Abstract
Hepatitis C virus (HCV) subverts the cellular DEAD-box RNA helicase DDX6 to promote virus infection. Using polysome gradient analysis and the subgenomic HCV Renilla reporter replicon genome, we determined that DDX6 does not affect HCV translation. Rather expression of the subgenomic HCV Renilla luciferase reporter at late times, as well as labeling of newly synthesized viral RNA with 4-thiouridine showed that DDX6 modulates replication. Because DDX6 is an effector protein of the microRNA pathway, we also investigated its role in miR-122-directed HCV gene expression. Similar to sequestering miR-122, depletion of DDX6 modulated HCV RNA stability. Interestingly, miR-122-HCV RNA interaction assays with mutant HCV genomes sites and compensatory exogenous miR-122 showed that DDX6 affects the function of miR-122 at one particular binding site. We propose that DDX6 facilitates the miR-122 interaction with HCV 5′ UTR, which is necessary for stabilizing the viral genome and the switch between translation and replication.
Keywords: DEAD-box RNA helicase, DDX6, Hepatitis C virus, miR-122, RNA stability, Replication
1. Introduction
Hepatitis C virus (HCV) significantly impacts the quality of life for the estimated 200 million chronically infected individuals worldwide (El-Serag, 2012). While mortality rates associated with HCV infection are relatively low at 1–5%, liver fibrosis and cirrhosis resulting from chronic HCV infection remains a leading cause of liver transplantation and hepatocellular carcinoma in the United States. Despite the recent success of several new direct-acting antiviral agents targeting some of the most common genotypes of HCV, the widespread prevalence of hepatitis C remains a significant public health concern (Bartenschlager et al., 2013).
The single-stranded positive-sense RNA genome of HCV comprises 5′ and 3′ untranslated regions (UTRs) and a single open reading frame that encodes three structural (core, envelope 1 and 2) and seven nonstructural proteins (p7, and NS2 through NS5B) (Lindenbach and Rice, 2013). Translation and replication of the HCV genome is directed by RNA elements within both UTRs. Interestingly, specific interactions of the liver-specific microRNA miR-122 with two binding sites in the 5′ UTR mask the 5′ end of the genome from Xrn1 and Xrn2 5′-to-3′ exonuclease digestion (Li et al., 2013, 2015; Sedano and Sarnow, 2014; Thibault et al., 2015), and thus contribute to maintaining HCV RNA abundance. While the binding of miR-122 to both sites 1 and 2 is critical (Jopling et al., 2008), the exact nature of these interactions remains elusive (Garcia-Sastre and Evans, 2013).
DDX6 (RCK/p54) is a member of the conserved family of DExD/H RNA helicases that contain conserved sequence motifs associated with RNA binding, ATP binding and hydrolysis, and coordination between RNA and ATP binding (helicase) activities (Presnyak and Coller, 2013; Weston and Sommerville, 2006). DEAD-box proteins are components of large ribonucleoprotein (RNP) complexes that remodel short RNA-RNA or RNA-protein interactions to facilitate a broad range of RNA functions (Linder and Jankowsky, 2011). DDX6 and its orthologs (Drosophila Me31B, Saccharomyces Dhh1, and Xenopus Xp54) repress translation to promote silencing of maternal mRNAs (Minshall et al., 2001; Nakamura et al., 2001), or the storage and/or degradation of cellular mRNAs in cytoplasmic RNA granules such as processing bodies (P-bodies) and stress granules (Buchan et al., 2008; Kedersha et al., 2005; Presnyak and Coller, 2013; Sweet et al., 2012). More recently DDX6 was shown to interact with Ago2 and CNOT1 (key components of the microRNA Induced Silencing Complex (miRISC) and deadenylation complex, respectively) to function as an effector of microRNA-mediated gene regulation (Chiu and Rana, 2002; Mathys et al., 2014; Su et al., 2011).
Given the important role of DDX6 in RNA metabolism, it is not surprising that misregulation of DDX6 is linked with cancer. Indeed DDX6 is overexpressed in colorectal adenocarcinomas (Akao et al., 1995; Lin et al., 2008; Nakagawa et al., 1999), and depletion of DDX6 contributes to the proliferation of specific cancer cell lines. Interestingly in human liver samples taken from patients with HCV-related chronic hepatitis and hepatocellular carcinoma (HCC) DDX6 was overexpressed, and treatment of patients with interferon-α decreased DDX6 expression (Miyanari et al., 2007). Because of the role of DDX6 in microRNA-mediated gene regulation, and misregulation in cancer we sought to dissect the mode by which DDX6 might modulate miR-122-directed HCV gene expression.
Our earlier studies showed that during HCV infection, P-bodies are dispersed likely as a result of the re-localization of DDX6 and other P-body components to HCV assembly sites on lipid droplets (Pager et al., 2013). In this study we investigated the role of DDX6 on key steps in the HCV infectious cycle. Our data show that depletion of endogenous DDX6 using target-specific siRNAs decreases HCV protein and RNA levels that were rescued by overexpression of siRNA-resistant 3xFlag-DDX6. DDX6 does not affect HCV translation, but modulates replication and the stability of HCV RNA. Furthermore, the effect of depleting DDX6 on HCV gene expression may be rescued with increasing concentration of exogenous miR-122. Importantly, we find that DDX6 modulates the interaction of miR-122 with a specific binding site in the 5′ UTR, which may facilitate the switch between translation and replication.
2. Results
2.1. Depletion of DDX6 decreases HCV protein and RNA abundance
To examine the effect of DDX6 on HCV gene expression, we used siRNAs to deplete endogenous DDX6 in Huh7 cells, infected with JFH-1 virus and examined HCV protein and RNA abundance by western and northern blot analysis. Decreasing DDX6 abundance resulted in a decrease in HCV core protein, and HCV RNA abundance (Fig. 1). To determine whether HCV gene expression could be rescued we used siRNAs to deplete endogenous DDX6, and overexpressed either 3xFlag-BAP (Bacterial Alkaline Phosphatase) or 3xFlag-RCKΔsi, that contained a mutated siRNA-binding site and thus was resistant to siRNA depletion. Overexpression of 3xFlag-RCKΔsi, but not 3xFlag-BAP, successfully rescued HCV gene expression (Fig. 1).
Fig. 1. Effect of depletion of DDX6 on HCV protein and RNA abundance.
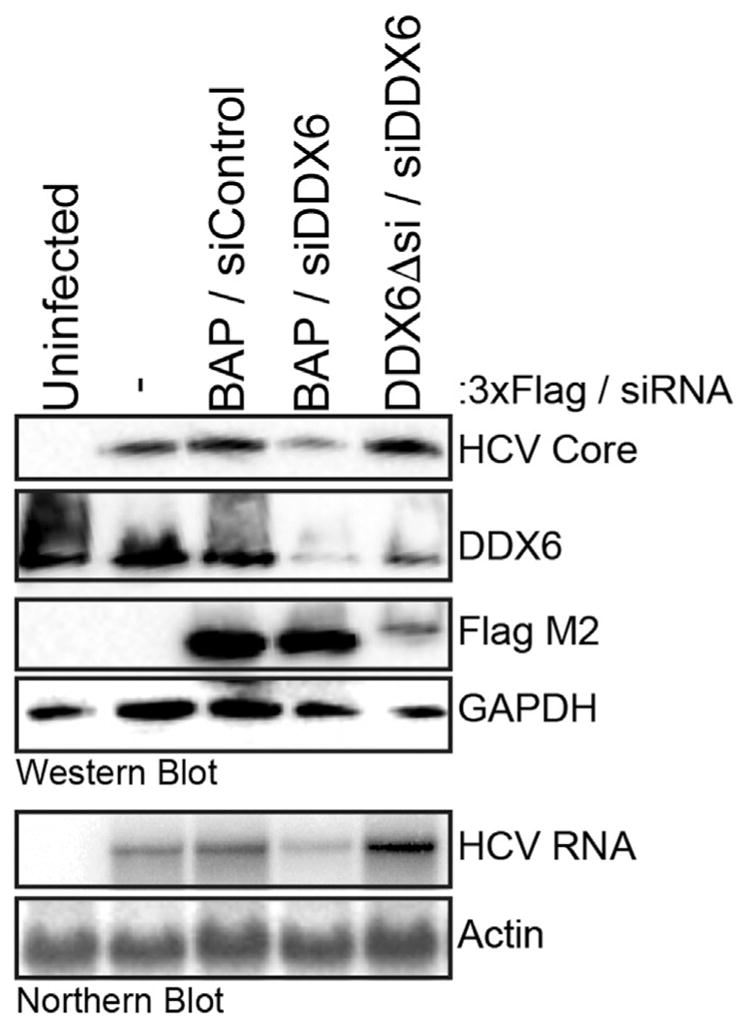
Huh7 cells were co-transfected with control and DDX6 specific siRNAs, and p3xFlag-BAP or p3xFlag-DDX6Δsi plasmids, and 24 h post-transfection infected with JFH-1 at a moi of 0.01. Three days p.i. cells were harvested, and viral and cellular proteins and RNAs analyzed by (A) western and northern blot.
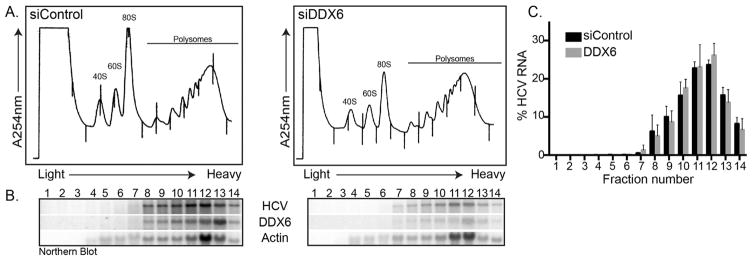
2.2. DDX6 does not change the distribution of HCV RNA in polysomes
In uninfected cells DDX6 and orthologs have been shown to modulate translation (Presnyak and Coller, 2013). To investigate whether DDX6 might similarly affect translation of the HCV poly-protein, we transfected Huh7 cells with control and DDX6 siRNAs, infected with JFH-1 and separated cell lysates in a polysome sucrose gradient. By measuring the absorbance at 254 nm we determined the profile of ribosomal RNA (40S, 60S, 80S and polysomes) across the gradients (Fig. 2A). Although the 80S peak (relative to the polysomes) was higher in JFH-1-infected cells transfected with the control siRNAs compared to the DDX6 siRNAs (Fig. 2A), we did not observe a significant change in the distribution of the 40S, 60S, 80S or polysome peaks. We next isolated RNA from each fraction of the gradient and analyzed the distribution of HCV and cellular RNAs across the gradient by northern blot (Fig. 2B). Our data show β-actin mRNA in both control and DDX6 siRNA transfected cells distributed in the polysome fractions indicating active translation of this cellular mRNA. Although siRNA depletion of DDX6 similarly decreased the abundance of HCV RNA, the distribution of HCV RNA across the gradient, including in the heavy translating polysome fractions, did not change in DDX6 compared to control siRNA transfected cells (Fig. 2C). Treatment of cell lysates with puromycin, a compound that results in early termination of the polypeptide chain, dissociates translating ribosomes, and results in the redistribution of translating mRNAs from heavy fractions into lighter fractions (Blobel and Sabatini, 1971). Puromycin treatment of HCV infected cell lysates from control and DDX6 siRNA transfected cells resulted in a similar redistribution of HCV RNA (data not shown), indicating that the viral RNA in polysome fractions was being actively translated. Together these data indicate that depletion of DDX6 does not change the distribution of HCV RNA on polysomes, suggesting that DDX6 does not affect translation of the viral polyprotein.
Fig. 2. Effect of decreased DDX6 abundance on HCV translation.
Control and DDX6-specific siRNA transfected Huh7 cells were infected with JFH-1 at a moi of 0.01. Three days p.i. cells were harvested, and cell lysates layered onto a 10–60% sucrose polysome gradient. Protein and RNA complexes were separated in the gradient by ultracentrifugation and fractionation. (A) Polysome profile of Huh7 cells transfected with either control or DDX6 siRNAs, and infected with JFH-1. (B) Northern blot of the distribution of HCV RNA, and DDX6 and β-actin mRNAs in the polysome gradient. (C) Percent of HCV RNA distributed across the gradient. Data are from three independent experiments. Error bars display the standard error of the mean.
Additionally, Huh7 5B cells that autonomously express the HCV 1b replicon were transfected with control and DDX6-specific siRNAs, and forty-eight hours post-transfection cells were metabolically labeled with 35S-[Met/Cys] for 30 min, and HCV NS5A was immunoprecipitated and examined by SDS-PAGE and phosphorimager analysis. In accord with the polysome analysis depletion of DDX6 did not affect translation or processing of the HCV polyprotein (data not shown).
2.3. Depleting DDX6 decreases luciferase activity from a subgenomic reporter genome
To further confirm that DDX6 does not affect translation we examined the effects of depleting DDX6 abundance on translation and replication, using the subgenomic JFH-1 Renilla luciferase reporter replicon (Berger et al., 2009). The sgJFH-1-RLuc reporter replicon contains the HCV 5′ UTR and 3′ UTRs, the Renilla luciferase gene, an EMCV internal ribosome entry site (IRES) and the HCV NS3-NS5B genes. The IRES in the HCV 5′ UTR mediates translation of the Renilla luciferase, while the EMCV IRES mediates translation of those HCV proteins (NS3-NS5B) critical for replication. Renilla activity measured at early time points (4 and 8 h) post-transfection of the wild-type (WT) replication-competent reporter replicon directly measures translation of the transfected RNA, while the Renilla activity at later time points (24 and 48 h) is the result of translation of the accumulated newly synthesized HCV genomes (Fig. 3). In contrast, Renilla activity from the replication-incompetent (GND) reporter replicon measures only translation of the transfected RNA and not replication, due to the absence of accumulating replicating HCV genomes (and subsequent RNA decay) at later time points (Fig. 3). We did not observe a significant difference in Renilla luciferase activity at 8 h post-transfection in Huh7 cells transfected with either control or DDX6-specific siRNAs and expressing either the wild-type and GND reporter replicon genomes, suggesting that depletion of DDX6 does not affect translation (Fig. 3). Between 8 and 24 h we noted a significant decrease in the decay of Renilla luciferase activity from both the replication incompetent (GND) and replication competent (WT) reporter genomes between cells transfected with DDX6-specific or control siRNAs (Fig. 3), indicating that a decrease in DDX6 abundance affects the stability of HCV RNA. At 48 h post-transfection we also observed a significant decrease in Renilla luciferase activity of the replication competent reporter genome in cells depleted of DDX6 compared to control (Fig. 3). As expected, at 48 h post-transfection minimal luciferase activity from the replication-incompetent replicon was observed. These data show that a decrease in the abundance of DDX6 may affect the stability of the HCV reporter RNA genomes and/or HCV replication.
Fig. 3.
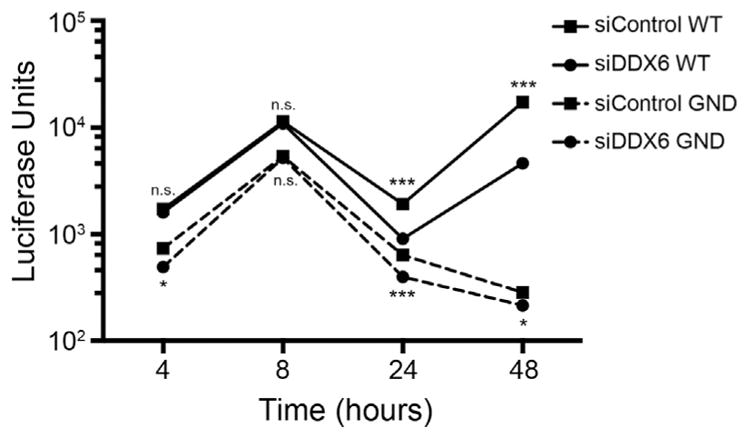
Examination of translation and replication of the subgenomic Renilla luciferase reporter JFH-1 replicon following depletion of DDX6. Replication competent (WT) and incompetent (GND) subgenomic reporter replicons were transfected and expressed in Huh7 cells transfected with either control or DDX6-specific siRNAs. Cells were harvested at 4, 8, 24 and 48 h post-transfection of the replicon genomes. Data are a representative of three experiments showing units of Renilla luciferase activity (presented on a log10 scale) following transfection of the subgenomic replicons. Data from each independent experiment was performed in duplicate and are presented as standard deviation (SD) of the mean. n.s. not significant, *p < 0.05 and ***p < 0.0001 by unpaired Student’s t-test.
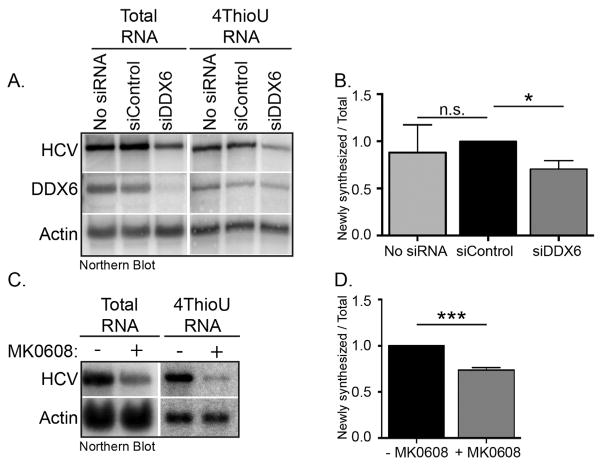
2.4. DDX6 affects the rate of synthesis of new HCV RNA
To confirm the role of DDX6 in HCV replication, we labeled and isolated newly synthesized RNA in HCV-infected cells transfected. To show that this 4-thiouridine assay would measure HCV replication, we incubated JFH-1 infected Huh7 cells with MK-0608, a nucleoside analog shown to inhibit NS5B, the HCV RNA-dependent RNA polymerase (Olsen et al., 2004).
Analysis by northern blotting detected both positive and negative-strand HCV RNA (Fig. 4C), and we determined the effect of MK-0608 on the rate of HCV replication by examining the ratio of newly synthesized HCV RNA to total HCV RNA. Treatment of JFH-1 infected cells for 8 h with MK-0608 significantly decreased the rate of HCV replication (Fig. 4D). We next investigated the effect on the rate of synthesis of new HCV RNA in Huh7 cells transfected with control and DDX6 siRNAs. Similar to our previous observation (Fig. 1) we observed that decreased DDX6 abundance reduced both total and newly synthesized HCV RNA (Fig. 4A). In determining the rate of 4-thiouridine incorporation, observed a 30% reduction when DDX6 abundance is decreased (Fig. 4B), supporting a role for DDX6 in HCV replication.
Fig. 4. Effect of decreased DDX6 abundance on replication of HCV RNA.
Huh7 cells were transfected with either control or DDX6-specific siRNAs. The following day untransfected and cells transfected with siRNAs were infected with JFH-1 at a moi of 0.01. Two days p.i. untransfected cells were incubated with 25 μM MK-0608 for 6 h. Untransfected cells and siRNA transfected cells were incubated with 700 μM 4-thiouridine for 2 h at 37 °C. TRIzol extracted RNA was incubated with MTSEA-biotin XX, and newly synthesized RNA separated from total RNA by streptavidin affinity purification. (A) Northern blot of total and newly synthesized HCV and cellular RNA. (B) Quantification of northern blot (Figure 4A) is from two independent experiments. Data were normalized to control siRNA. (C) Northern blot of total and newly synthesized HCV and cellular RNA of untreated orMK-0608 treated JFH-1 infected cells. (D) Quantification of northern blot (Figure 4C) is from three independent experiments. Data were normalized to the untreated cells. Data are represented as the fraction of newly synthesized RNA per amount of total RNA relative to the control siRNA. Error bars display the mean ± SD. Data were analyzed using an unpaired Student’s t-Test; n.s. not significant, *p < 0.05 and ***p < 0.001. 4-ThioU RNA, 4-thiouridine labeled RNA.
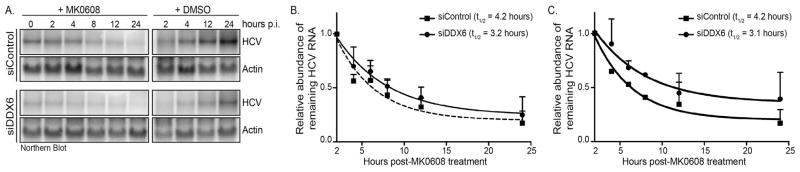
2.5. DDX6 affects stability of HCV RNA
The JFH-1 subgenomic Renilla reporter replicon data showed that depletion of DDX6 dramatically affected HCV replication (Fig. 3). Indeed, labeling of newly synthesized viral RNA showed that DDX6 decreased the rate of HCV replication by 30% (Fig. 4B). In Fig. 3 we also observed a significant change in the decay of luciferase activity from the GND replication-incompetent Renilla reporter replicon between cells transfected with the control siRNA versus DDX6-specific suggesting depletion of DDX6 decreases the stability of the viral genome. We therefore next examined whether the difference in the decrease in luciferase activity of the replication-incompetent reporter replicon, and the decrease in HCV replication observed with replication-competent subgenomic Renilla reporter replicon in the DDX6-depleted cells, were a result of a change in the stability of HCV RNA. Specifically, mock- or JFH-1-infected Huh7 cells transfected with control and DDX6-specific siRNAs, were incubated with the nucleoside analog MK-0608 to inhibit HCV replication, and total RNA at specific times post-drug treatment was isolated. HCV RNA abundance in control and DDX6 siRNA-transfected cells was compared by northern blot analysis (Fig. 5A) and RNA decay curves were generated to determine the half-life of HCV RNA (Fig. 5B). In two different sets of experiments (comprising four and two biological replicates, respectively) we observed that in HCV-infected cells transfected with the control siRNA the half-life of HCV RNA was 4.2 h (Fig. 5B and 5C). Similarly, depleting DDX6 in HCV-infected cells the half-life of HCV RNA was 3.2 h (Fig. 5B) and 3.1 h (Fig. 5C). In generating the RNA decay curves we noted that the effect on the stability of the HCV RNA correlated with the efficiency of the siRNA depletion of DDX6, which contributed to variability between assays (Fig. 5B and 5C). While the viral RNA half-life of HCV-infected cells transfected with the control or DDX6-specific siRNAs between the two sets of experiments did not achieve statistical significance (as determined by an unpaired Student’s t-test), the difference between the two sets of assays were reproducible. These data together with the increased rate in the decay of Renilla luciferase activity from the replication incompetent GND reporter genome (Fig. 3) suggest a role for DDX6 in HCV RNA stability. This change in HCV RNA stability combined with the replication defect likely account for the dramatic loss of HCV RNA.
Fig. 5. Effect of decreased DDX6 abundance on the stability of the HCV RNA.
JFH-1 infected Huh7 cells transfected with either control or DDX6-specific siRNAs were incubated with DMSO or 25 μM MK0608. Cells were harvested at the time of addition of the inhibitor, and 2, 4, 6, 8 and 24 h post-MK0806 treatment. (A) Northern blot shows HCV RNA and actin mRNA abundance over time. (B) Quantification of HCV RNA decay over time. The data presented are from four independent experiments. (C) Quantification of a second set of experiments showing HCV RNA decay over the same time period. The data presented are from two independent experiments. (B–C) HCV RNA was normalized to 1 at the 2 h time point. The graphical representation of the data was generated using a nonlinear regression (curve fit), and one phase decay analyses using the GraphPad Prism6 software. GraphPad software was also used to calculate of RNA half-life from the data presented in (B) and (C). The error bars display the mean ± SD.
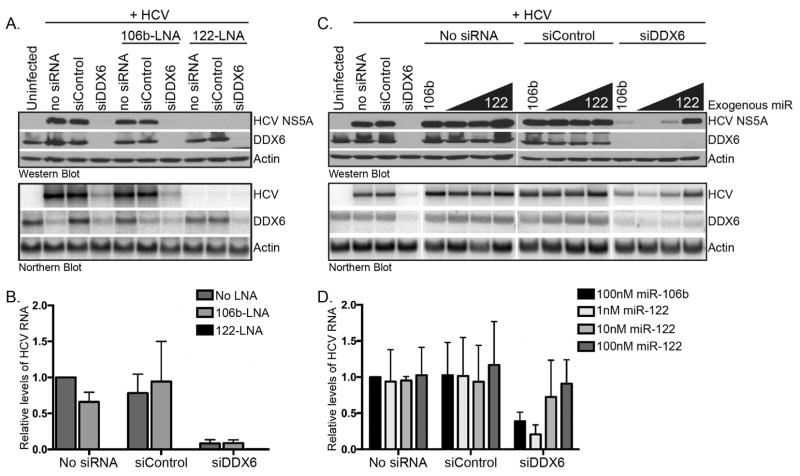
2.6. miR-122 modulates HCV RNA abundance independent of DDX6
Because of the known role of DDX6 in microRNA-mediated gene regulation, we investigated whether DDX6 and miR-122 functioned together to maintain HCV RNA abundance. To this end we first examined whether HCV gene expression required miR-122 when DDX6 was depleted. Huh7 cells were transfected with control and DDX6-specific siRNAs and locked nucleic acid (LNA) antisense oligonucleotides to sequester miR-106b or miR-122, and infected with JFH-1. Sequestration of an unrelated microRNA miR-106b did not affect HCV gene expression in cells transfected with the control siRNA; however in contrast depletion of DDX6 did decrease HCV protein and RNA levels (Fig. 6A and 6B). Sequestration of miR-122 dramatically decreased HCV protein and RNA in mock-transfected cells, and cells transfected with control and DDX6-specific siRNAs (Fig. 6A and 6B). Thus, HCV RNA levels require miR-122 regardless of the abundance of DDX6, and supports other reports of the critical role of miR-122 in HCV infection (Jangra et al., 2010b; Jopling et al., 2008, 2005).
Fig. 6. Effect on HCV gene expression following depletion of DDX6 and miR-122, or addition of exogenous miR-122.
(A) Huh7 cells were transfected with control or DDX6-specific siRNAs alone, or in combination with antisense LNA oligonucleotides targeting miR-122 or miR-106b. Transfected Huh7 cells were infected with JFH-1 at a moi of 0.01. HCV RNA and protein abundance was examined 3 days post-infection by western and northern blot analysis. (B) Quantitation of the relative levels of HCV RNA. HCV RNA abundance was normalized to cells transfected in the absence of siRNA and LNA antisense oligonucleotides. Data are representative of two independent experiments. (C) Huh7 transfected with control or DDX6-specific siRNAs alone, or in combination with 10 nM, 50 nM or 100 nM miR-122 (and 100 nM miR-106b), were infected with JFH-1, and HCV gene expression examined by western and northern blot. (D) Quantitation of the relative levels of HCV RNA. HCV RNA abundance was normalized to JFH-1-infected cells transfected with no siRNA and 100 nM miR-106b. Data are representative of two independent experiments. Error bars display the mean ± SD.
We next investigated whether the decrease in HCV gene expression following depletion of DDX6 was rescued by overexpression of miR-122. Specifically, Huh7 cells were transfected with control and DDX6-specific siRNAs, and exogenous miR-106b or increasing concentration of exogenous miR-122. Huh7 cells transfected with reagent alone (no siRNA) or control siRNA, and increasing concentrations of exogenous miR-122 resulted in increased HCV protein and RNA abundance (Fig. 6C and 6D). Interestingly, increasing concentration of exogenous miR-122 rescued HCV gene expression in Huh7 cells depleted of DDX6 (Fig. 6C and 6D). These data suggest that depletion of DDX6 does not limit the ability of miR-122 to promote the accumulation of HCV RNA.
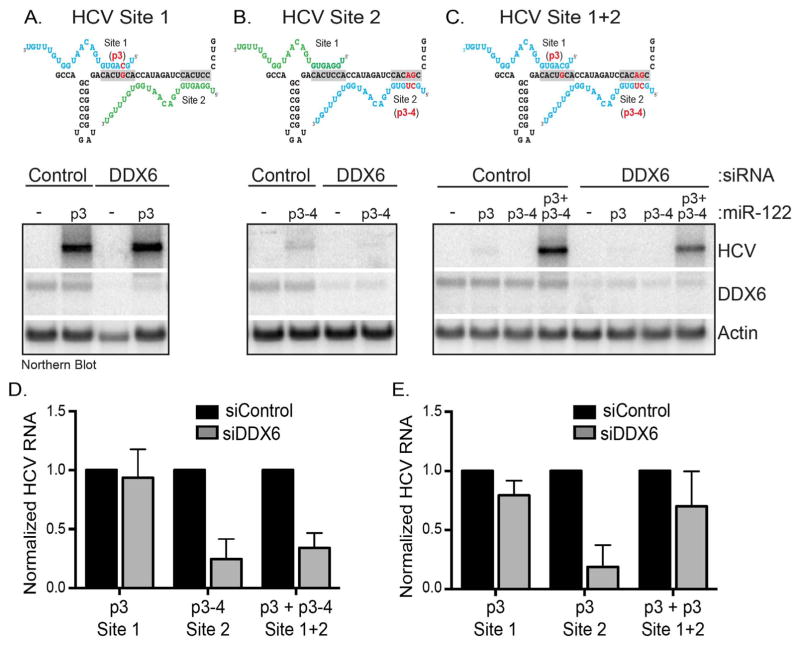
2.7. DDX6 facilitates a site-specific interaction of miR-122 with the HCV 5′ UTR
Binding of different cellular factors to the HCV 5′ UTR promote translation and replication. Interestingly, miR-122 binding sites overlap with the region to which PCBP2 and hnRNP K bind the 5′ UTR (Fan et al., 2014; Flynn et al., 2015), and the alternate binding of either PCBP2 or miR-122 was proposed to facilitate the transition between translation and replication (Masaki et al., 2015). Because DDX6 is a putative helicase we posited that DDX6 might modulate the interaction of miR-122 with the 5′ UTR. We therefore used the previously described miR-122-HCV RNA interaction assay to investigate the effect of DDX6 on miR-122 binding to site 1, site 2 or site 1+2 (Jopling et al., 2008). In this assay viral RNA does not accumulate if HCV genomes have mutations in either miR-122 binding site. Mutant viral RNA abundance is however rescued when synthetic miR-122 with complementary mutations is co-expressed. We used three different mutant HCV genomes (Fig. 7A–7C) that prevented wild-type (WT) endogenous miR-122 binding to HCV RNA at either site 1 (Fig. 7A), site 2 (Fig. 7B) or site 1+2 (Fig. 7C). Huh7 cells were first transfected with control or DDX6-specific siRNAs and synthetic mutant miR-122, then electroporated with mutant HCV genomes, and 5 days post-electroporation HCV RNA abundance was examined by northern blot. For HCV RNA to accumulate, miR-122 must bind site 1 and 2 (Fig. 7) (Jopling et al., 2008). In the absence of complementary mutant miR-122, endogenous miR-122 can only bind to one site (site 1 or site 2) or neither site. Therefore HCV RNA does not accumulate in control siRNA-transfected cells expressing p3 (site 1), p3-4 (site 2) or p3+p3-4 (site 1+2) mutant HCV genomes. However, when complementary mutant p3, p3-4 or p3+p3-4 miR-122 was transfected both sites were occupied with endogenous and/or mutant miR-122, and HCV RNA accumulated (Fig. 7A–7C). Although depletion of DDX6 dramatically reduced HCV gene expression (Fig. 1), depletion of DDX6 did not affect rescue of HCV RNA with p3 miR-122 targeted to site 1 (Fig. 7A and 7D). In contrast, miR-122 binding at site 2 was not rescued by p3-4 miR-122 when DDX6 was depleted (Fig. 7B and 7D). Interestingly, we observed an increased amount of rescue of HCV RNA following DDX6 depletion and when exogenous miR-122 was bound to site 1+2 (Fig. 7C and 7D). Because the rescue of HCV gene expression from the p3-4 site 2 mutant genome in control siRNA-cells was not as robust as the p3 site 1 HCV mutant, we repeated the miR-122-HCV interaction assay using H77 ΔE1/p7 genomes with a single p3 mutation in site 1, site 2 and site 1+2. We similarly observed that depletion of DDX6 limited the rescue of HCV gene expression with complementary p3 miR-122 targeted to site 2, compared to site 1 (Fig. 7E). These data suggest that DDX6 modulates the interaction of miR-122 specifically with site 2 in the 5′ UTR.
Fig. 7. Effect of decreased DDX6 abundance on the interaction of miR-122 with HCV RNA at site 1, site 2 and site 1+2 in the 5′ UTR.
Huh7 cells were transfected with control or DDX6-specific siRNAs and exogenous miR-122 duplexed RNA. These cells were electroporated with in vitro transcribed H77ΔE1-p7 RNA with mutations inhibiting the interaction of endogenous miR-122 with site 1 (p3), site 2 (p3-4) or site 1+2 (p3+p3-4). H77 RNA was analyzed by northern blot 5 days post-electroporation. (A–C) Schematic and northern blots of mutant HCV genomes bound by endogenous (green) and/or exogenous miR-122 (blue) at site 1 (A; p3), site 2 (B; p3-4) and site 1+2 (C; p3+p3-4). (D) Quantitation of each northern blot (Fig. 7A–C) from three independent experiments. (E) Quantitation of relative HCV RNA abundance following transfection of control and DDX6-specific siRNAs and p3 complementary miR-122, and electroporation of mutant H77ΔE1-p7 RNA with p3 mutations at site 1, site 2 and site 1+2. Effects of DDX6 depletion on each mutant genome were normalized HCV RNA abundance of the mutant HCV genomes in cells transfected with control siRNAs. Error bars display the mean ± SD.
3. Discussion
RNA viruses co-opt numerous cellular RNA binding proteins such as the DEAD-box RNA helicase DDX6 to aid different steps in the infectious cycle. Brome mosaic virus, for example relocalizes translating viral RNA to replication complexes by subverting Dhh1, an ortholog of RCK/54, and other proteins associated with the mRNA decapping complex (Mas et al., 2006). Retroviruses such as foamy virus and HIV in contrast, subvert DDX6 for capsid assembly (Reed et al., 2012; Yu et al., 2011). We previously observed DDX6 co-localizing with HCV core protein on lipid droplets, and depletion of DDX6 decreased cell-associated and extracellular virus titers, even though the amount of core protein released between control and DDX6-siRNA depleted cells was similar. These data together supported a role for DDX6 in HCV assembly (Pager et al., 2013). However, the dramatic decrease in HCV protein and RNA abundance, suggested that DDX6 might function at additional steps in the HCV infectious cycle. We now show that DDX6 also affects HCV RNA stability and replication, likely by modulating the interaction of miR-122 with the 5′ UTR.
There are numerous steps in the infectious pathway at which this cellular DEAD-box helicase might act to modulate HCV gene expression. We first investigated the effect of DDX6 on HCV translation by polysome gradient analysis (Fig. 2). Although the abundance of HCV RNA was decreased in cells depleted of DDX6 (Fig. 1 and 2B), the distribution of HCV RNA on polysomes between control and DDX6 siRNA-transfected cells did not change (Fig. 2C), suggesting that DDX6 did not modulate translation of the HCV polyprotein. After treating lysates from DDX6-depleted cells with puromycin, an elongation inhibitor (Blobel and Sabatini, 1971), translating ribosomes were disassembled into 40 S and 60 S subunits, and cellular and HCV RNA mRNA was released from the polysomes (data not shown), showing HCV RNA in the polysome fractions was indeed being translated. We also investigated the role of DDX6 on HCV translation using the replication competent and incompetent subgenomic luciferase replicons, and found no significant difference in Renilla luciferase activity during the period of protein synthesis early post-transfection between control and DDX6 depleted Huh7 cells. Together these data show that DDX6 does not modulate HCV translation. Although our data contrast previous studies showing an affect of DDX6 on HCV translation using HCV luciferase replicon genomes (Huys et al., 2013; Scheller et al., 2009), it is possible that replication efficiency and permissiveness between different cell passages and Huh7 lines (Lohmann et al., 2001; Sainz et al., 2009) contributes to these different effects on HCV translation that we observed when DDX6 was depleted. In agreement with this interpretation, we performed a third assay namely 35S-metabolic labeling (data not shown), which supports an effect downstream of translation. In the absence of the full-length genome and viral proteins, DDX6 was shown not to affect HCV IRES-directed translation of a dicistronic RNA (Jangra et al., 2010a).
Data from the expression of the subgenomic Renilla reporter replicons suggested a role for DDX6 in stabilizing the viral RNA and HCV replication (Fig. 3). We previously observed some DDX6 foci localizing adjacent to replication complexes (Pager et al., 2013). We confirmed the role of DDX6 in HCV replication by labeling and isolating newly synthesized HCV RNA with 4-thiouridine and streptavidin beads (Fig. 4). By northern blot we determined a 30% decrease in the rate of HCV RNA synthesis (Fig. 4B). Jangra et al. previously reported DDX6 affected HCV replication, the study however only showed a decrease in HCV protein and RNA abundance, and viral titers following siRNA depletion of DDX6 (Jangra et al., 2010a). Although Huys et al., similarly showed that depletion of DDX6 decreased replication of the subgenomic and full-length luciferase HCV reporter genomes, our study of labeling newly synthesized HCV RNA is the first to show that DDX6 impairs the rate of replication (Fig. 4B).
Because decreased HCV replication cannot completely account for the effect of depletion of DDX6, and that we also observed a change in the decay of luciferase activity from the replication-incompetent (GND) replicon between control and DDX6-specific siRNA transfected cells between 8 and 24 h, we speculated that DDX6 might modulate the stability of HCV RNA. Indeed by examining the abundance of HCV RNA at specific time points following inhibition of HCV replication with the nucleoside analog MK-0608 (Fig. 5) we determined that the half-life of HCV RNA decreased from 4.2 h to 3.1 h in Huh7 cells depleted of DDX6 (Fig. 5). Our study with the full length HCV genome, contrasts that of Scheller et al. who reported that siRNA depletion of DDX6 had no effect on the putative stability of a reporter mRNA containing the HCV 5′ and 3′ UTRs flanking the luciferase gene at 4 h post-transfection (Scheller et al., 2009). The ~25% decrease in HCV RNA stability in DDX6-depleted cells is in the range of the ~ 16% decrease in the half-life of replication incompetent H77S/GLuc22A genome (21.9 h to 18.2 h) following sequestration of miR-122 with an antisense 2′-O-methylated oligonucleotide (Shimakami et al., 2012).
DDX6 facilitates microRNA-mediated gene regulation (Chen et al., 2014; Chu and Rana, 2006; Mathys et al., 2014; Rouya et al., 2014). Thus we also examined how DDX6 might contribute to miR-122 regulation of HCV gene expression. Our data show that HCV RNA requires miR-122, even in the absence of DDX6 (Fig. 6A), and that overexpression of miR-122 can rescue HCV RNA levels in DDX6-depleted cells (Fig. 6C). While miR-122 and DDX6 might function independently to modulate HCV RNA, it is also possible that DDX6 acts before miR-122 to facilitate the interaction with the 5′ UTR and accumulation of HCV RNA. Our data show that depletion of DDX6 dramatically decreases HCV protein and RNA abundance (Fig. 1). Using the HCV-miR-122 interaction assay we were surprised when exogenous miR-122 targeted to interact with site 1, rescued HCV gene expression in both control and DDX6 siRNA-transfected cells (Fig. 7A). In contrast, exogenous miR-122 targeting site 2 poorly rescued HCV RNA levels in DDX6 depleted cells (Fig. 7B). Exogenous miR-122 binding site 1 and 2 rescued HCV RNA more than at just site 2 in DDX6 depleted cells, but not to HCV RNA levels in cells transfected with control siRNAs (Fig. 7C). Together these data suggest that the binding of miR-122 to site 2 and accumulation of HCV RNA are modulated by DDX6.
The ability of miR-122 to bind two sites presents unique points of regulation, where binding of miR-122 at individual or both sites, sequentially or cooperatively might modulate distinct steps in the HCV infectious cycle (Garcia-Sastre and Evans, 2013; Thibault et al., 2015). Jangra et al. showed that HJ3-5 with a p6 mutation at site 2 was rescued by exogenous miR-124, wild-type miR-122 or miR-122 p6, while the p6 mutation at site 1 was rescued only by the corresponding exogenous miR-122 p6 (Jangra et al., 2010b). Sequestration of miR-122 with a locked nucleic acid (LNA) antisense antagomir did not dramatically affect viral gene expression and titers of a chimeric HCV with a stem loop from the U3 snRNA at site 1 and intact site 2 (Li et al., 2011). More recently Thibault et al., showed in Xrn1-depleted cells, that binding of miR-122 at site 1 increased HCV gene expression more than miR-122 bound at site 2 (Thibault et al., 2015). Thibault et al. noted that the differential rescue might be a result of the p3 point mutation at site 2. Rather than examining luciferase activity as a proxy for HCV gene expression (Thibault et al., 2015), in our miR-122-HCV RNA interactions assays we measured rescue of HCV RNA abundance by northern blot (Fig. 7). We similarly observed that rescue of the H77 ΔE1/p7 genome containing the p3-4 mutation at site 2, in Huh7 cells transfected with a control siRNA was reduced. However in comparing the rescue of HCV RNA in cells depleted of DDX6, the data were normalized to the expression of the particular HCV mutant genome in cells transfected with the control siRNA. As such any effects we observed in DDX6-depleted cells integrated changes in the replicative capacity resulting from the mutation in the site 2. We also determined that a single p3 mutation in site 2 limited the rescue of HCV RNA in DDX6-depleted cells. While HCV infection assays have shown site 1 as the dominant miR-122 binding site in modulation of HCV RNA (Jangra et al., 2010b; Li et al., 2011; Thibault et al., 2015), in vitro assays show two miR-122 molecules binding sites 1 and 2 with different binding affinities (Mortimer and Doudna, 2013). Site 2 was shown to have a higher affinity for miR-122 compared to site 1 likely a result of additional base pair interactions downstream of the seed-sequence binding site (Mortimer and Doudna, 2013). The significance of these additional base-pairing interactions in infected cells is unclear however it is possible that DDX6 ensures the correct interaction of miR-122 within the site 2 seed sequence and additional binding site at nucleotides 30 and 31, such that miR-122 is also able to efficiently bind site 1.
The RNA genome is pivotal in the life of positive-sense RNA viruses as it functions as a mRNA that is translated to produce viral proteins, a template during replication for negative-strand and subsequent positive-strand synthesis, and the nascent viral genome that is assembled into virions. In different RNA viruses the key switch from translation to replication and initiation of negative-strand synthesis, is governed by long-range RNA-RNA interactions and circularization of the viral RNA, either by complementary sequences in the 5′ and 3′ UTRs or RNA-protein interactions (Nicholson and White, 2014). In HCV the interaction of miR-122 with the 5′ UTR has been proposed to act as the switch between translation and replication (Masaki et al., 2015). In particular, the cellular RNA binding protein PCBP2 was shown to bind both the 5′ and 3′ UTR of HCV (Fig. 8A), and facilitate circularization of the HCV genome, and viral translation and replication (Flynn et al., 2015; Masaki et al., 2015; Wang et al., 2011). Interestingly the binding of miR-122 to either site 2 or site 1+2 hindered the interaction of PCBP2 with the 5′ UTR, and stimulated replication of HCV RNA (Fig. 8B) (Masaki et al., 2015). We propose that DDX6 is key to the miR-122-directed translation-to-replication switch. In particular cellular DEAD-box RNA helicases remodel RNA-RNA interactions, RNA-protein interactions, and facilitate the loading of protein complexes on RNA (Putnam and Jankowsky, 2013), and the helicase activity of DDX6 has been shown to be important for HCV gene expression (Jangra et al., 2010a). To this end DDX6 binding HCV RNA might aid the displacement of PCBP2 and the loading of miR-122-Ago2 complexes, to affect the switch between translation and replication. In our model depletion of DDX6 would decrease the association of miR-122 with the 5′ UTR (Fig. 7), thus increasing the susceptibility of the viral RNA to degradation by cellular 5′-to-3′ exonucleases (Fig. 5). Thereby the translation-to-replication switch would be effectively hindered, delaying the initiation of negative-strand synthesis (Fig. 4) and thus the accumulation of genomes available for replication (Fig. 3). The mechanism by which DDX6 acts is unclear and future studies will determine whether DDX6 displaces PCBP2 from the HCV 5′ UTR, remodels the 5′ UTR RNA structure, or facilitates a specific interaction of miR-122-Ago2 complexes with site 2 in the 5′ UTR.
Fig. 8. A model for the interaction of DDX6 and miR-122 with the HCV genome.
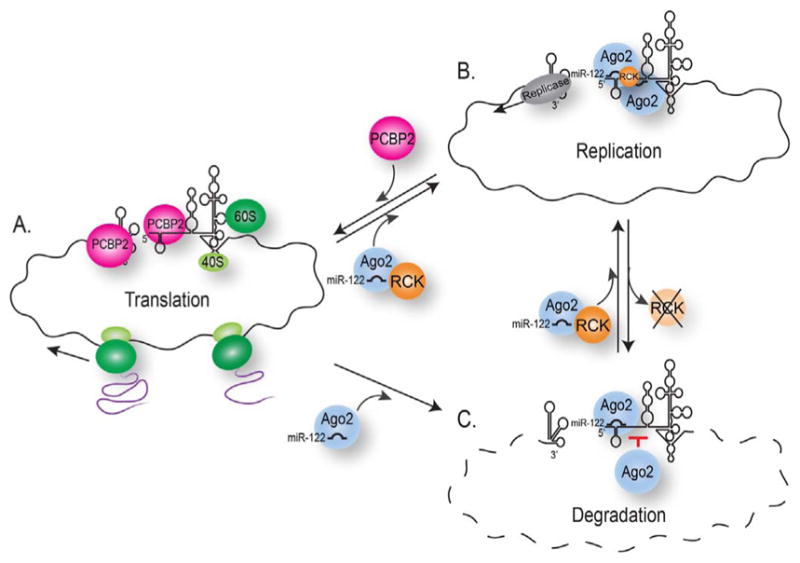
Early during HCV infection the positive-sense RNA genome acts as transcript for translation of viral proteins, and later is used as a template for replication of the viral genome. (A–B) The transition from translation to replication is closely regulated by components within viral RNA secondary structure, displacement of PCBP2, and binding of host miR-122, and host and viral proteins. (C) In this study we also show that depletion of DDX6 affects the interaction of miR-122 with site 2 in the 5′ UTR to decrease HCV RNA stability, and that by modulating the interaction of miR-122 with the 5′ UTR, DDX6 is a key regulator in the switch between translation and replication, where depletion of DDX6 delays the transition to and initiation of replication.
More recently, HCV was shown to replicate independent of miR-122 (Hopcraft et al., 2016; Thibault et al., 2013), and specific mutations in the HCV 5′ UTR not only facilitate HCV gene expression when miR-122 abundance is decreased but also in the absence of miR-122 (Hopcraft et al., 2016; Israelow et al., 2014). Interestingly depletion of DDX6 affects both miR-122 dependent and independent HCV gene expression. The mode by which DDX6 affects miR-122 independent gene expression remains to be determined; however it is possible that DDX6 facilitates the displacement of cellular factors such as PCBP2 from the 5′ UTR, where this displacement by DDX6 is the switch between translation and replication, and that the interaction of miR-122 increases the efficiency of the switch.
4. Materials and methods
4.1. Cell culture and reagents
Huh-7 cells were cultured in DMEM supplemented with 10% fetal bovine serum, 1% non-essential amino acids and 200 μM L-glutamine at 37 °C with 5% CO2.
MK-0608 (7-deaza-2′-C-methyladenosine; sc-284810) purchased from Santa Cruz Biotechnologies, was resuspended to 100 μM in DMSO. A working stock of 25 μM was prepared in cell culture media.
4.2. Cloning and site-directed mutagenesis
p3xFlag-DDX6 was created using standard molecular biology techniques. Specifically DDX6 was PCR amplified from pmRFP-DDX6 (a generous gift from Nancy Kedersha, Brigham and Women’s Hospital) and cloned into pCR2.1-Topo. DDX6 was subcloned from pCR2.1-Topo into p3xFlag-CMV 7.1 (Sigma) using the HindIII and XbaI restriction enzyme sites. We then used site-directed mutagenesis with the QuikChange XLII to create an siRNA-resistant gene of DDX6 (3xFlag-RCKΔsi). In particular six nucleotides in the siRNA binding were mutated, while maintaining the correct amino acid sequence (from gcagaaaccctatgagatt to tcagaagccttacgaaatc). These mutations were introduced with the following primers: prCTP34 (5′-CTTTCCCTCTTAGTGTACAGAAGTTCATGAATTCCCATCTTCAGAAGCCTTACGAAATCAACCTGATGGAGGAACTAAC-3′) and prCTP35 (5′-GTTAGTTCCTCCATCAGGTTGATTTCGTAAGGCTTCTGAAGATGGGAATTCATGAACTTCTGTACACTAAGAGGGAAAG-3′). p3xFlag-BAP (Bacterial Alkaline Phosphatase, Sigma) was used as a control plasmid.
4.3. siRNA and plasmid transfections
Commercially synthesized siRNA oligonucleotides (IDT) were annealed to a final concentration of 20 μM (Pager et al., 2013). The following siRNA oligonucleotides were used: the RISC-free siRNA (Dharmacon, D-001220-01-05), siGL2 sense-strand 5′-CGUACGCGGAAUACUUCGAUU-3′ and siGL2 antisense-strand 5′-UCGAAGUAUUCCGCGUACGUU-3′, and siDDX6 sense-strand 5′-GCAGAAACCCUAUGAGAUUUU-3′ and siDDX6 antisense-strand 5′-AAUCUCAUAGGGUUUCUGCUU-3′. Huh7 were seeded in a 60 mm or 100 mm cell culture dish, and 24 h later transfected with siRNAs (25 nM or 50 nM) alone using either Dharmafect I (Dharmacon) or Lipofectamine 2000 (Invitrogen), or with 25 nM siRNAs and 1 μg plasmid DNA using Lipofectamine 2000 (Invitrogen). Transfection reactions were prepared according to the manufacturer’s specifications.
4.4. JFH-1 virus infections
Huh7 cells seeded in 60 mm tissue culture plates and transfected with siRNAs (or siRNAs and p3xFlag-RCKΔsi or -BAP) were infected with the JFH-1 virus at a multiplicity of infection (moi) of 0.01 for 5 h at 37 °C. Following incubation, infected cells were trypsinized, replated into 100 mm tissue culture plates and incubated at 37 °C. Three days post-infection (dpi) cells were harvested for polysome sucrose gradients, western blot and northern blot analysis. For polysome gradients cells in 100 mm tissue culture plates were transfected and re-plated into two 150 mm tissue culture plates following JFH-1 infection.
4.5. Polysome sucrose gradient analysis
Media on uninfected and JFH-1-infected Huh7 cells was replaced 2 dpi, and cells were incubated for a further 18 h at 37 °C. Two hours before harvesting, the media on the cells was replaced again. Immediately prior harvesting, cells were incubated with 0.1 mg/ml cycloheximide (Sigma) for 3 min at 37 °C. Cells were placed on ice and washed twice with cold PBS containing 0.1 mg/ml cycloheximide. Excess PBS was aspirated, and cells were scraped and collected. Cells pooled from two 150 mm plates were pelleted at 14,000 rpm for 5 s, and excess PBS removed. Each cell pellet was lysed for 10 min on ice in 600 μl lysis buffer (150 mM NaCl, 15 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 0.1 mg/ml cycloheximide, 1 mg/ml heparin [Sigma] and 1% Triton X-100). The chilled lysate was then centrifuged at 8400 rpm for 5 min at 4 °C. The cleared supernatant was layered on to a 10–60% sucrose gradient containing 150 mM NaCl, 15 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 1 mg/ml heparin and 0.1 mg/ml cycloheximide. To fractionate lysates in puromycin gradients, cells were lysed in buffer (500 mM KCl, 15 mM Tris-HCl (pH 7.5) and 15 mM MgCl2) containing 1% Triton X-100 in the absence or presence of 2 mM puromycin (Sigma). To facilitate release of ribosomes from mRNAs, lysates were first incubated on ice for 15 min and then incubated at 37 °C 10 min. Hereafter lysates were clarified as before. Clarified supernatants were separated in 10–60% sucrose gradients containing puromycin buffer. Polysome gradients were centrifuged in a SW41 rotor at 35,000 rpm for 2 h 45 min at 4 °C. The Isco fractionation system (Brandel) was used to collect fourteen 750 μl fractions. The distribution of ribosomal RNA in the gradient was monitored at 254 nm. RNA was extracted from each fraction (Wehner et al., 2010).
4.6. Protein isolation and western blot analysis
Cells were washed once with cold PBS, scraped, collected, and pelleted, following a brief high-speed spin. Excess PBS was removed, and cells were lysed for 20 min on ice in RIPA buffer (100 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% sodium deoxycholic acid, 1% Triton-X100 and 0.1% SDS), containing the EDTA-free Complete protease inhibitors cocktail (Roche). Lysates were centrifuges at 14,000 rpm for 20 min at 4 °C, and clarified supernatants collected. Protein concentration was determined with Bradford reagent (BioRad). Proteins (20 μg) were separated in 10% polyacrylamide/SDS gels at 100 V, and transferred to PVDF membranes at 100 V at 4 °C for 1 h. Membranes were first blocked for 1 h at room temperature with 5% skim milk powder in PBS containing 0.1% Tween-20 (PBS/T), and then incubated with primary antibodies overnight at 4 °C. Membranes were washed with PBS/T and incubated with HRP-conjugated secondary antibodies (Donkey anti-Mouse and Donkey anti-Rabbit; Santa Cruz Biotechnologies) at 1:10,000 for 1 h at room temperature. Proteins were detected with ECL blotting reagent (Pierce, Thermo Scientific) and Biomax light film (Kodak) or BioRad ChemiDoc XRS & MP imager. The following primary antibodies were used for immunoblotting: rabbit anti-DDX6 (DDX6) (1:5000; Bethyl Laboratories A300–460A), rabbit anti-β-Actin (Sigma A2066); mouse anti-HCV Core C7-50 (1:1000; Abcam ab2740), and anti-HCV NS5A (1:10,000; a generous gift from Dr. Charles Rice, Rockefeller University).
4.7. RNA extractions and Northern blot analysis
RNA from uninfected and JFH-1 infected cells was extracted with TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. An amount of 10 μg total RNA was analyzed by northern blot analysis. To isolate RNA from sucrose gradients, input lysate and isolated fractions were first incubated with 0.5% SDS and 0.25 mg/ml proteinase K (New England Biolabs) at 37 °C for 30 min. An equal volume of acid-phenol: chloroform (Ambion) was used for the first extraction, and an equal volume of chloroform for the second extraction. Isolated RNA was precipitated overnight at −20 °C with 40 μg glycogen (Roche), 1/10 vol sodium acetate and 2.5 volumes ethanol. Total RNA isolated from each fraction was examined by northern blot analysis. Isolated RNA was resuspended in loading buffer (1% MOPS-EDTA-sodium acetate (MESA), 67% formaldehyde, and 50% formamide), denatured for 15 min at 65 °C and separated in 1.2% agarose/6.7% formaldehyde gels. RNA was separated in gels in MESA buffer, containing 18% formaldehyde, at 100 V, and then transferred overnight and UV cross-linked to a Zeta probe membrane (BioRad). HCV and β-actin RNA was detected with [α–32P] dATP-RadPrime DNA labeled (Invitrogen) probes and ExpressHyb hybridization buffer (Clontech) according to manufacturer’s protocols.
4.8. 4-Thiouridine (4ThioU) labeling and isolation of newly synthesized RNA
Newly synthesized HCV RNA was labeled and isolated following a previously described protocol (Duffy et al., 2015; Duffy and Simon, 2016; Norman and Sarnow, 2010). In brief, Huh7 cells transfected with siRNAs were infected with JFH-1 at a moi of 0.01. Two dpi the media was removed, the cells washed twice with PBS and then incubated at 37 °C for 8 h in OptiMEM (Invitrogen) without or with MK0608. After 6 h the cells were labeled with 700 μM 4-thiouridine (4-ThioU; Sigma) for 2 h. 4-ThioU is readily taken up by Huh7 cells and converted 4-thiouridine-triphosphate; cellular RNA polymerase II and HCV 5B RNA-dependent RNA polymerase (RdRp) incorporate the 4SU-triphosphate into newly synthesized RNA transcripts (Cleary et al., 2005). Following incubation with 4ThioU, cells were harvested and RNA extracted with TRIzol. To remove any remaining contaminants from the TRIzol-isolated RNA, the RNA was re-extracted with acid phenol/chloroform. Twenty μg of RNA was used for further isolation. To modify the thiol group on the 4ThioU molecules, the RNA was incubated at room temperature and in the dark for 30 min with 16.4 μM MTSEA-biotin-XX (Biotium Catalog no. 89139-636). To remove unreacted MTS-biotin from the RNA samples the biotinylated RNA was incubated with chloroform and centrifuged for 10 min at 12,000×g at 4 °C. Hereafter the biotinylated RNA was precipitated at room temperature for 10 min with 1/10 vol of 5 M NaCl and 1 vol of isopropanol. The RNA was pelleted in a microfuge at high speed at 4 °C for 10 min and then resuspended in RNase-free water. The biotinylated RNA was further isolated with streptavidin beads. Dynabeads MyOne Streptavidin C1 magnetic beads (Life Technologies) were prepared by a series of washes in a high-salt wash buffer (1 M NaCl, 10 mM EDTA and 100 mM Tris-HCl (pH 7.4) and 0.05% Tween 20) and then blocked with 40 ng/μl glycogen for 1 h minutes at room temperature. Biotinylated RNA was incubated with the streptavidin beads for 15 min at room temperature and then washed with MPG buffer. The biotinylated RNA was eluted from the beads with elution buffer (100 mM dithiothreitol (DTT), 1 M NaCl, 10 mM EDTA and 100 mM Tris-HCl (pH 7.4) and 0.05% Tween 20). Eluted biotinylated RNA was precipitated and then analyzed by northern blot.
4.9. Subgenomic JFH1 Renilla luciferase (sgJFH-1-RLuc) assay
Replication competent and incompetent subgenomic Renilla luciferase replicon plasmids (a kind gift from Dr. Glenn Randall, University of Chicago) (Berger et al., 2009) were linearized with XbaI, and subgenomic replicon RNA was in vitro transcribed using the MegaScript T7 kit (Ambion) according to the manufacturer’s protocol. Huh7 cells were transfected with control and DDX6-specific siRNAs and 24 h post-transfection were seeded into 24 well plates. Cells were transfected 24 h later with 0.8 μg of replicon RNA using TransMessenger reagent (Qiagen). Media was changed after 1 h, and lysates prepared in Passive Lysis Buffer (according to the manufacturer’s instructions; Promega) at 4, 8, 24, and 48 h post-transfection of replicon RNA. Renilla luciferase activity was measured in three technical replicates using the Renilla Luciferase Assay System (Promega) with a 96-well luminometer (Synergy H1, BioTek).
4.10. RNA stability assay
Huh7 cells were seeded in 100 mm tissue culture plates and transfected with control and DDX6-targeting siRNAs using DharmaFect I (Dharmacon). The following day transfected cells were infected with JFH-1 at a moi of 0.01 for 6 h at 37 °C. Cells were incubated overnight in new media. The following day cells were trypsinized and re-plated into 60 mm cell culture plates and incubated at 37 °C. Twenty-four hours later, media was removed and replaced with media containing DMSO or 25 μM MK-0608. At 0, 2, 4, 6, 8, 12, and 24 h post-treatment, the media was removed and TRIzol added to the cells. RNA was extracted as before, and HCV and β-actin RNA abundance examined by northern blot and phosphorimager analysis. DDX6 depletion was confirmed by western blot analysis from untreated JFH-1 infected cells harvested at the 24 h time point. One phase decay analysis and half-life of HCV RNA was determined using GraphPad Prism software.
4.11. miR-122-HCV interaction assay
Mutant H77ΔE1/p7 genomes containing mutation at the miR122 site, site 2 and site 1+2 (Fig. 7A) have been previously described (Jopling et al., 2008; Machlin et al., 2011). HCV RNA was in vitro transcribed using the MegaScript kit (Ambion) according to the manufacturer’s protocol. Similar to the preparation of siRNA duplexes (Pager et al., 2013), exogenous miR-122 was prepared by annealing p3 and p3-4 miR-122 oligonucleotides with the miR-122* oligonucleotide. Huh7 cells in 100 mm tissue culture plates were transfected with 25 nM siRNAs and 50 nM of duplexed exogenous miR-122. The following day cells were trypsinized and centrifuged at 1000 rpm for 5 min. The cells were first washed once with PBS and then twice with Cytomix buffer (120 mM KCl, 150 mM CaCl2, 10mMK2HPO4, 25mM Hepes, 2 mM EDTA and 2 mM MgCl2; pH 7.6). Huh7 cells in the Cytomix buffer were placed in a 0.4 cm BioRad cuvette and electroporated with 10 μg in vitro transcribed mutant H77ΔE1/p7 RNA using the BioRad GenePulser Xcell (exponential decay, ∞ resistance, 270 V, 950 μF). Following electroporation the cells were allowed to recover at room temperature for 10 min and then plated into 100 mm tissue culture dishes and incubated at 37 °C. Electroporated cells were further transfected with 50 nM miR-122 duplexes at 1 and 3 days postelectroporation. The sequences of the miR-122 oligonucleotides are as follows: miR-122 p3: 5′-UGCAGUGUGACAAUGGUGUUUGU-3′; miR-122 p3-4: 5′-UGCUGUGUGACAAUGGUGUUUGU-3′; and miR-122*: 5′-AAACGCCAUUAUCACACUAAAUA-3′. The following mutant H77ΔE1/p7 genomes were used: p3 mutation at site 1, a p3-4 mutation at site 2, and a p3+p3-4 mutation at sites 1+2 (Fig. 7A), and p3 mutation at site 1, a p3 mutation at site 2, and a p3 mutation at both sites 1+2.
Acknowledgments
Funding
This work was supported in part from grants from the NIH (AI069000, AI47365), the American Association for the Study of Liver Diseases (AASLD), and from start-up funds and Faculty Research Awards Program from the University at Albany-SUNY and the State of New York.
We would like to thank Dr. Nancy Kedersha (Brigham and Women’s Hospital, Harvard University), Dr. Glenn Randall (University of Chicago), Dr. Charles Rice (Rockefeller University), and Dr. Takaji Wakita (National Institute of Infectious Diseases, Tokyo) for valuable reagents. We especially thank Marlene Belfort, Ing-Nang Wang, Gabriele Fuchs, Kouacou Konan and members of the Pager lab for helpful comments on the manuscript.
References
- Akao Y, Marukawa O, Morikawa H, Nakao K, Kamei M, Hachiya T, Tsujimoto Y. The rck/p54 candidate proto-oncogene product is a 54-kilodalton D-E-A-D box protein differentially expressed in human and mouse tissues. Cancer Res. 1995;55:3444–3449. [PubMed] [Google Scholar]
- Bartenschlager R, Lohmann V, Penin F. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat Rev Microbiol. 2013;11:482–496. doi: 10.1038/nrmicro3046. [DOI] [PubMed] [Google Scholar]
- Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci USA. 2009;106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G, Sabatini D. Dissociation of mammalian polyribosomes into subunits by puromycin. Proc Natl Acad Sci USA. 1971;68:390–394. doi: 10.1073/pnas.68.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Boland A, Kuzuoglu-Ozturk D, Bawankar P, Loh B, Chang CT, Weichenrieder O, Izaurralde E. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol Cell. 2014;54:737–750. doi: 10.1016/j.molcel.2014.03.034. [DOI] [PubMed] [Google Scholar]
- Chiu YL, Rana TM. RNAi in human cells: basic structural and functional features of small interfering RNA. Mol Cell. 2002;10:549–561. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary MD, Meiering CD, Jan E, Guymon R, Boothroyd JC. Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nat Biotechnol. 2005;23:232–237. doi: 10.1038/nbt1061. [DOI] [PubMed] [Google Scholar]
- Duffy EE, Simon MD. Enriching s4 U-RNA using Methane Thiosulfonate (MTS) chemistry. Curr Protoc Chem Biol. 2016;8:234–250. doi: 10.1002/cpch.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy EE, Rutenberg-Schoenberg M, Stark CD, Kitchen RR, Gerstein MB, Simon MD. Tracking Distinct RNA Populations Using Efficient and Reversible Covalent Chemistry. Mol Cell. 2015;59:858–866. doi: 10.1016/j.molcel.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–1273. e1261. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan B, Lu KY, Reymond Sutandy FX, Chen YW, Konan K, Zhu H, Kao CC, Chen CS. A human proteome microarray identifies that the heterogeneous nuclear ribonucleoprotein K (hnRNP K) recognizes the 5′ terminal sequence of the hepatitis C virus RNA. Mol Cell Proteom: MCP. 2014;13:84–92. doi: 10.1074/mcp.M113.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RA, Martin L, Spitale RC, Do BT, Sagan SM, Zarnegar B, Qu K, Khavari PA, Quake SR, Sarnow P, Chang HY. Dissecting noncoding and pathogen RNA-protein interactomes. RNA. 2015;21:135–143. doi: 10.1261/rna.047803.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A, Evans MJ. miR-122 is more than a shield for the hepatitis C virus genome. Proc Natl Acad Sci USA. 2013;110:1571–1572. doi: 10.1073/pnas.1220841110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopcraft SE, Azarm KD, Israelow B, Lévêque N, Schwarz MC, Hsu TH, Chambers MT, Sourisseau M, Semler BL, Evans MJ, Coyne CB. Viral determinants of miR-122-independent hepatitis C virus replication. mSphere. 2016;1:e00009–e00015. doi: 10.1128/mSphere.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huys A, Thibault PA, Wilson JA. Modulation of hepatitis C Virus RNA accumulation and translation by DDX6 and miR-122 are mediated by separate mechanisms. PLoS One. 2013;8:e67437. doi: 10.1371/journal.pone.0067437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelow B, Mullokandov G, Agudo J, Sourisseau M, Bashir A, Maldonado AY, Dar AC, Brown BD, Evans MJ. Hepatitis C virus genetics affects miR-122 requirements and response to miR-122 inhibitors. Nat Commun. 2014;5:5408. doi: 10.1038/ncomms6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra RK, Yi M, Lemon SM. DDX6 (Rck/p54) is required for efficient hepatitis C virus replication but not for internal ribosome entry site-directed translation. J Virol. 2010a;84:6810–6824. doi: 10.1128/JVI.00397-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra RK, Yi M, Lemon SM. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J Virol. 2010b;84:6615–6625. doi: 10.1128/JVI.00417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Jopling CL, Schutz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Masaki T, Yamane D, McGivern DR, Lemon SM. Competing and noncompeting activities of miR-122 and the 5′ exonuclease Xrn1 in regulation of hepatitis C virus replication. Proc Natl Acad Sci USA. 2013;110:1881–1886. doi: 10.1073/pnas.1213515110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yamane D, Lemon SM. Dissecting the roles of the 5′ exoribonucleases xrn1 and xrn2 in restricting hepatitis C virus replication. J Virol. 2015;89:4857–4865. doi: 10.1128/JVI.03692-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YP, Gottwein JM, Scheel TK, Jensen TB, Bukh J. MicroRNA-122 antagonism against hepatitis C virus genotypes 1–6 and reduced efficacy by host RNA insertion or mutations in the HCV 5′ UTR. Proc Natl Acad Sci USA. 2011;108:4991–4996. doi: 10.1073/pnas.1016606108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Wang R, Shen JJ, Wang X, Gao P, Dong K, Zhang HZ. Knockdown of RCK/p54 expression by RNAi inhibits proliferation of human colorectal cancer cells in vitro and in vivo. Cancer Biol Ther. 2008;7:1669–1676. doi: 10.4161/cbt.7.10.6660. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Flaviviridae: The Viruses and Their Replication. 6. Lippincott-Williams & Williams Publishers; Philadelphia, Pennsylvania: 2013. [Google Scholar]
- Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- Lohmann V, Korner F, Dobierzewska A, Bartenschlager R. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J Virol. 2001;75:1437–1449. doi: 10.1128/JVI.75.3.1437-1449.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machlin ES, Sarnow P, Sagan SM. Masking the 5′ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc Natl Acad Sci USA. 2011;108:3193–3198. doi: 10.1073/pnas.1012464108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas A, Alves-Rodrigues I, Noueiry A, Ahlquist P, Diez J. Host deadenylation-dependent mRNA decapping factors are required for a key step in brome mosaic virus RNA replication. J Virol. 2006;80:246–251. doi: 10.1128/JVI.80.1.246-251.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki T, Arend KC, Li Y, Yamane D, McGivern DR, Kato T, Wakita T, Moorman NJ, Lemon SM. miR-122 stimulates hepatitis C virus RNA synthesis by altering the balance of viral RNAs engaged in replication versus translation. Cell Host Microbe. 2015;17:217–228. doi: 10.1016/j.chom.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathys H, Basquin J, Ozgur S, Czarnocki-Cieciura M, Bonneau F, Aartse A, Dziembowski A, Nowotny M, Conti E, Filipowicz W. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol Cell. 2014;54:751–765. doi: 10.1016/j.molcel.2014.03.036. [DOI] [PubMed] [Google Scholar]
- Minshall N, Thom G, Standart N. A conserved role of a DEAD box helicase in mRNA masking. RNA. 2001;7:1728–1742. doi: 10.1017/s135583820101158x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- Mortimer SA, Doudna JA. Unconventional miR-122 binding stabilizes the HCV genome by forming a trimolecular RNA structure. Nucleic Acids Res. 2013;41:4230–4240. doi: 10.1093/nar/gkt075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa Y, Morikawa H, Hirata I, Shiozaki M, Matsumoto A, Maemura K, Nishikawa T, Niki M, Tanigawa N, Ikegami M, Katsu K, Akao Y. Overexpression of rck/p54, a DEAD box protein, in human colorectal tumours. Br J Cancer. 1999;80:914–917. doi: 10.1038/sj.bjc.6690441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Amikura R, Hanyu K, Kobayashi S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development. 2001;128:3233–3242. doi: 10.1242/dev.128.17.3233. [DOI] [PubMed] [Google Scholar]
- Nicholson BL, White KA. Functional long-range RNA-RNA interactions in positive-strand RNA viruses. Nat Rev Microbiol. 2014;12:493–504. doi: 10.1038/nrmicro3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman KL, Sarnow P. Modulation of hepatitis C virus RNA abundance and the isoprenoid biosynthesis pathway by microRNA miR-122 involves distinct mechanisms. J Virol. 2010;84:666–670. doi: 10.1128/JVI.01156-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen DB, Eldrup AB, Bartholomew L, Bhat B, Bosserman MR, Ceccacci A, Colwell LF, Fay JF, Flores OA, Getty KL, Grobler JA, LaFemina RL, Markel EJ, Migliaccio G, Prhavc M, Stahlhut MW, Tomassini JE, MacCoss M, Hazuda DJ, Carroll SS. A 7-deaza-adenosine analog is a potent and selective inhibitor of hepatitis C virus replication with excellent pharmacokinetic properties. Antimicrob Agents Chemother. 2004;48:3944–3953. doi: 10.1128/AAC.48.10.3944-3953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pager CT, Schutz S, Abraham TM, Luo G, Sarnow P. Modulation of hepatitis C virus RNA abundance and virus release by dispersion of processing bodies and enrichment of stress granules. Virology. 2013;435:472–484. doi: 10.1016/j.virol.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnyak V, Coller J. The DHH1/RCKp54 family of helicases: an ancient family of proteins that promote translational silencing. Biochim Biophys Acta. 2013;1829:817–823. doi: 10.1016/j.bbagrm.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam AA, Jankowsky E. DEAD-box helicases as integrators of RNA, nucleotide and protein binding. Biochim Biophys Acta. 2013;1829:884–893. doi: 10.1016/j.bbagrm.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JC, Molter B, Geary CD, McNevin J, McElrath J, Giri S, Klein KC, Lingappa JR. HIV-1 Gag co-opts a cellular complex containing DDX6, a helicase that facilitates capsid assembly. J Cell Biol. 2012;198:439–456. doi: 10.1083/jcb.201111012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouya C, Siddiqui N, Morita M, Duchaine TF, Fabian MR, Sonenberg N. Human DDX6 effects miRNA-mediated gene silencing via direct binding to CNOT1. RNA. 2014;20:1398–1409. doi: 10.1261/rna.045302.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz B, Jr, Barretto N, Uprichard SL. Hepatitis C virus infection in phenotypically distinct Huh7 cell lines. PLoS One. 2009;4:e6561. doi: 10.1371/journal.pone.0006561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller N, Mina LB, Galao RP, Chari A, Gimenez-Barcons M, Noueiry A, Fischer U, Meyerhans A, Diez J. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc Natl Acad Sci USA. 2009;106:13517–13522. doi: 10.1073/pnas.0906413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedano CD, Sarnow P. Hepatitis C virus subverts liver-specific miR-122 to protect the viral genome from exoribonuclease Xrn2. Cell Host Microbe. 2014;16:257–264. doi: 10.1016/j.chom.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimakami T, Yamane D, Jangra RK, Kempf BJ, Spaniel C, Barton DJ, Lemon SM. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc Natl Acad Sci USA. 2012;109:941–946. doi: 10.1073/pnas.1112263109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Meng S, Lu Y, Trombly MI, Chen J, Lin C, Turk A, Wang X. Mammalian hyperplastic discs homolog EDD regulates miRNA-mediated gene silencing. Mol Cell. 2011;43:97–109. doi: 10.1016/j.molcel.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet T, Kovalak C, Coller J. The DEAD-box protein Dhh1 promotes decapping by slowing ribosome movement. PLoS Biol. 2012;10:e1001342. doi: 10.1371/journal.pbio.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault PA, Huys A, Dhillon P, Wilson JA. MicroRNA-122-dependent and -independent replication of Hepatitis C Virus in Hep3B human hepatoma cells. Virology. 2013;436:179–190. doi: 10.1016/j.virol.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Thibault PA, Huys A, Amador-Canizares Y, Gailius JE, Pinel DE, Wilson JA. Regulation of hepatitis C virus genome replication by Xrn1 and MicroRNA-122 binding to individual sites in the 5′ untranslated region. J Virol. 2015;89:6294–6311. doi: 10.1128/JVI.03631-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Jeng KS, Lai MM. Poly(C)-binding protein 2 interacts with sequences required for viral replication in the hepatitis C virus (HCV) 5′ untranslated region and directs HCV RNA replication through circularizing the viral genome. J Virol. 2011;85:7954–7964. doi: 10.1128/JVI.00339-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehner KA, Schutz S, Sarnow P. OGFOD1, a novel modulator of eukaryotic translation initiation factor 2alpha phosphorylation and the cellular response to stress. Mol Cell Biol. 2010;30:2006–2016. doi: 10.1128/MCB.01350-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston A, Sommerville J. Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic Acids Res. 2006;34:3082–3094. doi: 10.1093/nar/gkl409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SF, Lujan P, Jackson DL, Emerman M, Linial ML. The DEAD-box RNA helicase DDX6 is required for efficient encapsidation of a retroviral genome. PLoS Pathog. 2011;7:e1002303. doi: 10.1371/journal.ppat.1002303. [DOI] [PMC free article] [PubMed] [Google Scholar]