Abstract
Hepatocellular carcinoma (HCC) is one of the most prevalent and life-threatening malignancies worldwide. There are few diagnostic and prognostic biomarkers and druggable targets for HCC. Aldehyde dehydrogenase 1 (ALDH1) is a marker of stem cells in a variety of cancers, but the mRNA levels and prognostic value of ALDH1 isoforms in HCC patients remain unknown. In the present study, gene ontology annotation of the ALDH1 family was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID), and the gene pathway analsis was performed using GeneMANIA software. The initial prognostic value of ALDH1 expression in 360 HCC patients was assessed using the OncoLnc database. The expression levels of ALDH1 isoforms in normal liver tissues and clinical specimens of cancer vs. normal control datasets were determined using the GTEx and Oncomine databases, respectively. We then analyzed the prognostic value of ALDH1 expression in 212 hepatitis B virus (HBV)–related HCC patients using the GEO database. We found that the ALDH1 isoform showed high aldehyde dehydrogenase activity. The ALDH1A1, ALDH1B1, and ALDH1L1 genes encoded for the ALDH1 enzyme. High ALDH1B1 expression had protective qualities in HCC patients. Moreover, HBV–related HCC patients who showed high ALDH1L1 gene expression had a better clinical outcomes. In addition, high ALDH1A1 expression was associated with a 57–month recurrence–free survival in HBV-related HCC patients. High ALDH1B1 expression was protective for HCCs with multiple nodules and high serum alpha–fetoprotein (AFP) level. Furthermore, high serum AFP levels contributed to lower ALDH1L1. ALDH1A1, ALDH1B1, and ALDH1L1, all of which were considered promising diagnostic and prognostic markers as well as potential drug targets.
Introduction
Liver cancer is the fifth most common cancer and the second leading cause of cancer–related death in males worldwide, with hepatocellular carcinoma (HCC) representing the largest proportion (70%–90%) of primary liver cancers [1, 2]. Although most HCC cases (> 80%) occur in sub–Saharan Africa or Eastern Asia, China alone accounts for more than 50% of cases worldwide [3, 4]. Although early diagnostic methods, surgical resection, liver transplantation, and ablation by radiofrequency or ethanol injection have provided considerable advancements, the 5-year survival rate ranges from 50%–70% at early disease stages, and the prognosis of HCC patients remains poor due to the high postoperative recurrence rate and metastasis [5, 6]. Thus, the elucidation of mechanisms of initiation and progression of HCC and the identification of diagnostic and prognostic markers as well as potential drug targets are critical to provide better prognosis and individualized treatments for HCC patients.
Aldehyde dehydrogenases (ALDHs) are a family of intracellular enzymes that are involved in cellular detoxification, differentiation, and drug resistance by oxidation of cellular aldehydes [7]. The major function of ALDHs is aldehyde detoxification, which serves to protect stem cells against the destructive properties of oxidative aldehydes. It has been shown that both human and murine hematopoietic and neural stem cells, as well as related progenitor cells, exhibit high ALDH activity [8–11], and that increased cell differentiation leads to decreased ALDH activity [11]. As a marker of stem cells in a variety of cancers, ALDH1 acts as a modulator for cell proliferation, stem cell differentiation, and resistance to chemotherapeutic agents [12]. Tanei et al. reported that tumors from breast cancer with ALDH1–positive cells displayed higher rates of chemotherapy resistance [13]. Furthermore, because ALDH1–positive cells exhibited tumorigenic ability with prognostic significance, the ALDH1–positive status may be involved not only in proliferation of the primary tumor but also in the formation of metastases [9]. However, of the ALDH1 isoforms (ALDH1A1, ALDH1A2, ALDH1A3, ALDH1B1, ALDH1L1, and ALDH1L2) that alone or in combination contribute to ALDH1 activity have not been determined. Previous studies [14–16] have demonstrated distinct prognostic values of ALDH1 isoforms in breast cancer, gastric cancer, and non–small cell lung cancer using an online Kaplan–Meier plotted database. Nonetheless, the prognostic value of individual ALDH1 isoforms in HCC is not yet clear. In the current study, we determined the mRNA level and prognostic value of ALDH1 isoforms in HCC patients.
Materials and methods
This study was approved by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University. The gene ontology of the ALDH1 was annotated using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/), which provides resources to understand the biological functions of a large group of genes [17]. Genes’ pathway analysis was performed by a GeneMANIA Software (www.genemania.org) [18]. The prognostic value of mRNA expression of ALDH1 isoforms in HCC was initially assessed using the OncoLnc database (www.oncolnc.org). OncoLnc is a tool for interactively exploring survival correlations, and it contains survival data for 8,647 patients from 21 cancer studies performed by The Cancer Genome Atlas (TCGA), along with RNA–SEQ expression for mRNAs from TCGA cases [19]. TCGA contains microarray-based and RNA sequencing–based mRNA expression variants. The survival analyses were performed using cutoff values of median or quartile of ALDH1 family expression in 360 HCC patients. mRNA expression levels above or below the cutoff value divided the cases into high expression and low expression groups, respectively. However, since the different background and type of liver cancer was analyzed in TCGA, OncLnc was subsequently used to evaluate the initial prognostic value of ALDH1 isoforms with different cutoff values. Considering the complexity of tumorigenesis and tumor progression in HCC cases, we further analyzed the association of the ALDH1 isoforms mRNA expressions with a total of 445 hepatitis B virus (HBV)–related HCC and paramalignant samples from the Gene Expression Omnibus (GEO accession: GSE14520) using the median cutoff value. Sample mRNA expression was measured using the Affymetrix HT Human Genome U133A Array (HT_HG-U133A). To explore the relationships between the ALDH1 isoforms and clinical features, we downloaded clinical data coupled to expression data for the ALDH1 isoform mRNA expressions from OncoLnc and GEO. In GSE14520, we identified 212 HBV-related HCC patients with ontology features and clinical survival information.
The mRNA expression data for the ALDH1 isoforms in normal human tissues were obtained from the Genotype Tissue Expression (GTEx) database (www.gtexportal.org). The GTEx database provides a resource to study human gene expression and regulation and their relationships to genetic variation [20]. This release of GTEx includes genotype data from approximately 450 donors and over 9600 RNA–Seq samples across 51 tissue sites and 2 cell lines as well as adequate power to detect expression quantitative trait loci in 44 tissues. Moreover, Oncomine gene expression array datasets (www.oncomine.org) were used to analyze the mRNA levels of different ALDH1 isoforms in different cancers. Oncomine is an online cancer microarray database used to facilitate the discovery of genome-wide expression analyses [21]. In this study, we compared the clinical specimens of cancer vs. normal control datasets, using the Student’s t–test to generate a P value. A P value of < 0.01 was considered statistically significant, and fold change was defined as 2, whereas the data type was restricted to mRNA.
The correlation of ALDH1 isoform expression was analyzed by Pearson’s correlation coefficient. Kaplan–Meier survival analysis was used to calculate survival percentages, and the differences in survival percentages were then estimated using a generalized log–rank test. A Cox proportional hazard regression model was used to calculate the hazard ratio (HR) with 95% confidence interval (CI). All statistical analyses were two–sided and were performed using SPSS version 17.0 (SPSS, Chicago, IL, USA). A P value of < 0.05 was considered statistically significant. The survival curves and correlation plots were depicted by GraphPad 5.01 (www.graphpad.com) and R 3.3.2 (www.r-project.org), respectively.
Results
The DAVID gene ontology terms associated with ALDH1 isoforms are presented in S1 Table. All numbers for the ALDH1 isoforms were enriched in aldehyde dehydrogenase activity. In addition, the ALDH1 isoforms also catalyzed oxidation–reduction reactions and were involved in retinol metabolism.
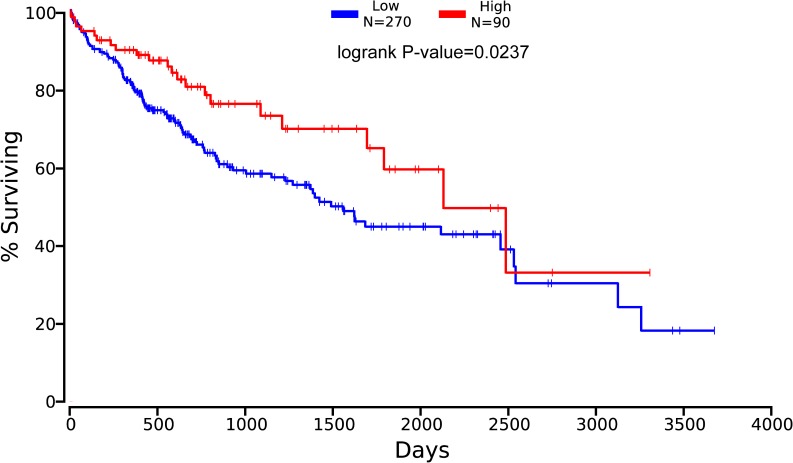
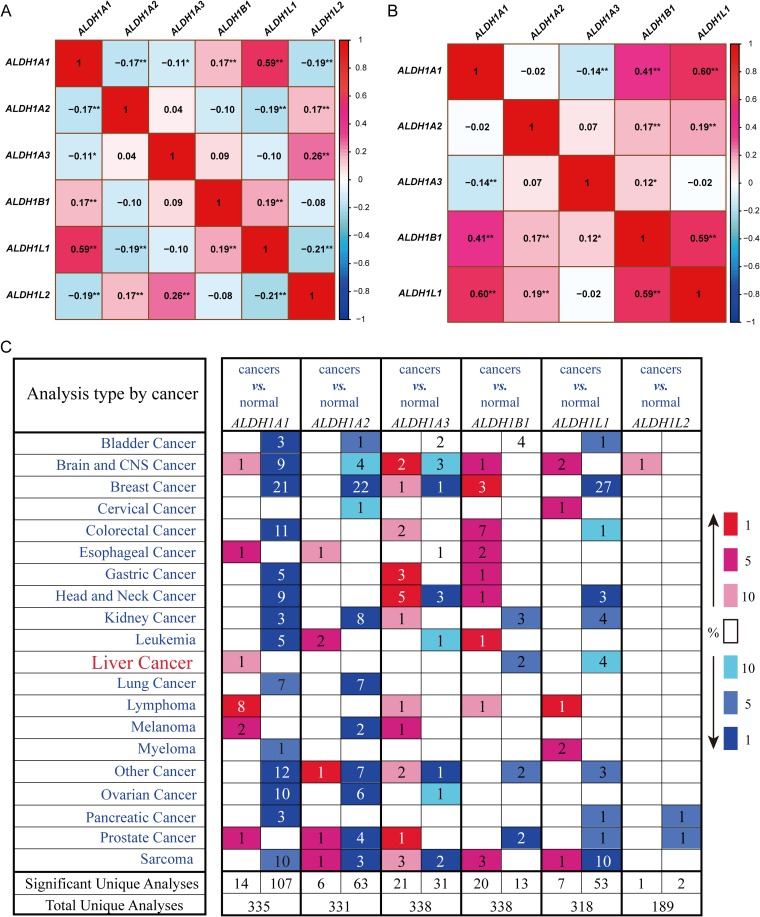
With the cutoff value set at the median or quartile, we determined that ALDH1B1 was the only isoform associated with the clinical outcomes of 360 liver cancer patients in the OncoLnc database (Fig 1). There were no significant finding from analyses of the other five ALDH1 isoforms. The high expression of ALDH1B1 correlated with a favorable clinical outcome in liver cancer patients (log–rank, P = 0.0237). The baseline data of the 360 liver cancer patients are reported in S2 Table. Moreover, Tumor, Node, Metastasis (TNM) stage and associated with clinical outcomes of liver cancer patients are also reported in S2 Table. The univariate Cox proportional hazard regression model revealed a lower hazard risk in the liver cancer patients with high ALDH1B1 expression levels (S3 Table). To further define the positive relationships among the ALDH1 isoforms, the expression levels of mRNAs were analyzed and the correlations determined using the OncLnc database. As shown in Fig 2A, we found that ALDH1A1, ALDH1B1, and ALDH1L1 had positive relationships.
Fig 1. The prognostic value of ALDH1B1 mRNA expression in liver cancer patients, and the survival curve as plotted using the OncoLnc database.
ALDH = aldehyde dehydrogenase.
Fig 2.
Correlation analyses of ALDH1 isoform mRNA expressions from the OncoLnc (A) and GEO databases (B, accession: GSE14520). The figure shows the association of mRNA expressions of aldehyde dehydrogenase1 (ALDH1) isoforms with a positive correlation (red) or negative correlation (blue). The numbers of cells with statistically significant correlation coefficients are denoted by *P < 0.05 and **P < 0.01. (C) ALDH1 isoform mRNA expressions in different tumor types. The figure shows the number of datasets with statistically significant mRNA overexpression (red) or downregulation (blue) of the target gene (cancer vs. normal tissue). The P value threshold is 0.01. The number of datasets that met the threshold set for each experiment and the cancer types are shown. Genes were ranked by the percentile of the target genes in the top of all genes as measured in each experiment.The best gene rank percentile are denoted in color.
Furthermore, we downloaded the clinical data of 445 HCC and paramalignant samples with HBV infection in the GEO database. Clinicopathological characteristics of HBV-related HCC patients were shown in S4 Table (only 212 samples have entries for baseline data). Most characteristics were associated with prognosis of HBV-related HCC patients, except for age and alanine aminotransferase level. A correlation with the ALDH1 family was detected, except for ALDH1L2, which was not found in the GSE14520 (i.e., probably did not contain the probe for ALDH1L2). The results also demonstrated that ALDH1A1, ALDH1B1, and ALDH1L1 were positively related (Fig 2B). Oncomine analyses of six ALDH1 family submembers in cancer vs. normal samples showed that ALDH1A1 was significantly upregulated in liver cancer in the different datasets. In contrast, downregulation of ALDH1B1 and ALDH1L1 was found in liver cancer compared with normal tissues (Fig 2C). In normal tissues, the relatively higher expression levels of ALDH1A1, ALDH1B1, and ALDH1L1 were analyzed using the GTEx database (S1 Fig). Considering these results, we speculated that ALDH1A1/B1/L1 expression levels contributed to ALDH1 activity.
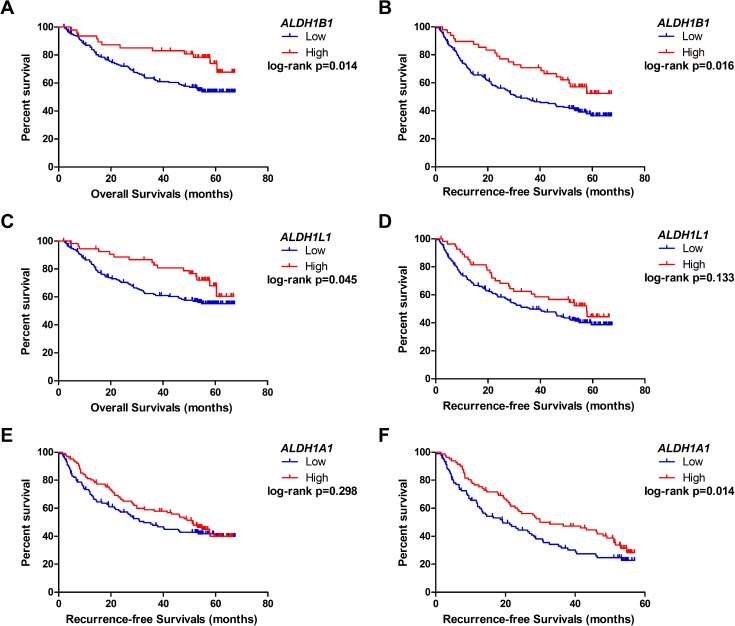
Moreover, survival analyses have shown that high expression of ALDH1B1 in 212 HBV-related HCC patients had a favorable outcome in terms of overall survival (OS) (Fig 3A) and recurrence–free survival (RFS) (Fig 3B). After adjusting for age and sex, high ALDH1B1 expression had a lower risk in OS and RFS of the HBV–HCC patients (HROS = 0.46; HRRFS = 0.53, respectively; Table 1). Likewise, high ALDH1L1 mRNA expression was associated with a lower hazard in clinical outcomes of HBV–HCC patients (HROS = 0.54; HRRFS = 0.65, respectively; Table 1). Using Kaplan–Meier survival analyses, high ALDH1L1-expressing patients had a longer OS time (Fig 3C), whereas there was no statistical difference between the two groups in RFS (Fig 3D). Although we did not find that ALDH1A1 expression was associated with long–term clinical outcome (Fig 3E), high ALDH1A1 mRNA expression was significantly associated with a 57–month RFS (Fig 3F) and had a lower hazard risk (HRRFS = 0.67; Table 1). However, there was no statistical significance in the associations of ALDH1A2/A3 expression levels with clinical outcomes of HBV-related HCC patients.
Fig 3. The prognostic value of ALDH1A1/B1/L1 mRNA expression in HBV-related HCC patients [data from the GEO database (accession: GSE14520)].
A and C: The Kaplan–Meier survival analyses of aldehyde dehydrogenase (ALDH)1B1 and ALDH1L1 expression of the overall survival time of HBV-related HCC patients. B and D: The Kaplan–Meier survival analyses of ALDH1B1 and ALDH1L1 expression in recurrence–free survival time of HBV-related HCC patients. E and F: The Kaplan–Meier survival analyses of ALDH1A1 expression in recurrence–free survival time and 57–month recurrence–free survival time of HBV related HCC patients, respectively. HCC = hepatocellular carcinoma.
Table 1. Association between ALDH1A1/B1/L1 mRNA expression and clinical outcomes in HBV-related HCC patients.
| OS | RFS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | number | MST | P value | HR a | 95%CI | MRT | P value | HR a | 95%CI | |
| ALDH1A1 | Low | 119 | >67.4 | 0.230 | Ref. | 32.6 | 0.298 | Ref. | ||
| High | 102 | >67.0 | 0.75 | 0.49–1.16 | 51.1 | 0.78 | 0.55–1.13 | |||
| ALDH1A2 | Low | 138 | >67.4 | 0.549 | Ref. | 45.9 | 0.970 | Ref. | ||
| High | 83 | >66.6 | 0.85 | 0.55–1.33 | 41.6 | 0.98 | 0.68–1.41 | |||
| ALDH1A3 | Low | 153 | >67.4 | 0.184 | Ref. | 45.9 | 0.927 | Ref. | ||
| High | 68 | >67.3 | 0.73 | 0.44–1.20 | 40.1 | 1.02 | 0.69–1.51 | |||
| ALDH1B1 | Low | 173 | >67.4 | 0.014 | Ref. | 30.9 | 0.016 | Ref. | ||
| High | 48 | >67.3 | 0.46 | 0.25–0.84 | >67.3 | 0.53 | 0.33–0.86 | |||
| ALDH1L1 | Low | 165 | >67.4 | 0.045 | Ref. | 36.0 | 0.133 | Ref. | ||
| High | 56 | >67.0 | 0.54 | 0.31–0.94 | 57.7 | 0.65 | 0.42–0.996 | |||
| ALDH1A1b | Low | 85 | 32.6 | 0.093 | Ref. | 19.2 | 0.039 | Ref. | ||
| High | 71 | 52.7 | 0.69 | 0.44–1.07 | 29.9 | 0.67 | 0.47–0.97 | |||
Note
a HR adjusted for age and sex for the Cox proportional hazard model.
b The association of aldehyde dehydrogenase (ALDH) 1A1 expression with the 57-month survival time and 57-month recurrence–free survival time. The bold terms are statistically significant.
Abbreviations: OS, overall survival; RFS, recurrence–free survival; MST, median survival time; MRT, median recurrence time; HR, hazard ratio; 95% CI, 95% confidence intervals; Ref., reference.
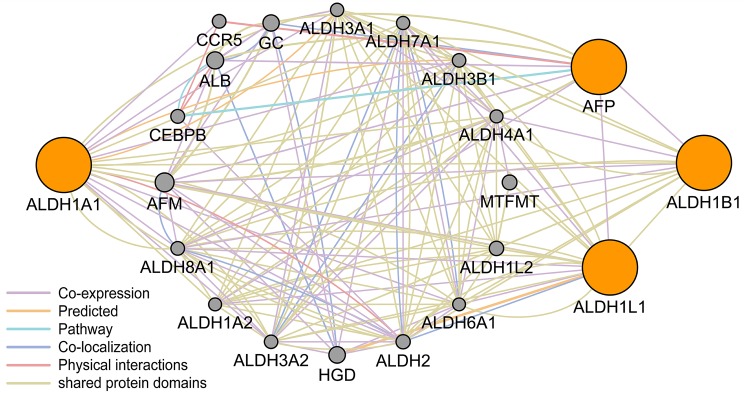
We also performed logistical regression analysis adjusted for age and sex to evaluate the relationship between ALDH1A1/B1/L1 expression and clinical features of HBV-related HCC patients (Table 2). The results demonstrated that high ALDH1B1-expression was a protective factor for multiple nodules (OR = 0.28, 95% CI = 0.10–0.85, P = 0.024). In addition, lower expression of ALDH1B1/L1 was associated with high serum alpha–fetoprotein (AFP) levels (≥ 300 ng/mL) (OR = 0.22 and 0.39, respectively). We performed pathway analysis using GeneMANIA software, and we found, upon co–expression, that AFP was associated with ALDH1B1/L1 (Fig 4).
Table 2. Association between ALDH1A1/B1/L1 mRNA expression and the tumor biological features in HBV-related HCC patients.
| Variables | ALDH1A1 | ALDH1B1 | ALDH1L1 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | OR (95%CI) | P value | Low | High | OR (95%CI) | P value | Low | High | OR(95%CI) | P value | ||
| Tumor size | <5 | 70 | 70 | Ref. | 108 | 32 | Ref. | 101 | 39 | Ref. | |||
| (cm) | ≥5 | 49 | 31 | 0.61 (0.34–1.07) | 0.085 | 65 | 15 | 0.75 (0.38–1.50) | 0.418 | 64 | 16 | 0.59 (0.30–1.16) | 0.126 |
| Nodule | single | 92 | 84 | Ref. | 132 | 44 | Ref. | 129 | 47 | Ref. | |||
| multiple | 27 | 18 | 0.76 (0.39–1.49) | 0.427 | 41 | 4 | 0.28 (0.10–0.85) | 0.024 | 36 | 9 | 0.74 (0.33–1.69) | 0.476 | |
| Cirrhosis | no | 13 | 5 | Ref. | 14 | 4 | Ref. | 16 | 2 | Ref. | |||
| yes | 106 | 97 | 2.20 (0.75–6.48) | 0.152 | 159 | 44 | 0.90 (0.28–2.94) | 0.866 | 149 | 54 | 2.41 (0.53–11.03) | 0.257 | |
| BCLC | 0 or A | 87 | 81 | Ref. | 132 | 36 | Ref. | 126 | 42 | Ref. | |||
| B or C | 31 | 20 | 0.72 (0.38–1.36) | 0.308 | 39 | 12 | 1.16 (0.55–2.54) | 0.698 | 38 | 13 | 1.12 (0.54–2.34) | 0.757 | |
| AFP (ng/mL) | <300 | 87 | 81 | Ref. | 124 | 44 | Ref. | 120 | 48 | Ref. | |||
| ≥300 | 31 | 20 | 0.69 (0.36–1.32) | 0.262 | 47 | 4 | 0.22 (0.07–0.66) | 0.007 | 44 | 7 | 0.39 (0.16–0.93) | 0.033 | |
Note: OR adjusted age and sex for logistic regression, the bold terms are statistically significant. Abbreviations: OR, odds ratio; 95% CI, 95% confidence intervals; Ref, reference; BCLC, Barcelona Clinic Liver Cancer; AFP, alpha–fetoprotein; ALDH, aldehyde dehydrogenase; HCC, hepatocellular carcinoma.
Fig 4. The gene pathway analysis between ALDH1A1/B1/L1 and AFP using GeneMANIA software.
ALDH = aldehyde dehydrogenase; AFP = alpha–fetoprotein.
Discussion
Using online data resources, we showed that the expression of ALDH1 family members in normal tissues was significantly different from that in liver cancer tissues. High ALDH1B1 expression was found to play a protective role in liver cancer patients. Moreover, HBV-related HCC patients who expressed high levels of ALDH1L1 had a better clinical outcomes than expressed in other ALDH1 isoforms. In addition, high ALDH1A1 expression was associated with a 57–month RFS for HBV-related HCC patients. Multiple nodules and high serum AFP levels were considered risk factors for ALDH1B1 expression. Furthermore, high serum AFP level may contribute to lower ALDH1L1 expression. Therefore, they could potentially serve as diagnostic and prognostic markers as well as potential drug targets.
Epidemiological studies have unequivocally identified chronic alcohol consumption as an important risk factor for the development of various types of cancers [22]. Ethanol is absorbed by the small intestine and then metabolized, primarily by the liver. Ethanol is oxidized by cytosolic alcohol dehydrogenase (ADH) to form acetaldehyde, which is subsequently oxidized by mitochondrial aldehyde dehydrogenases, mainly ALDH2, to produce acetate [23]. Among the known human ALDHs, cytosolic ALDH1A1 has been shown to play a major role in acetaldehyde oxidation and elimination [24]. In a rodent study, cytosolic ALDH1A1 has been shown to be involved in acetaldehyde metabolism and alcohol preference [25]. A study using crude lysate from Huh7 hepatoma cells reported that ALDH1B1 contributed to the oxidation of short-chain aldehydes including acetaldehyde and propionaldehyde, which suggested a role for ALDH1B1 in ethanol metabolism [26]. Stagos et al. [27] observed the punctate positive staining of human ALDH1B1 in the human liver and pancreas using immunohistochemical staining. As Stagos et al. found a similarly higher level of ALDH1B1 and ALDH2 mRNAs in mouse liver relative to other tissues examined, these results further suggested an important role for the two mitochondrial ALDHs in ethanol metabolism. Furthermore, the ALDH1L1 enzyme appeared to be a chief regulator of cellular metabolism, as it is strongly downregulated under certain physiological and pathological conditions [28, 29], while its upregulation can produce drastic antiproliferative effects [28]. Hwang et al. [30] proposed a hypothesis that upregulating ALDH1L1 expression in order to decrease acetaldehyde concentrations and promote DNA stability would thereby decrease cancer susceptibility.
Several markers have been identified for the selection of human (cancer) stem cells, of which ALDH1 is among the most widely studied. ALDH1 consists of detoxifying enzymes that are responsible for the oxidation of retinaldehydes to retinoids. Previous reports of the liver evaluated by flow cytometry suggested that high ALDH activity could be a marker of liver progenitor cells in normal liver [31] and cancer stem cells in HCC cases [32]. In humans, ALDH1 has six subfamily members, which exhibit > 60% amino acid identity, with ALDH1A1 as the predominant isoform [33]. Previous studies showed that upregulation of ALDH1A1 expression was associated with enhanced invasiveness in acute myeloid leukemia[34], nasopharyngeal carcinoma [35], bladder cancer [36], and pancreatic cancer [37]. Moreb et al. [38] reported that disrupting ALDH1A1 using small interfering (si) RNAs in lung cancer cell lines reduced neoplastic activity. However, we found that high ALDH1A1 expression was associated with a better 57–month RFS in HBV-related HCC patients. The results were consistent with those of Suzuki et al.[39], which suggested that high ALDH1A1 was a favorable prognostic factor in RFS of HCC patients. Compared to the normal tissues in the Oncomine database, ALDH1A1 expression was upregulated in HCC tissues. The result came from Mas et al. [40] study, enrolling 88 distinct patients with hepatitis C viral (HCV) infection (41 HCV-cirrhotic tissues,17 HCV-cirrhotic tissues from patients with HCC, and 47 HCV-HCC tissues). Differentially expressed genes were studied by use of high-density oligonucleotide arrays, including hybridized to HG-U133A and HG-U133A 2.0 GeneChips. We therefore speculated that ALDH1A1 expression was involved in cancer heterogeneity and the differences in prognoses for different virus infection backgrounds and types of cancer.
ALDH1B1, a mitochondrial ALDH, is another promising good prognostic marker for cancer, sharing 65% homology in peptide sequence with cytosolic ALDH1A1 in humans [41]. ALH1B1 catalytically metabolizes a wide range of aldehyde substrates, including acetaldehyde and products of lipid peroxidation, and is activated in ethanol metabolism [27]. In our gene ontology annotation studies, the main functions of ALDH1B1 were aldehyde dehydrogenase activity and oxidoreductase activity. Singh et al. [42] suggested that ALDH1B1, a significant contributor to ALDH activity as measured by the Aldefluor™ assay, could promote tumor formation by modulating the Wnt/β–catenin, Notch, and PI3K/Akt signaling pathways. However, ALDH1B1 is a relatively uncharacterized member of the ALDH1 superfamily in the context of HCC. In the present study, we found that high ALDH1B1 expression in HCC patients had a favorable clinical survival prediction for OS and RFS. From the Oncomine database, we found that ALDH1B1 expression was downregulated in liver cancer tissues compared to that of the normal tissues, which suggested ALDH1B1 as a potential tumor suppressor gene. Furthermore, multiple nodules and high AFP expression could lead to the downregulation of ALDH1B1 expression. A relationship of co–repression between AFP and ALDH1B1was identified via the GeneMANIA software. Although ALDH1B1 could be a potential prognostic marker for HCC patients, the mechanism is still undetermined.
ALDH1L1, another member of the ALDH gene superfamily, is also called FDH [10–formyl tetrahydrofolate (THF) dehydrogenase] and is involved in nervous system development and reduced proliferation. These findings imply that ALDH1L1 may have tumor suppressor properties [43]. Oleinik et al. found that ALDH1L1 suppressed cancer cell proliferation by depleting intracellular 10–formyl THF, which is essential for de novo purine biosynthesis. This effect could be reversed by excess extracellular folate [44]. Future investigations regarding the critical role of ALDH1L1 in cancer cell survival and induction of folate stress will provide important insights into the malignant process. Furthermore, these studies could link deregulation of key metabolic pathways to cancer, as well as establish new targets for diagnostics of the malignant transformation process. Microarray data showed that mRNA levels of ALDH1L1 were remarkably reduced in HCC [45]. A previous study [46] assessed ALDH1L1 expression in HCC tissues using real–time quantitative RT–PCR, western blotting, and immunohistochemistry staining, and further findings showed that low expression of ALDH1L1 in HCC was significantly associated with pathology grade, hepatitis B surface antigen status, and serum AFP. The studies also suggested that decreased expression of ALDH1L1 was associated with a poor prognosis in HCC patients. Using gene pathway analysis, we found that AFP was associated with the ALDH1 family. Concerning the results of our study, high ALDH1L1 expression was associated with prognosis and serum AFP levels, which was consistent with previous studies. Thus, ALDH1L1 may be a novel prognostic marker for HCC, but the specific mechanism remains unclear.
Several limitations of this study warrant discussion. First, our sample size was limited and a small fraction of the clinical data was missing. As such, additional studies with larger sample sizes will be needed to clarify our results. Second, the subjects evaluation collected in this study included background and type of tumor, which may introduce background heterogeneity and may also represent a major limitation of the study. Third, we specifically used median mRNA expression to evaluate the prognostic value in HBV-related HCC patients, the appropriate cut-off value of ALDH1 isoform mRNA expression for HCC patients remains unclear.
In conclusion, our findings demonstrated that ALDH1A1, ALDH1B1, and ALDH1L1 contributed to the ALDH1 family activity. The expressions of ALDH1B1 and ALDH1L1 were associated with clinical features and survival of HBV-related HCC patients. Moreover, high ALDH1A1 expression in HBV-related HCC patients had a favorable 57–month RFS time. The results provided a better understanding of the heterogeneity and complexity of HCC at the molecular level, and further provided a foundation of more accurate prognoses and the development of new treatment targets.
Supporting information
Red frame is normal liver tissue. ALDH = aldehyde dehydrogenase.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All data are available from the public database and software (i.e. OncLnc; DAVID; GEO(GEO accession: GSE14520); Oncomine; GTEx; GeneMania).
Funding Statement
This work was supported in part by the National Nature Science Foundation of China (No: 81560535, 81072321, 30760243, 30460143 and 30560133), 2009 Program for New Century Excellent Talents in University (NCET), Guangxi Nature Sciences Foundation (No: GuiKeGong 1104003A-7), and Guangxi Health Ministry Medicine Grant (Key-Scientific Research-Grant Z201018).
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. Ca A Cancer Journal for Clinicians. 2015;65(2):87 doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. International Journal of Cancer Journal International Du Cancer. 2001;94(2):153–6. [DOI] [PubMed] [Google Scholar]
- 3.Elserag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132(7):2557 doi: 10.1053/j.gastro.2007.04.061 [DOI] [PubMed] [Google Scholar]
- 4.Venook AP, Papandreou C, Furuse J, Guevara LLD. The Incidence and Epidemiology of Hepatocellular Carcinoma: A Global and Regional Perspective. Oncologist. 2010;15 Suppl 4(Supplement 4):5. [DOI] [PubMed] [Google Scholar]
- 5.Bruix J, Llovet JM. Major achievements in hepatocellular carcinoma. Lancet. 2009;373(9664):614 doi: 10.1016/S0140-6736(09)60381-0 [DOI] [PubMed] [Google Scholar]
- 6.Shimozawa N, Hanazaki K. Longterm prognosis after hepatic resection for small hepatocellular carcinoma. Journal of the American College of Surgeons. 2004;198(3):356 doi: 10.1016/j.jamcollsurg.2003.10.017 [DOI] [PubMed] [Google Scholar]
- 7.Moreb J, Schweder M, Suresh A, Zucali JR. Overexpression of the human aldehyde dehydrogenase class I results in increased resistance to 4-hydroperoxycyclophosphamide. Cancer Gene Therapy. 1996;3(1):24–30. [PubMed] [Google Scholar]
- 8.Balicki D. Moving forward in human mammary stem cell biology and breast cancer prognostication using ALDH1. Cell Stem Cell. 2007;1(5):485–7. doi: 10.1016/j.stem.2007.10.015 [DOI] [PubMed] [Google Scholar]
- 9.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2011;1(5):555–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corti S, Locatelli F, Papadimitriou D, Donadoni C, Salani S, Del BR, et al. Identification of a primitive brain-derived neural stem cell population based on aldehyde dehydrogenase activity. Stem Cells. 2006;24(4):975–85. doi: 10.1634/stemcells.2005-0217 [DOI] [PubMed] [Google Scholar]
- 11.Armstrong L, Stojkovic M, Dimmick I, Ahmad S, Stojkovic P, Hole N, et al. Phenotypic characterization of murine primitive hematopoietic progenitor cells isolated on basis of aldehyde dehydrogenase activity. Stem Cells. 2004;22(7):1142–51. doi: 10.1634/stemcells.2004-0170 [DOI] [PubMed] [Google Scholar]
- 12.Nishikawa S, Konno M, Hamabe A, Hasegawa S, Kano Y, Ohta K, et al. Aldehyde dehydrogenase high gastric cancer stem cells are resistant to chemotherapy. International Journal of Oncology. 2013;42(4):1437 doi: 10.3892/ijo.2013.1837 [DOI] [PubMed] [Google Scholar]
- 13.Tanei T, Morimoto K, Shimazu K, Kim SJ, Tanji Y, Taguchi T, et al. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clinical Cancer Research. 2009;15(12):4234–41. doi: 10.1158/1078-0432.CCR-08-1479 [DOI] [PubMed] [Google Scholar]
- 14.Wu S, Xue W, Huang X, Yu X, Luo M, Huang Y, et al. Distinct prognostic values of ALDH1 isoenzymes in breast cancer. Tumor Biology. 2015;36(4):2421–6. doi: 10.1007/s13277-014-2852-6 [DOI] [PubMed] [Google Scholar]
- 15.Shen JX, Liu J, Li GW, Huang YT, Wu HT. Mining distinct aldehyde dehydrogenase 1 (ALDH1) isoenzymes in gastric cancer. Oncotarget. 2016;7(18):25340–9. doi: 10.18632/oncotarget.8294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.You Q, Guo H, Xu D. Distinct prognostic values and potential drug targets of ALDH1 isoenzymes in non-small-cell lung cancer. Drug Design Development & Therapy. 2014;9(default):5087–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols Erecipes for Researchers. 2009;4(1):44. [DOI] [PubMed] [Google Scholar]
- 18.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–7. doi: 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anaya J. OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. 2016;2(2):e67. [Google Scholar]
- 20.Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The Genotype-Tissue Expression (GTEx) project. Biopreservation & Biobanking. 2013;13(5):307–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: A Cancer Microarray Database and Integrated Data-Mining Platform. Neoplasia. 2004;6(1):1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bagnardi V, Blangiardo M, La VC, Corrao G. Alcohol consumption and the risk of cancer: a meta-analysis. Alcohol Research & Health the Journal of the National Institute on Alcohol Abuse & Alcoholism. 2001;25(4):263. [PMC free article] [PubMed] [Google Scholar]
- 23.SZ, Li TK. Determinants of alcohol use and abuse: Impact of quantity and frequency patterns on liver disease †. Hepatology. 2007;46(6):2032–9. doi: 10.1002/hep.22010 [DOI] [PubMed] [Google Scholar]
- 24.Vasiliou V, Pappa A. Polymorphisms of human aldehyde dehydrogenases. Consequences for drug metabolism and disease. Pharmacology. 2000;61(3):192 [DOI] [PubMed] [Google Scholar]
- 25.Singh SM. Studies with cDNA probes on the in vivo effect of ethanol on expression of the genes of alcohol metabolism. Alcohol Alcohol. 1990;25(4):385–94. [PubMed] [Google Scholar]
- 26.Stewart MJ, Malek K, Crabb DW. Distribution of messenger RNAs for aldehyde dehydrogenase 1, aldehyde dehydrogenase 2, and aldehyde dehydrogenase 5 in human tissues. Journal of investigative medicine: the official publication of the American Federation for Clinical Research. 1996;44(2):42–6. [PubMed] [Google Scholar]
- 27.Stagos D, Chen Y, Brocker C, Donald E, Jackson BC, Orlicky DJ, et al. Aldehyde Dehydrogenase 1B1: Molecular Cloning and Characterization of a Novel Mitochondrial Acetaldehyde-Metabolizing Enzyme. Drug Metabolism & Disposition the Biological Fate of Chemicals. 2010;38(10):1679–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krupenko SA, Oleinik NV. 10-formyltetrahydrofolate dehydrogenase, one of the major folate enzymes, is down-regulated in tumor tissues and possesses suppressor effects on cancer cells. Cell Growth Differ. 2002;13(5):227–36. [PubMed] [Google Scholar]
- 29.Epperson LE, Dahl TA, Martin SL. Quantitative analysis of liver protein expression during hibernation in the golden-mantled ground squirrel. Mol Cell Proteomics. 2004;3(9):920–33. doi: 10.1074/mcp.M400042-MCP200 [DOI] [PubMed] [Google Scholar]
- 30.Hwang PH, Lian L, Zavras AI. Alcohol intake and folate antagonism via CYP2E1 and ALDH1: effects on oral carcinogenesis. Med Hypotheses. 2012;78(2):197–202. doi: 10.1016/j.mehy.2011.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dollé L, Best J, Empsen C, Mei J, Rossen EV, Roelandt P, et al. Successful isolation of liver progenitor cells by aldehyde dehydrogenase activity in naïve mice. Hepatology. 2012;55(2):540–52. doi: 10.1002/hep.24693 [DOI] [PubMed] [Google Scholar]
- 32.Ma S, Chan KW, Lee TK, Tang KH, Wo JY, Zheng BJ, et al. Aldehyde dehydrogenase discriminates the CD133 liver cancer stem cell populations. Molecular Cancer Research. 2008;6(7):1146 doi: 10.1158/1541-7786.MCR-08-0035 [DOI] [PubMed] [Google Scholar]
- 33.Wang K, Chen X, Zhan Y, Jiang W, Liu X, Wang X, et al. Increased expression of ALDH1A1 protein is associated with poor prognosis in clear cell renal cell carcinoma. Medical Oncology. 2013;30(2):574 doi: 10.1007/s12032-013-0574-z [DOI] [PubMed] [Google Scholar]
- 34.Pearce DJ, Taussig D, Simpson C, Allen K, Rohatiner AZ, Lister TA, et al. Characterization of cells with a high aldehyde dehydrogenase activity from cord blood and acute myeloid leukemia samples. Stem Cells. 2005;23(6):752 doi: 10.1634/stemcells.2004-0292 [DOI] [PubMed] [Google Scholar]
- 35.Hou W, He W, Li Y, Ma R, Wang Z, Zhu X, et al. Increased expression of aldehyde dehydrogenase 1 A1 in nasopharyngeal carcinoma is associated with enhanced invasiveness. European Archives of Oto-Rhino-Laryngology. 2014;271(1):171–9. doi: 10.1007/s00405-013-2517-x [DOI] [PubMed] [Google Scholar]
- 36.Su Y, Qiu Q, Zhang X, Jiang Z, Leng Q, Liu Z, et al. Aldehyde Dehydrogenase 1 A1-Positive Cell Population Is Enriched in Tumor-Initiating Cells and Associated with Progression of Bladder Cancer. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2010;19(2):327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasheed ZA, Yang J, Wang Q, Kowalski J, Freed I, Murter C, et al. Prognostic Significance of Tumorigenic Cells With Mesenchymal Features in Pancreatic Adenocarcinoma. Journal of the National Cancer Institute. 2010;102(5):340 doi: 10.1093/jnci/djp535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moreb JS, Mohuczy D, Ostmark B, Zucali JR. RNAi-mediated knockdown of aldehyde dehydrogenase class-1A1 and class-3A1 is specific and reveals that each contributes equally to the resistance against 4. Cancer Chemotherapy and Pharmacology. 2007;59(1):127–36. doi: 10.1007/s00280-006-0233-6 [DOI] [PubMed] [Google Scholar]
- 39.Suzuki E, Chiba T, Zen Y, Miyagi S, Tada M, Kanai F, et al. Aldehyde dehydrogenase 1 is associated with recurrence‐free survival but not stem cell‐like properties in hepatocellular carcinoma. Hepatology Research. 2012;42(11):1100–11. doi: 10.1111/j.1872-034X.2012.01028.x [DOI] [PubMed] [Google Scholar]
- 40.Mas V, Maluf D, Kj, Yanek K, Kong X, Kulik L, Freise C, et al. Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma. Mol Med. 2009;15(4):85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y, Orlicky DJ, Matsumoto A, Singh S, Thompson DC, Vasiliou V. Aldehyde dehydrogenase 1B1 (ALDH1B1) is a potential biomarker for human colon cancer. Biochem Biophys Res Commun. 2011;405(2):173–9. doi: 10.1016/j.bbrc.2011.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh S, Arcaroli J, Chen Y, Thompson DC, Messersmith W, Jimeno A, et al. ALDH1B1 Is Crucial for Colon Tumorigenesis by Modulating Wnt/β-Catenin, Notch and PI3K/Akt Signaling Pathways. Plos One. 2015;10(5):e0121648 doi: 10.1371/journal.pone.0121648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anthony TE, Heintz N. The folate metabolic enzyme ALDH1L1 is restricted to the midline of the early CNS, suggesting a role in human neural tube defects. Journal of Comparative Neurology. 2007;500(2):368–83. doi: 10.1002/cne.21179 [DOI] [PubMed] [Google Scholar]
- 44.Krupenko SA, Oleinik NV. 10-formyltetrahydrofolate dehydrogenase, one of the major folate enzymes, is down-regulated in tumor tissues and possesses suppressor effects on cancer cells. Cell Growth & Differentiation the Molecular Biology Journal of the American Association for Cancer Research. 2002;13(5):227–36. [PubMed] [Google Scholar]
- 45.Tackelshorne D, Goodman MD, Williams AJ, Wilson DJ, Eskandari T, Vogt LM, et al. Identification of differentially expressed genes in hepatocellular carcinoma and metastatic liver tumors by oligonucleotide expression profiling. Cancer. 2001;92(2):395–405. [DOI] [PubMed] [Google Scholar]
- 46.Chen XQ, He JR, Wang HY. Decreased expression of ALDH1L1 is associated with a poor prognosis in hepatocellular carcinoma. Medical Oncology. 2012;29(3):1843–9. doi: 10.1007/s12032-011-0075-x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Red frame is normal liver tissue. ALDH = aldehyde dehydrogenase.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All data are available from the public database and software (i.e. OncLnc; DAVID; GEO(GEO accession: GSE14520); Oncomine; GTEx; GeneMania).