Abstract
Adverse in utero events can alter the development and function of numerous physiological systems, giving rise to lasting neurodevelopmental deficits. In particular, data have shown that prenatal alcohol exposure can reprogram neurobiological systems, altering developmental trajectories and resulting in increased vulnerability to adverse neurobiological, behavioral and health outcomes. Increasing evidence suggests that epigenetic mechanisms are potential mediators for the reprogramming of neurobiological systems, as they may provide a link between the genome, environmental conditions and neurodevelopmental outcomes. This review outlines the current state of epigenetic research in fetal alcohol spectrum disorder, highlighting the role of epigenetic mechanisms in the reprogramming of neurobiological systems by alcohol and as potential diagnostic tools for fetal alcohol spectrum disorder. We also present an assessment of the current limitations in studies of prenatal alcohol exposure, and highlight the future steps needed in the field.
Keywords: : animal models, clinical cohorts, development, epigenetics, FASD, fetal programming, prenatal alcohol exposure
Prenatal alcohol exposure reprograms physiological systems
Adverse early-life conditions have the potential to permanently imprint or program physiological and behavioral systems during development and lead to long-term consequences in offspring [1,2]. In particular, epigenetic mechanisms are emerging as potential mediators for the biological embedding of these experiences, as they provide a link between in utero conditions and the genome in the modulation of subsequent developmental trajectories [3,4]. Prenatal alcohol exposure can result in an adverse in utero environment that causes numerous adverse developmental consequences. Fetal alcohol spectrum disorder (FASD) refers to the broad spectrum of structural, neurocognitive, physiological and behavioral abnormalities or deficits that can occur following prenatal alcohol exposure (PAE) [5,6]. At the most severe end of the spectrum is fetal alcohol syndrome (FAS), which can occur with chronic exposure to high doses of alcohol [7]. The diagnostic criteria for FAS consist of pre- and postnatal growth retardation, a characteristic facial dysmorphology and central nervous system (CNS) alterations, including neurological abnormalities, developmental delays and intellectual impairment [6]. Exposure to alcohol at levels that do not produce full FAS can result in either partial FAS, where only some of the diagnostic features occur, or in numerous alcohol-related effects that can be primarily physical (alcohol-related birth defects) or primarily neurobehavioral (alcohol-related neurodevelopmental disorder). Of note, alcohol-related birth defects and alcohol-related neurodevelopmental disorder are not mutually exclusive and both may occur in an individual exposed to alcohol in utero [6].
The degree to which alcohol affects development depends on a variety of factors such as timing and level of alcohol exposure, overall maternal health and nutrition, and genetic background [8]. However, neurobehavioral/neurodevelopmental deficits are consistently seen across the spectrum, and include neurocognitive impairment (cognitive function, learning and memory, executive function), impairment in self-regulation (attention, impulsivity, behavioral regulation, stress responsiveness, mood/affect, sleep abnormalities) and deficits in adaptive function (communication, social behavior, activities of daily living) [9–14]. Importantly, the adverse neurodevelopmental outcomes of children with FASD often persist well into adulthood, including metabolic changes, immune dysfunction, and altered stress responsitivity (reviewed in [15]).
Animal models of PAE were first developed in response to the skepticism that greeted the first description of FAS by Jones and Smith (1973) [16]. These were particularly important to the field, as they provided a level of control not possible in the clinical setting and allowed for the analysis of deficits associated with timing, pattern (acute vs chronic) and dose of alcohol, genetic factors, environment, nutrition and interactions with other drugs. An additional important strength of animal models is the ability to make direct correlations between central and peripheral tissues, as clinical studies do not have ready access to critical tissues such as the brain and other organs, except through biopsy, and changes in clinically accessible tissues do not always reflect alterations in the brain. Furthermore, animal models can provide critical insight into the molecular mechanisms underlying effects of PAE, and can thus pave the way for identification of novel biomarkers. Important recent studies have made significant progress in characterizing the neurodevelopmental, physiological, and behavioral alterations associated with PAE, as well as elucidating molecular mechanisms through which these alterations occur at different doses and patterns of alcohol exposure. In vitro studies have provided further vital insights into the molecular mechanisms by which alcohol affects cellular functions, allowing for the dissection of molecular pathways in highly specific and controlled environments [17–22]. These different strategies have provided key insights into the altered neurodevelopmental profiles resulting from PAE and highlight the complex and long-term programming effects of alcohol on numerous developmental processes. Taken together, these studies provide support for the suggestion that the underlying adverse effects of PAE on development may involve fetal programming. This concept suggests that early environmental or nongenetic factors, including maternal undernutrition, stress, and exposure to drugs or other toxic agents, can permanently organize or imprint physiological and neurobiological systems to increase adverse cognitive, adaptive and behavioral outcomes, as well as vulnerability to diseases or disorders later in life [1,2,23]. Of particular relevance, research has shown that alcohol is an early life insult that programs developing neurobiological systems and markedly increases risk for adverse outcomes [24]. Although the exact molecular mechanisms underlying these effects remain unknown, epigenetics is a prime candidate for the programming effects of environmental factors on physiological systems, as it might serve as a link between environmental stimuli and neurodevelopmental outcomes to influence health and behavior well into adulthood [25–27].
Epigenetic reprogramming by developmental alcohol exposure
Epigenetics refers to modifications of DNA and/or its regulatory factors, including chromatin and noncoding RNA, that alter the accessibility of DNA to modulate gene expression and cellular functions without changes to underlying genomic sequences [28]. Patterns of epigenetic modifications, in general, have been closely associated with cell fate specification and differentiation, suggesting a crucial role for epigenetics in the regulation of cellular functions [29]. Notably, cell specificity is the major driver of global epigenetic patterns, as each cell type is associated with a unique epigenomic landscape. In addition to the stability of the epigenome in regulating cellular identity, epigenetic patterns can also be influenced by environmental stimuli, although these effects tend to be subtler. Importantly, epigenetic mechanisms exist in a seeming paradox between the stability of cellular identity and plasticity of environmental responses, modulating cellular functions through both short- and long-term responses to stimuli [4].
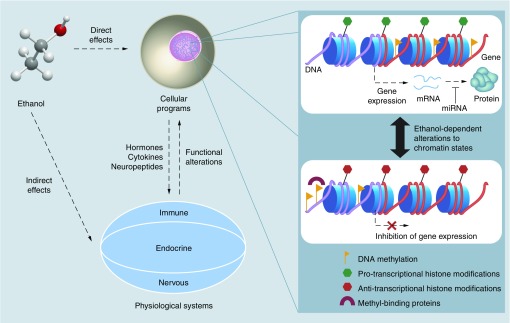
Given the close relationship between gene expression and epigenetic patterns, it is not surprising that initial evidence of the programming effects of alcohol on the genome were identified through changes in transcription. A number of key studies have demonstrated that alcohol exposure during gestation leads to persistent genome-wide alterations to the transcriptome [30–34]. Indeed, PAE causes widespread changes to gene expression levels in the brain of fetal, neonatal and adult rodents [18,31–33,35,36]. Importantly, we have shown that changes in gene expression occur not only under basal conditions, but also in the transcriptomic responses to an immune challenge in PAE compared with control animals, highlighting the robust long-term reprogramming of transcriptomic profiles in the brains of PAE animals [34]. Although the relationship between gene expression and epigenetic patterns remains extremely complex, these findings suggest that PAE-induced deficits may be associated with reprogramming of the epigenome during development [27]. Indeed, numerous lines of evidence now point to epigenetic alterations in the etiology of FASD, including evidence from cell culture, various animal models of prenatal and neonatal alcohol exposure, as well as a few recent clinical studies of FASD. Figure 1 provides an overview of our take on fetal programming of physiological systems by developmental alcohol exposure. Of note, many studies have used either FASD phenotypes or exposure paradigms as their main focus when assessing epigenetic changes. Here, we delineate the current state of epigenetic research in FASD, which includes investigation of DNA modifications, chromatin alterations and ncRNA expression profiles, highlighting the role of epigenetics in the reprogramming of biological systems by PAE.
Figure 1. . Overview of the mechanisms underlying fetal programming by prenatal alcohol exposure.
Ethanol exposure can have both direct effects on cellular programs through intracellular pathways, and indirect effects through the stimulation of various physiological systems, which release signaling molecules that can affect cellular functions. Cell death may play a role in the reprogramming of physiological systems, but is not depicted here. Together, these likely play a role in the biological embedding of PAE, with long-term consequences on health and behavior. Importantly, epigenetic mechanisms may mediate these effects through ethanol-dependent alterations in chromatin states. Chromatin, located in the cell’s nucleus, dynamically influences gene expression to modulate cellular functions. Transcriptionally active epigenetic states involve the presence of pro-transcriptional histone modifications and gene body DNA methylation. Micro-RNA (miRNA) provide an additional layer of regulation by modulating protein levels through translational inhibition. By contrast, transcriptionally inactive chromatin landscapes are associated with promoter DNA methylation, which can inhibit transcription factor binding and recruit methyl-binding proteins that promote anti-transcriptional histone modifications through their association with histone-modifying complexes. Nevertheless, the relationship between epigenetic states and transcription remains extremely complex, and this figure is merely a simplified representation of epigenetic regulation (expanded from our previous review [27]).
PAE is correlated with altered DNA modification programs throughout the lifespan
Covalent modifications of DNA nucleotides have long been established as a form of epigenetic regulation, and include both DNA methylation and hydroxymethylation. DNA methylation is perhaps the most studied epigenetic modification and involves the covalent attachment of a methyl group to the 5′ position of cytosine, typically occurring at cytosine-guanine dinucleotide (CpG) sites [37]. As CpG dinucleotides are palindromic, both DNA strands are generally methylated at CpGs, allowing for conservation of DNA methylation patterns during cell division [22]. Additionally, recent evidence suggests that DNA methylation also occurs at low levels at CpH sites (where H = A, C or T), which is found at high levels in the brain, particularly within neurons [39,40]. DNA methylation is associated with the regulation of gene expression, although its effects on transcription are highly dependent on genomic context [38]. For example, DNA methylation at gene promoters generally represses gene expression, while its role is more variable within gene bodies and intergenic regions [38]. DNA methylation can also directly control the binding of transcription factors and repressors to gene regulatory regions, such as promoters and enhancers, to modulate gene expression patterns [41]. In addition to its role in transcriptional control, DNA methylation within exon–intron boundaries has been associated with altered mRNA splicing, and its presence within certain exons potentially regulates alternative transcriptional start sites [42–44]. Although DNA methylation patterns at promoters within an individual tend to negatively correlate with gene expression, emerging evidence shows that when comparing a single gene across a population, the association between DNA methylation and gene expression can be negative, positive or nonexistent, highlighting the complex relationship between DNA methylation and transcription [45–47]. In contrast to DNA methylation, hydroxymethylation of cytosine residues is thought to act as an intermediate in the active demethylation cycle of DNA by the TET family of enzymes. However, recent studies have shown that DNA hydroxymethylation may also play a role in the regulation of gene expression through the recruitment of various chromatin modifiers [48]. Moreover, given its high levels in pluripotent cells and the adult brain, DNA hydroxymethylation may also potentially have a functional role in neurodevelopment and various cognitive processes [39,49–53]. Perhaps most importantly, in addition to its role in the regulation of developmental programs, DNA methylation is also emerging as a potential biomarker for early-life exposures due to its stability over time and malleability in response to environmental cues [54].
A large number of studies have identified changes in DNA modifications in response to PAE, and the current review will present a snapshot of the different approaches to assess these alterations, which range from ‘bulk’ levels to candidate gene approaches and genome-wide investigations (here, we define bulk levels as measures of epigenetic patterns that do not delineate specific regions, but rather represent the total levels within a given tissue or cell population). For a detailed overview of all current studies of DNA modifications and developmental alcohol exposure, please refer to Table 1. The first evidence of alcohol-induced changes to DNA methylation programs was generated in a mouse model, where embryos were exposed to alcohol during gestational days (GD) 9–11. This study demonstrated that alcohol reduced bulk levels of DNA methylation in the genome, potentially by inhibiting DNMT1 activity, and opened the door for future studies of epigenetic mechanisms in FASD [55]. Several studies have extended this line of evidence by studying the effects of alcohol exposure during various stages of development and identifying alterations to bulk levels of DNA methylation in different brain regions under basal and intervention conditions (Table 1) [56–61] For instance, PAE throughout gestation delays the accumulation of DNA methylation in neural stem cells, and increases DNA methylation levels in the mouse hippocampus, a brain region involved in learning and memory [58]. This same study assessed bulk DNA hydroxymethylation in parallel, identifying a decrease in the neural progenitor cells of the hippocampus, which suggests widespread alterations to DNA methylation programs [58]. In addition to assessing the impact of PAE on bulk DNA methylation levels, a number of studies have used bulk DNA methylation levels as a measurable outcome for dietary or therapeutic interventions in combination with different behavioral tasks. For example, choline supplementation has been proposed as a potential intervention due to its role as a methyl donor, and has been associated with the partial rescue of behavioral alterations and increased DNA methylation levels in the hippocampus and prefrontal cortex of PAE rats [56,62]. Similar outcomes are also observed in embryos and neural stem cells treated with alcohol or 5-azacytidine, a potent inhibitor of DNA methylation, suggesting that alcohol-induced deficits are likely related to altered epigenomic profiles and functions [18]. Overall, these findings demonstrate that developmental alcohol exposure tends to impair the establishment of typical DNA methylation levels, which may reprogram downstream cellular and biological functions.
Table 1. . DNA modification studies of developmental alcohol exposure.
| Type |
Exposure paradigm |
Model |
Effects |
Ref. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Level | Treatment | Time | Species | Tissue | Culture | Age | Sex | Dir. | Target | |||
| Bulk | A | 3 g/kg (2×/day) | Gavage | GD 9–11 | Mouse | Whole fetus | N | GD 11 | ND | ↓ | DNA methylation | [55] |
| ↓ | DNMT1 activity | |||||||||||
| A | 3 g/kg (1×/day) | Intragastric intubation | PN 2–10 | Rat | Hippocampus/prefrontal cortex | N | PN 21 | Male/female | ↑ | DNA methylation | [56] | |
| A | 4.5 g/kg | Gavage | GD 1–22 | Rat | Hippocampus | N | PN 21 | Male/female | ↑ | DNMT activity | [57] | |
| M | 4% v/v ethanol | Liquid diet | GD 1–17 | Mouse | Hippocampus | N | GD 17 | ND | ↑ | DNAm/DNAhm of CA1 cells, intermediate zone (DNAm only) | [58] | |
| ↓ | DNAm/DNAhm of NE | |||||||||||
| PN 7 | ND | ↑ | DNAm of outer granule layer | |||||||||
| ↓ | DNAhm of outer and inner granule layer cells | |||||||||||
| A | ||||||||||||
| 200 mM | 2 days | NA | Mouse | Embryonic fibroblast (NIH/3T3) | Y | GD 13.5 | ND | ↓ | DNA methylation | [59] | ||
| ↓ | Dnmt1, Dnmt3a, Dnmt3b levels | |||||||||||
| A | ||||||||||||
| 2.5 g/kg (2× at 0 h and 2 h) | Subcut. injection | PN 7 | Mouse | Hippocampus and neocortex | N | PN 7 | Male/female | ↓ | DNA methylation | [60] | ||
| ↓ | Dnmt1, Dnmt3a levels | |||||||||||
| |
A |
70 mM |
8 days |
GD 14.5 |
Mouse |
Neural stem cells |
Y |
NA |
NA |
↑ |
DNA methylation |
[61] |
| Targeted | M | 5.0 g/kg | Liquid diet | GD 1–18 | Rat | Astrocyte | Y | GD 21 | Male/female | ↑ | Gfap DNA methylation | [65] |
| A | 6.0 g/kg | Intragastric intubation | GD 1–20 | Rat | Olfactory bulb | N | PN 10 | Male/female | ↑ | Bdnf DNA methylation | [66] | |
| M | 10% v/v ethanol | Free drinking | GD 0.5–8.5 | Agouti mouse | Tail | N | PN 21 | Male/female | ↑ | Avy IAP DNA methylation | [64] | |
| A | 5.8 g/kg | Intragastric intubation | GD 9 | Mouse | Embryo | N | GD 17 | Male | ↓ | Igf2 DNA methylation | [67] | |
| ↑ | Igf2 expression | |||||||||||
| M | 6.7% v/v ethanol | Liquid diet | GD 7–21 | Rat | Mediobasal hypothalamus | N | PN 60–65 | Male | ↑ | Pomc DNA methylation | [68] | |
| M | 10% v/v ethanol | Free drinking | GD 0.5–8.5 | Mouse | Hippocampus | N | PN 87 | Male | ↓ | Vglut2 DNA methylation | [69] | |
| M | 10% v/v ethanol | Free drinking | GD 0.5–8.5 | Mouse | Hippocampus | N | PN 28 | Male | ↑ | DNAm Vmn2r64, Olfr110 (2 CpG) | [71] | |
| ↓ | DNAm Olfr601, Olfr110 (1 CpG) | |||||||||||
| A | 70 mM | 8 days | GD 14.5 | Mouse | Neural stem cells | Y | NA | NA | DNAm MeCP2 | [61] | ||
| |
M |
35.5% ethanol-derived calories |
Liquid diet |
GD 1–22 |
Rat |
Hippocampus |
N |
PN 55 |
Male/female |
↑ |
DNAm Slc6a4 |
[70] |
| Genome wide | A | 400 mg/dl | 44 h | GD 8.25 | Mouse | Embryo | Y | GD 10 | ND | ↑ | 1028 genes DNA methylation | [17] |
| ↓ | 1136 genes DNA methylation | |||||||||||
| A | 400 mg/dl | 2 days | NA | Mouse | NS-5 neural stem cells | Y | NA | ND | ↑ | DNAm in 83 cell cycle genes | [19] | |
| A | 400 mg/dl | 6 h | NA | Rat | DRG-derived neural stem cells | Y | NA | ND | ↓ | NSC gene DNAm | [72] | |
| M | 20 mM | 4 days | NA | Human | Embryonic stem cells | Y | NA | ND | No changes in DNA methylation at 27,000 CpGs | [74] | ||
| M | 10% v/v ethanol | Free drinking | GD 1- PN10 | Mouse | Whole brain | N | PN 70 | Male | 6660 differentially methylated promoters | [73] | ||
| M or A | 20 mM or 50 mM | 1 or 2 days | NA | Human | Embryonic stem cells | Y | NA | Male/female | DNA hypermethylation at TSS and CpG islands, but inconsistent across individual regions | [75] | ||
| A | 2.5 g/kg (2× at 0 and 2 h) | Subcut. injection | PN 4,7 | Mouse | Hippocampus | N | PN 70 | Male | 4640 differentially methylated regions in gene promoters and 459 in CpG islands | [76] | ||
| Confirmed FASD | Human | Buccal epithelial cells | N | 3–6 years | Male | 269 differentially methylated CpGs | [77] | |||||
| Confirmed PAE | Human | Buccal epithelial cells | N | 5–18 years | Male/female | 356 hypomethylated CpGs 302 hypermethylated CpGs | [78] | |||||
A: Acute; DNAm: DNA methylation; DNAhm: DNA hydroxymethylation; GD: Gestational day; M: Moderate; NA: Not applicable; ND: Not disclosed; NE: Neuroepithelium; PN: Postnatal day.
Proof of principle of alcohol’s programming effects was further exemplified using the agouti viable (Avy) yellow mouse model, which contains a DNA methylation-sensitive element within the Avy locus that regulates coat color [63]. In this model, PAE increased the incidence of pseudo-agouti animals, indicating that specific loci are responsive to the effects of alcohol during development and can influence phenotypic outcomes [64]. As such, more recent studies have sought to identify specific gene targets of PAE-induced epigenetic effects, either through hypothesis- or discovery-driven approaches. An initial study using cultured cells showed that, rather than a global demethylation of the genome, specific regions become more methylated and others less methylated in response to alcohol exposure, suggesting that some regions may be differentially sensitive to alcohol-induced reprogramming effects [17]. Consequently, numerous groups have invested in targeted analyses of epigenetic patterns in genes associated with the deficits observed in individuals with FASD (e.g., immune, stress, cognitive and otherwise-related; Table 1) [61,65–71]. For instance, in mice, the expression of Igf2, an imprinted gene involved in growth, is decreased in the embryo and placenta following PAE, concomitant with increased DNA methylation of the differentially methylated region 1 in its promoter and growth deficits in offspring. Interestingly, choline supplementation during gestation partially rescues the effects of PAE on growth and DNA methylation within this locus, further highlighting a potential role for dietary supplements in the attenuation of alcohol-induced deficits [67]. PAE also results in increased DNA methylation and decreased expression of proopiomelanocortin in the hypothalamus, which is a key regulator of the stress response [68]. Moreover, we have shown that Slc6a4, an important serotonin transporter, also displays sex-dependent alterations to DNA methylation and gene expression patterns in the hypothalamus of adult PAE rats [70]. While the hypothesis-driven approach has proven fruitful in many regard, it relies heavily on previously identified biological pathways and has not been very successful in identifying novel targets of developmental alcohol exposure.
More recently, researchers have begun using genome-wide tools to study the effects of alcohol exposure beyond classical candidate pathways (Table 1) [17,19,72–78]. For example, widespread changes in DNA methylation patterns were identified in the brains of adult male mice, with some alterations overlapping with changes in gene expression profiles [73,76]. In addition to the identification of new genes potentially associated with alcohol exposure, these findings provide evidence for the lasting effects of developmental alcohol exposure on the DNA methylome. Moreover, these PAE-related changes in the DNA methylome may, in turn, alter transcriptional profiles and reprogram physiological systems. Two recent studies have also characterized the DNA methylation profile of buccal epithelial cells (BECs) of children with FASD, identifying widespread alterations to the epigenome, and have provided preliminary evidence of a DNA methylation ‘signature’ of FASD [77,78]. Specifically, this study identified alterations in novel genes, as well as in genes previously associated with alcohol exposure, including several imprinted genes and the protocadherin clusters [78]. While the use of a peripheral tissue, BECs, makes it difficult to readily interpret these findings in the context of FASD-associated deficits, these studies provide important insight into potential biomarkers of PAE in human populations. Nevertheless, additional cohorts from different cultural backgrounds are necessary to validate these findings and solidify their usefulness in a clinical context.
Taken together, these studies highlight the widespread effects of developmental alcohol exposure on DNA methylation patterns, although the direction of change varies depending on the model of alcohol exposure, the tissue analyzed and the specific genes assessed. While most studies of bulk DNA methylation identify a decrease in methylation levels, potentially due to lower activity of DNMTs and the inhibition of 1-carbon metabolism by alcohol, results have varied across models due to a number of factors, including differences in levels and timing of alcohol exposure, developmental stage, analyzed tissue and analysis methods. These findings highlight the importance of using different models to assess the molecular mechanisms underlying the effects of ethanol at different stages, doses, etc. Furthermore, the analysis of DNA hydroxymethylation in the context of FASD remains an elusive topic of research, only investigated in a single study of PAE [58]. Given its seemingly key role in neurons, it could potentially play an important role in the etiology of FASD. As a whole, multiple lines of evidence support a role for DNA methylation in the fetal programming of biological systems by PAE and represent an important avenue for the discovery of biomarkers of FASD.
Chromatin modifications associated with PAE
In eukaryotic genomes, DNA is found in nucleoprotein polymers known as chromatin, which compact DNA within the nucleus and modulate access to genetic material. More specifically, DNA is wrapped around nucleosomes, protein octamers containing two molecules each of core histone proteins H2A, H2B, H3 and H4 [79,80]. The flexible N-terminal tails of these proteins are the main targets for histone modifications, which control chromatin structure and transcription by modulating the accessibility of the local chromatin structure. Different histone modifications may regulate transcription in their vicinity by serving as a docking platform to recruit various effector proteins – such as chromatin remodeling complexes – or by preventing various proteins from binding to chromatin [81]. The different types of histone marks include acetylation, methylation, phosphorylation, ubiquitination, sumoylation and ADP-ribosylation, which play various functional roles depending on their cellular context and location on the histone tail [82–84]. For instance, histone acetylation and histone H3 lysine 4 (H3K4) methylation are typically associated with increased transcription, while methylation of histone H3 lysine 9 (H3K9) and 27 (H3K27) is associated with transcriptional repression. Importantly, histone modifications play a crucial role in mediating the crosstalk between transcriptional regulation and chromatin-modifying enzymes to dynamically regulate chromatin structure, downstream transcriptional activity and cellular programs [83]. Additional relevant chromatin-associated proteins include methylated DNA-binding proteins, such as MeCP2, and the methyl CpG-binding (MBD) domain family of proteins. These can act as intermediaries between DNA and histone modifications, recruiting histone-modifying enzymes to modulate the chromatin landscape [38,85]. In particular, they have been tightly associated with development, and mutations in MeCP2 lead to a severe neurodevelopmental disorder, Rett syndrome, suggesting that they may play a role in mediating the effects of PAE [86,87].
To date, the effects of developmental alcohol on chromatin structure have been relatively understudied. Again, we present a broad overview of the different approaches to studying histone modifications following PAE, including the investigation of bulk levels, targeted genes and genome-wide associations. Table 2 delineates the different studies assessing chromatin alterations following developmental alcohol exposure. The earliest evidence of alcohol-induced changes to chromatin proteins came from a sheep model of chronic PAE, where fetal sheep display lower levels of H2A family proteins compared with controls [88]. Furthermore, several genome-wide studies of PAE have identified differences in the expression of histone modifying enzymes and enrichment of gene ontology terms associated with chromatin proteins [32–34]. Together, these findings suggest that alcohol exposure during development could potentially impact the abundance of different chromatin proteins, leading to fundamental alterations to chromatin structure and function. More recent studies have focused primarily on the bulk levels of histone modifications following PAE (Table 2) [68,89–96]. For instance, exposure to high levels of alcohol during the ‘third-trimester equivalent’ (PN 7) rat model causes reductions in bulk levels of acetylated histone H3 and H4 in the cerebellum, suggesting a global decrease in transcriptional activity [89]. Similar results were found using a prenatal exposure model, where a more repressed chromatin state was found in the brains of adult male rats exposed throughout gestation (equivalent to the first two trimesters of human pregnancy). These animals show decreased bulk levels of total H3K4 di- and trimethylation and H3K9 acetylation, as well as an increase in H3K9 dimethylation, concomitant with altered expression profiles of the chromatin-modifying enzymes, Set7/9, Setdb1, Hdac2 and G9a, which modulate the levels of these marks. Together, these findings support a role for alcohol in long-lasting effects on the regulation of chromatin states [91].
Table 2. . Studies of chromatin alterations in response to developmental alcohol exposure.
| Type |
Exposure paradigm |
Model |
Effects |
Ref. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Level | Treatment | Time | Species | Tissue | Culture | Age | Sex | Dir. | Target | |||
| Bulk | M | 4 h/day | Inhalation chamber | PN 2–12 |
Rat | Cerebellum | N | PN 2, 4, 6, 8, 10, 12 | Male/female | ↓ | H3ace, H4ace, H3K23ace | [89] |
| ↑ | Cbp expression (not PN10,12) | |||||||||||
| M or A | 50 or 200 mM ethanol | 24 h | NA | Mouse | Cardiac progenitor cells | Y | NA | ND | ↑ | H3K9ace | [90] | |
| M | 6.7% v/v ethanol | Liquid diet | GD 7–21 |
Rat | POMC neurons of the arcuate nucleus | N | PN 60–80 | Male/female | ↑ | H3K9me2, G9a, Hdac2, Setdb1 expression | [91] | |
| ↓ | H3K4me2/3, H3K9ace Set7/9 expression |
|||||||||||
| A | 262.5 mM | 24 h | 48–72 hpf | Zebrafish | Embryo | N | 72 hpf | Male/female | ↑ | Phosphorylated histone H3 | [96] | |
| M | 6.7% v/v ethanol | Liquid diet | GD 7–21 |
Rat | β-endorphin neurons of the arcuate nucleus | N | PN 60–65 | Male | ↑↓ | H3K9me2 H3K4me2/3, H3K9ace, H3S10phospho | [68] | |
| A | 2.5 g/kg (2× at 0 and 2h) | Subcut. injection | PN 7 | Mouse | Hippocampus & Neocortex Neocortex | N | PN 7 | Male/female | ↑ | H3K9me2, H3K27me2 | [92] | |
| ↑ | G9a expression | |||||||||||
| M | 1.0 g/kg (2× at 0 and 2 h) | Subcut. injection | PN 7 | Mouse | Hippocampus & Neocortex | N | PN 7 | ND | ↑ | H3K9me2, H3K27me2 | [93] | |
| A | 6 g/kg | Intragastric intubation | GD 7–14.5 |
Mouse | Fetal heart | N | GD 14.5 | Male/female | ↑ | H3K14ace | [94] | |
| A | 5 g/kg | Intragastric intubation | GD 8.5–16.5 | Mouse | Fetal heart | N | GD 14.5 PN 7 |
Male/Female | ↑ | H3K9ace | [95] | |
| |
A |
5.0 g/kg (2× at 0 and 2h) |
Subcut. injection |
PN 7 |
Mouse |
Cerebral cortex |
N |
PN 7 |
Male |
↑ |
H2AX serine 139 phosphorylation |
[97] |
| Methyl-binding proteins | A | 200 mM | 2 days | NA | Mouse | Embryonic fibroblast (NIH/3T3) | Y | GD 13.5 | ND | ↓ | Mbd3 level | [59] |
| A | 6 g/kg (2× at 0 and 6h) | Intragastric intubation | GD 6–15 | Mouse | Cortex & Striatum | N | 5 weeks | Male/female | ↓ | MeCP2 levels | [100] | |
| A | 70 mM | 8 days | GD 14.5 | Mouse | Neural stem cells | Y | NA | NA | ↑ | MeCP2 levels | [61] | |
| |
A |
300 mM |
6 days |
0–6 dpf |
Oryzias latipes |
Brain |
N |
6 dpf |
Male/female |
↑ |
Mecp2, Mbd1b, Mbd3a, Mbd3b |
[101] |
| Targeted | A | 320 mg/dl | 5 days | GD 12.5 | Mouse | Fetal cerebral cortical neuroepithelial stem cells | Y | NA | ND | ↓ | H3K4me3, H3K27me3 of NSC/hemeobox genes and transposable elements | [20] |
| M | 1.0 g/kg (2× at 0 and 2h) | Subcut. injection | PN 7 | Mouse | Hippocampus and neocortex | N | PN 7 | ND | ↑ | H3K14ace of G9a exon 1 | [93] | |
| A | 5 g/kg | Intragastric intubation | GD 8.5–16.5 | Mouse | Fetal heart | N | GD 14.5 PN 7 |
Male/female | ↑ | H3K9ace of Gata4 promoter | [95] | |
| A | 2.9 g/kg (2× at 0 and 4 h) | IP injection | GD 7 | Mouse | Cortex | N | GD 17 | ND | ↑ | H3K9ace of NSC/hemeobox genes | [21] | |
| ↓ | H3K27me3 of NSC/hemeobox genes | |||||||||||
| A | 160 or 240 mg/dl | 3 days | GD 12.5 | Mouse | Fetal cerebral cortical neuroepithelial stem cells (GD 12.5) | Y | NA | ND | ↑↓ | Dose-dependent alterations to H3K4me3, H3K9ace, H3K9me2, H3K27me3 in NSC genes | [21] | |
| A | 2.5 g/kg (2× at 0 and 2 h) | Subcut. injection | PN 7 | Mouse | Hippocampus and neocortex | N | PN 7 | ND | ↑↓ | H4K8ace of Cb1r exon 1 H3K9me2 of Cb1r exon 1 | [99] | |
| |
M |
10% v/v ethanol |
Free drinking |
GD 0.5–8.5 |
Mouse |
Hippocampus |
N |
PN 87 |
Male |
↑ |
Vglut2 H3K4me3 |
[69] |
| Genome-wide | A | 2.5 g/kg (2× at 0 and 2 h) | Subcut. injection | PN 7 | Mouse | Hippocampus | N | PN 70 | Male | Widespread alterations to H3K4me3 (798 genes) and H3K27me3 (223 genes) – little overlap | [76] | |
A: Acute; Dpf: Day post-fertilization; GD: Gestational day; G-W: Genome-wide; Hpf: Hours past fertilization; M: Moderate; NA: Not applicable; ND: Not disclosed; PN: Postnatal day.
In addition to their roles in the regulation of transcriptional programs, histone marks can also serve as a proxy for DNA damage, as in the case of phosphorylation of serine 129 on histone H2AX (γH2AX), which is associated with double-stranded breaks and DNA fragmentation, a mark of apoptosis. Of note, the levels of this modification increase in the mouse cortex 7 h after alcohol exposure and may serve as a quantitative indicator of ethanol-induced DNA damage [97]. Several other histone modifications have also been associated with the high levels of neuroapoptosis observed after acute ethanol exposure. Extensive work has been performed related to the effects of developmental alcohol on H3K9 dimethylation and its association with ethanol-induced neurodegeneration, another mechanism through which PAE might reprogram physiological systems. Indeed, third-trimester equivalent exposure to alcohol increases the expression of G9a and induces a concomitant rise in bulk levels of dimethylated H3K9 and H3K27 in the neocortex and hippocampus. In turn, these are associated with caspase-3 activation and neuronal apoptosis in the postnatal day 7 brain of mice, potentially leading to long-term deficits in synaptic plasticity, learning, and memory [92,98]. Interestingly, inhibition of G9a reduces the levels of neurodegeneration in these animals and may represent a potential intervention target for PAE [92,98]. These results highlight the complex interplay among different mechanisms of fetal programming following alcohol exposure, placing epigenetic mechanisms at the nexus of different exposure paradigms and neurodevelopmental outcomes.
Similar to DNA methylation, the study of chromatin has also begun moving toward more specific target genes (Table 2) [20,21,69,93,95,99]. For example, primary neurosphere cultures exposed to high levels of ethanol show alterations in H3K4 and H3K27 trimethylation within the regulatory regions of genes involved in differentiation and neural cell identity, as well as transposable elements [20]. Furthermore, in primary cultures of neural stem cells from mice, levels of H3K4 trimethylation, H3K27 trimethylation, H3K9 acetylation, and H3K9 methylation display dose-dependent alterations in response to ethanol exposure within the promoters of several genes involved in neuronal patterning [21]. These results highlight both the global effects of alcohol exposure on chromatin, and the importance of reconciling differences among the various models of alcohol exposure. Depending on the model, different genes exhibit differential histone modification profiles, which also shift between the immediate response to ethanol and 96 hours post-exposure, suggesting that the effects of alcohol are dependent on both the gene affected and the timing of exposure [21]. While the direction of alcohol-induced changes is variable across all modifications at different doses in this in vitro model, H3K9 dimethylation is consistently depleted compared with that in controls, contrasting studies of global H3K9 dimethylation levels. Furthermore, the same study also showed that exposure to alcohol on GD 7 alters histone modifications profiles of the same subset of neural patterning genes in the mouse cortex [21]. This finding supports the value of cell culture models in providing data relevant to those in in vivo models, and suggests that cell culture models could prove useful in the initial assessment of therapeutic interventions. Finally, genome-wide analysis of H3K4 and H3K27 trimethylation in the adult mouse brain revealed widespread differences in the enrichment profiles of ethanol-exposed animals compared with controls [76]. Although few overlaps between the enrichment patterns of the two modifications were identified, these findings nevertheless indicate long-term alterations to histone modification programs in the brain following PAE, which may have important ramifications in the assessment of functional deficits in FASD. Of note, this same study also investigated DNA methylation patterns, and similarly, found very few overlaps between alterations in the enrichment of histone marks and DNA methylation in response to alcohol exposure [76].
Overall, these findings suggest that alcohol exposure reprograms histone modifications both genome-wide and in a locus-specific manner, with different genes displaying unique patterns of epigenetic programming in response to alcohol. Overall, it appears that developmental alcohol exposure may generally promote a repressive epigenomic state at the bulk histone modification level, though the causative link between epigenetics states and gene expression remains unclear. However, as a vast number of different combinations exist in the histone code, the interpretation of these findings is extremely complex, especially given the number of different models of developmental alcohol exposure. More research is needed to assess additional histone modifications and their overlap throughout the genome, as well as their intersection with DNA modifications. Alcohol exposure also appears to influence the levels of methylated DNA-binding proteins, including MeCP2, MBD1 and MBD3, though the results vary across exposure paradigms [59,61,100,101]. These findings reflect the complex interplay between DNA modifications and higher level chromatin structure, further highlighting the importance of integrating multiple levels of epigenetic mechanisms. Furthermore, variants of canonical histones may also play a role in the etiology of alcohol-induced deficits, and represent an intriguing avenue of research. Future studies should also strive to assess alterations to chromatin in human populations, as human research is lacking in this area, likely due to the difficulty of obtaining primary samples from human populations and the need for immediate sample fixation following collection for the targeted analysis of histone modifications. Although evidence of long-term effects on histone modification programs remains sparse, the data to date support a role for histone modifications in the reprogramming of biological systems by alcohol.
Noncoding RNA are associated with FASD phenotypes
Another layer of epigenetic regulation is mediated through ncRNAs, which are distinct from messenger RNA (mRNA) in that they are not translated into protein. These perform a wide variety of regulatory functions, ranging from the regulation of mRNA and protein levels to the repression of repetitive and transposable elements. For instance, micro-RNAs (miRNA) inhibit the translation of mRNA into protein, modulating the levels of different factors in the cell. By contrast, small nuclear RNA and small nucleolar (snoRNA) are involved in the post-transcriptional control of mRNA, mediating alternative splicing events and promoting post-transcriptional modifications of different RNA species. Finally, long non-coding RNAs (lncRNA) are emerging as key regulators of the cell, as they provide an essential layer of epigenetic control for several different processes. Specifically, lncRNAs act as guides for epigenetic repression or activation, decoys for chromatin modifying enzymes or RNAi pathways, and regulators of post-transcriptional RNA processing [102–106]. These transcripts also play important roles in the integration of developmental, spatial, temporal and stimulus-specific cues necessary for the integration of complex gene expression and epigenetic patterns required during development [106]. In the CNS, this additional level of epigenetic control appears to play a crucial role, as neurons express very high levels of ncRNAs, and the abnormal expression of several ncRNA species has been associated with various neurological disorders, including autism, Fragile X, Rett syndrome, schizophrenia and anxiety-like disorder [107–116].
Emerging evidence places ncRNA signaling at the nexus of brain development and alcohol-induced neurodevelopmental deficits. In particular, miRNAs have recently become an active area of research, particularly in the context of alcohol-induced neuroapoptosis and potential biomarkers of alcohol exposure [22,69,73,117–125]. Table 3 outlines the studies assessing miRNA expression patterns in response to developmental alcohol exposure. In contrast to DNA methylation and histone modifications, the first study of ncRNA was performed in vitro, using cultured neuroepithelial cells isolated from second-trimester mouse embryos [117]. These showed several differentially expressed miRNAs in response to ethanol treatment, including the suppression of miR-9, a crucial regulator of neurogenesis [117]. High doses of ethanol also cause levels of miR-9, miR-29a and miR-29b to decrease in primary cultures of cerebellar granule neurons isolated from PN 7 mice [125]. Importantly, miR-29b appears to mediate ethanol-induced apoptosis in this model and as a developmentally-regulated gene, could be involved in the cerebellum’s period of alcohol sensitivity [125]. Furthermore, the decrease in miR-9 expression was replicated in murine neural stem cells, where it is associated with increased CpG dinucleotide methylation at the miR-9–2 locus, highlighting the interplay among the different layers of epigenomic regulation [121]. Chronic intermittent exposure of ethanol also causes widespread alterations in miRNA expression profiles in primary neuronal cells from GD 15 mice [126]. Interestingly, removal of ethanol in this model altered the expression of miRNAs involved in neuron differentiation, embryonic development and synaptic plasticity, suggesting that alcohol exposure can permanently reprogram the miRNome, and importantly, that withdrawal might also profoundly alter the epigenome of the cell [126].
Table 3. . Micro-RNA studies of developmental alcohol exposure.
|
Exposure paradigm |
Model |
Effects |
Ref. |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Level | Treatment | Time | Species | Tissue | Culture | Age | Sex | Dir. | Target (miR-) | ||
| A |
320 mg/dl |
5 days |
GD 12.5 |
Mouse |
Cerebral cortical NPC |
Y |
NA |
ND |
↓ |
9, 21, 153, 335 |
[117] |
| L to A |
2.0, 4.0 or 6.0 g/kg (2× at 0 and 2 h) |
Gavage |
GD 6–15 |
Mouse |
Fetal brain |
N |
GD 17.5 |
ND |
↑ |
9, 10a, 10b, 30a-3p, 145, 152 |
[118] |
| |
|
|
|
|
|
|
|
|
↓ |
29c, 30e-5p, 154, 200a, 296, 339, 362, 496 |
|
| A |
1 or 1.5% ethanol |
NA |
4–24 hpf |
Zebrafish |
Embryo |
N |
24 hpf |
ND |
↑ |
30d, 153a, 183, 736 |
[119] |
| |
|
|
|
|
|
|
|
|
↓ |
23a |
|
| A |
320 mg/dl |
5 days |
GD 12.5 |
Mouse |
Cerebral cortical NPC |
Y |
NA |
ND |
↓ |
9, 21, 153, 335, 140–3p |
[22] |
| A |
2.5 g/kg (2× at 0 and 2 h) |
Subcut. injection |
PN 4, 7 |
Mouse |
Whole brain |
N |
PN 60 |
Male |
↑ |
26b |
[120] |
| A |
430 mM |
NA |
3.5–7.5 hpf |
Zebrafish |
Embryo |
N |
24, 48, 72 hpf |
Male/female |
↑ |
pre-9–3 (precursor to 9) |
[121] |
| |
|
|
|
|
|
|
|
|
↓ |
9 |
|
| M |
10% v/v ethanol |
Free drinking |
GD 1 - PN 10 |
Mouse |
Whole brain |
N |
PN 70 |
Male |
↑ |
679–5p |
[73] |
| A |
400 mg/dl |
24 or 48 h |
PN 7 |
Mouse |
Cerebellar granule neurons |
Y |
PN 7 |
ND |
↑ |
34a, 181a |
[125] |
| |
|
|
|
|
|
|
|
|
↓ |
9, 29a, 29b, 133 |
|
| A |
2.5 g/kg (2× at 0 and 2 h) |
Subcut injection |
GD 14, 16 |
Mouse |
Whole brain |
N |
PN 70 |
Male |
↑ |
302c |
[122] |
| A |
2.9 g/kg at 0 h 1.45 g/kg at 2 h |
IP injection |
GD 12 |
Rat |
Amygdala and ventral striatum |
N |
PN 42 |
Male/female |
↑ |
34c, 155, 542–1, let-7c-1, let7c-2–3p |
[123] |
| |
|
|
|
|
|
|
|
|
↓ |
9a-2, 29c-3p, 191–1, 138–2, 221–5p, 322–2, 384–5p, 412–3p, 496, 874–5p 1843a-3p |
|
| A |
1.75 g/kg (1 h/day × 3 days/week) |
Intravenous catheter |
GD 4–132 |
Sheep |
Plasma |
N |
GD 147 |
Male/female |
↑ |
34b |
[124] |
| |
|
|
|
|
|
|
|
|
↓ |
9, 15b, 17–92, 572, 720 |
|
| M |
10% v/v ethanol |
Free drinking |
GD 0.5–8.5 |
Mouse |
Hippocampus |
N |
PN 87 |
Male |
↑ |
135a, 135b, 467b-5p, 487b |
[69] |
| Heavily prenatally exposed and affected children | Human | Maternal plasma | N | NA | Female | ↑ | 187–5p, 204–5p, 222–5p, 299–3p, 449, 491–3p, 518f-3p, 519a-3p, 671–5-. 760, 885–3p | [129] | |||
A: Acute; GD: Gestational day; L: Low; Hpf: hours past fertilization; M: Moderate; NA: Not applicable; ND: Not disclosed; NPC: Neural progenitor cell; PN: Postnatal day.
In vivo rodent models of PAE have validated many of these findings and identified additional miRNAs associated with alcohol exposure. For instance, exposure to high levels of alcohol during early postnatal life decreases miR-29b expression in the mouse cerebellum, consistent with the observations from cultured cerebellar granule neurons [125]. Additionally, alcohol exposure from the mid-first to mid-second trimesters leads to widespread changes in miRNA levels in E17.5 mouse brains, including the upregulation of miR-10a and -10b, which regulate Hoxa1, a key factor during embryonic development [118]. This finding was partially replicated in adult mice, as acute alcohol exposure on GD14 and 16 also causes alterations to miRNA expression in the PN70 mouse brain, including miR-10b. This suggests that PAE may cause long-term reprogramming of ncRNA patterns, which may play a role in the functional deficits induced by alcohol [122].
Perhaps the best characterized miRNA in the context of alcohol exposure, miR-9 displays differential expression following developmental alcohol exposure across multiple models and ages. In addition to the in vitro models mentioned above, this finding has been replicated in both zebrafish embryos and newborn lambs [121,124]. As miR-9 plays a key role in both the developing and adult brain as a vital regulator of neural function and development, its altered expression patterns could potentially have lasting effects on physiological functions [127]. In vivo knockout of miR-9 in zebrafish results in the same juvenile swimming phenotype as that in alcohol-treated animals [121,128]. Of note, knockdown of miR-153c also results in this phenotype, and thus it is possible that this miRNA is implicated in the facial dysmorphisms observed in FASD due to its role in the development of the craniofacial skeleton [128]. Finally, the ovine model of alcohol exposure also identified a number of plasma miRNA in pregnant ewes and newborn lambs exposed to alcohol, including miR-9, miR-15b, miR-19b and miR-20a [124]. Given their high sensitivity and specificity for alcohol exposure, these miRNAs may represent potential biomarkers of alcohol exposure or key mediators of alcohol-induced epigenetic reprogramming. Indeed, a recent study showed that changes in plasma miRNAs in alcohol-exposed pregnant mothers could predict infant outcomes either alone or in conjunction with other clinical variables [129]. These findings suggest that maternal plasma miRNAs may be useful to classify difficult-to-diagnose FASD subpopulations, although further validation in independent cohorts is required.
In contrast to miRNA, few other ncRNA have been assessed in different models of developmental alcohol exposure. It was shown that first, second or third trimester equivalent exposure to alcohol increases MBII-52-specific snoRNA expression in the PN70 mouse brain [73]. This finding represents the only evidence of PAE-induced alterations to snoRNA expression to date and represents a promising avenue of research. Moreover, only a single differentially expressed lncRNA following developmental alcohol exposure, linc1354, has been identified to date. Associated with neural stem cell differentiation, this lncRNA shows decreased levels in neurosphere stem cells following ethanol exposure, supporting a role for lncRNAs in FASD [20]. While a wide variety of additional ncRNA species have not yet been assessed in PAE models, they may provide important functional insight given their key role in neural functions, and represent an important avenue of research in the study of epigenetic mechanisms in FASD. Finally, covalent modifications to mRNA and different ncRNA species are emerging as important regulatory mechanisms in the brain. In particular, levels of N6-methyladenosine increase in the prefrontal cortex of mice following behavioral training, suggesting a potential role in cognitive functions [130]. While the functional implications of these modifications remain unclear, they may prove important in alcohol-induced deficits given their abundance in the brain. Overall, ncRNA appears to play important roles in the development of deficits associated with PAE, including facial dysmorphisms, neuroapoptosis and altered neurodevelopment, supporting a vital role in the etiology of deficits associated with FASD.
Important considerations for epigenetic studies of FASD
Although much headway has been made in characterizing the epigenetic patterns associated with developmental alcohol exposure and their role in fetal programming, a number of key considerations will be crucial for the next wave of epigenetic studies in FASD. First, most studies of alcohol exposure focus exclusively on male animals or do not highlight sex-specific differences, an issue found throughout many research fields and recently highlighted by the new funding guidelines from the National Institutes of Health [131]. Since epigenetic patterns are highly associated with sex, this further reduces the generalizability and applicability of findings from animal models of PAE to clinical settings [132]. This is particularly relevant to the domain of FASD, as males typically display different cognitive and behavioral phenotypes, as well as differential susceptibilities to stressors and mental health disorders compared with their female counterparts [133–135]. As such, the paucity of data on females in the FASD research field must be addressed in order to fully assess the role of epigenetics in the etiology of alcohol-induced deficits. Given the wide variety of PAE models, we must also begin to integrate findings from different models of exposure, which vary in terms of dosage (low to high), pattern of exposure (acute or chronic), trimester of exposure and type of ethanol administration, to identify the most robust epigenetic signatures of PAE (for a review on the different models of developmental alcohol exposure, please refer to [136]).
Additionally, a large portion of whole-genome analyses of epigenomic patterns have been performed either in cell culture or whole brains, which does not necessarily reflect the downstream functional implications of alcohol-induced alterations. Future studies should begin to assess changes within specific brain structures and primary tissues to further dissect the role of epigenetics in the various deficits associated with developmental alcohol exposure. Cell type differences must also be taken into account when analyzing epigenetic data, as cell type composition is a major driver of epigenetic patterns and may be altered by alcohol exposure. Various strategies can be used to address this issue, including the isolation of single cell types prior to epigenetic analyses, the inclusion of cell type proportions in statistical models, or bioinformatic methods such as cell type deconvolution and surrogate variable analysis. In addition, robust statistical methods must be used in the analysis of genome-wide alterations to prevent spurious associations with alcohol exposure. These considerations include the use of multiple-test correction and other methods to correct for discrepancies between groups (age, ethnicity, smoking, etc.), which tend to occur frequently in population studies of epigenetic mechanisms. Of note, the phenotypes associated with FASD have been rather heterogeneous in human studies, and possibly confounded with genetic ancestry, highlighting our need for large and diverse cohorts to tease apart the often subtle influences of PAE on the epigenome and identify critical periods of vulnerability.
Finally, to fully assess the role of epigenetic mechanisms in PAE-induced associated physiological functions, we must begin to integrate the multiple layers of epigenetic machinery, from chromatin alterations and DNA methylation to miRNA and lncRNA expression [39]. Future studies should also assess the concordance of these changes with mRNA expression, as the relationship between epigenetic patterns and transcription is highly complex and has yet to be fully elucidated. Although some studies of PAE have already begun to fill this niche, much work is needed before we can successfully integrate the multiple layers of genome-wide epigenomic regulation in the etiology of FASD [68,69].
Future perspective
Although the study of epigenetic patterns following PAE is progressing at a rapid rate, a number of key issues remain elusive in regard to both mechanisms of fetal programming and biomarkers of FASD. For one, early evidence from some groups suggests that developmental alcohol exposure could potentially have lasting impacts on the epigenome of future generations, suggesting a possible role for inter- or transgenerational epigenetic inheritance [91]. While these data are certainly intriguing and raise important ethical considerations in the study and prevention of FASD, they must be interpreted with relative caution due to severe limitations in studying such effects. First, the interpretation of these results must take into consideration the number of generations to determine whether they are considered inter or transgenerational, which are commonly confounded due to the presence of cells for the F2 generation in the pregnant F0 female [137]. Second, these studies were largely performed in rodent models, which have not yet been shown to display the same inheritance patterns as humans. Third, no cohorts are currently available for the study of transgenerational inheritance in humans, and the current evidence remains tenuous at best. Nevertheless, although much work must be done to fully assess the implications of inter- or transgenerational epigenetic inheritance in FASD, this remains an intriguing and important area of research that certainly warrants further investigation.
Another issue facing the field is that as of yet, and perhaps not surprisingly at this time, the vast majority of epigenetic studies rely on correlation, rather than causation. More specifically, even though epigenetic mechanisms are correlated with PAE in cross-sectional clinical cohorts and animal models, it is not yet clear whether their reversal would dampen PAE phenotypes, which would indicate a more causal role. Moreover, although epigenetic patterns appear to associate with changes in transcription, the majority of these relationships show inconsistent results across studies and the functional implications of epigenetic changes have yet to be fully established [45,138]. As such, prior to making inferences concerning cognitive, behavioral or physiological outcomes from alcohol-induced epigenetic alterations, we must first establish a direct line of evidence between epigenetic patterns, gene expression profiles, and the phenotype in question, either through genetic manipulation of model organisms or therapeutic interventions. Given that different environmental factors have been shown to modulate PAE-induced deficits, including stress, immune challenges, or early-life adversity, futures studies must also begin to address the differences and similarities between basal and inducible alterations to gene expression and epigenetic patterns. Model organisms, such as mice, rats, zebrafish, and others will play a crucial role in addressing this issue, as they allow for finer manipulations of biological systems and tighter control of environmental conditions. Perhaps most importantly, we must begin to position epigenetic mechanisms at the nexus of exposure paradigms and phenotypic outcomes to provide better insight into the etiology of FASD. Furthermore, analysis of both central and peripheral tissues in animal models will be vital before we can begin to make functional inferences in clinical models, as human epigenetic studies mainly rely on peripheral tissues such as BEC and blood.
Although the degree to which peripheral alterations are linked to the mechanisms underlying FASD remains unknown, they may present a unique opportunity to develop accurate epigenetic biomarkers of PAE. In many cases, the deficits associated with FASD only become evident long after exposure, highlighting the importance of early biomarkers as tools to identify at-risk children and mitigate the long-term effects of alcohol. More recent studies in animal models and clinical populations of individuals with FASD are beginning to provide a solid foundation for biomarker discovery with hopes for definitive markers in the relatively near future. Of the utmost importance in this line of research are additional studies to validate current findings and to begin to assess the accuracy and specificity of these types of markers. While a characteristic epigenomic signature appears to occur in the buccal cells of children with FASD, this finding requires additional validation and testing in a clinical setting [78]. Furthermore, strong correlations have been identified between genetic background and epigenetic patterns, particularly in the case of gene by environment (GxE) interactions [139–141]. Given the challenges in obtaining cohorts of children with homogenous ethnicities, it will be vital to assess the relevance and implications of methylation quantitative trait loci or allelic variants correlating with nearby CpG methylation levels in the context of FASD. Longitudinal studies will also be integral to the identification of PAE-associated alterations to epigenetic profiles, as cross-sectional studies may not fully reflect the diversity of individuals with FASD across development and aging. Importantly, the field must also begin to move beyond early life outcomes and extend its focus into adolescence and adulthood, as data on adolescents and adults with FASD remains sparse. These studies will further develop a role for altered epigenetic programming in FASD and long-term health outcomes, be they immune, neurological or stress-related [15]. In addition, these may prove crucial to our understanding of the etiology of FASD, particularly given the relationship between aging, disease, and DNA methylation [142]. These longitudinal cohorts will also be necessary to assess the persistence of epigenetic reprogramming by PAE and the potential validity of biomarkers over time. Epigenetic profiles may also serve as a better marker of FASD if they are developed in conjunction with different stratification tools, such as MRI, eye tracking, physical and mental health diagnostics, and immune markers, to parse out the wide range of deficits associated with FASD and create more accurate diagnostic tools. Finally, we must also begin to assess the overlaps, or lack thereof, in epigenetic patterns among different neurodevelopmental disorders, as they may display similar deficits and share common or overlapping molecular etiologies [143]. The integration of these findings will provide important insight into the root cause of these disorders and may provide additional strategies for both diagnostic tools and therapeutic interventions.
Conclusion
Despite the recognition of FAS over 40 years ago, PAE remains a leading cause of developmental disability in the developed world, as recent North American estimates place the incidence between 2 and 5% [144–148]. However, early identification of individuals with FASD remains difficult, limiting the effectiveness of current interventions, which still lack specific molecular or neurobiological targets [149]. Although the study of epigenetic patterns in FASD remains a emerging field, it has provided important contributions to our understanding of the molecular underpinnings of FASD. To date, epigenetic research has identified numerous alterations in DNA methylation patterns, chromatin states and ncRNA expression levels, which provide important biological insight into the deficits associated with FASD, while also potentially uncovering targets for therapeutic intervention. This work has begun to lay the groundwork for the development of epigenetic biomarkers of PAE, which may be the key to identifying children at risk for FASD. In turn, the identification of valid biomarkers will eventually support the creation of strategies for earlier diagnoses and targeted interventions to improve the lives of children and families affected by FASD.
Executive summary.
Prenatal alcohol exposure (PAE) reprograms physiological systems, which may increase vulnerability to adverse behavioral and health outcomes.
The fetal programming effects of PAE could be explained by alterations to epigenetic mechanisms, such as DNA methylation, chromatin modifications and noncoding RNA.
Considerations for future epigenetic studies of fetal alcohol spectrum disorder (FASD) include the assessment of sex differences, reconciliation of different exposure paradigm and integration of multiple layers of epigenetic regulation.
Longitudinal studies of individuals with FASD will be imperative to identify potential inter- and trans-generational inheritance patterns, to elucidate the persistence of deficits in adolescents or adults and to develop of diagnostic tools for FASD.
Animal models will be crucial to establishing a causal link between epigenetic alterations, gene expression and the deficits caused by PAE.
In conjunction with different stratification tools, such as MRI, eye tracking, physical and mental health diagnostics, and immune markers, epigenetic mechanisms may prove key to the development of accurate diagnostic tools for FASD.
Acknowledgements
The authors thank NeuroDevNet for continuous encouragement and support for this work. They would also like to thank N Lan, P Holman and V Lam from J Weinberg’s lab; M Jones, S Moore and S Islam from MS Kobor’s lab; and P Lu for their input on this article.
Footnotes
Financial & competing interests disclosure
The work in J Weinberg’s laboratory is supported by grants from NIAAA (R37 AA007789, ROI AA022460, 5U24AA014811 [CIFASD subaward]) and NeuroDevNet (Canadian Networks of Centres of Excellence). MS Kobor is a Senior Fellow of the Canadian Institute For Advanced Research and the Canada Research Chair in Social Epigenetics. AA Lussier is supported by a Developmental Neurosciences Research Training Award from Brain Canada and NeuroDevNet. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Godfrey K, Robinson S. Maternal nutrition, placental growth and fetal programming. Proc. Nutr. Soc. 1998;57(1):105–111. doi: 10.1079/pns19980016. [DOI] [PubMed] [Google Scholar]
- 2.Hanson MA, Gluckman PD. Developmental origins of health and disease: new insights. Basic Clin. Pharmacol. Toxicol. 2008;102(2):90–93. doi: 10.1111/j.1742-7843.2007.00186.x. [DOI] [PubMed] [Google Scholar]
- 3.Meaney MJ. Epigenetics and the biological definition of gene X environment interactions. Child Dev. 2010;81(1):41–79. doi: 10.1111/j.1467-8624.2009.01381.x. [DOI] [PubMed] [Google Scholar]
- 4.Boyce WT, Kobor MS. Development and the epigenome: the “synapse” of gene-environment interplay. Dev. Sci. 2015;18(1):1–23. doi: 10.1111/desc.12282. [DOI] [PubMed] [Google Scholar]; • Provides an overview of the epigenetics mechanisms and their potential interactions with the environment.
- 5.Hoyme HE, Kalberg WO, Elliott AJ, et al. Updated clinical guidelines for diagnosing fetal alcohol spectrum disorders. Pediatrics. 2016;138(2) doi: 10.1542/peds.2015-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stratton K, Howe C, Battaglia F. Fetal Alcohol Syndrome: Diagnosis, Epidemiology, Prevention and Treatment. National Academy Press; Washington, DC: [Google Scholar]
- 7.Jones KL, Smith DW. Recognition of the fetal alcohol syndrome in early infancy. Lancet. 1973;302(7836):999–1001. doi: 10.1016/s0140-6736(73)91092-1. [DOI] [PubMed] [Google Scholar]
- 8.Pollard I. Neuropharmacology of drugs and alcohol in mother and fetus. Semin. Fetal Neonatal Med. 2007;12(2):106–113. doi: 10.1016/j.siny.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Carter RC, Jacobson JL, Molteno CD, et al. Fetal alcohol growth restriction and cognitive impairment. Pediatrics. 2016;138(2):176192. doi: 10.1542/peds.2016-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynch ME, Kable JA, Coles CD. Prenatal alcohol exposure, adaptive function, and entry into adult roles in a prospective study of young adults. Neurotoxicol. Teratol. 2015;51:52–60. doi: 10.1016/j.ntt.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panczakiewicz AL, Glass L, Coles CD, et al. Neurobehavioral deficits consistent across age and sex in youth with prenatal alcohol Exposure. Alcohol. Clin. Exp. Res. 2016;40(9):1971–1981. doi: 10.1111/acer.13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doyle LR, Mattson SN. Neurobehavioral disorder associated with prenatal alcohol exposure (ND-PAE): review of evidence and guidelines for assessment. Curr. Dev. Disord. Rep. 2015;2(3):175–186. doi: 10.1007/s40474-015-0054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Astley SJ, Olson HC, Kerns K, et al. Neuropyschological and behavioral outcomes from a comprehensive magnetic resonance study of children with fetal alcohol spectrum disorders. Can. J. Clin. Pharmacol. 2009;16(1):e178–e201. [PMC free article] [PubMed] [Google Scholar]
- 14.Streissguth AP, O’Malley K. Neuropsychiatric implications and long-term consequences of fetal alcohol spectrum disorders. Semin. Clin. Neuropsychiatry. 2000;5(3):177–190. doi: 10.1053/scnp.2000.6729. [DOI] [PubMed] [Google Scholar]
- 15.Moore EM, Riley EP. What happens when children with fetal alcohol spectrum disorders become adults? Curr. Dev. Disord. Rep. 2015;2(3):219–227. doi: 10.1007/s40474-015-0053-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Provides important insight into the necessity to investigate fetal alcohol spectrum disorder (FASD) in adults.
- 16.Jones KL, Smith DW, Ulleland CN, Streissguth P. Pattern of malformation in offspring of chronic alcoholic mothers. Lancet. 1973;1(7815):1267–1271. doi: 10.1016/s0140-6736(73)91291-9. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Balaraman Y, Wang G, Nephew KP, Zhou FC. Alcohol exposure alters DNA methylation profiles in mouse embryos at early neurulation. Epigenetics. 2009;4(7):500–511. doi: 10.4161/epi.4.7.9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou FC, Zhao Q, Liu Y, et al. Alteration of gene expression by alcohol exposure at early neurulation. BMC Genomics. 2011;12:124. doi: 10.1186/1471-2164-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hicks SD, Middleton FA, Miller MW. Ethanol-induced methylation of cell cycle genes in neural stem cells. J. Neurochem. 2010;114(6):1767–1780. doi: 10.1111/j.1471-4159.2010.06886.x. [DOI] [PubMed] [Google Scholar]
- 20.Veazey KJ, Carnahan MN, Muller D, Miranda RC, Golding MC. Alcohol-induced epigenetic alterations to developmentally crucial genes regulating neural stemness and differentiation. Alcohol. Clin. Exp. Res. 2013;37(7):1111–1122. doi: 10.1111/acer.12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veazey KJ, Parnell SE, Miranda RC, Golding MC. Dose-dependent alcohol-induced alterations in chromatin structure persist beyond the window of exposure and correlate with fetal alcohol syndrome birth defects. Epigenetics Chromatin. 2015;8(1):39. doi: 10.1186/s13072-015-0031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balaraman S, Winzer-Serhan UH, Miranda RC. Opposing actions of ethanol and nicotine on microRNAs are mediated by nicotinic acetylcholine receptors in fetal cerebral cortical-derived neural progenitor cells. Alcohol. Clin. Exp. Res. 2012;36(10):1669–1677. doi: 10.1111/j.1530-0277.2012.01793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swanson JM, Entringer S, Buss C, Wadhwa PD. Developmental origins of health and disease: environmental exposures. Semin. Reprod. Med. 2009;27(5):391–402. doi: 10.1055/s-0029-1237427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hellemans KGC, Sliwowska JH, Verma P, Weinberg J. Prenatal alcohol exposure: fetal programming and later life vulnerability to stress, depression and anxiety disorders. Neurosci. Biobehav. Rev. 2010;34(6):791–807. doi: 10.1016/j.neubiorev.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Outlines the concept of fetal programming in the context of FASD, providing a framework for the interpretation of long-term alcohol-induced deficits.
- 25.Yuen RKC, Neumann SMA, Fok AK, et al. Extensive epigenetic reprogramming in human somatic tissues between fetus and adult. Epigenetics Chromatin. 2011;4:7. doi: 10.1186/1756-8935-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shulha HP, Cheung I, Guo Y, Akbarian S, Weng Z. Coordinated cell type-specific epigenetic remodeling in prefrontal cortex begins before birth and continues into early adulthood. PLoS Genet. 2013;9(4):e1003433. doi: 10.1371/journal.pgen.1003433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobor MS, Weinberg J. Focus on: epigenetics and fetal alcohol spectrum disorders. Alcohol Res. Health. 2011;34(1):29–37. [PMC free article] [PubMed] [Google Scholar]
- 28.Bird A. Perceptions of epigenetics. Nature. 2007;447(7143):396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 29.Ziller MJ, Gu H, Müller F, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500(7463):477–81. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleiber ML, Laufer BI, Stringer RL, Singh SM. Third trimester-equivalent ethanol exposure is characterized by an acute cellular stress response and an ontogenetic disruption of genes critical for synaptic establishment and function in mice. Dev. Neurosci. 2014;36(6):499–519. doi: 10.1159/000365549. [DOI] [PubMed] [Google Scholar]
- 31.Kleiber ML, Laufer BI, Wright E, Diehl EJ, Singh SM. Long-term alterations to the brain transcriptome in a maternal voluntary consumption model of fetal alcohol spectrum disorders. Brain Res. 2012;1458:18–33. doi: 10.1016/j.brainres.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 32.Kleiber ML, Mantha K, Stringer RL, Singh SM. Neurodevelopmental alcohol exposure elicits long-term changes to gene expression that alter distinct molecular pathways dependent on timing of exposure. J. Neurodev. Disord. 2013;5(1):6. doi: 10.1186/1866-1955-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Downing C, Flink S, Florez-McClure ML, Johnson TE, Tabakoff B, Kechris KJ. Gene expression changes in C57BL/6J and DBA/2J mice following prenatal alcohol exposure. Alcohol. Clin. Exp. Res. 2012;36(9):1519–1529. doi: 10.1111/j.1530-0277.2012.01757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lussier AA, Stepien KA, Neumann SM, Pavlidis P, Kobor MS, Weinberg J. Prenatal alcohol exposure alters steady-state and activated gene expression in the adult rat brain. Alcohol. Clin. Exp. Res. 2015;39(2):251–261. doi: 10.1111/acer.12622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Green ML, Singh AV, Zhang Y, Nemeth KA, Sulik KK, Knudsen TB. Reprogramming of genetic networks during initiation of the fetal alcohol syndrome. Dev. Dyn. 2007;236(2):613–631. doi: 10.1002/dvdy.21048. [DOI] [PubMed] [Google Scholar]
- 36.Hard ML, Abdolell M, Robinson BH, Koren G. Gene-expression analysis after alcohol exposure in the developing mouse. J. Lab. Clin. Med. 2005;145(1):47–54. doi: 10.1016/j.lab.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 37.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293(5532):1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 38.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 39.Lister R, Mukamel EA, Nery JR, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341(6146):1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Assesses epigenetic changes during brain development (mouse and human), and is a prime example of integrating multiple layers of epigenomic regulation.
- 40.Guo JU, Su Y, Shin JH, et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2014;17(2):215–222. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr. Opin. Genet. Dev. 1993;3(2):226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 42.Shukla S, Kavak E, Gregory M, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479(7371):74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013;23(11):1256–1269. doi: 10.1038/cr.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maunakea AK, Nagarajan RP, Bilenky M, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466(7303):253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lam LL, Emberly E, Fraser HB, et al. Factors underlying variable DNA methylation in a human community cohort. Proc. Natl Acad. Sci. USA. 2012;109(Suppl. 2):17253–17260. doi: 10.1073/pnas.1121249109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gutierrez-Arcelus M, Lappalainen T, Montgomery SB, et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife. 2013;2(2):e00523. doi: 10.7554/eLife.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones MJ, Fejes AP, Kobor MS. DNA methylation, genotype and gene expression: who is driving and who is along for the ride? Genome Biol. 2013;14(7):126. doi: 10.1186/gb-2013-14-7-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sadakierska-Chudy A, Kostrzewa RM, Filip M. A comprehensive view of the epigenetic landscape part I: DNA methylation, passive and active DNA demethylation pathways and histone variants. Neurotox. Res. 2014;27(1):84–97. doi: 10.1007/s12640-014-9497-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Santiago M, Antunes C, Guedes M, Sousa N, Marques CJ. TET enzymes and DNA hydroxymethylation in neural development and function – how critical are they? Genomics. 2014;104(5):334–340. doi: 10.1016/j.ygeno.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 52.Alaghband Y, Bredy TW, Wood MA. The role of active DNA demethylation and Tet enzyme function in memory formation and cocaine action. Neurosci. Lett. 2016;625:40–46. doi: 10.1016/j.neulet.2016.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang T, Pan Q, Lin L, et al. Genome-wide DNA hydroxymethylation changes are associated with neurodevelopmental genes in the developing human cerebellum. Hum. Mol. Genet. 2012;21(26):5500–5510. doi: 10.1093/hmg/dds394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bock C. Epigenetic biomarker development. Epigenomics. 2009;1(1):99–110. doi: 10.2217/epi.09.6. [DOI] [PubMed] [Google Scholar]
- 55.Garro AJ, McBeth DL, Lima V, Lieber CS. Ethanol consumption inhibits fetal DNA methylation in mice: implications for the fetal alcohol syndrome. Alcohol. Clin. Exp. Res. 1991;15(3):395–398. doi: 10.1111/j.1530-0277.1991.tb00536.x. [DOI] [PubMed] [Google Scholar]
- 56.Otero NKH, Thomas JD, Saski CA, Xia X, Kelly SJ. Choline supplementation and DNA methylation in the hippocampus and prefrontal cortex of rats exposed to alcohol during development. Alcohol. Clin. Exp. Res. 2012;36(10):1701–1709. doi: 10.1111/j.1530-0277.2012.01784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perkins A, Lehmann C, Lawrence RC, Kelly SJ. Alcohol exposure during development: Impact on the epigenome. Int. J. Dev. Neurosci. 2013;31(6):391–397. doi: 10.1016/j.ijdevneu.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y, Ozturk NC, Zhou FC. DNA methylation program in developing hippocampus and its alteration by alcohol. PLoS ONE. 2013;8(3):1–11. doi: 10.1371/journal.pone.0060503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mukhopadhyay P, Rezzoug F, Kaikaus J, Greene RM, Pisano MM. Alcohol modulates expression of DNA methyltranferases and methyl CpG-/CpG domain-binding proteins in murine embryonic fibroblasts. Reprod. Toxicol. 2013;37:40–48. doi: 10.1016/j.reprotox.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagre NN, Subbanna S, Shivakumar M, Psychoyos D, Basavarajappa BS. CB1-receptor knockout neonatal mice are protected against ethanol-induced impairments of DNMT1, DNMT3A, and DNA methylation. J. Neurochem. 2015;132(4):429–442. doi: 10.1111/jnc.13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liyanage VRB, Zachariah RM, Davie JR, Rastegar M. Ethanol deregulates Mecp2/MeCP2 in differentiating neural stem cells via interplay between 5-methylcytosine and 5-hydroxymethylcytosine at the Mecp2 regulatory elements. Exp. Neurol. 2015;265:102–117. doi: 10.1016/j.expneurol.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas JD, Biane JS, O’Bryan KA, O’Neill TM, Dominguez HD. Choline supplementation following third-trimester-equivalent alcohol exposure attenuates behavioral alterations in rats. Behav. Neurosci. 2007;121(1):120–130. doi: 10.1037/0735-7044.121.1.120. [DOI] [PubMed] [Google Scholar]
- 63.Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12(11):949–957. [PubMed] [Google Scholar]
- 64.Kaminen-Ahola N, Ahola A, Maga M, et al. Maternal ethanol consumption alters the epigenotype and the phenotype of offspring in a mouse model. PLoS Genet. 2010;6(1):e1000811. doi: 10.1371/journal.pgen.1000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vallés S, Pitarch J, Renau-Piqueras J, Guerri C. Ethanol exposure affects glial fibrillary acidic protein gene expression and transcription during rat brain development. J. Neurochem. 1997;69:2484–2493. doi: 10.1046/j.1471-4159.1997.69062484.x. [DOI] [PubMed] [Google Scholar]
- 66.Maier SE, Cramer JA, West JR, Sohrabji F. Alcohol exposure during the first two trimesters equivalent alters granule cell number and neurotrophin expression in the developing rat olfactory bulb. J. Neurobiol. 1999;41(3):414–423. doi: 10.1002/(sici)1097-4695(19991115)41:3<414::aid-neu9>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 67.Downing C, Johnson TE, Larson C, et al. Subtle decreases in DNA methylation and gene expression at the mouse Igf2 locus following prenatal alcohol exposure: effects of a methyl-supplemented diet. Alcoholism. 2011;45(1):65–71. doi: 10.1016/j.alcohol.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bekdash RA, Zhang C, Sarkar DK. Gestational choline supplementation normalized fetal alcohol-induced alterations in histone modifications, DNA methylation, and proopiomelanocortin (POMC) gene expression in??-endorphin-producing POMC neurons of the hypothalamus. Alcohol. Clin. Exp. Res. 2013;37(7):1133–1142. doi: 10.1111/acer.12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang CR, Ho M-F, Vega MCS, Burne THJ, Chong S. Prenatal ethanol exposure alters adult hippocampal VGLUT2 expression with concomitant changes in promoter DNA methylation, H3K4 trimethylation and miR-467b-5p levels. Epigenetics Chromatin. 2015;8(1):40. doi: 10.1186/s13072-015-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ngai YF, Sulistyoningrum DC, O’Neill R, Innis SM, Weinberg J, Devlin AM. Prenatal alcohol exposure alters methyl metabolism and programs serotonin transporter and glucocorticoid receptor expression in brain. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015;309(5):R613–R622. doi: 10.1152/ajpregu.00075.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marjonen H, Sierra A, Nyman A, et al. Early maternal alcohol consumption alters hippocampal DNA methylation, gene expression and volume in a mouse model. PLoS ONE. 2015;10(5):e0124931. doi: 10.1371/journal.pone.0124931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou FC, Chen Y, Love A. Cellular DNA methylation program during neurulation and its alteration by alcohol exposure. Birth Defects Res. Part A - Clin. Mol. Teratol. 2011;91(8):703–715. doi: 10.1002/bdra.20820. [DOI] [PubMed] [Google Scholar]
- 73.Laufer BI, Mantha K, Kleiber ML, Diehl EJ, Addison SMF, Singh SM. Long-lasting alterations to DNA methylation and ncRNAs could underlie the effects of fetal alcohol exposure in mice. Dis. Model. Mech. 2013;6(4):977–92. doi: 10.1242/dmm.010975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krishnamoorthy M, Gerwe BA, Scharer CD, et al. Ethanol alters proliferation and differentiation of normal and chromosomally abnormal human embryonic stem cell-derived neurospheres. Birth Defects Res. Part B Dev. Reprod. Toxicol. 2013;98(3):283–295. doi: 10.1002/bdrb.21063. [DOI] [PubMed] [Google Scholar]
- 75.Khalid O, Kim JJ, Kim HS, et al. Gene expression signatures affected by alcohol-induced DNA methylomic deregulation in human embryonic stem cells. Stem Cell Res. 2014;12(3):791–806. doi: 10.1016/j.scr.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chater-Diehl EJ, Laufer BI, Castellani CA, Alberry BL, Singh SM. Alteration of gene expression, DNA methylation, and histone methylation in free radical scavenging networks in adult mouse hippocampus following fetal alcohol exposure. PLoS ONE. 2016;11(5):e0154836. doi: 10.1371/journal.pone.0154836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Laufer BI, Kapalanga J, Castellani CA, Diehl EJ, Yan L, Singh SM. Associative DNA methylation changes in children with prenatal alcohol exposure. Epigenomics. 2015;7(8):1259–1274. doi: 10.2217/epi.15.60. [DOI] [PubMed] [Google Scholar]
- 78.Portales-Casamar E, Lussier AA, Jones MJ, et al. DNA methylation signature of human fetal alcohol spectrum disorder. Epigenetics Chromatin. 2016;9(25):81–101. doi: 10.1186/s13072-016-0074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Identifies a characteristic DNA methylation signature of prenatal alcohol exposure from the largest cohort of children with FASD to date.
- 79.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 80.Luger K, Mäder AW, Richmond RK, Sargent DF. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 81.Venkatesh S, Smolle M, Li H, et al. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2013;489(7416):452–455. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
- 82.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 83.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bowman GD, Poirier MG. Post-translational modifications of histones that influence nucleosome dynamics. Chem. Rev. 2015;115(6):2274–2295. doi: 10.1021/cr500350x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Du Q, Luu P-L, Stirzaker C, Clark SJ. Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics. 2015;7(6):1051–1073. doi: 10.2217/epi.15.39. [DOI] [PubMed] [Google Scholar]
- 86.Bogdanović O, Veenstra GJC. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 2009;118(5):549–565. doi: 10.1007/s00412-009-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 88.Ramadoss J, Liao WX, Chen DB, Magness RR. High-throughput caveolar proteomic signature profile for maternal binge alcohol consumption. Alcoholism. 2010;44(7–8):691–697. doi: 10.1016/j.alcohol.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guo W, Crossey EL, Zhang L, et al. Alcohol exposure decreases CREB binding protein expression and histone acetylation in the developing cerebellum. PLoS ONE. 2011;6(5) doi: 10.1371/journal.pone.0019351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhong L, Zhu J, Lv T, et al. Ethanol and its metabolites induce histone lysine 9 acetylation and an alteration of the expression of heart development-related genes in cardiac progenitor cells. Cardiovasc. Toxicol. 2010;10(4):268–274. doi: 10.1007/s12012-010-9081-z. [DOI] [PubMed] [Google Scholar]
- 91.Govorko D, Bekdash RA, Zhang C, Sarkar DK. Male germline transmits fetal alcohol adverse effect on hypothalamic proopiomelanocortin gene across generations. Biol. Psychiatry. 2012;72(5):378–388. doi: 10.1016/j.biopsych.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Subbanna S, Shivakumar M, Umapathy NS, et al. G9a-mediated histone methylation regulates ethanol-induced neurodegeneration in the neonatal mouse brain. Neurobiol. Dis. 2013;54:475–485. doi: 10.1016/j.nbd.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Subbanna S, Nagre NN, Shivakumar M, Umapathy NS, Psychoyos D, Basavarajappa BS. Ethanol induced acetylation of histone at G9a exon1 and G9a-mediated histone H3 dimethylation leads to neurodegeneration in neonatal mice. Neuroscience. 2014;258:422–432. doi: 10.1016/j.neuroscience.2013.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang W, Peng C, Zheng M, et al. Prenatal alcohol exposure causes the over-expression of DHAND and EHAND by increasing histone H3K14 acetylation in C57 BL/6 mice. Toxicol. Lett. 2014;228(3):140–146. doi: 10.1016/j.toxlet.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 95.Peng C, Zhu J, Sun HC, et al. Inhibition of histone H3K9 acetylation by anacardic acid can correct the over-expression of Gata4 in the hearts of fetal mice exposed to alcohol during pregnancy. PLoS ONE. 2014;9(8):e104135. doi: 10.1371/journal.pone.0104135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chung H-Y, Chang C-T, Young H-W, Hu SP, Tzou W-S, Hu C-H. Ethanol inhibits retinal and CNS differentiation due to failure of cell cycle exit via an apoptosis-independent pathway. Neurotoxicol. Teratol. 2013;38:92–103. doi: 10.1016/j.ntt.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 97.Goldowitz D, Lussier AA, Boyle JK, et al. Molecular pathways underpinning ethanol-induced neurodegeneration. Front. Genet. 2014;5:203. doi: 10.3389/fgene.2014.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Subbanna S, Basavarajappa BS. Pre-administration of G9a/GLP inhibitor during synaptogenesis prevents postnatal ethanol-induced LTP deficits and neurobehavioral abnormalities in adult mice. Exp. Neurol. 2014;261:34–43. doi: 10.1016/j.expneurol.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Subbanna S, Nagre NN, Umapathy NS, Pace BS, Basavarajappa BS. Ethanol exposure induces neonatal neurodegeneration by enhancing CB1R Exon1 Histone H4K8 acetylation and up-regulating CB1R function causing neurobehavioral abnormalities in adult mice. Int. J. Neuropsychopharmacol. 2015;18(5):1–15. doi: 10.1093/ijnp/pyu028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim P, Park JH, Choi CS, et al. Effects of ethanol exposure during early pregnancy in hyperactive, inattentive and impulsive behaviors and MeCP2 expression in rodent offspring. Neurochem. Res. 2013;38(3):620–631. doi: 10.1007/s11064-012-0960-5. [DOI] [PubMed] [Google Scholar]
- 101.Dasmahapatra AK, Khan IA. Modulation of DNA methylation machineries in Japanese rice fish (Oryzias latipes) embryogenesis by ethanol and 5-azacytidine. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2016;179:174–183. doi: 10.1016/j.cbpc.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 102.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013;14(11):699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Orom UA, Derrien T, Beringer M, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in Human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 2011;12(8):542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- 106.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kapranov P, St Laurent G, Raz T, et al. The majority of total nuclear-encoded non-ribosomal RNA in a human cell is “dark matter” un-annotated RNA. BMC Biol. 2010;8:149. doi: 10.1186/1741-7007-8-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gustincich S, Sandelin A, Plessy C, et al. The complexity of the mammalian transcriptome. J. Physiol. 2006;575(2):321–332. doi: 10.1113/jphysiol.2006.115568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Spadaro PA, Bredy TW. Emerging role of non-coding RNA in neural plasticity, cognitive function, and neuropsychiatric disorders. Front. Genet. 2012;3:1–16. doi: 10.3389/fgene.2012.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Williams JM, Beck TF, Pearson DM, Proud MB, Sau WC, Scott DA. A 1q42 deletion involving DISC1, DISC2, and TSNAX in an autism spectrum disorder. Am. J. Med. Genet. Part A. 2009;149(8):1758–1762. doi: 10.1002/ajmg.a.32941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ziats MN, Rennert OM. Aberrant expression of long noncoding RNAs in autistic brain. J. Mol. Neurosci. 2013;49(3):589–593. doi: 10.1007/s12031-012-9880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pastori C, Peschansky VJ, Barbouth D, Mehta A, Silva JP, Wahlestedt C. Comprehensive analysis of the transcriptional landscape of the human FMR1 gene reveals two new long noncoding RNAs differentially expressed in Fragile X syndrome and Fragile X-associated tremor/ataxia syndrome. Hum. Genet. 2014;133(1):59–67. doi: 10.1007/s00439-013-1356-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Petazzi P, Sandoval J, Szczesna K, et al. Dysregulation of the long non-coding RNA transcriptome in a Rett syndrome mouse model. RNA Biol. 2013;10(7):1197–1203. doi: 10.4161/rna.24286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Millar JK, Wilson-Annan JC, Anderson S, et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 2000;9(9):1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 115.Barry G, Briggs J, Vanichkina D, et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry. 2013;19:486–494. doi: 10.1038/mp.2013.45. [DOI] [PubMed] [Google Scholar]
- 116.Spadaro PA, Flavell CR, Widagdo J, et al. Long noncoding RNA-directed epigenetic regulation of gene expression is associated with anxiety-like behavior in mice. Biol. Psychiatry. 2015;78(12):848–859. doi: 10.1016/j.biopsych.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sathyan P, Golden HB, Miranda RC. Competing interactions between micro-RNAs determine neural progenitor survival and proliferation after ethanol exposure: evidence from an ex vivo model of the fetal cerebral cortical neuroepithelium. J. Neurosci. 2007;27(32):8546–8557. doi: 10.1523/JNEUROSCI.1269-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang L-L, Zhang Z, Li Q, et al. Ethanol exposure induces differential microRNA and target gene expression and teratogenic effects which can be suppressed by folic acid supplementation. Hum. Reprod. 2009;24(3):562–579. doi: 10.1093/humrep/den439. [DOI] [PubMed] [Google Scholar]
- 119.Soares AR, Pereira PM, Ferreira V, et al. Ethanol exposure induces upregulation of specific microRNAs in zebrafish embryos. Toxicol. Sci. 2012;127(1):18–28. doi: 10.1093/toxsci/kfs068. [DOI] [PubMed] [Google Scholar]
- 120.Stringer RL, Laufer BI, Kleiber ML, Singh SM. Reduced expression of brain cannabinoid receptor 1 (Cnr1) is coupled with an increased complementary micro-RNA (miR-26b) in a mouse model of fetal alcohol spectrum disorders. Clin. Epigenetics. 2013;5(1):14. doi: 10.1186/1868-7083-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pappalardo-Carter DL, Balaraman S, Sathyan P, Carter ES, Chen WJA, Miranda RC. Suppression and epigenetic regulation of MiR-9 contributes to ethanol teratology: Evidence from zebrafish and murine fetal neural stem cell models. Alcohol. Clin. Exp. Res. 2013;37(10):1657–1667. doi: 10.1111/acer.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mantha K, Laufer BI, Singh SM. Molecular changes during neurodevelopment following second-trimester binge ethanol exposure in a mouse model of fetal alcohol spectrum disorder: From immediate effects to long-term adaptation. Dev. Neurosci. 2014;36(1):29–43. doi: 10.1159/000357496. [DOI] [PubMed] [Google Scholar]
- 123.Ignacio C, Mooney SM, Middleton FA. Effects of acute prenatal exposure to ethanol on microRNA expression are ameliorated by social enrichment. Front. Pediatr. 2014;2:103. doi: 10.3389/fped.2014.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Balaraman S, Lunde ER, Sawant O, Cudd TA, Washburn SE, Miranda RC. Maternal and neonatal plasma microrna biomarkers for fetal alcohol exposure in an ovine model. Alcohol. Clin. Exp. Res. 2014;38(5):1390–1400. doi: 10.1111/acer.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Qi Y, Zhang M, Li H, et al. MicroRNA-29b regulates ethanol-induced neuronal apoptosis in the developing cerebellum through SP1/RAX/PKR Cascade. J. Biol. Chem. 2014;289(14):10201–10210. doi: 10.1074/jbc.M113.535195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Guo Y, Chen Y, Carreon S, Qiang M. Chronic intermittent ethanol exposure and its removal induce a different miRNA expression pattern in primary cortical neuronal cultures. Alcohol. Clin. Exp. Res. 2012;36(6):1058–1066. doi: 10.1111/j.1530-0277.2011.01689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Miranda RC. MicroRNAs and ethanol toxicity. Int. Rev. Neurobiol. 2014;115:245–284. doi: 10.1016/B978-0-12-801311-3.00007-X. [DOI] [PubMed] [Google Scholar]
- 128.Tal TL, Franzosa JA, Tilton SC, et al. MicroRNAs control neurobehavioral development and function in zebrafish. FASEB J. 2012;26(4):1452–1461. doi: 10.1096/fj.11-194464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Balaraman S, Schafer JJ, Tseng AM, et al. Plasma miRNA profiles in pregnant women predict infant outcomes following prenatal alcohol exposure. PLoS ONE. 2016;11(11):e0165081. doi: 10.1371/journal.pone.0165081. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Identifies a miRNA signature in the plasma of pregnant women that could potentially predict deficits in children with heavy prenatal alcohol exposure.
- 130.Widagdo J, Zhao Q, Kempen M, et al. Experience-dependent accumulation of N 6-methyladenosine in the prefrontal cortex is associated with memory processes in mice. J. Neurosci. 2016;36(25):6771–6777. doi: 10.1523/JNEUROSCI.4053-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Clayton JA, Collins FS. NIH to balance sex in cell and animal studies. Nature. 2014;509(7500):282–283. doi: 10.1038/509282a. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This commentary outlines the establishment of new funding guidelines by the National Institutes of Health to support research assessing sex differences in health and disease.
- 132.Zhang FF, Cardarelli R, Carroll J, et al. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics. 2011;6(5):623–629. doi: 10.4161/epi.6.5.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hellemans KGC, Verma P, Yoon E, Yu W, Weinberg J. Prenatal alcohol exposure increases vulnerability to stress and anxiety-like disorders in adulthood. Ann. N. Y. Acad. Sci. 2008;1144:154–175. doi: 10.1196/annals.1418.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bale TL, Epperson CN. Sex differences and stress across the lifespan. Nat. Neurosci. 2015;18(10):1413–1420. doi: 10.1038/nn.4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Oldehinkel AJ, Bouma EMC. Sensitivity to the depressogenic effect of stress and HPA-axis reactivity in adolescence: a review of gender differences. Neurosci. Biobehav. Rev. 2011;35(8):1757–1770. doi: 10.1016/j.neubiorev.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 136.Patten AR, Fontaine CJ, Christie BR. A comparison of the different animal models of fetal alcohol spectrum disorders and their use in studying complex behaviors. Front. Pediatr. 2014;2:93. doi: 10.3389/fped.2014.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.van Otterdijk SD, Michels KB. Transgenerational epigenetic inheritance in mammals: how good is the evidence? FASEB J. 2016;30:1–9. doi: 10.1096/fj.201500083. [DOI] [PubMed] [Google Scholar]; •• Reviews the current evidence for transgenerational inheritance, providing insight into the current limitations and future direction for the field.
- 138.Gutierrez-Arcelus M, Lappalainen T, Montgomery SB, et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife. 2013;2:e00523–e00523. doi: 10.7554/eLife.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fraser HB, Lam LL, Neumann SM, Kobor MS. Population-specificity of human DNA methylation. Genome Biol. 2012;13(2):R8. doi: 10.1186/gb-2012-13-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Heyn H, Moran S, Hernando-Herraez I, et al. DNA methylation contributes to natural human variation. Genome Res. 2013;23(9):1363–1372. doi: 10.1101/gr.154187.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Moen EL, Zhang X, Mu W, et al. Genome-wide variation of cytosine modifications between European and African populations and the implications for complex traits. Genetics. 2013;194(4):987–996. doi: 10.1534/genetics.113.151381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jones MJ, Goodman SJ, Kobor MS. DNA methylation and healthy human aging. Aging Cell. 2015;14(6):924–932. doi: 10.1111/acel.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kelleher E, Corvin A. The Genetics of Neurodevelopmental Disorders. John Wiley & Sons, Inc.; NJ, USA: 2015. Overlapping etiology of neurodevelopmental disorders; pp. 29–48. [Google Scholar]
- 144.Jones KL, Smith DW. Recognition of the fetal alcohol syndrome in early infancy. Lancet. 1973;302(7836):999–1001. doi: 10.1016/s0140-6736(73)91092-1. [DOI] [PubMed] [Google Scholar]
- 145.Lemoine P, Harousseau H, Borteyru J, Menuet J. Les enfants des parents alcoholiques: anomolies observées a propos de 127 cas. Ouest Med. 1968;8:476–482. [Google Scholar]
- 146.May PA, Gossage JP. Estimating the prevalence of fetal alcohol syndrome: a summary. Alcohol Res. Health. 2001;25(3):159–167. [PMC free article] [PubMed] [Google Scholar]
- 147.May PA, Baete A, Russo J, et al. Prevalence and characteristics of fetal alcohol spectrum disorders. Pediatrics. 2014;134(5):855–866. doi: 10.1542/peds.2013-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.May PA, Keaster C, Bozeman R, et al. Prevalence and characteristics of fetal alcohol syndrome and partial fetal alcohol syndrome in a Rocky Mountain Region City. Drug Alcohol Depend. 2015;155:118–127. doi: 10.1016/j.drugalcdep.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Murawski N, Moore E, Thomas J, Riley E. Advances in diagnosis and treatment of fetal alcohol spectrum disorders from animal models to human studies. Alcohol. Res. 2015;37(1):97–108. [PMC free article] [PubMed] [Google Scholar]