The use of biomarkers as an objective measure of pain has received substantial attention in the recent literature, with proponents arguing that brain-derived markers in particular may some day surpass, or even replace, self-report in the characterization of pain.1 Several empirical studies regarding the use of pain biomarkers for diagnosis and classification have been published in recent years.3,7,14,22 However, to our knowledge, no such studies have considered the critical effect of previous probabilities on the diagnostic utility of biomarkers for pain. In other words, it is unclear whether these biomarkers provide diagnostic benefit gained over simply assuming that all patients reporting to the clinic have chronic pain conditions or that all people in the general population do not have chronic pain.
Bayes’ theorem provides a useful context for the understanding of potential pitfalls related to the use of clinical tests (eg, pain biomarkers) by presenting a mathematical framework for calculating the likelihood of a particular event occurring given its previous probability or base rate.12 This principle is expressed in the following equation, where A and B are hypothetical events:
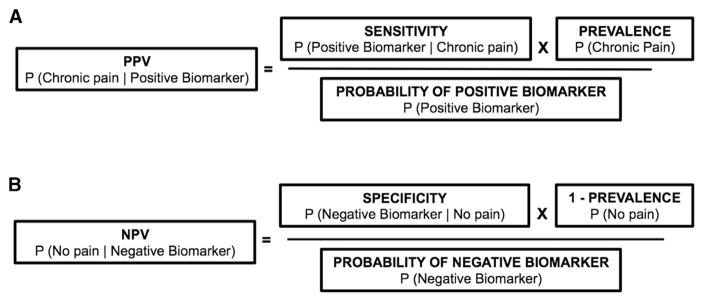
Bayes’ theorem can inform the diagnostic utility of pain biomarkers, specifically in calculating positive predictive value (PPV; ie, the probability that an individual with a positive brain biomarker result actually has chronic pain) and negative predictive value (NPV; ie, the probability that an individual with a negative brain biomarker result actually does not have chronic pain). The PPV and NPV estimates are commonly reported in studies proposing brain biomarkers of pain in which the study uses an equal number of patients and control subjects in their sample (eg, 50% patients and 50% control subjects). This estimate assumes that there are an equal number of individual with and without the condition in a given population. No studies have used ecologically-valid base rates (eg, 12% of the general population for chronic low back pain [cLBP]) in calculating these values.13 As a result, PPV and NPV reported in these studies do not allow for a valid, real-world application. Figure 1 shows the application of Bayes’ theorem in calculating PPV and NPV, to account for empirically-supported base rates. Critically, unlike sensitivity and specificity, PPV and NPV depend on the prevalence of the condition in question.
Figure 1.
Use of Bayes’ theorem to calculate (A) PPV and (B) NPV for chronic pain biomarkers. Note that prevalence directly modulates PPV and NPV, with PPV tending to increase as prevalence increases, and NPV decreasing as prevalence increases.
The primary aim of this article is to illustrate the effect of base rates on the diagnostic utility of proposed brain biomarkers. We selected 4 representative investigations that used magnetic resonance imaging (MRI) data to either differentiate chronic pain patients from healthy control subjects or distinguish from among multiple conditions.3,7,14,22 They were selected for their combined use of multiple neuroimaging modalities, statistical methods, and inclusion of a variety of patient groups. As a group, these studies took advantage of functional and structural MRI, applied multivariate classification techniques, and attempted to differentiate among a number of distinct pain conditions (ie, back pain, irritable bowel syndrome, osteoarthritis, and complex regional pain syndrome). Our purpose was not to provide an exhaustive review of the existing literature on biomarkers of chronic pain, and as such we chose only a subset of the available studies. Furthermore, although other similar studies exist, the general principles illustrated here remain the same across studies.2,17,21 For each of these studies, we performed 3 sets of calculations considering population base rate, clinic base rate, and study sample base rate. Because biomarkers are often proposed on the basis of their presumed clinical utility, PPV and NPV were also recalculated on the basis of a conservative 90% base rate for each condition in a clinic setting. We note, however, that the base rate in the clinic for each condition is likely closer to 100%.
Across studies examined, PPV was substantially reduced when epidemiological base rates were used in place of each study’s unrepresentative sample, suggesting that the application of the proposed biomarkers in the general population would result in a high probability that a person with the biomarker did not actually have chronic pain. For the least common condition examined in the included studies, complex regional pain syndrome, PPV decreased from 88% in the study population to 5% in the general population. NPV was substantially reduced when clinical base rates were used in place of each study’s initial sample, suggesting that the application of the proposed biomarkers in clinical settings would result in a high probability of the biomarker mis-identifying patients who actually have chronic pain as not having pain.
For example, Ung et al developed their proposed neural marker of cLBP using MRI gray matter density values in a support vector machine analysis.22 Their sample consisted of 47 patients with cLBP and 47 individuals without cLBP, resulting in a base rate of 50%. The authors reported sensitivity (76%) and specificity (75%) for this marker, concluding that the marker was useful for discriminating between individuals with and without cLBP (PPV = 75%, NPV = 76%).
The epidemiological base rate of cLBP has been reported as approximately 12% of the population.13 Applying this base rate via Bayes’ theorem to the reported sensitivity and specificity values by Ung et al, the probability that the marker will correctly identify someone with cLBP (PPV) in the general population decreases to 29%. The probability that the marker correctly identifies someone who does not have the condition (NPV) is 96%. Coupled together, these values suggest that in the general population, this marker would perform well at identifying individuals who do not have cLBP. However, it will have a high rate of false positive results (71%).
When a 90% base rate for cLBP (intended as a conservative estimate of prevalence in a clinical setting) is applied to the sensitivity and specificity reported by Ung et al, PPV becomes 96% and NPV becomes 26%. This suggests that using a neural marker of cLBP in the clinic will be likely to correctly identify individuals who do have the condition, but will perform poorly on correctly classifying individuals who do not have the condition. Additional results, as well as relevant specifics regarding each of the included studies, are described in detail in Table 1.
Table 1.
PPV and NPV of Biomarkers in Example Studies
| Study | Pain Condition |
Sensitivity, % | Specificity, % | Manuscript N | Biomarker Type |
Manuscript Base Rate, % |
Manuscript PPV, % |
Manuscript NPV, % |
Epi Base Rate, % |
Epi PPV, % |
Epi NPV, % |
Clinical PPV, % |
Clinical NPV, % |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Labus et al14 | IBS | 65 | 75 | 160 | Structural MRI | 50 | 72 | 68 | 3–208 | 7–39 | 90–98.5 | 96 | 19 |
| Ung et al22 | cLBP | 76 | 75 | 94 | Structural MRI | 50 | 75 | 76 | 1213 | 29 | 96 | 96 | 26 |
| Baliki et al3 | cLBP | 91 | 92 | 130 | Structural MRI | 28 | 82 | 96 | 1213 | 61 | 99 | 99 | 54 |
| CRPS | 94 | 96 | 22 | 88 | 98 | .214 | 5 | 99 | 99 | 64 | |||
| OA | 75 | 95 | 22 | 81 | 93 | 1210 | 67 | 97 | 99 | 30 | |||
| Callan et al7 | cLBP | 92 | 92 | 26 | Functional MRI | 50 | 92 | 92 | 1213 | 61 | 99 | 99 | 57 |
Abbreviations: Epi, epidemiological; IBS, irritable bowel syndrome; CRPS, complex regional pain syndrome; OA, osteoarthritis.
NOTE. The bold data represent the markers’ predictive value with the epidemiological and clinical base rates.
Discussion
For this report, we reanalyzed PPV and NPV for several studies intended to identify biomarkers for chronic pain using epidemiologically derived base rates for prevalence in the general population and rationally derived (yet conservative) base rates for prevalence in a clinical setting. Results strongly suggest that despite appearing promising in laboratory samples with low ecological validity, each proposed marker would, in fact, perform quite poorly when realistic base rates are taken into account. One previous study applied Bayes’ theorem to a proposed neural biomarker of autism spectrum disorders and had findings and conclusions similar to those in the present study.11
Another major concern is that biomarker development studies at present assume that the main purpose of the biomarker is determining whether a person has chronic pain or does not. In a clinical setting, however, the application would be much more complicated, with differential diagnosis among chronic pain conditions being the main purpose of assessment. Furthermore, in a clinical scenario in which a physician desires to confirm a patient is undertreated for their pain to justify additional treatment, or to detect cases in which a patient is not in need of additional pain control, the high rate of false negative results would be a significant impediment. For these reasons, the diagnostic value of these biomarkers surpassing an assumption that all patients will have the pain condition is unsupported.
Biomarkers evaluated to be sufficiently valid and relevant may prove to be useful for understanding mechanisms of pain, as well as for potential individualization of treatment.13,19 It is not the intention of the authors to imply that the studies of neural classifiers for chronic pain conditions discussed in this editorial are without scientific merit. We believe strongly that these studies contribute substantially to scientific understanding of the neural correlates of these conditions. It is their clinical utility for diagnostic purposes, and their justification using measures such as sensitivity and specificity, that we question.
Suggestions for Future Biomarker Development
The clinical value of a biomarker for chronic pain (or indeed, the diagnosis of any condition) is more nuanced than simply considering the sensitivity and specificity. It is also imperative to consider base rates of the condition, in the particular context or setting in which the biomarker is intended for use. Other factors also bear consideration. In a recent commentary, Woo and Wager proposed desirable characteristics of neuroimaging biomarkers: diagnosticity, interpretability, deployability, and generalizability.23 They emphasized the importance of: 1) diagnosticity as adequate sensitivity and specificity, 2) interpretability as scientifically meaningful, 3) deployability as clinically practical and useful, and 4) generalizability as replication of results across sites and testing conditions.
Although these criteria provide a basis for future biomarker development, we believe they should be expanded. First, our results indicate that the reported diagnosticity of current neural pain markers is inflated because of unrepresentative samples used to derive the markers, and critical failure to take into account base rates of the diagnoses in designing studies. As discussed previously and illustrated in Table 1, even a test that performed very well in the general population, using epidemiological base rates, may not be adequate to significantly outperform the base rate in a clinical setting. Future studies might avoid this issue by proactively reporting the performance of the tested marker under a range of ecologically appropriate base rates.
Deployability of diagnostic markers in the clinic is also questionable. For demonstrative purposes, we made a conservative assumption that 90% of individuals tested in a clinic would actually have a given chronic pain condition. Although the PPV of the biomarkers examined in this report are generally quite high using this presumed base rate, they are not necessarily high enough that the added patient burden and financial cost is justified. Were a clinician to simply assume that every patient actually had a given chronic pain condition, they would be correct at least 90% of the time; a biomarker-based classifier would have to be extremely accurate and low-cost (in terms of patient burden and financially) to justify its use. In situations of high or low previous probability, biomarkers should be used with caution because unexpected results are likely to be false positive or negative.12 Additional concerns of deployability are related to how these markers would be practically implemented in a clinical setting. Algorithms used to derive the markers reported on in this article require extensive knowledge of sophisticated statistics and data analysis software packages. Therefore, future studies should describe logistically how clinics might use their marker.
Our final concern is regarding generalizability. Before any assumptions can be made about the generalizability of these markers, it is imperative that we establish their ability to be reproduced across time points. We previously examined test-retest reliability of functional MRI data compared with self-report in a highly-controlled, experimental design and found that these data did not outperform the reliability of participants’ pain ratings.15,16 This finding suggests that inherent assumptions about the reproducibility of neuroimaging findings over time are inadequate. Future studies should examine test-retest reliability and specificity of particular brain regions and connections of proposed markers to determine their robustness and relationship to clinical end points over time. Another key limitation for implementing neuroimaging markers of pain is the lack of convergence among reported markers. Although it should be acknowledged that structural and functional neuroimaging results are not expected to perfectly overlap,9 reported biomarkers for the same population that implement the same imaging technique presently show poor convergence.
There may be situations in which a biomarker-based diagnostic tool could have utility. In certain cases, self-report may not be available to aid in the diagnosis and treatment of an individual, and in these scenarios, bio-markers may prove helpful in facilitating diagnosis and treatment.7 Base rates of the diagnosis remain critical in these decisions as well, and are unknown. Furthermore, because biomarkers of chronic pain are necessarily validated against self-report, and patterns of brain activation related to pain sensation are likely to be altered in nonverbal or cognitively impaired patients, use of a biomarker classifier in these patients will be a challenging proposition. Signatures validated in healthy individuals may not accurately map onto pain-related brain activity in impaired patients. The characterization of brain-based markers for chronic pain has also been described as an avenue for mechanism-based treatment development; however, to our knowledge this translation has not yet been successfully performed.5,6 Finally, in addition to the obvious cost of MRI scanning, ethical dilemmas may result in the event that there is a conflict between self-report and test results from brain-based markers.20
Even if the proposed criteria for pain biomarkers are met, it is still unclear what diagnostic value they have compared to current clinical methods. We must stress that pain biomarkers are always provisional and cannot fully replace variables that represent the patient perspective of health, such as self-report.18,19
Footnotes
The authors have no conflicts of interest to declare.
Contributor Information
Michael Robinson, Department of Clinical and Health Psychology, University of Florida, Gainesville, Florida.
Jeff Boissoneault, Department of Clinical and Health Psychology, University of Florida, Gainesville, Florida.
Landrew Sevel, Department of Clinical and Health Psychology, University of Florida, Gainesville, Florida.
Janelle Letzen, Department of Clinical and Health Psychology, University of Florida, Gainesville, Florida.
Roland Staud, Department of Medicine, University of Florida, Gainesville, Florida.
References
- 1.Apkarian AV, Hashmi JA, Baliki MN. Pain and the brain: Specificity and plasticity of the brain in clinical chronic pain. Pain. 2011;152:S49–S64. doi: 10.1016/j.pain.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagarinao E, Johnson KA, Martucci KT, Ichesco E, Farmer MA, Labus J, Ness TJ, Harris R, Deutsch G, Apkarian AV, Mayer EA, Clauw DJ, Mackey S. Preliminary structural MRI based brain classification of chronic pelvic pain: A MAPP network study. Pain. 2014;155:2502–2509. doi: 10.1016/j.pain.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baliki MN, Schnitzer TJ, Bauer WR, Apkarian AV. Brain morphological signatures for chronic pain. PLoS One. 2011;6:e26010. doi: 10.1371/journal.pone.0026010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borchers AT, Gershwin ME. Complex regional pain syndrome: A comprehensive and critical review. Autoimmun Rev. 2014;13:242–265. doi: 10.1016/j.autrev.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Borsook D, Becerra L, Hargreaves R. Biomarkers for chronic pain and analgesia. Part 1: The need, reality, challenges, and solutions. Discov Med. 2011;11:197–207. [PubMed] [Google Scholar]
- 6.Borsook D, Becerra L, Hargreaves R. Biomarkers for chronic pain and analgesia. Part 2: How, where, and what to look for using functional imaging. Discov Med. 2011;11:209–219. [PubMed] [Google Scholar]
- 7.Callan D, Mills L, Nott C, England R, England S. A tool for classifying individuals with chronic back pain: Using multivariate pattern analysis with functional magnetic resonance imaging data. PLoS One. 2014;9:e98007. doi: 10.1371/journal.pone.0098007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canavan C, West J, Card T. The epidemiology of irritable bowel syndrome. Clin Epidemiol. 2014;6:71–80. doi: 10.2147/CLEP.S40245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damoiseaux JS, Greicius MD. Greater than the sum of its parts: A review of studies combining structural connectivity and resting-state functional connectivity. Brain Struct Funct. 2009;213:525–533. doi: 10.1007/s00429-009-0208-6. [DOI] [PubMed] [Google Scholar]
- 10.Dillon CF, Rasch EK, Gu Q, Hirsch R. Prevalence of knee osteoarthritis in the United States: Arthritis data from the Third National Health and Nutrition Examination Survey 1991–94. J Rheumatol. 2006;33:2271–2279. [PubMed] [Google Scholar]
- 11.Griffin R, Westbury C. Infant EEG activity as a biomarker for autism: A promising approach or a false promise? BMC Med. 2011;9:61. doi: 10.1186/1741-7015-9-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grimes DA, Schulz KF. Refining clinical diagnosis with likelihood ratios. Lancet. 2005;365:1500–1505. doi: 10.1016/S0140-6736(05)66422-7. [DOI] [PubMed] [Google Scholar]
- 13.Hoy D, Bain C, Williams G, March L, Brooks P, Blyth F, Woolf A, Vos T, Buchbinder R. A systematic review of the global prevalence of low back pain. Arthritis Rheum. 2012;64:2028–2037. doi: 10.1002/art.34347. [DOI] [PubMed] [Google Scholar]
- 14.Labus JS, Van Horn JD, Gupta A, Alaverdyan M, Torgerson C, Ashe-McNalley C, Irimia A, Hong JY, Naliboff B, Tillisch K, Mayer EA. Multivariate morphological brain signatures predict patients with chronic abdominal pain from healthy control subjects. Pain. 2015;156:1545–1554. doi: 10.1097/j.pain.0000000000000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Letzen JE, Boissoneault J, Sevel LS, Robinson ME. Test-retest reliability of pain-related functional brain connectivity compared to pain self-report. Pain. 2016;157:546–551. doi: 10.1097/j.pain.0000000000000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letzen JE, Sevel LS, Gay CW, O’Shea AM, Craggs JG, Price DD, Robinson ME. Test-retest reliability of pain-related brain activity in healthy controls undergoing experimental thermal pain. J Pain. 2014;15:1008–1014. doi: 10.1016/j.jpain.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson ME, O’Shea AM, Craggs JG, Price DD, Letzen JE, Staud R. Comparison of machine classification algorithms for fibromyalgia: Neuroimages versus self-report. J Pain. 2015;16:472–477. doi: 10.1016/j.jpain.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson ME, Staud R, Price DD. Pain measurement and brain activity: Will neuroimages replace pain ratings? J Pain. 2013;14:323–327. doi: 10.1016/j.jpain.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strimbu K, Tavel JA. What are biomarkers? Curr Opin HIV AIDS. 2010;5:463–466. doi: 10.1097/COH.0b013e32833ed177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan MD, Cahana A, Derbyshire S, Loeser JD. What does it mean to call chronic pain a brain disease? J Pain. 2013;14:317–322. doi: 10.1016/j.jpain.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 21.Sundermann B, Burgmer M, Pogatzki-Zahn E, Gaubitz M, Stüber C, Wessolleck E, Heuft G, Pfleiderer B. Diagnostic classification based on functional connectivity in chronic pain: Model optimization in fibromyalgia and rheumatoid arthritis. Acad Radiol. 2014;21:369–377. doi: 10.1016/j.acra.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Ung H, Brown JE, Johnson KA, Younger J, Hush J, Mackey S. Multivariate classification of structural MRI datadetects chronic low back pain. Cereb Cortex. 2014;24:1037–1044. doi: 10.1093/cercor/bhs378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woo CW, Wager TD. Neuroimaging-based biomarker discovery and validation. Pain. 2015;156:1379–1381. doi: 10.1097/j.pain.0000000000000223. [DOI] [PMC free article] [PubMed] [Google Scholar]