because it is a well characterized method that provides the best compromise between speed and the number of identified proteins/proteoforms.21 Because HCD is limited in the depth of sequence coverage that can be obtained and may cause the loss of labile PTMs,21, 25 alternative fragmentation methods have been developed to both increase sequence coverage and minimize PTM loss, including ultraviolet photodissociation [UVPD]26; electron transfer dissociation [ETD]27; and hybrid electron transfer dissociation/higher-energy collisional dissociation [EThcD]28). Each of these alternative methods has shown increasing promise for top-down analysis of proteins.9, 10, 26, 29, 30
We have focused our efforts on the development and application of UVPD, especially using 193 nm photons, because in many cases UVPD provides unprecedented sequence coverage for peptides and proteins26, 31 while also allowing retention of post-translational modifications.32, 33 UVPD exhibits impressive performance for both denatured proteins and native-like proteins, the latter typically generated in low charge states which have proven more challenging to characterize by collision- and electron-based methods.32–36 To date, UVPD has only had limited usage for high-throughput top-down analysis of complex protein mixtures, such as cell lysates.10 Previously, Cannon et al.10 reported the detection of the majority of Escherichia coli ribosomal proteins, in addition to 215 proteins and 292 proteoforms from a Saccharomyces cerevisiae lysate after online UPLC separation of the proteins on a C4 column and fragmentation by UVPD. This study comprised the first attempt at high-throughput, top-down proteomics using UVPD, but was limited by the bioinformatic platforms available at the time (i.e., prior to optimization of high-throughput database searching for UVPD).
Our present study combined recent advances in proteome fractionation, developments in top-down informatics, and the latest in high performance instrumentation to provide the most extensive identification and characterization to date of human proteins and proteoforms using 193 nm UVPD-MS. We also compared the performance and attributes of 193 nm UVPD to HCD, demonstrating the complementarity of these methods and the ability of UVPD to increase confidence in characterization compared to HCD for overlapping proteoforms.
Materials and Methods
Samples and Cell Lysis
Washed, pelleted, and frozen HeLa cells (1 × 109 cells/pellet) were purchased from Biovest International/National Cell Culture. Cells were lysed on ice for 20 minutes in 20 mM Tris, pH 7.5, containing 100 mM NaCl, 1% (w/v) N-lauroylsarcosine, and 1X final concentration of HALT EDTA-free protease inhibitor cocktail (ThermoFisher Scientific), using a modified version of the method in DeHart et al.37 After lysis, 1 M MgCl2 was added to a final concentration of 1 mM, followed by digestion of all DNA and RNA with 750 U of benzonase nuclease at 37°C for 20 min. After pelleting debris at 13,200 rpm for 15 min at 4°C, lysate protein concentration was measured using the Pierce BCA Protein Assay Kit (ThermoFisher Scientific). Following the BCA assay, a 400 μg quantity was precipitated overnight in 6 volumes of acetone at −80°C. Precipitated lysate proteins were pelleted by centrifugation at 13,200 rpm for 10 minutes at 4°C, followed by decanting of the supernatant and washing of the pellet in an additional 6 volumes of acetone, with repeated centrifugation and decanting.
GELFrEE Protein fractionation
The acetone precipitated pellets were subsequently resuspended in 100 μL of 1% SDS by vigorous pipetting. After adding 8 μL of 1 M dithiothreitol, 12 μL of Optima-grade water (ThermoFisher Scientific), and 30 μL of 5X Tris-Acetate sample buffer (Expedeon Inc.) for a final volume of 150 μL, the proteins were denatured for 10 min at 95°C. Following denaturation, all samples were pelleted at 13,200 rpm for 10 min at 4°C. Each sample was separated into 12 MW-based fractions on a 10%T gel-eluted liquid-fraction entrapment electrophoresis (GELFrEE) cartridge, following manufacturer’s instructions (GELFrEE 8100 Fractionation System, Expedeon Inc.). For each fraction, a 10 μL sample was taken to visualize protein content and resolution by SDS-PAGE and subsequent silver nitrate stain.38 Immediately prior to LC-MS analysis, fractions were precipitated using the chloroform/methanol/water method.39 Precipitated pellets were washed with an additional 4 volumes of methanol to maximize SDS removal. After a short drying period (1–5 min), all pellets were resuspended in 5% ACN/0.1% formic acid using repeated pipetting aspirations. Resuspended pellets were further diluted 1:3–1:8 times depending on initial protein amount (as determined from Figure S1).
LC-MS
Fractions 1–7 were separated on in-house packed PLRP-S (5 μm particle size, 1000 Å pore size; Agilent Technologies) columns (trap column: 3 cm × 100 μm i.d.; analytical column: 40 cm × 75 μm i.d.) using a Dionex Ultimate 3000 UHPLC system (ThermoFisher Scientific). Mobile phases consisted of A: 0.1% formic acid and B: 99% acetonitrile, 0.1% formic acid. An initial gradient was applied as follows at a flow rate of 300 nl/min: 0–8 min at 2% B; 8–10 min increasing from 2–15% B; 10–37 min increasing from 15–55% B; 37–40 min rapidly increasing from 55–95% B; 40–43 min at 95% B; 43–45 min decreasing from 95–2% B; 45–60 min at 2%B). 100–500 ng of protein was injected on-column from each fraction. After this initial injection, the gradient was optimized using the gradient optimization and analysis tool (GOAT; https://proteomics.swmed.edu/goat/).40 All GOAT gradients are provided in Table S1. The nanoLC system was coupled to a Fusion Lumos Orbitrap mass spectrometer (Thermo Scientific Instruments) modified for 193 nm UVPD as described earlier for a Fusion Orbitrap mass spectrometer.41 UVPD was performed in the high-pressure trap using a single pulse (1.4 mJ, 5 ns) from a 193 nm excimer laser (Coherent Existar XS). HCD was performed in the ion routing multipole at 15% NCE during a 0.1 ms period. The Orbitrap mass spectrometer was run using the following parameters regardless of fragmentation type: MS1 at 120,000 FTRP, 4 μscans averaged per spectrum, 1 × 105 AGC target, 15 V source fragmentation, intact protein monoisotopic precursor selection; and MS2 at 120,000 FTRP with top speed mode enabled for 7 s, 6 μscans averaged per spectrum, and 1 × 105 AGC target. (“Top speed mode” affords a variable number of MS/MS spectra per scan cycle, typically 3–4 MS2 spectra in the present study). For fractions 1–4, charge state targets were set to 10–25+, and for fractions 5–7, charge state targets were set to 10–25+ and undetermined charge states.
Data analysis
Raw data were uploaded to the National Resource for Translational and Developmental Proteomics (NRTDP, Northwestern University, Evanston, IL) TDPortal13 high-performance computing environment for analysis of high-throughput top-down proteomics data (available for academic collaborators at: http://nrtdp.northwestern.edu/tdportal-request/). Intact protein MS1 spectra were first averaged using the cRAWler algorithm (developed in-house), followed by deconvolution to monoisotopic masses by means of the Xtract algorithm (Thermo Fisher Scientific). Processed data were then searched against a database generated from the SwissProt 2016_04 release of the human proteome comprising 1 × 107 total candidate proteoforms. All searches entailed a three-pronged strategy, each mode of which was first defined for ProSight PTM 2.0.42 The first stage entailed a narrow absolute mass search (with a 2.2 Da tolerance for MS1 and a 10 ppm tolerance for MS2) to confidently identify well-matching previously detected proteoforms. An MS1 tolerance of 2.2 Da is used to be tolerant of isotoping errors in the Xtract deconvolution algorithm. The second stage involved a biomarker search (equivalent to a no-enzyme search in peptide analysis, with a 10 ppm tolerance for both MS1 and MS2) which enabled detection of previously unknown truncations. The strategy concluded with a wide absolute mass search using a 200 Da tolerance for MS1, a 10 ppm tolerance for MS2, and Δm mode enabled to accommodate unexpected post-translational modifications. The 200 Da search tolerance allows inclusion of unexpected modifications which requires a large precursor tolerance. The choice of 200 Da is meant to encompass the possible incorporation of up to two phosphorylation modifications. Data derived from each fragmentation type (HCD or UVPD) were analyzed separately.
False discovery rate (FDR) and instantaneous q-value estimation at the protein and proteoform level were accomplished by means of a recently-developed in-house target-decoy method. First, local FDR calculations were performed by searching against a scrambled database created from both MS1 and MS2 data with all resulting hits assumed to be incorrect, using a method first described in a 2011 top-down proteomics study.7 The distribution of these initial incorrect hits was then used to calculate the null distribution for the given scoring metrics, which was in turn used to determine the probability of a forward hit being assigned the observed score (or better) due to chance alone. Local FDRs were then accurately determined with minimal outside influence from unknown dependencies by applying conservative corrections from multiple tests to each of these probabilities.43 Global FDR calculations were performed at the protein and proteoform level by pooling the local FDR results from multiple searches and applying the “pick best” approach.44 Only those proteoform entries mapping to a protein entry at 1% FDR or better were reported, to increase overall accuracy of the results.
Post-search results were visualized by TDViewer version 0.9.1.1 (available for download at: (http://topdownviewer.northwestern.edu). Protein and proteoform identifications at 1% FDR or better were exported in list form into Microsoft Excel 2013, for direct comparison by histogram or collation into Venn diagrams with Venny 2.1 (http://bioinfogp.cnb.csic.es/tools/venny/). The .raw files analyzed in the searches described above, the .txt file used for search database creation, and all resulting .tdReport files, which show all identified protein entries and proteoforms, as well as other statistics such as observed sequence coverage or calculated q-values and C-scores, are available for download here: ftp://massive.ucsd.edu/MSV000080432 (access: ftp://MSV000080432@massive.ucsd.edu/). The evaluation of HCD and UVPD C-score statistical differences were determined using analysis of variance in SAS 9.4 (SAS Institute).45 Other statistical measures were performed in Microsoft Excel 2013.
Results and Discussion
For a systematic comparison of UVPD and HCD for large-scale high-throughput top-down proteomics, HeLa whole-cell lysate proteins were fractionated by molecular weight via gel-eluted liquid-fraction entrapment electrophoresis (GELFrEE)46 prior to LCMS/MS analysis on a high-performance Orbitrap mass spectrometer. Each of seven GELFrEE fractions, ranging in MW from ~5–45 kDa (Fig. S1), was analyzed in triplicate using UVPD or HCD in back-to-back series to allow direct comparison of MS/MS modes, for a total of 42 LC-MS runs. Raw data files were analyzed via the TDPortal high-performance computing environment at Northwestern University, with an average processing time of 16 wall-clock hours, and specific metrics were used to evaluate the performance of HCD and UVPD as summarized in the following sections. Specifically, key metrics included the number of proteins and proteoforms identified by each method, as well as the sequence coverages, q-values, and C-scores within each set of identified proteoforms. Sequence coverage is a standard parameter used to evaluate the percentage of backbone sites cleaved in a protein. The q-value47 is the instantaneous false discovery rate associated with the proteoform identification, and represents the false discovery rate the study would have if the proteoform in question was taken as the worst result to be considered an identification (e.g. a q value of 0.01 means that the overall FDR of the study would be 1% if the proteoform in question where accepted as correctly identified). The q-values are derived from the distribution of Poisson scores48 for the forward hits compared with a distribution of scrambled results. In the present study, the q-values range from 9.8×10−3 to 4.6×10−150, with lower scores being more favorable. The characterization score (C-score) is a more recently introduced concept20 used to provide a metric for estimating the ability to confidently differentiate and assign proteoforms. C-scores range from zero to greater than 500. Proteoforms assigned C-scores below 3 have been neither confidently identified nor characterized, while proteoforms assigned C-scores between 3 and 40 have been confidently identified but not fully characterized, and those proteoforms assigned C-scores above 40 have been confidently identified and extensively characterized.
UVPD vs HCD metrics
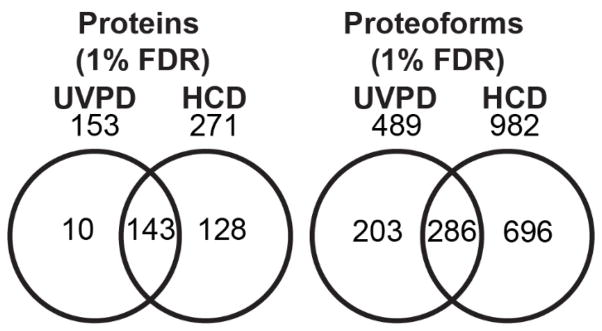
While high-throughput UVPD-MS has primarily been limited by bioinformatic capabilities, the recently developed TDPortal platform has allowed the first direct comparison of high-throughput UVPD and HCD top-down proteomics data acquired on an Orbitrap Lumos mass spectrometer. Pooling results from seven HeLa fractions analyzed in triplicate resulted in the identification of 153 proteins (Table S2; defined as UniProt accession numbers) and 489 proteoforms (Table S3; defined as a Proteoform Record, PFR; Consortium for Top-Down Proteomics Proteoform Repository http://repository.topdownproteomics.org/) at 1% FDR using UVPD and 271 proteins (Table S4) and 982 proteoforms (Table S5) at 1% FDR using HCD. The overlapping and unique proteins and proteoforms identified by UVPD and HCD are summarized in Venn diagram format in Figure 1. While HCD resulted in the identification of a greater number of proteins and proteoforms overall, the UVPD and HCD datasets still had 143 proteins (~51%) and 286 proteoforms (~24%) in common (Figure 1), thus giving ample overlap to facilitate a more detailed comparison of the MS/MS performance metrics for specific proteoforms, as described below. Based solely on the number of identifications, HCD outperforms UVPD; however, the confidence in the identifications vary considerably for UVPD compared to HCD, as shown in Figures 2 to 4. Interestingly, 203 proteoforms (17.1%) were uniquely identified by UVPD, suggesting a high level of complementarity of UVPD and HCD.
Figure 1.
Proteins (left) and proteoforms (right) detected by high-throughput top-down proteomic analysis of seven HeLa GELFrEE fractions using HCD or UVPD fragmentation.
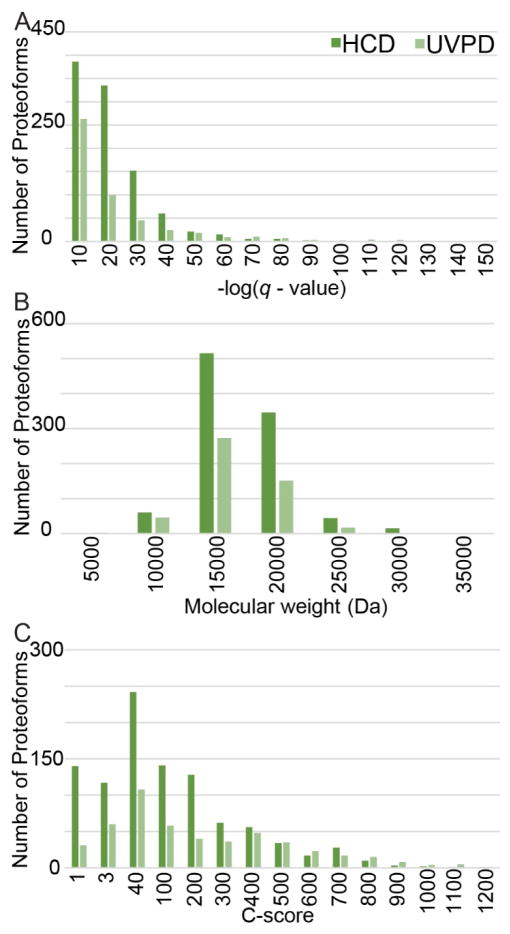
Figure 2.
(A) -log(q-value), (B) molecular weight, and (C) C-score distributions for all detected proteoforms within the HCD (dark green) and UVPD (light green) datasets.
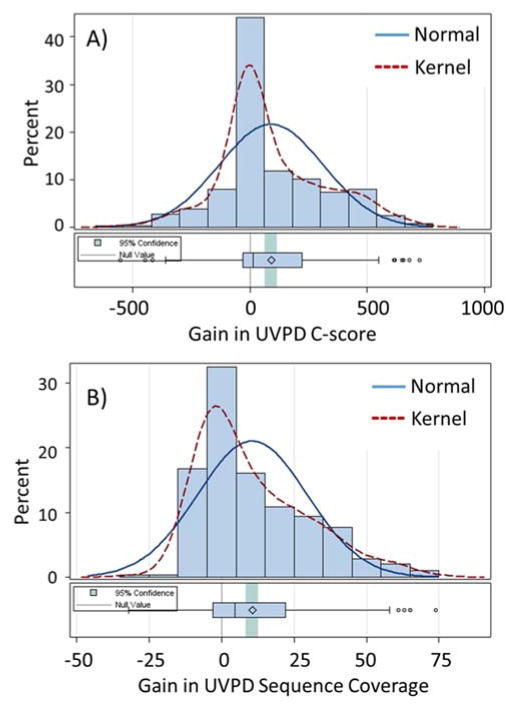
Figure 4.
Distribution of gains in UVPD compared to HCD for (A) C-scores and (B) sequence coverage for characterization of proteoforms. Overlays of normal distributions and kernel distributions are given for each metric.
Figure 2 displays the number of proteoforms identified for HCD and UVPD in histogram format according to q-value, molecular weight, and C-scores in order to show the range of values and clustering of proteoform parameters. The total distribution of q-values (Figure 2A), molecular weights (Figure 2B), and C-scores (Figure 2C) were similar for the UVPD and HCD datasets. The average molecular weight of proteins identified by HCD was slightly higher (UVPD: 13.4 ± 3.0 kDa; HCD: 14.4 ± 3.4 kDa; p<0.001) than that identified by UVPD for all detected proteoforms (Figure 2B). UVPD afforded a significantly higher average C-score (UVPD: 215 ± 260; HCD: 126 ± 187; p<0.001) and trend toward higher average -log(q-value) (UVPD: 18 ± 22; HCD: 16 ± 13; p=0.07) than HCD. This outcome is rationalized by an increase in the average degree of sequence coverage (UVPD: 25 ± 18%; HCD: 13 ± 7%; p < 0.001) as well as the greater number of diagnostic fragment types associated with UVPD (Figure 2A and 2C). Furthermore, UVPD resulted in 59% of identified proteoforms being assigned a C-score of 40 or higher, whereas 49% of the proteoforms identified by HCD fell into the same highest C-score category (Figure 2C). The results from Figure 2A (q-values) and Figure 2C (C-scores) recapitulate that the richer MS/MS spectra afforded by UVPD provides a boost in scoring metrics for proteoform identification compared to HCD.
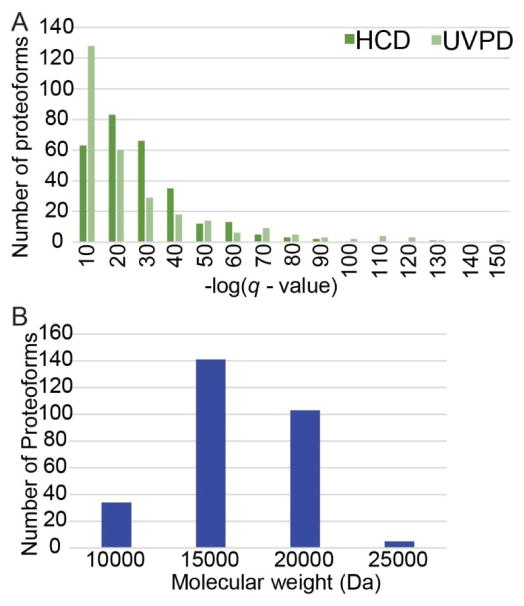
The 286 proteoforms that overlapped for UVPD and HCD were re-grouped in histogram format according to q-value and molecular weight in Figure 3. UVPD afforded a significantly higher average C-score (mean UVPD gain: 89.8, 95% CI 64.0 to 115.6, t(285 d.f.)=6.86; p<0.001) and higher average percent of sequence coverage (mean UVPD increase in fragment coverage: 10.6%, 95% CI 8.38% to 12.8%, t(285 d.f.)=9.45; p < 0.001). This increase in C-score is gained with no significant change in q-value (t(285 d.f.)=−0.7, p=0.4847; Figure 3A). The average molecular weight for these 286 proteoforms (13.4 kDa; Figure 3B) was representative of the molecular weight range of all of the UVPD proteoforms (p=0.98); however, this average molecular weight was smaller than that of all proteoforms identified by HCD (p<0.001). This discrepancy in molecular size arises from the greater number of proteoforms falling in the 25–30 kDa range identified by HCD; however, the 286 overlapping proteoforms provide a direct means of evaluating differences in C-score (Fig. 4A) and sequence coverage (Fig. 4B) distributions between the UVPD and HCD datasets. The comparisons of C-scores and sequence coverages obtained by UVPD and HCD of individual proteoforms are illustrated as difference plots in Figure S2, in which the 286 proteoforms are displayed across the x-axis, and the y-axis is used to convey the increase or decrease in the C-score or sequence coverage for each proteoform (UVPD relative to HCD). The benchmark HCD values are tracked on the green line, and the bars that extend above the green line signify those proteoforms for which UVPD outperformed HCD.
Figure 3.
(A) -log(q-value) and (B) molecular weight distributions of the 286 overlapping proteoforms between the HCD (dark green) and UVPD (light green) datasets.
As illustrated in Figure 4A, C-scores generated from the UVPD mass spectra were approximately 9% higher overall than those C-scores generated from HCD mass spectra (UVPD: 68% greater than C-score 40; HCD: 63% greater than C-score 40) for the 286 overlapping proteoforms, with 62% of overlapping proteoforms having a higher UVPD C-score than HCD C-score. UVPD also resulted in a significant increase in average sequence coverage (28% ± 20%, p<0.0001) compared to HCD (17% ± 8%) with 59% of the overlapping proteoforms having greater UVPD sequence coverage (Figure 4B). On average, UVPD increased sequence coverage by 11% compared to HCD; however, five of the proteoforms displayed a sequence coverage that increased by over 60% compared to HCD. For example, there was a 74% increase in sequence coverage for small EDRK-rich factor 2 (P84101-1; PFR21392) for UVPD compared to HCD (Figure 5A). These combined results suggest that UVPD provided increased confidence in PTM localization and proteoform characterization compared to HCD, owing to the larger number of fragment ions produced, and consistent with a previous report10 of the promising merits of UVPD for top-down proteomics.
Figure 5.
Fragmentation maps of A) EDRK-rich factor 2 (PFR21392) for HCD (12+) and UVPD (12+), and B) high mobility group protein HMG-I with three post-translational modifications (PFR17157) for HCD (14+) and UVPD (17+). Specific residues or sites are shaded as follows: blue box = phosphorylation, red box = acetylation, green box = methylation. Backbone fragmentation markers are shown along the sequence as colored flags: a,x: green; b,y: blue; c,z: red.
Localization of Post-translational Modifications
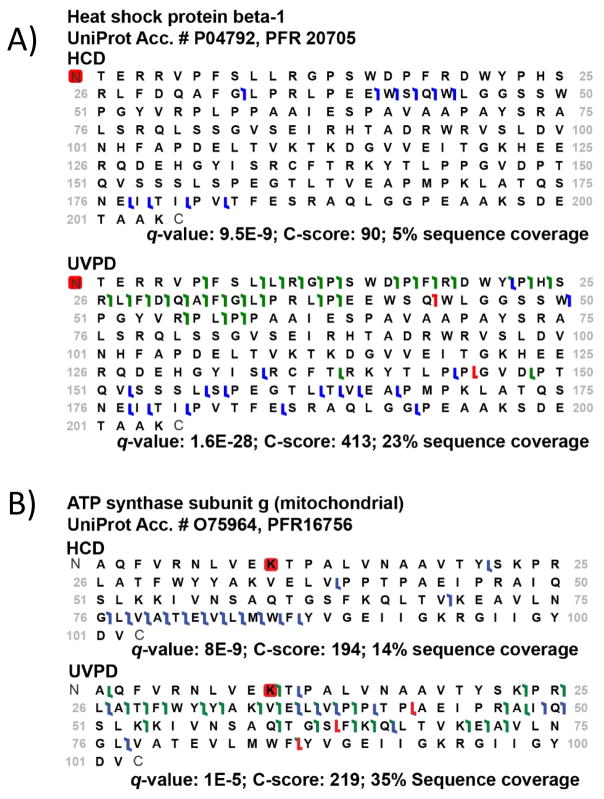
Based on the increase in sequence coverage and C-score observed for UVPD, the MS/MS results for six of the 286 overlapping proteoforms were examined in greater detail to evaluate differences in PTM localization reflected by the changes in C-scores (Fig. S2A, blue circles) and sequence coverage variation (Fig. S2B, blue circles). Among the group of 286 proteoforms, up to five PTMs, including N-terminal acetylation, lysine acetylation, and arginine methylation, were localized on a single protein. For the high mobility group protein HMG-I (P17096-1; 106 residues; Figure 5B and Figure S3), at least two overlapping proteoforms possessing three (PFR17157) or four (PFR13815) PTMs were characterized. For PFR17157 (Figure 5B), UVPD localized acetylated Lys6 and yielded limited coverage of phosphorylation of Ser101 and Ser102 (C-score: 368). A similar level of localization was observed for the two phosphorylations by HCD; however, acetylation of the N-terminus or Lys6 could not be confirmed, reducing confidence in characterization (C-score: 256) (Fig. 5B). For PFR 13815, the methylation of Arg25 was localized by UVPD (Figure S3), and there was limited localization of the acetylation of Lys14. The phosphorylation of Ser101 and Ser102 were pinpointed by appropriate fragment ions to these two positions by UVPD (Figure S3). For HCD of this same proteoform, the phosphorylation sites were not localized, and only limited localization of the acetylation at Lys14 and methylation at Arg25 was obtained, causing the C-score to plummet to 0.1 for HCD (from 149 for UVPD). For singly modified proteoforms, a large number of N-terminal acetylations (e.g., Small EDRK-rich factor 2 (Fig. 5A); Heat shock protein beta-1 [Fig. 6A]) were mapped in addition to lysine acetylations that UVPD successfully differentiated from the alternative N-terminal forms, as exemplified by ATP synthase subunit g (PFR16756) in Figure 6B.
Figure 6.
Fragmentation maps of A) heat shock protein beta-1 (PFR20705) for HCD (29+) and UVPD (26+), and B) ATP synthase subunit g (mitochondrial) (PFR16756) for HCD (12+) and UVPD (12+). UVPD allows localization of the acetylation (red box) of lysine-10, but HCD does not. Specific residues or sites are shaded as follows: blue box = phosphorylation, red box = acetylation, green box = methylation. Backbone fragmentation markers are shown along the sequence as colored flags: a,x: green; b,y: blue; c,z: red.Specific residues or sites are shaded as follows: blue box = phosphorylation, red box = acetylation, green box = methylation. Backbone fragmentation markers are shown along the sequence as colored flags: a,x: green; b,y: blue; c,z: red.
In some cases, HCD outperformed UVPD, as illustrated for ATP synthase delta (P30049; 146 residues; PFR1028), a proteoform which contained no PTMs. HCD returned greater sequence coverage and C-score than UVPD (Figure S4). This outcome reinforces the complementarity of HCD and UVPD, as not all proteoforms will fragment as extensively with UVPD. Some proteins are clearly better suited for HCD fragmentation than UVPD fragmentation, possibly owing to the favorable distribution of mobile protons or low frequency of amino acids susceptible to directing preferential cleavages; two factors that might otherwise suppress appropriate fragmentation upon collisional activation.
Conclusions
The use of UVPD fragmentation for the analysis of human proteins by high-throughput top-down proteomics resulted in both increased overall confidence in proteoform identification and improved degree of PTM localization in comparison to the more typical HCD fragmentation, as evaluated by scoring metrics provided by a new and more advanced bioinformatics platform. From these comparative analyses of HeLa GELFrEE fractions, UVPD was observed to increase sequence coverage up to 74% and the level of proteoform characterization by up to 9% compared to HCD, reflected in a particular increase in confident PTM localization. The comprehensive results obtained by the present study recapitulate the complementary nature of HCD and UVPD fragmentation for more extensive protein profiling and proteoform characterization in high-throughput top-down proteomics studies on an Orbitrap Fusion Lumos platform.
Supplementary Material
Acknowledgments
Funding from the NSF (CHE-1402753), the Welch Foundation (F-1155) and NIH 1K12GM102745 (fellowship to TPC) is acknowledged. Funding from the UT System for support of the UT System Proteomics Core Facility Network is gratefully acknowledged. This work was supported in part by the National Institute of General Medical Sciences P41GM108569 for the National Resource for Translational and Developmental Proteomics (NRTDP) based at Northwestern University (to NLK). Further support was provided by the computational resources and staff for the Quest high performance computing facility at Northwestern University, which is jointly supported by the Office of the Provost, the Office for Research, and Northwestern University Information Technology.
References
- 1.Zhang Y, Fonslow BR, Shan B, Baek MC, Yates JR. Protein Analysis by Shotgun/Bottom-up Proteomics. Chemical Reviews. 2013;113(4):2343–2394. doi: 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gillet LC, Leitner A, Aebersold R. Mass Spectrometry Applied to Bottom-Up Proteomics: Entering the High-Throughput Era for Hypothesis Testing. Annual Review of Analytical Chemistry. 2016;9(1):449–472. doi: 10.1146/annurev-anchem-071015-041535. [DOI] [PubMed] [Google Scholar]
- 3.Toby TK, Fornelli L, Kelleher NL. Progress in Top-Down Proteomics and the Analysis of Proteoforms. Annual Review of Analytical Chemistry. 2016;9:499–519. doi: 10.1146/annurev-anchem-071015-041550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gregorich ZR, Ge Y. Top-down proteomics in health and disease: Challenges and opportunities. PROTEOMICS. 2014;14(10):1195–1210. doi: 10.1002/pmic.201300432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith LM, Kelleher NL, Proteomics CTD. Proteoform: a single term describing protein complexity. Nature Methods. 2013;10(3):186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ntai I, Kim K, Fellers RT, Skinner OS, Smith AD, Early BP, Savaryn JP, LeDuc RD, Thomas PM, Kelleher NL. Applying Label-Free Quantitation to Top Down Proteomics. Analytical Chemistry. 2014;86(10):4961–4968. doi: 10.1021/ac500395k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tran JC, Zamdborg L, Ahlf DR, Lee JE, Catherman AD, Durbin KR, Tipton JD, Vellaichamy A, Kellie JF, Li M, Wu C, Sweet SMM, Early BP, Siuti N, LeDuc RD, Compton PD, Thomas PM, Kelleher NL. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature. 2011;480:254–258. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durbin KR, Fornelli L, Fellers RT, Doubleday PF, Narita M, Kelleher NL. Quantitation and Identification of Thousands of Human Proteoforms below 30 kDa. J Proteome Res. 2016;15(3):976–82. doi: 10.1021/acs.jproteome.5b00997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catherman AD, Durbin KR, Ahlf DR, Early BP, Fellers RT, Tran JC, Thomas PM, Kelleher NL. Large-scale top-down proteomics of the human proteome: membrane proteins, mitochondria, and senescence. Mol Cell Proteomics. 2013;12(12):3465–73. doi: 10.1074/mcp.M113.030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannon JR, Cammarata MB, Robotham SA, Cotham VC, Shaw JB, Fellers RT, Early BP, Thomas PM, Kelleher NL, Brodbelt JS. Ultraviolet Photodissociation for Characterization of Whole Proteins on a Chromatographic Time Scale. Analytical Chemistry. 2014;86(4):2185–2192. doi: 10.1021/ac403859a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valeja SG, Xiu L, Gregorich ZR, Guner H, Jin S, Ge Y. Three Dimensional Liquid Chromatography Coupling Ion Exchange Chromatography/Hydrophobic Interaction Chromatography/Reverse Phase Chromatography for Effective Protein Separation in Top-Down Proteomics. Analytical Chemistry. 2015;87(10):5363–5371. doi: 10.1021/acs.analchem.5b00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y, Sun L, Zhu G, Dovichi NJ. Coupling Capillary Zone Electrophoresis to a Q Exactive HF Mass Spectrometer for Top-down Proteomics: 580 Proteoform Identifications from Yeast. Journal of Proteome Research. 2016;15(10):3679–3685. doi: 10.1021/acs.jproteome.6b00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fornelli L, Durbin KR, Fellers RT, Early BP, Greer JB, LeDuc RD, Compton PD, Kelleher NL. Advancing top-down analysis of the human proteome using a benchtop quadrupole-Orbitrap mass spectrometer. Journal of Proteome Research. 2017;16(2):609–618. doi: 10.1021/acs.jproteome.6b00698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bunger MK, Cargile BJ, Ngunjiri A, Bundy JL, Stephenson JL., Jr Automated proteomics of E. coli via top-down electron-transfer dissociation mass spectrometry. Anal Chem. 2008;80(5):1459–67. doi: 10.1021/ac7018409. [DOI] [PubMed] [Google Scholar]
- 15.Roth MJ, Parks BA, Ferguson JT, Boyne MT, Kelleher NL. “Proteotyping”: Population proteomics of human leukocytes using top down mass spectrometry. Analytical Chemistry. 2008;80(8):2857–2866. doi: 10.1021/ac800141g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JE, Kellie JF, Tran JC, Tipton JD, Catherman AD, Thomas HM, Ahlf DR, Durbin KR, Vellaichamy A, Ntai I, Marshall AG, Kelleher NL. A Robust Two-Dimensional Separation for Top-Down Tandem Mass Spectrometry of the Low-Mass Proteome. Journal of the American Society for Mass Spectrometry. 2009;20(12):2183–2191. doi: 10.1016/j.jasms.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parks BA, Jiang L, Thomas PM, Wenger CD, Roth MJ, Boyne MT, Burke PV, Kwast KE, Kelleher NL. Top-down proteomics on a chromatographic time scale using linear ion trap Fourier transform hybrid mass spectrometers. Analytical Chemistry. 2007;79(21):7984–7991. doi: 10.1021/ac070553t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patrie SM, Ferguson JT, Robinson DE, Whipple D, Rother M, Metcalf WW, Kelleher NL. Top down mass spectrometry of < 60-kDa proteins from Methanosarcina acetivorans using quadrupole FRMS with automated octopole collisionally activated dissociation. Mol Cell Proteomics. 2006;5(1):14–25. doi: 10.1074/mcp.M500219-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Kellie JF, Catherman AD, Durbin KR, Tran JC, Tipton JD, Norris JL, Witkowski CE, Thomas PM, Kelleher NL. Robust Analysis of the Yeast Proteome under 50 kDa by Molecular-Mass-Based Fractionation and Top-Down Mass Spectrometry. Analytical Chemistry. 2012;84(1):209–215. doi: 10.1021/ac202384v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LeDuc RD, Fellers RT, Early BP, Greer JB, Thomas PM, Kelleher NL. The C-score: a Bayesian framework to sharply improve proteoform scoring in high-throughput top down proteomics. J Proteome Res. 2014;13(7):3231–40. doi: 10.1021/pr401277r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahlf DR, Compton PD, Tran JC, Early BP, Thomas PM, Kelleher NL. Evaluation of the Compact High-Field Orbitrap for Top-Down Proteomics of Human Cells. Journal of Proteome Research. 2012;11(8):4308–4314. doi: 10.1021/pr3004216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Catherman AD, Li M, Tran JC, Durbin KR, Compton PD, Early BP, Thomas PM, Kelleher NL. Top Down Proteomics of Human Membrane Proteins from Enriched Mitochondrial Fractions. Analytical Chemistry. 2013;85(3):1880–1888. doi: 10.1021/ac3031527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Compton PD, Tran JC, Ntai I, Kelleher NL. Optimizing capillary electrophoresis for top-down proteomics of 30–80 kDa proteins. PROTEOMICS. 2014;14(10):1158–1164. doi: 10.1002/pmic.201300381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ntai I, LeDuc RD, Fellers RT, Erdmann-Gilmore P, Davies SR, Rumsey J, Early BP, Thomas PM, Li S, Compton PD, Ellis MJC, Ruggles KV, Fenyö D, Boja ES, Rodriguez H, Townsend RR, Kelleher NL. Integrated Bottom-Up and Top-Down Proteomics of Patient-Derived Breast Tumor Xenografts. Molecular & Cellular Proteomics. 2016;15(1):45–56. doi: 10.1074/mcp.M114.047480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michalski A, Neuhauser N, Cox J, Mann M. A Systematic Investigation into the Nature of Tryptic HCD Spectra. Journal of Proteome Research. 2012;11(11):5479–5491. doi: 10.1021/pr3007045. [DOI] [PubMed] [Google Scholar]
- 26.Shaw JB, Li WZ, Holden DD, Zhang Y, Griep-Raming J, Fellers RT, Early BP, Thomas PM, Kelleher NL, Brodbelt JS. Complete Protein Characterization Using Top-Down Mass Spectrometry and Ultraviolet Photodissociation. Journal of the American Chemical Society. 2013;135(34):12646–12651. doi: 10.1021/ja4029654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101(26):9528–33. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunner AM, Lossl P, Liu F, Huguet R, Mullen C, Yamashita M, Zabrouskov V, Makarov A, Altelaar AF, Heck AJ. Benchmarking multiple fragmentation methods on an orbitrap fusion for top-down phospho-proteoform characterization. Anal Chem. 2015;87(8):4152–8. doi: 10.1021/acs.analchem.5b00162. [DOI] [PubMed] [Google Scholar]
- 29.Zheng Y, Fornelli L, Compton PD, Sharma S, Canterbury J, Mullen C, Zabrouskov V, Fellers RT, Thomas PM, Licht JD, Senko MW, Kelleher NL. Unabridged Analysis of Human Histone H3 by Differential Top-Down Mass Spectrometry Reveals Hypermethylated Proteoforms from MMSET/NSD2 Overexpression. Mol Cell Proteomics. 2016;15(3):776–90. doi: 10.1074/mcp.M115.053819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ansong C, Wu S, Meng D, Liu X, Brewer HM, Deatherage Kaiser BL, Nakayasu ES, Cort JR, Pevzner P, Smith RD, Heffron F, Adkins JN, Paša-Tolić L. Top-down proteomics reveals a unique protein S-thiolation switch in Salmonella Typhimurium in response to infection-like conditions. Proceedings of the National Academy of Sciences. 2013;110(25):10153–10158. doi: 10.1073/pnas.1221210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw JB, Madsen JA, Xu H, Brodbelt JS. Systematic comparison of ultraviolet photodissociation and electron transfer dissociation for peptide anion characterization. J Am Soc Mass Spectrom. 2012;23(10):1707–15. doi: 10.1007/s13361-012-0424-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson MR, Taliaferro JM, Dalby KN, Brodbelt JS. 193 nm Ultraviolet Photodissociation Mass Spectrometry for Phosphopeptide Characterization in the Positive and Negative Ion Modes. J Proteome Res. 2016;15(8):2739–48. doi: 10.1021/acs.jproteome.6b00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fort KL, Dyachenko A, Potel CM, Corradini E, Marino F, Barendregt A, Makarov AA, Scheltema RA, Heck AJ. Implementation of Ultraviolet Photodissociation on a Benchtop Q Exactive Mass Spectrometer and Its Application to Phosphoproteomics. Anal Chem. 2016;88(4):2303–10. doi: 10.1021/acs.analchem.5b04162. [DOI] [PubMed] [Google Scholar]
- 34.Morrison LJ, Brodbelt JS. Charge site assignment in native proteins by ultraviolet photodissociation (UVPD) mass spectrometry. Analyst. 2016;141(1):166–76. doi: 10.1039/c5an01819f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cammarata MB, Brodbelt JS. Structural characterization of holo- and apo-myoglobin in the gas phase by ultraviolet photodissociation mass spectrometry. Chemical Science. 2015;6(2):1324–1333. doi: 10.1039/c4sc03200d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cammarata MB, Thyer R, Rosenberg J, Ellington A, Brodbelt JS. Structural Characterization of Dihydrofolate Reductase Complexes by Top-Down Ultraviolet Photodissociation Mass Spectrometry. J Am Chem Soc. 2015;137(28):9128–35. doi: 10.1021/jacs.5b04628. [DOI] [PubMed] [Google Scholar]
- 37.DeHart CJ, Chahal JS, Flint SJ, Perlman DH. Extensive Post-translational Modification of Active and Inactivated Forms of Endogenous p53. Molecular & Cellular Proteomics. 2014;13(1):1–17. doi: 10.1074/mcp.M113.030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shevchenko A, Wilm M, Vorm O, Mann M. Mass Spectrometric Sequencing of Proteins from Silver-Stained Polyacrylamide Gels. Analytical Chemistry. 1996;68(5):850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 39.Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138(1):141–3. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 40.Trudgian DC, Fischer R, Guo X, Kessler BM, Mirzaei H. GOAT--a simple LC-MS/MS gradient optimization tool. Proteomics. 2014;14(12):1467–71. doi: 10.1002/pmic.201300524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klein DR, Holden DD, Brodbelt JS. Shotgun Analysis of Rough-Type Lipopolysaccharides Using Ultraviolet Photodissociation Mass Spectrometry. Analytical Chemistry. 2016;88(1):1044–1051. doi: 10.1021/acs.analchem.5b04218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zamdborg L, LeDuc RD, Glowacz KJ, Kim YB, Viswanathan V, Spaulding IT, Early BP, Bluhm EJ, Babai S, Kelleher NL. ProSight PTM 2.0: improved protein identification and characterization for top down mass spectrometry. Nucl Acids Res. 2007;35(suppl_2):W701–706. doi: 10.1093/nar/gkm371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Annals of Statistics. 2001;29(4):1165–1188. [Google Scholar]
- 44.Higdon R, Haynes W, Kolker E. Meta-analysis for Protein Identification: A Case Study on Yeast Data. Omics-a Journal of Integrative Biology. 2010;14(3):309–314. doi: 10.1089/omi.2010.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ntai I, Toby TK, LeDuc RD, Kelleher NL. A Method for Label-Free, Differential Top-Down Proteomics. In: Sechi S, editor. Quantitative Proteomics by Mass Spectrometry. Springer New York; New York, NY: 2016. pp. 121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tran JC, Doucette AA. Gel-eluted liquid fraction entrapment electrophoresis: an electrophoretic method for broad molecular weight range proteome separation. Anal Chem. 2008;80(5):1568–73. doi: 10.1021/ac702197w. [DOI] [PubMed] [Google Scholar]
- 47.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meng F, Cargile BJ, Miller LM, Forbes AJ, Johnson JR, Kelleher NL. Informatics and multiplexing of intact protein identification in bacteria and the archaea. Nat Biotechnol. 2001;19(10):952–7. doi: 10.1038/nbt1001-952. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.