Abstract
Rheumatoid arthritis is an immune-mediated disease that primarily affects diarthrodial joints. Susceptibility and severity of this disease are influenced by nongenetic factors, such as environmental stress, suggesting an important role of epigenetic changes. In this review, we summarize the epigenetic changes (DNA methylation, histone modification and miRNA expression) in fibroblast-like synoviocytes, which are the joint-lining mesenchymal cells that play an important role in joint inflammation and damage. We also review the effects of these epigenetic changes on rheumatoid arthritis pathogenesis and discuss their therapeutic potential.
Keywords: : epigenetics, fibroblast-like synoviocytes, histone, methylation, microRNA, rheumatoid arthritis
The pathogenesis of autoimmune diseases is dependent on the complex interplay between genetics and environmental triggers. While the contribution of genetics is unquestioned, studies in identical twins suggest that environmental stress makes a substantial contribution to heritability and disease risk. The wide range of symptom severity, remission and relapse rates, response to therapy, as well as the progression of disease over time and the global increase of disease prevalence over the past several years suggest a strong contribution of environmental triggers. Epigenetic marks are key to integrating these stochastic influences and regulating the genomic response, and recent studies have begun to shed light on how these changes can alter cell behavior. This is well documented in rheumatoid arthritis (RA), where studies on peripheral blood cells and synovial fibroblast-like synoviocytes (FLS) have been particularly illuminating.
RA is a chronic, systemic immune-mediated disease that is primarily characterized by inflammation of the joint lining (synovium) and destruction of cartilage and underlying bone. Twin and familiar aggregation studies showed that in RA genetic factors explain up to ∼60% of the variance; disease concordance between identical twins is only 12–15% suggesting that other influences are also important [1,2]. The many known RA-associated SNPs currently explain only a limited amount of the expected disease variance [3], and the relative contribution of each new SNP is reaching the point of diminishing returns [4]. Furthermore, a recent study of mono- and dizygotic twins has suggested that shared and nonshared environmental effects are actually greater than that of genetic effects in regards to RA susceptibility [5]. This observation highlights the importance of researching the role of nongenetic factors in RA. RA is greatly dependent on age [6], environmental factors such as smoking [6,7] and microbiome composition [8], and certain early life factors such as birth weight [9] and breastfeeding [10]. Epigenetic factors are another promising area of investigation since they are both heritable and acquired throughout life and are ideal candidates to bridge environmental and genetic contribution to risk of disease.
While many epigenetic studies in autoimmune diseases focus on marks in immune cells, perhaps the most information in RA relates to the stromal element, primarily FLS that form the innermost layer of the synovial membrane surrounding joints. In RA they acquire an aggressive phenotype and form a hyperproliferative intimal lining layer (pannus), propagate joint inflammation through the production of cytokines and small molecule mediators and are the primary mediators of cartilage destruction in the joint [11]. While FLS are able to respond to existing inflammatory mediators from immune cells, they are not merely ‘passive responders’ [12]. RA FLS become imprinted and their abnormal behavior continues independently of the inflammatory milieu of the RA joint [12]. For example, FLS isolated from RA patients retain their ability to degrade cartilage in an SCID mouse model months after isolation from the rheumatoid synovium [13]. Such imprinted phenotypes of FLS suggest epigenetic alterations of these cells, which could partly explain the variability in RA severity.
The biology of RA FLS remains incompletely understood and currently the RA treatment armamentarium does not include any FLS-targeted agents. Understanding the epigenetic signature of RA FLS could be an important way to identify novel diagnostic markers or therapeutic targets. In this review, we describe epigenetic alterations in RA FLS including methylation, histone modification and microRNA, and discuss the implications of these alterations on disease pathogenesis and future therapies for RA.
DNA methylation in RA FLS
DNA methylation is perhaps the most prevalent epigenetic alteration in RA. Methylation occurs through enzymatic modification of deoxycytidine through the transfer of a methyl group to the fifth carbon on cytosine pyrimidine rings (5-methylcytosine), which almost exclusively exist in CpG dinucleotides [14]. While most CpGs are methylated, those occurring in CpG islands including those within gene promoters are largely nonmethylated [15]. Methylation of promoter CpGs results in inactivation of expression of its cognate gene, although methylation in other regions, such as gene bodies, introns and enhancers, can have variable effects [15].
Methylation can occur as a way of maintaining methylation patterns during DNA replication or can occur de novo in response to external stimuli [16], and is carried out by a family of enzymes called dinucleotide methyltransferases [17]. Aberrant global methylation has been described for several families of diseases such as cancer [18], neurodegenerative disease [19] and autoimmune disease [20]. In RA, many DNA methylation studies have been performed on immune cells, including peripheral blood mononuclear cells and T cells, and have been recently reviewed [21,22].
A DNA methylation signature in RA FLS
Early investigations into global DNA methylation levels in RA synovium using HPLC suggested no difference between RA patients and that from OA patients when whole synovial tissue was assessed, as opposed to peripheral blood mononuclear cells (PBMCs), which had lower methylation in RA patients [23]. Altered methylation of RA FLS was first described in 2009 when investigators observed reduced 5-methylcytosine levels in RA FLS compared with osteoarthritis (OA) FLS using immunohistochemistry and flow cytometry [24]. Hypomethylation was also observed in the promoter of long interspersed nuclear element 1 (L1; LINE-1), a mammalian retrotransposon that comprises approximately 17% of the human genome [25] and is therefore used as a surrogate for measuring global genomic methylation [26]. Further, normal FLS treated with 5-azacytidine, a DNA methylation inhibitor, showed decreased global methylation and displayed a more aggressive RA FLS-like phenotype [24].
When whole genomic methylation patterns in FLS were studied using the chip technology, global genomic hypomethylation was not observed in RA FLS compared with OA FLS [27], although it should be noted that methylation of repetitive elements such as LINE-1 are not well represented on such chips and could, therefore, be underestimated. Instead, a more complex pattern of altered methylation in RA emerged. Interestingly, unbiased assessment of methylation levels at individual loci in RA and OA FLS revealed an RA ‘methylome’ signature, in which specific regions were hypermethylated while others were hypomethylated [27]. These regions corresponded to pathways involved in FLS-matrix interactions, inflammation and cell trafficking, giving insight to how altered methylation in RA FLS could be linked to their aggressive phenotype. A follow-up study with additional RA and OA FLS confirmed a nonrandom DNA methylation pattern in RA FLS and, importantly, showed that this signature is stable across several passages and between replicates [28]. This study also identified a set of differentially methylated genes that are involved in RA pathways (including matrix regulation, cytokine regulation and cell adhesion), and confirmed that their expression is correlated to the methylation status at that gene promoter [28]. This stable methylation signature, and its relationship to RA phenotypes and pathways, suggest that RA FLS are imprinted in the rheumatoid joints and retain this signature ex vivo.
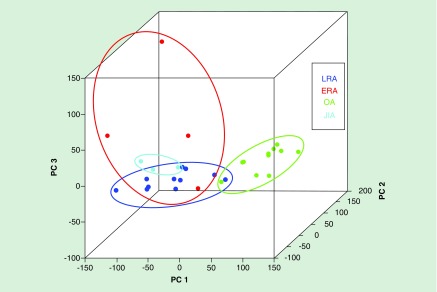
The existence of a stable RA signature for DNA methylation in FLS is intriguing in regards to addressing the distinct behavior of RA FLS compared with FLS in normal or noninflamed joints, yet it has proven to be much more informative. A recent study has shown that this ‘methylome signature’ appears to change as the duration of disease increases. FLS from patients with early RA differ somewhat from longstanding RA [29] in genes related to cell differentiation and proliferation (Figure 1). The data suggest early plasticity in the RA methylome and that changes can become fixed over time. This study also demonstrated that juvenile idiopathic arthritis FLS methylation was also more similar to RA than OA FLS and that they formed a subgroup of the ‘inflammatory methylotype’ [29]. This precise fine-tuning of methylation patterns, rather than a nonspecific alteration of global methylation levels, may represent a possible diagnostic and prognostic tool for RA patients in the future.
Figure 1. . Unique DNA methylation signatures in rheumatoid arthritis fibroblast-like synoviocytes from early and long-standing disease.
Principal component analysis shows differentially methylated loci in ERA, LRA, JIA and OA FLS lines. Methylation patterns in RA FLS lines segregate from OA FLS lines, while ERA FLS lines further segregate from LRA FLS lines.
ERA: Early rheumatoid arthritis; FLS: Fibroblast-like synoviocytes; JIA: Juvenile idiopathic arthritis; LRA: Longstanding rheumatoid arthritis; OA: Osteoarthritis; RA: Rheumatoid arthritis.
Adapted with permission from [29].
Another recent study has shown that the RA FLS methylome signature also displays spatial specificity. The assessment of FLS isolated from human hip and knee joints revealed an expanded methylome signature that differed between RA and OA FLS [30]. Interestingly, both RA and OA FLS showed unique methylation patterns between hip and knee location, especially in developmental genes like HOX and WNT families. These genes likely determine how a cell in a particular joint should behave, and it is not clear if these epigenetic marks are preprogrammed or whether they are acquired after they arrive at each joint. Perhaps even more interesting, a group of epigenetic marks and transcriptome differences remained between RA hip and knee FLS even when ‘disease-independent’ variances were removed [30]. Pathways critical to disease pathogenesis, such as JAK-STAT, were distinctly different in the two joint locations and provide a possible explanation for the asynchronous responses to targeted therapeutic agents. The methylome signatures of more commonly affected joints in RA, such as wrists and the small joints of hands and feet remain to be determined, as hip and knee joints in this study were used due to their accessibility during routine surgeries. Another recent study looked at RA fibroblasts isolated from rheumatoid synovial fluid, which can be obtained more easily and less invasively from patients (although it is not clear if these are a separate population from traditionally defined FLS). The authors show that the previously described RA FLS methylome signature was also found in the synovial fluid fibroblasts [31], and if verified, might serve as a readily available source of patient samples for assessing methylation signatures. A better understanding of how the methylome signature is established based on environmental factors, and how joint location affects this signature, will be crucial to understanding and predicting disease development.
Mechanisms of altering DNA methylation in RA FLS
While it is clear that a DNA methylation signature exists for RA FLS, it is not yet clear exactly how and when RA FLS are imprinted. One explanation is that the methyl donor S-adenosyl methionine is consumed in other pathways in RA FLS, and this results in reduced methylation [32]; however, the RA FLS signature is much more complex than a global hypomethylation, and therefore other mechanisms must also play a role. Another explanation is that the inflammatory milieu, in which RA FLS reside, influences methylation of these cells. Indeed, methylation levels appear to be sensitive to inflammatory cytokines. In one study, 5-methylcytosine levels were increased upon stimulation with TNF and IL-1ß, with RA FLS maintaining a lower methylation level than OA FLS [24]. Another study has shown that proinflammatory cytokines, especially IL-1ß, decrease dinucleotide methyltransferase expression in FLS [33]. In this regard, treatment of FLS with IL-1ß decreases not only global methylation but also selectively decreases methylation at specific loci that are typically hypomethylated in the RA signature [33]. While these results support a role for inflammation in the imprinting of FLS in RA, it is important to note that the effects of cytokines such as IL-1ß are mostly reversible when the cytokine is removed from the FLS [33] and, therefore, cannot fully explain the stable methylome signature that persists in RA FLS ex vivo.
Clues to RA pathogenesis from the methylome signature
RA FLS possess a unique, nonrandom DNA methylome pattern that is finely tuned during disease progression and varies with joint location. While it is still not clear exactly how this fine-tuning occurs, a general pattern of differential methylation persists and must in some way reflect the genes and pathways responsible for the aggressive phenotype acquired by FLS during RA. Indeed, differential methylation at specific loci in FLS has helped identify novel mediators of RA pathogenesis. For example, the promoter of the gene for the chemokine CXCL12 is normally highly methylated, but is hypomethylated in RA FLS [34]. This decrease in promoter methylation results in increased expression and production of CXCL12, which is highly abundant in RA joints and is thought to promote chronic inflammation [34]. Another study identified the transcription factor T-box transcription factor 5 as having differential methylation at its promoter in RA FLS compared with OA FLS [35]. Further, T-box transcription factor 5 expression was linked to the expression of several downstream genes including the chemokine CXCL12 [35], demonstrating how differential methylation of select genes as described in the RA FLS methylome signature may cause widespread effects resulting in the dramatic phenotype of RA FLS.
Recently, an approach for probing the methylome signature along with other types of data obtained from RA and OA FLS was proposed as a way to identify novel therapeutic targets for RA. Whitaker et al. have described a method combining data from genome-wide association studies, differential expression studies comparing RA versus OA FLS, as well as differential DNA methylation studies comparing RA versus OA FLS [36]. The result is the identification of so-called ‘multievidence genes’ [36] that can then be individually explored in FLS in the context of RA pathogenesis.
This integrative approach has led to the recent elucidation of important participants in RA pathology and possible therapeutic targets such as ELMO1 [36], LBH [37,38] and PTPN11 [39]. The first multievidence gene reported using this approach was ELMO1, which encodes a protein involved in cellular motility and engulfment, and whose promoter was found to have increased methylation in RA FLS [36]. Knockdown of ELMO1 suppressed FLS migration and invasion by decreasing activation of the GTPase RAC1 [36]. These results show how the integration of methylation and expression data can identify a protein previously unrelated to RA that plays a critical role in FLS-mediated RA pathology.
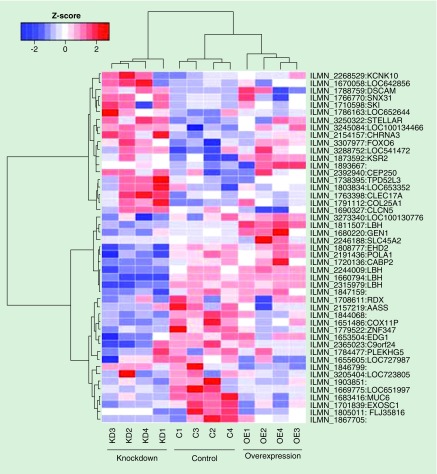
This integrated ‘-omics’ approach can be expanded beyond promoter analysis to include enhancers, on which the effect of methylation is largely unknown. This led to the recent identification of new loci in RA FLS. One example is LBH, a gene encoding a protein previously thought to be only involved in embryonic development. LBH was first identified as a multievidence gene based on hypomethylation of its promoter in RA FLS and, using knockdown experiments in RA FLS, LBH was found to influence the transcriptome in these cells (Figure 2) including cell growth and proliferation networks [37]. A follow-up study, which included putative enhancer loci in the integrative analysis, again identified LBH as a multievidence gene, this time due to hypomethylation of an enhancer region [38]. Interestingly, this enhancer region also includes an RA-associated SNP, and the combination of SNP genotype and methylation levels at the enhancer was shown to influence enhancer activity and thus LBH expression [38].
Figure 2. . LBH regulates the fibroblast-like synoviocyte transcriptome.
Heatmap showing gene expression in control (C) FLS and FLS in which LBH has been KD or OE. Probes include genes with significant (p < 0.05) differential expression between any two of the three FLS populations (control, KD and OE). Systems biology showed that cell cycle genes were enriched in the FLS with modulated LBH expression. Z scores represent gene expression levels and were calculated for each gene.
FLS: Fibroblast-like synoviocyte; KD: Knocked-down; OE: Overexpressed.
Adapted with permission from [37].
Another gene identified in this study was PTPN11, which encodes the protein tyrosine phosphatase SHP2 and is overexpressed in RA FLS [40]. Analysis of an enhancer region in PTPN11 in RA FLS revealed hypermethylation that increased sensitivity of the cells to glucocorticoids and increased their aggressiveness [39]. This study not only helped explain the mechanism for PTPN11 overexpression in RA FLS but also showed that SHP2 is a potential therapeutic target in RA using genetic deficiency studies and pharmacological inhibition in mouse models of arthritis [39].
In summary, the establishment of a DNA methylome signature in RA FLS will undoubtedly improve our understanding of RA pathogenesis. While it is known that the local cytokine environment can potentially influence this signature, other environmental factors likely come into play in order to establish the stable imprinted phenotype observed in RA FLS. Identifying these environmental factors will help shed light on RA susceptibility and disease progression. In addition, exploring the DNA methylome in RA FLS using new integrated ‘-omics’ involving other types of epigenetic marks approaches that are helping identify novel mediators of RA pathogenesis could lead to new classes of therapeutic targets for RA.
Histone modification in RA FLS
Histone modification represents a second critical epigenetic mark that can profoundly influence cell phenotype and gene expression. Histone proteins associate with DNA and regulate the accessibility of gene promoters to transcriptional machinery. Multiple mechanisms can modify the histone epigenetic landscape including acetylation, methylation, citrullination, phosphorylation, ubiquition and sumoylation [41]. Recently, alteration of histone citrullination was reported in RA FLS [42]; however, most studies have focused on histone acetylation.
Histone acetylation involves the transfer of acetyl groups from acetyl-CoA to histone lysine residues, and is catalyzed by enzymes called histone acetyl transferases. Acetylation is reversible, and such deacetylation is catalyzed by a family of histone deacetylases (HDACs) which have been extensively studied in various cancers due to anticancer properties of HDAC inhibitors [43], and more recently in RA [21]. HDACs can be divided into four groups: Class I (HDAC1,-2,-3,-8), Class II (HDAC4,-5,-6,-7,-9,-10), Class III (Sirt1,-2,-3,-4,-5,-6,-7) and Class IV (HDAC11) [44]. HDACs can differ based on tissue expression, subcellular localization and function.
Histone acetylation in RA FLS
Measurement of total histone acetylation levels using whole synovial tissue from RA patients has revealed variable alterations in acetylation. One study showed an overall increase in acetylation associated with decreased HDAC activity as well as decreased expression of HDAC1 and -2 [45]. Another report indicated that HDAC activity and HDAC1 expression in RA synovial tissue was increased [46]. HDAC1–11 RNA transcripts in cultured FLS showed that HDAC1 and -2 have higher expression in RA compared with OA FLS, and knockdown suggested that these HDACs have a modest effect on proliferation [47]. While histone acetylation appears to be altered in RA FLS, the degree to which and the means by which this change takes place remain uncertain.
Inflammation-induced alteration of HDACs in RA FLS
Because RA FLS become imprinted in the background of an inflammatory milieu, it is not surprising that, like methylation, histone acetylation of these cells is affected by inflammatory cytokines. For instance, TNF expression levels in synovial tissue positively correlate with HDAC activity in synovial tissue and treatment of RA FLS with TNF increases overall HDAC activity [46]. A more direct link between TNF and histone acetylation was observed in TNF-stimulated FLS, with increased histone H4 acetylation [48]. The study also showed that this was accompanied by increased chromatin accessibility, and a prolonged inflammatory response by these cells which is not present in macrophages, which are another abundant cell in the synovial lining [48]. This idea of TNF-induced imprinting of FLS via histone acetylation was also addressed by Sohn et al. who found that chronic exposure of FLS primes these cells leading to enhanced inflammatory response upon secondary exposure to cytokines such as interferon-gamma [49]. This effect was demonstrated to be due to an overall increase in histone acetylation [49].
While there is a relationship between inflammation and HDAC expression, not all members of the HDAC family are affected similarly. HDAC1 was the only HDAC with increased expression in synovial tissue from RA patients in one study, while HDAC4 expression was decreased [46]. A more recent study showed that TNF expression correlates with HDAC1 in synovial tissues from RA patients, while IL-6 inversely correlates with HDAC expression. Clinical parameters such as CRP levels inversely correlate with HDAC5 expression [50]. Like methylation, histone acetylation is likely influenced be several factors that remain to be determined and likely contribute to the etiology of RA.
HDAC inhibition in RA FLS & in arthritis
Transformed cells have a multitude of epigenetic alterations, which often include reduced acetylation, and hence silencing, of tumor suppressor genes [51]. HDAC inhibitors have been extensively studied in the context of cancer therapy, and can be considered for their use in RA in light of the aggressive behavior of FLS. One of the most commonly used HDAC inhibitors to target FLS is trichostatin A, a natural product used as an antifungal agent with HDAC inhibitor properties [52]. Treatment of RA FLS with trichostatin A increases apoptosis [53–56] and decreases the inflammatory response [57] and invasiveness [56]. One study showed that trichostatin A suppresses arthritis in the antigen-induced arthritis mouse model [58].
Givinostat (ITF2357) is another HDAC inhibitor with activity against multiple HDACs and is currently in clinical trials for several diseases. Givinostat decreases the inflammatory response in RA FLS [57] and a preclinical study showed a therapeutic effect of givinostat in multiple rodent models of RA [59]. Givinostat is also the only HDAC inhibitor to enter clinical trials for an inflammatory arthritis. A small Phase II open-label study in 2011 demonstrated that givinostat was safe and potentially beneficial in the treatment of juvenile RA [60]. It should be noted, however, that cell-based studies indicate that some anti-inflammatory effects of givinostat, as well as the trichostatin A, might be due to inhibition of nonhistone HDAC substrates [57].
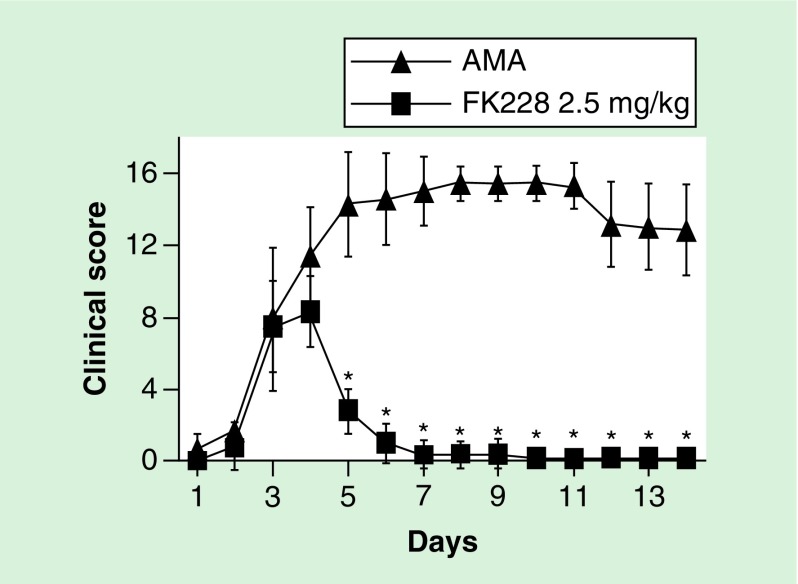
Other general inhibitors, some of which are in clinical trials for various cancers, decrease RA FLS proliferation or disease severity in mouse models of RA and include phenylbutyrate [58], romidepsin (FK228) (Figure 3) [61], entinostat (MS-275) [62], zolinza (suberoylanilide hydroxamic acid) [63] and 10-hydroxy-2-decanoic acid [64]. Although promising, HDACs have different patterns of expression and function in RA FLS as previously discussed, and so a specific HDAC inhibitor profile could be important. One example is NK-HDAC1, an HDAC inhibitor specific for HDAC1 [46,47] that has a modest effect on proliferation [47]. When NK-HDAC1 was used to treat RA FLS it reduced proliferation, induced apoptosis and decreased cytokine production [65]. NK-HDAC1 also reduced severity of joint disease in a mouse model of RA [65]. Another inhibitor tested on RA FLS is tubastatin A, a selective HDAC6 inhibitor that decreases the expression of IL-6 [66], a key cytokine in the pathogenesis of RA. Tubastatin A was also shown to decrease joint destruction and overall disease severity in a mouse model of RA [66].
Figure 3. . Effect of histone deacetylase in a mouse model of autoantibody-mediated arthritis.
Male DBA/1 mice were injected with 2 mg of an anti-type II collagen monoclonal antibody cocktail on Day 0, and boosted with lipopolysaccharide on Day 2 to induce autoantibody-mediated arthritis. Mice were treated with 2.5 mg/kg of the HDAC inhibitor FK228 (n = 10) or saline control (n = 9) on Day 4. Graphs show the clinical score of arthritis severity over the disease course. FK228 has an immediate and dramatic effect of reducing arthritis.
*p < 0.01.
HDAC: Histone deacetylase.
Adapted with permission from [61].
Overall, it appears that histone acetylation patterns are altered in RA FLS, and that inhibition of HDACs reduces inflammation and joint damage in preclinical models. Although each HDAC could control different genes and functions in RA FLS, and HDAC inhibitors often target multiple HDACs, HDAC inhibition could be a promising option for the treatment of RA. Finally, inhibitors targeting bromodomains, domains that allow a protein to ‘read’ histone acetylation marks, have also been explored for the treatment of cancer and inflammation [67]. More recent data showed that bromodomain inhibition reduces arthritis severity in mouse models of RA [68,69]. The effects of bromodomain inhibition on RA FLS include decreased inflammatory response, proliferation and matrix degradation [70].
MicroRNA in RA FLS
Like DNA methylation and histone modification, microRNAs represent another mode of epigenetic control of gene expression; however, they are unique in that they are encoded by the genome. MicroRNAs form a large group of noncoding RNAs that are transcribed and processed to mature miRNAs approximately 22 nucleotides in length. The endonucleases Drosha and Dicer are responsible for the processing of primary miRNAs, which results in production of a mature miRNA strand as well as a ‘star’ or ‘asterisk’ strand that is often degraded [71]. MicroRNAs contain sequences with complementarity to specific mRNAs, and with assistance of the RNA-induced silencing complex (RISC) complex binds these target mRNAs resulting in their cleavage or inhibition of translation [71].
MicroRNAs are implicated in many biological processes and diseases including cancer [72]. Their important role in the immune system has led to the identification of several microRNAs that are involved in various autoimmune diseases, including RA [73]. While many microRNAs have aberrant expression in immune cells in RA [74], RA FLS express a distinct miRNA profile [75–77]. This profile can be reproduced in FLS from a mouse model of RA [76] which suggests that microRNAs are important regulators of FLS phenotype. In addition, it appears that the DNA methylation and miRNA profiles of RA FLS are not mutually exclusive, and that integrative analysis of these and other types of modifications are needed in the future in order to fully appreciate the epigenetic landscape in RA FLS [77]. Here, we focus on microRNAs that have been identified as upregulated or downregulated in RA FLS in studies that give insight to their function in RA pathogenesis and possible therapeutic potential. A full list of microRNAs identified in RA FLS can be found in Table 1.
Table 1. . MicroRNAs with altered expression in rheumatoid arthritis fibroblast-like synoviocytes.
| MicroRNA ID | Expression in RA FLSs | Putative function in RA FLSs | Ref. |
|---|---|---|---|
| miR-10a |
Decreased |
Anti-inflammatory; regulation of NF-κB pathway and production of inflammatory cytokines |
[90] |
| miR-18a |
Increased in response to TNF |
Overexpression increases inflammatory cytokines, MMP-1 and NF-κB signaling |
[91] |
| miR-19 |
Decreased in response to lipopolysaccharide |
Anti-inflammatory; regulation of TLR2, IL-6, IL-8 and MMP3 |
[92,93] |
| miR-20a |
Decreased in response to lipopolysaccharide |
Anti-inflammatory; regulation of IL-6, TNF and IL-1β |
[94] |
| miR-23b |
Decreased |
Anti-inflammatory; overexpression reduces arthritis in mouse model |
[87] |
| miR-34a* |
Decreased |
Regulation of FLS survival |
[86] |
| miR-124a |
Decreased |
Regulation of FLS proliferation; overexpression reduces arthritis in mouse model |
[75,84,85] |
| miR-155 |
Increased |
Protective; inhibition of MMP3 expression, proliferation and survival of FLS |
[76,81,82] |
| miR-146a |
Increased |
Unknown |
[75,81] |
| miR-203 |
Increased |
Promotion of MMP1 and IL-6 production |
[79] |
| miR-221 |
Increased |
Promotion of inflammatory cytokines, survival, migration and invasion of FLS |
[76,78] |
| miR-222 |
Increased |
Unknown |
[76] |
| miR-223 |
Increased |
Unknown |
[81] |
| miR-323–3p |
Increased |
Promotes Wnt/cadherin pathway |
[76] |
| miR-346 |
Increased in response to lipopolysaccharide |
Inhibition of Bruton tyrosine kinase expression; decreases IL-18 mRNA expression and TNF mRNA stability |
[95,96] |
| miR-663 | Increased | Regulation of FLS proliferation and IL-6 production | [80] |
FLS: Fibroblast-like synoviocyte; MMP: Matrix metalloproteinase; RA: Rheumatoid arthritis.
MicroRNAs increased in RA FLS
Profiling of RA FLS has revealed several microRNAs to be upregulated in RA FLS compared with OA FLS or FLS from healthy controls. MiR-221 was first identified as a highly expressed microRNA in a deep sequencing screen of RA FLS as well as FLS from a mouse model of RA [76]. MiR-221 expression was confirmed in synovial tissue from RA patients, and is induced upon stimulation of RA FLS with lipopolysaccharide (LPS) [78]. Inhibition of miR-221 was shown to decrease inflammatory cytokine production by RA FLS upon LPS stimulation, as well as increase apoptosis and decrease migration and invasion of these cells [78]. MiR-203 is also highly expressed in RA, and overexpression of this microRNA increases matrix metalloproteinase (MMP) MMP1 and IL-6 production [79]. Interestingly, the high expression of miR-203 in RA FLS was linked to hypomethylation at its promoter [79], suggesting miR-203 may be part of the ‘methylome signature’ of RA FLS discussed above.
Another noteworthy example of an overexpressed microRNA is miR-663, which is highly expressed in RA FLS compared with FLS from healthy controls and regulates FLS proliferation and IL-6 production [80]. MiR-663 regulates these pathogenic behaviors by decreasing expression of the tumor suppressor gene APC [80], ultimately suppressing Wnt signaling, which mediates its role in conferring a transformed-like phenotype to RA FLS.
While overexpression may suggest a positive role in RA pathogenesis, other miRNAs appear to be induced in RA FLS that have a protective role. One such microRNA is miR-155, which was found in three studies to be overexpressed in RA FLS [76,81,82] and whose expression is increased in FLS upon exposure to inflammatory cytokines or TLR ligands [81]. While miR-155 deficiency was protective in one mouse model of RA [83], much of its proinflammatory activity was attributed to its expression in monocytes. A subsequent functional study in FLS showed that overexpression of miR-155 decreased expression of the matrix metalloproteinase MMP3 and decreased RA FLS proliferation and invasion, while inhibition of miR-155 had the opposite effect, suggesting a protective role for miR-155 in FLS [82].
MicroRNAs decreased in RA FLS
Expression of some microRNAs is low in RA, and are therefore attractive targets for gene therapy. MiR-124a was especially low in a screen of RA versus OA FLS [75], and its overexpression decreased proliferation of FLS from RA patients [75,84], but not those of healthy controls [75]. The cell cycle regulator CDK2 was one of the targets identified for miR-124a [75,84], supporting its role in controlling FLS proliferation. MiR-124a overexpression in a rat model of RA decreased disease severity and suppressed FLS proliferation [85].
Another example of a microRNA with suppressed expression in RA FLS is miR-34a* [86]. MiR-34a* is an example of a ‘star’ or ‘asterisk’ strand described above, which occur during processing of a mature microRNA strand and are often degraded with unknown biological function [71]. While increased methylation of the miR-34a/34a* promoter was observed, only miR-34a* had reduced expression in RA FLS [86]. Overexpression of miR-34a* induced apoptosis in RA FLS, and the apoptosis inhibitor XIAP was identified as a putative target [86].
Finally, miR-23b was recently identified as decreased in RA patients as well as other autoimmune diseases [87]. In RA FLS, expression of miR-23b was suppressed by IL-17, a key cytokine in RA pathogenesis [87]. Overexpression of miR-23b reduced the expression of inflammatory cytokines in stimulated FLS, and its transgenic expression decreased joint disease in a mouse model of RA [87], suggesting that re-expression of miR-23b in FLS could be of therapeutic benefit in RA. Thus, patterns of miRNA expression could influence the function of RA FLS. These patterns could provide insight into RA pathogenesis, and could be leveraged with therapeutic strategies that remodel the miRNA profile.
Although we have focused on microRNAs in RA FLS in this review as they are direct epigenetic modifiers, other noncoding RNAs that may affect epigenetic changes include long noncoding RNA (lncRNA). While knowledge of the role of lncRNA in RA FLS is limited, a recent study showed that RA FLS may express a unique lncRNA signature that correlates with certain clinical parameters [88].
Future perspective
RA FLS are a relevant model for studying epigenetics in chronic disease since they acquire a stable imprinted phenotype through the disease process. While great strides have been made in understanding how FLS become imprinted in RA, there are several areas that should be addressed. First, identification of environmental factors that affect the epigenetic signature in RA FLS need to be defined. Recent studies have begun to address the impacts of environmental factors on epigenetic modifications. For example, cigarette smoking (which is the best defined environmental influence on RA susceptibility) also affects DNA methylation at certain loci [89]; however, how environmental effects such as smoking specifically alter FLS in RA is not known.
Second, rapid advances in omics technology are expected to improve the resolution of the RA FLS epigenetic signature. The ability to examine modifications such as DNA methylation at the single cell level will greatly enhance our understanding of specific effects of epigenetic changes in RA FLS. In addition, deconvoluting data from mixed cell populations will help tease apart cell-specific effects. Furthermore, the assembly of a definitive RA FLS signature through genome-wide studies such as ChIP-seq, RNA-seq, ATAC-seq and whole genome bisulfite sequencing, will be important to define the rheumatoid epigenomic landscape.
An additional point for future study is understanding whether the RA FLS epigenetic signature can be used as a diagnostic and prognostic tool. As it emerges, the RA FLS epigenetic signature can be envisioned as a set of distinct markers that can be paired with other diagnostic tools such as ultrasound to provide staging of joint disease. Since RA FLS appear to be imprinted quite early in the disease process, this may allow earlier detection in people at risk for the disease. As well, the further validation of epigenetic RA signatures in FLS from different joints, and FLS from synovial fluid as opposed to tissue biopsies, may also lead to joint-targeted and easier sample collection protocols. In addition to its diagnostic potential, the epigenetic signature continues to identify novel therapeutic targets that may control RA FLS behavior and are expected to complement conventional immune-targeted therapies.
Conclusion
RA is an immune-mediated disease with a substantial nongenetic susceptibility component. Epigenetic abnormalities likely explain a significant component of the nongenetic risk of RA and might even underlie some of the unexplained genetic variance of disease. DNA methylation studies in RA FLS have revealed a methylome signature, which is evident early in the disease process and can be influenced by the inflammatory milieu characteristic of the RA joint. Likewise, histone modifications, in particular histone acetylation, also show a pattern unique to RA FLS. Such patterns are accompanied by altered expression and activity of HDACs, and HDAC inhibition might control RA FLS aggressiveness. Finally, miRNA expression is also altered in RA FLS, with some miRNAs being proinflammatory and some protective. While it is clear that all these different types of epigenetic anomalies are important in RA FLS, it remains to be seen how much interplay exists between these types of epigenetic modifications, and what other modifications affect these cells.
Executive summary.
DNA methylation in rheumatoid arthritis fibroblast-like synoviocytes
Rheumatoid arthritis fibroblast-like synoviocytes (RA FLS) exhibit a unique, stable methylome signature that is established early and changes with disease progression.
Methylation of RA FLS is influenced by inflammatory cytokines.
The combination of expression and methylation data have revealed new genes involved in the pathogenesis of RA.
Histone modification in RA FLS
Histone acetylation levels in RA FLS respond to inflammatory cytokines as a result of altered histone deacetylase expression.
Histone deacetylase inhibition is a potential approach to control RA FLS proliferation, inflammatory response and invasiveness, and decrease arthritis severity in some animal models.
MicroRNA in RA FLS
RA FLS express a distinct profile of microRNAs.
Most microRNAs that are increased in RA FLS influence cytokine production and proliferation, while some may be increased as a protective response.
Re-expression of microRNAs with low expression in RA FLS can improve disease severity in animal models of RA.
Conclusion
Epigenetic alterations explain a large component of nongenetic risk factors for RA.
Further research is needed to identify additional epigenetic alterations, as well as to define the interplay between specific types of epigenetic alterations.
Understanding the epigenetic landscape of RA FLS is expected to yield novel, cell-targeted and potentially personalized therapeutic strategies.
Footnotes
Financial & competing interests disclosure
This work was supported, in part, by grants from the Rheumatology Research Foundation, the Arthritis Foundation and the National Institute of Arthritis and Musculoskeletal and Skin Diseases R01 AR065466. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest
- 1.Aho K, Koskenvuo M, Tuominen J, Kaprio J. Occurrence of rheumatoid arthritis in a nationwide series of twins. J. Rheumatol. 1986;13(5):899–902. [PubMed] [Google Scholar]
- 2.Macgregor AJ, Snieder H, Rigby AS, et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 2000;43(1):30–37. doi: 10.1002/1529-0131(200001)43:1<30::AID-ANR5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 3.Okada Y, Wu D, Trynka G, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506(7488):376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stahl EA, Raychaudhuri S, Remmers EF, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 2010;42(6):508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Svendsen AJ, Kyvik KO, Houen G, et al. On the origin of rheumatoid arthritis: the impact of environment and genes – a population based twin study. PLoS ONE. 2013;8(2):e57304. doi: 10.1371/journal.pone.0057304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sparks JA, Chang SC, Deane KD, et al. Associations of smoking and age with inflammatory joint signs among first-degree relatives without rheumatoid arthritis: results from the Studies of the Etiology of RA. Arthritis Rheumatol. 2016;68(8):1828–1838. doi: 10.1002/art.39630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pedersen M, Jacobsen S, Garred P, et al. Strong combined gene-environment effects in anti-cyclic citrullinated peptide-positive rheumatoid arthritis: a nationwide case-control study in Denmark. Arthritis Rheum. 2007;56(5):1446–1453. doi: 10.1002/art.22597. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Zhang D, Jia H, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 2015;21(8):895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 9.Jacobsson LT, Jacobsson ME, Askling J, Knowler WC. Perinatal characteristics and risk of rheumatoid arthritis. BMJ. 2003;326(7398):1068–1069. doi: 10.1136/bmj.326.7398.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karlson EW, Mandl LA, Hankinson SE, Grodstein F. Do breast-feeding and other reproductive factors influence future risk of rheumatoid arthritis? Results from the Nurses’ Health Study. Arthritis Rheum. 2004;50(11):3458–3467. doi: 10.1002/art.20621. [DOI] [PubMed] [Google Scholar]
- 11.Bartok B, Firestein GS. Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunol. Rev. 2010;233(1):233–255. doi: 10.1111/j.0105-2896.2009.00859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bottini N, Firestein GS. Duality of fibroblast-like synoviocytes in RA: passive responders and imprinted aggressors. Nat. Rev. Rheumatol. 2013;9(1):24–33. doi: 10.1038/nrrheum.2012.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lefevre S, Knedla A, Tennie C, et al. Synovial fibroblasts spread rheumatoid arthritis to unaffected joints. Nat. Med. 2009;15(12):1414–1420. doi: 10.1038/nm.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson KD. DNA methylation and human disease. Nat. Rev. Genet. 2005;6(8):597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 15.Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer. 2011;2(6):607–617. doi: 10.1177/1947601910393957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schubeler D. Function and information content of DNA methylation. Nature. 2015;517(7534):321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 17.Subramaniam D, Thombre R, Dhar A, Anant S. DNA methyltransferases: a novel target for prevention and therapy. Front. Oncol. 2014;4:80. doi: 10.3389/fonc.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klutstein M, Nejman D, Greenfield R, Cedar H. DNA methylation in cancer and aging. Cancer Res. 2016;76(12):3446–3450. doi: 10.1158/0008-5472.CAN-15-3278. [DOI] [PubMed] [Google Scholar]
- 19.Klein HU, De Jager PL. Uncovering the role of the methylome in dementia and neurodegeneration. Trends Mol. Med. 2016;22(8):687–700. doi: 10.1016/j.molmed.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 20.Gupta B, Hawkins RD. Epigenomics of autoimmune diseases. Immunol. Cell Biol. 2015;93(3):271–276. doi: 10.1038/icb.2015.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bottini N, Firestein GS. Epigenetics in rheumatoid arthritis: a primer for rheumatologists. Curr. Rheumatol. Rep. 2013;15(11):372. doi: 10.1007/s11926-013-0372-9. [DOI] [PubMed] [Google Scholar]
- 22.Cribbs A, Feldmann M, Oppermann U. Towards an understanding of the role of DNA methylation in rheumatoid arthritis: therapeutic and diagnostic implications. Ther. Adv. Musculoskelet. Dis. 2015;7(5):206–219. doi: 10.1177/1759720X15598307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corvetta A, Della Bitta R, Luchetti MM, Pomponio G. 5-Methylcytosine content of DNA in blood, synovial mononuclear cells and synovial tissue from patients affected by autoimmune rheumatic diseases. J. Chromatogr. 1991;566(2):481–491. doi: 10.1016/0378-4347(91)80265-e. [DOI] [PubMed] [Google Scholar]
- 24.Karouzakis E, Gay RE, Michel BA, Gay S, Neidhart M. DNA hypomethylation in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 2009;60(12):3613–3622. doi: 10.1002/art.25018. [DOI] [PubMed] [Google Scholar]; • Showed that epigenetic modifications have a direct impact on rheumatoid arthritis fibroblast-like synoviocytes’ (RA FLS) pathogenic behavior.
- 25.Kazazian HH., Jr Genetics. L1 retrotransposons shape the mammalian genome. Science. 2000;289(5482):1152–1153. doi: 10.1126/science.289.5482.1152. [DOI] [PubMed] [Google Scholar]
- 26.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32(3):e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakano K, Whitaker JW, Boyle DL, Wang W, Firestein GS. DNA methylome signature in rheumatoid arthritis. Ann. Rheum. Dis. 2013;72(1):110–117. doi: 10.1136/annrheumdis-2012-201526. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The first study of DNA methylation levels at the whole genome level; demonstrated that DNA methylation patterns are stably imprinted in RA FLS.
- 28.Whitaker JW, Shoemaker R, Boyle DL, et al. An imprinted rheumatoid arthritis methylome signature reflects pathogenic phenotype. Genome Med. 2013;5(4):40. doi: 10.1186/gm444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ai R, Whitaker JW, Boyle DL, et al. DNA methylome signature in synoviocytes from patients with early rheumatoid arthritis compared to synoviocytes from patients with longstanding rheumatoid arthritis. Arthritis Rheumatol. 2015;67(7):1978–1980. doi: 10.1002/art.39123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ai R, Hammaker D, Boyle DL, et al. Joint-specific DNA methylation and transcriptome signatures in rheumatoid arthritis identify distinct pathogenic processes. Nat Commun. 2016;7:11849. doi: 10.1038/ncomms11849. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Demonstrates that the DNA methylome signature of RA FLS is dependent on joint location and helps explain joint preference in RA.
- 31.Glossop JR, Haworth KE, Emes RD, et al. DNA methylation profiling of synovial fluid FLS in rheumatoid arthritis reveals changes common with tissue-derived FLS. Epigenomics. 2015;7(4):539–551. doi: 10.2217/epi.15.15. [DOI] [PubMed] [Google Scholar]
- 32.Karouzakis E, Gay RE, Gay S, Neidhart M. Increased recycling of polyamines is associated with global DNA hypomethylation in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 2012;64(6):1809–1817. doi: 10.1002/art.34340. [DOI] [PubMed] [Google Scholar]
- 33.Nakano K, Boyle DL, Firestein GS. Regulation of DNA methylation in rheumatoid arthritis synoviocytes. J. Immunol. 2013;190(3):1297–1303. doi: 10.4049/jimmunol.1202572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karouzakis E, Rengel Y, Jungel A, et al. DNA methylation regulates the expression of CXCL12 in rheumatoid arthritis synovial fibroblasts. Genes Immun. 2011;12(8):643–652. doi: 10.1038/gene.2011.45. [DOI] [PubMed] [Google Scholar]
- 35.Karouzakis E, Trenkmann M, Gay RE, Michel BA, Gay S, Neidhart M. Epigenome analysis reveals TBX5 as a novel transcription factor involved in the activation of rheumatoid arthritis synovial fibroblasts. J. Immunol. 2014;193(10):4945–4951. doi: 10.4049/jimmunol.1400066. [DOI] [PubMed] [Google Scholar]
- 36.Whitaker JW, Boyle DL, Bartok B, et al. Integrative omics analysis of rheumatoid arthritis identifies non-obvious therapeutic targets. PLoS ONE. 2015;10(4):e0124254. doi: 10.1371/journal.pone.0124254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ekwall AK, Whitaker JW, Hammaker D, Bugbee WD, Wang W, Firestein GS. The rheumatoid arthritis risk gene LBH regulates growth in fibroblast-like synoviocytes. Arthritis Rheumatol. 2015;67(5):1193–1202. doi: 10.1002/art.39060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hammaker D, Whitaker JW, Maeshima K, et al. Limb bud and heart development gene transcription is regulated by the interplay of an enhancer risk allele and DNA methylation in rheumatoid arthritis. Arthritis Rheumatol. 2016;68(11):2637–2645. doi: 10.1002/art.39746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maeshima K, Stanford SM, Hammaker D, et al. Abnormal PTPN11 enhancer methylation promotes rheumatoid arthritis fibroblast-like synoviocyte aggressiveness and joint inflammation. JCI Insight. 2016;1(7):e86580. doi: 10.1172/jci.insight.86580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stanford SM, Maestre MF, Campbell AM, et al. Protein tyrosine phosphatase expression profile of rheumatoid arthritis fibroblast-like synoviocytes: a novel role of SH2 domain-containing phosphatase 2 as a modulator of invasion and survival. Arthritis Rheum. 2013;65(5):1171–1180. doi: 10.1002/art.37872. [DOI] [PMC free article] [PubMed] [Google Scholar]; • An example of how an integrated ‘-omic’ approach can identify a novel mediator in FLS behavior, and also lead to a potential theraputic target.
- 41.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 2014;15(11):703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 42.Wang F, Chen FF, Gao WB, et al. Identification of citrullinated peptides in the synovial fluid of patients with rheumatoid arthritis using LC-MALDI-TOF/TOF. Clin. Rheumatol. 2016;35(9):2185–2194. doi: 10.1007/s10067-016-3247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat. Rev. Drug Discov. 2014;13(9):673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 44.Chen HP, Zhao YT, Zhao TC. Histone deacetylases and mechanisms of regulation of gene expression. Crit. Rev. Oncog. 2015;20(1–2):35–47. doi: 10.1615/critrevoncog.2015012997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huber LC, Brock M, Hemmatazad H, et al. Histone deacetylase/acetylase activity in total synovial tissue derived from rheumatoid arthritis and osteoarthritis patients. Arthritis Rheum. 2007;56(4):1087–1093. doi: 10.1002/art.22512. [DOI] [PubMed] [Google Scholar]
- 46.Kawabata T, Nishida K, Takasugi K, et al. Increased activity and expression of histone deacetylase 1 in relation to tumor necrosis factor-alpha in synovial tissue of rheumatoid arthritis. Arthritis Res. Ther. 2010;12(4):R133. doi: 10.1186/ar3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horiuchi M, Morinobu A, Chin T, Sakai Y, Kurosaka M, Kumagai S. Expression and function of histone deacetylases in rheumatoid arthritis synovial fibroblasts. J. Rheumatol. 2009;36(8):1580–1589. doi: 10.3899/jrheum.081115. [DOI] [PubMed] [Google Scholar]
- 48.Lee A, Qiao Y, Grigoriev G, et al. Tumor necrosis factor alpha induces sustained signaling and a prolonged and unremitting inflammatory response in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 2013;65(4):928–938. doi: 10.1002/art.37853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sohn C, Lee A, Qiao Y, Loupasakis K, Ivashkiv LB, Kalliolias GD. Prolonged tumor necrosis factor alpha primes fibroblast-like synoviocytes in a gene-specific manner by altering chromatin. Arthritis Rheumatol. 2015;67(1):86–95. doi: 10.1002/art.38871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Angiolilli C, Grabiec AM, Ferguson BS, et al. Inflammatory cytokines epigenetically regulate rheumatoid arthritis fibroblast-like synoviocyte activation by suppressing HDAC5 expression. Ann. Rheum. Dis. 2016;75(2):430–438. doi: 10.1136/annrheumdis-2014-205635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26(37):5420–5432. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- 52.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 1990;265(28):17174–17179. [PubMed] [Google Scholar]
- 53.Jungel A, Baresova V, Ospelt C, et al. Trichostatin A sensitises rheumatoid arthritis synovial fibroblasts for TRAIL-induced apoptosis. Ann. Rheum. Dis. 2006;65(7):910–912. doi: 10.1136/ard.2005.044065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morinobu A, Wang B, Liu J, Yoshiya S, Kurosaka M, Kumagai S. Trichostatin A cooperates with FAS-mediated signal to induce apoptosis in rheumatoid arthritis synovial fibroblasts. J. Rheumatol. 2006;33(6):1052–1060. [PubMed] [Google Scholar]
- 55.Nakamura C, Matsushita I, Kosaka E, Kondo T, Kimura T. Anti-arthritic effects of combined treatment with histone deacetylase inhibitor and low-intensity ultrasound in the presence of microbubbles in human rheumatoid synovial cells. Rheumatology (Oxford) 2008;47(4):418–424. doi: 10.1093/rheumatology/ken003. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, Zhang B. Trichostatin A, an inhibitor of histone deacetylase, inhibits the viability and invasiveness of hypoxic rheumatoid arthritis fibroblast-like synoviocytes via PI3K/Akt signaling. J. Biochem. Mol. Toxicol. 2015;30(4) doi: 10.1002/jbt.21774. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 57.Grabiec AM, Korchynskyi O, Tak PP, Reedquist KA. Histone deacetylase inhibitors suppress rheumatoid arthritis fibroblast-like synoviocyte and macrophage IL-6 production by accelerating mRNA decay. Ann. Rheum. Dis. 2012;71(3):424–431. doi: 10.1136/ard.2011.154211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chung YL, Lee MY, Wang AJ, Yao LF. A therapeutic strategy uses histone deacetylase inhibitors to modulate the expression of genes involved in the pathogenesis of rheumatoid arthritis. Mol. Ther. 2003;8(5):707–717. doi: 10.1016/s1525-0016(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 59.Joosten LA, Leoni F, Meghji S, Mascagni P. Inhibition of HDAC activity by ITF2357 ameliorates joint inflammation and prevents cartilage and bone destruction in experimental arthritis. Mol. Med. 2011;17(5–6):391–396. doi: 10.2119/molmed.2011.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vojinovic J, Damjanov N, D'urzo C, et al. Safety and efficacy of an oral histone deacetylase inhibitor in systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. 2011;63(5):1452–1458. doi: 10.1002/art.30238. [DOI] [PubMed] [Google Scholar]; • The first clinical trial in which an histone deacetylase inhibitor has been tested in inflammatory arthritis. Givinostat was shown to be potentially beneficial in the treatment of juvenile rheumatoid arthritis.
- 61.Nishida K, Komiyama T, Miyazawa S, et al. Histone deacetylase inhibitor suppression of autoantibody-mediated arthritis in mice via regulation of p16INK4a and p21(WAF1/Cip1) expression. Arthritis Rheum. 2004;50(10):3365–3376. doi: 10.1002/art.20709. [DOI] [PubMed] [Google Scholar]
- 62.Choo QY, Ho PC, Tanaka Y, Lin HS. Histone deacetylase inhibitors MS-275 and SAHA induced growth arrest and suppressed lipopolysaccharide-stimulated NF-kappaB p65 nuclear accumulation in human rheumatoid arthritis synovial fibroblastic E11 cells. Rheumatology (Oxford) 2010;49(8):1447–1460. doi: 10.1093/rheumatology/keq108. [DOI] [PubMed] [Google Scholar]
- 63.Chen H, Pan J, Wang JD, Liao QM, Xia XR. Suberoylanilide hydroxamic acid, an inhibitor of histone deacetylase, induces apoptosis in rheumatoid arthritis fibroblast-like synoviocytes. Inflammation. 2016;39(1):39–46. doi: 10.1007/s10753-015-0220-3. [DOI] [PubMed] [Google Scholar]
- 64.Wang J, Zhang W, Zou H, et al. 10-Hydroxy-2-decenoic acid inhibiting the proliferation of fibroblast-like synoviocytes by PI3K-AKT pathway. Int. Immunopharmacol. 2015;28(1):97–104. doi: 10.1016/j.intimp.2015.05.036. [DOI] [PubMed] [Google Scholar]
- 65.Li M, Liu X, Sun X, et al. Therapeutic effects of NK-HDAC-1, a novel histone deacetylase inhibitor, on collagen-induced arthritis through the induction of apoptosis of fibroblast-like synoviocytes. Inflammation. 2013;36(4):888–896. doi: 10.1007/s10753-013-9616-0. [DOI] [PubMed] [Google Scholar]
- 66.Lee J, Hong EC, Jeong H, et al. A novel histone deacetylase 6-selective inhibitor suppresses synovial inflammation and joint destruction in a collagen antibody-induced arthritis mouse model. Int. J. Rheum. Dis. 2015;18(5):514–523. doi: 10.1111/1756-185X.12501. [DOI] [PubMed] [Google Scholar]
- 67.Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat. Rev. Drug Discov. 2014;13(5):337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 68.Park-Min KH, Lim E, Lee MJ, et al. Inhibition of osteoclastogenesis and inflammatory bone resorption by targeting BET proteins and epigenetic regulation. Nat. Commun. 2014;5:5418. doi: 10.1038/ncomms6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang QG, Qian J, Zhu YC. Targeting bromodomain-containing protein 4 (BRD4) benefits rheumatoid arthritis. Immunol. Lett. 2015;166(2):103–108. doi: 10.1016/j.imlet.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 70.Klein K, Kabala PA, Grabiec AM, et al. The bromodomain protein inhibitor I-BET151 suppresses expression of inflammatory genes and matrix degrading enzymes in rheumatoid arthritis synovial fibroblasts. Ann. Rheum. Dis. 2016;75(2):422–429. doi: 10.1136/annrheumdis-2014-205809. [DOI] [PubMed] [Google Scholar]
- 71.Hammond SM. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu. Rev. Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Simpson LJ, Ansel KM. MicroRNA regulation of lymphocyte tolerance and autoimmunity. J. Clin. Invest. 2015;125(6):2242–2249. doi: 10.1172/JCI78090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vicente R, Noel D, Pers YM, Apparailly F, Jorgensen C. Deregulation and therapeutic potential of microRNAs in arthritic diseases. Nat. Rev. Rheumatol. 2016;12(4):211–220. doi: 10.1038/nrrheum.2015.162. [DOI] [PubMed] [Google Scholar]
- 75.Nakamachi Y, Kawano S, Takenokuchi M, et al. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum. 2009;60(5):1294–1304. doi: 10.1002/art.24475. [DOI] [PubMed] [Google Scholar]
- 76.Pandis I, Ospelt C, Karagianni N, et al. Identification of microRNA-221/222 and microRNA-323–3p association with rheumatoid arthritis via predictions using the human tumour necrosis factor transgenic mouse model. Ann. Rheum. Dis. 2012;71(10):1716–1723. doi: 10.1136/annrheumdis-2011-200803. [DOI] [PubMed] [Google Scholar]
- 77.De La Rica L, Urquiza JM, Gomez-Cabrero D, et al. Identification of novel markers in rheumatoid arthritis through integrated analysis of DNA methylation and microRNA expression. J. Autoimmun. 2013;41:6–16. doi: 10.1016/j.jaut.2012.12.005. [DOI] [PubMed] [Google Scholar]; • Integrated analysis of DNA methylation and miRNA expression levels in RA FLS.
- 78.Yang S, Yang Y. Downregulation of microRNA221 decreases migration and invasion in fibroblastlike synoviocytes in rheumatoid arthritis. Mol. Med. Rep. 2015;12(2):2395–2401. doi: 10.3892/mmr.2015.3642. [DOI] [PubMed] [Google Scholar]
- 79.Stanczyk J, Ospelt C, Karouzakis E, et al. Altered expression of microRNA-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activation. Arthritis Rheum. 2011;63(2):373–381. doi: 10.1002/art.30115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miao CG, Shi WJ, Xiong YY, et al. MicroRNA-663 activates the canonical Wnt signaling through the adenomatous polyposis coli suppression. Immunol. Lett. 2015;166(1):45–54. doi: 10.1016/j.imlet.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 81.Stanczyk J, Pedrioli DM, Brentano F, et al. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58(4):1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 82.Long L, Yu P, Liu Y, et al. Upregulated microRNA-155 expression in peripheral blood mononuclear cells and fibroblast-like synoviocytes in rheumatoid arthritis. Clin. Dev. Immunol. 2013:296139. doi: 10.1155/2013/296139. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kurowska-Stolarska M, Alivernini S, Ballantine LE, et al. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc. Natl Acad. Sci. USA. 2011;108(27):11193–11198. doi: 10.1073/pnas.1019536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kawano S, Nakamachi Y. miR-124a as a key regulator of proliferation and MCP-1 secretion in synoviocytes from patients with rheumatoid arthritis. Ann. Rheum. Dis. 2011;70(Suppl. 1):i88–i91. doi: 10.1136/ard.2010.138669. [DOI] [PubMed] [Google Scholar]
- 85.Nakamachi Y, Ohnuma K, Uto K, Noguchi Y, Saegusa J, Kawano S. MicroRNA-124 inhibits the progression of adjuvant-induced arthritis in rats. Ann. Rheum. Dis. 2016;75(3):601–608. doi: 10.1136/annrheumdis-2014-206417. [DOI] [PubMed] [Google Scholar]
- 86.Niederer F, Trenkmann M, Ospelt C, et al. Down-regulation of microRNA-34a* in rheumatoid arthritis synovial fibroblasts promotes apoptosis resistance. Arthritis Rheum. 2012;64(6):1771–1779. doi: 10.1002/art.34334. [DOI] [PubMed] [Google Scholar]
- 87.Zhu S, Pan W, Song X, et al. The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-alpha. Nat. Med. 2012;18(7):1077–1086. doi: 10.1038/nm.2815. [DOI] [PubMed] [Google Scholar]
- 88.Zhang Y, Xu YZ, Sun N, et al. Long noncoding RNA expression profile in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Res. Ther. 2016;18(1):227. doi: 10.1186/s13075-016-1129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tsaprouni LG, Yang TP, Bell J, et al. Cigarette smoking reduces DNA methylation levels at multiple genomic loci but the effect is partially reversible upon cessation. Epigenetics. 2014;9(10):1382–1396. doi: 10.4161/15592294.2014.969637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mu N, Gu J, Huang T, et al. A novel NF-kappaB/YY1/microRNA-10a regulatory circuit in fibroblast-like synoviocytes regulates inflammation in rheumatoid arthritis. Sci. Rep. 2016;6:20059. doi: 10.1038/srep20059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Trenkmann M, Brock M, Gay RE, Michel BA, Gay S, Huber LC. Tumor necrosis factor alpha-induced microRNA-18a activates rheumatoid arthritis synovial fibroblasts through a feedback loop in NF-kappaB signaling. Arthritis Rheum. 2013;65(4):916–927. doi: 10.1002/art.37834. [DOI] [PubMed] [Google Scholar]
- 92.Philippe L, Alsaleh G, Suffert G, et al. TLR2 expression is regulated by microRNA miR-19 in rheumatoid fibroblast-like synoviocytes. J. Immunol. 2012;188(1):454–461. doi: 10.4049/jimmunol.1102348. [DOI] [PubMed] [Google Scholar]
- 93.Gantier MP, Stunden HJ, Mccoy CE, et al. A miR-19 regulon that controls NF-kappaB signaling. Nucleic Acids Res. 2012;40(16):8048–8058. doi: 10.1093/nar/gks521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Philippe L, Alsaleh G, Pichot A, et al. MiR-20a regulates ASK1 expression and TLR4-dependent cytokine release in rheumatoid fibroblast-like synoviocytes. Ann. Rheum. Dis. 2013;72(6):1071–1079. doi: 10.1136/annrheumdis-2012-201654. [DOI] [PubMed] [Google Scholar]
- 95.Alsaleh G, Suffert G, Semaan N, et al. Bruton's tyrosine kinase is involved in miR-346-related regulation of IL-18 release by lipopolysaccharide-activated rheumatoid fibroblast-like synoviocytes. J. Immunol. 2009;182(8):5088–5097. doi: 10.4049/jimmunol.0801613. [DOI] [PubMed] [Google Scholar]
- 96.Semaan N, Frenzel L, Alsaleh G, et al. miR-346 controls release of TNF-alpha protein and stability of its mRNA in rheumatoid arthritis via tristetraprolin stabilization. PLoS ONE. 2011;6(5):e19827. doi: 10.1371/journal.pone.0019827. [DOI] [PMC free article] [PubMed] [Google Scholar]