Abstract
Background
Reg4, a member of the Reg multigene family, is highly upregulated in many gastrointestinal cancers including gastric cancer (GC). The enhanced expression of Reg4 is associated with the resistance of GC to 5-fluorouracil (5-FU), while the underlying mechanism is not clear. The aim of the present study was to explore the resistant mechanism underlying 5-FU resistance.
Material/Methods
Reg4 expression was assessed by Western blot analysis for SGC-7901, BGC-823, AGS, MKN28, and MKN45. Synthetic short single strand RNA oligonucleotides and Flag-Reg4 plasmid were used to investigate the biological function of Reg4 in vitro. The cell viability assay was performed by MTT. Flow cytometry was carried out to measure the apoptosis caused by 5-FU. Reverse-transcriptase quantitative polymerase chain reaction (RT-qPCR) was used to examine the expression of 5-FU metabolism related enzymes. The effect of Reg4 on intracellular signaling was evaluated by Western blot.
Results
Western blot analysis of 5 GC cells showed that Reg4 was low or null in SGC-7901 and BGC-823, while high in AGS, MKN28, and MKN45. Over-expression of flag-Reg4 in SGC-7901 led to an increase in cell viability and lower apoptosis with 5-FU treatment. In contrast, siRNA knockdown of Reg4 enhanced 5-FU induced apoptosis. However, over-expression or knockdown of Reg4 had no significant influence on the expression of 5-FU metabolic enzymes. Further investigation revealed that Reg4 could activate Erk1/2-Bim-caspase3 cascade.
Conclusions
Reg4 inhibited apoptosis through regulating MAPK/Erk/Bim signaling pathway and thereby enhanced the resistance of GC to 5-FU.
MeSH Keywords: Apoptosis, Fluorouracil, Stomach Neoplasms
Background
Gastric cancer (GC) is the third leading cause of cancer-related mortality in China [1]. While early stage gastric cancer has a favorable prognosis [2]; most GC patients are diagnosed at advanced stages when effective therapeutics are not readily available [3]. As a result, the 5-year survival rate of late stage GC is less than 20%–25% and the median survival time is only 6–15 months [4,5].
The 5-fluorouracil (5-FU) based chemotherapy is a standard treatment for patients with advanced GC [6]. Although combining 5-FU with cisplatin, docetaxel, or oxaliplatin can improve treatment response [7], resistance to 5-FU in GC is still an intractable issue in the clinic. It was reported that upregulation of the enzymes associated with 5-FU metabolism such as thymidylate synthase (TS), orotate phosphoribosyl transferase (OPRT) and dihydropyrimidine dehydrogenase (DPD) were involved in the 5-FU-resistance [8]. Besides, alterations of apoptosis pathway [9] or proteins in response to DNA damage [10,11] can lead to resistance to 5-FU.
Our previous study [12] indicated that the mRNA levels of regenerating family member 4 (Reg4) were strongly related to the intrinsic resistance of GC cells to 5-FU. As a member of the REG gene family, Reg4 was originally identified by high-through put sequencing analysis of an inflammatory bowel disease library [13], and its overexpression was proven to be closely correlated to the carcinogenesis in some types of cancer [14–19]. In the present study, the role of Reg4 in promoting 5-FU-resistance in GC cells was confirmed and its mechanism was investigated.
Material and Methods
Cell lines and culture
Human gastric carcinoma cell lines SGC7901, AGS, MKN28, MKN45, and BGC823 were bought from the Cell Bank of the Chinese Academy of Sciences, Shanghai, China. AGS cells were maintained in F12 medium (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco); other cells were maintained in RPMI 1640 medium (Gibco) supplemented with 10% FBS. All cells were cultured in a 5% CO2 and 37°C incubator.
Western blot
Cells were harvested and lysed in NP40 buffer with protease inhibitor cocktails added. The concentration of total protein was measured using the Bradford calorimetric assay (Bio-Rad Laboratories, CA, USA). Thirty micrograms of protein were separated on 12% SDS-PAGE gel and transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, MA, USA). Then the membrane was blocked with 5% bovine serum albumin (Amresco, USA) at room temperature for two hours and then incubated with antibodies against Reg4 (1: 1,000, R&D Systems Inc., USA), P44/42 (1: 1,000, Cell Signaling Technology, USA), phosphorylated P44/42 (1: 1,000, Cell Signaling Technology, USA), Bim (1: 1,000, Cell Signaling Technology, USA), Bcl-2 (1: 1,000, Epitomics, USA), Bax (1: 1,000, Epitomics, USA), active-caspase3 (1: 1,000, Antibody Revolution Inc., USA) at 4°C overnight. In the next day, the membrane was covered with HRP-conjugated secondary antibody (1: 3,000, R&D Systems Inc., USA) at room temperature for two hours and detected using enhanced chemiluminescence regent (ECL, Millipore). Tubulin (1: 1,000, Antibody Revolution Inc., USA) was applied as a loading control.
Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR)
Total RNA were extracted from cells with the TRIZol reagent (Invitrogen). Five hundred nanograms of RNA were reverse transcribed to cDNA using PrimerScript RT Reagent (Takara Biotechnology, Dalian, China). The Quantitative PCR was carried out using SYBR Premix Ex Taq II kit (Takara Biotechnology) on ABI 7500 Real-time PCR System (Life Technology, Foster City, CA, USA). All the primers were obtained from Invitrogen and the sequences of primers were
5′-GAGTGTCAGTCTTACGGAAACG-3′ (Reg4 forward),
5′-TCGTGCAGGCCAATCCATATC-3′ (Reg4 reverse),
5′-GAAGGTGAAGGTCGGAGTC-3′ (GAPDH forward),
5′-GAAGATGGTGATGGGATTTC-3′ (GAPDH reverse),
5′-GCCTCGGTGTGCCTTTCA-3′ (TS forward),
5′-CCCGTGATGTGCGCAAT-3′ (TS reverse),
5′-TAGTGTTTTGGAAACTG TTGAGGTT-3′ (OPRT forward),
5′-TGGCATCAGTGACCTTCAAGCCC TCCT-3′ (OPRT reverse),
5′-AGGACGCAAGGAGGGTTTG-3′ (DPD forward),
5′-GTCCGCCGAGTCCTTACTGA-3′
(DPD reverse). Cycling conditions were 95°C for 10 minute, 40 repeats of 95 °C for 15 seconds followed by 60°C for one minute. The expression levels of mRNAs were calculated using the 2−ΔΔCt method which was described before [20].
Reg4 knockdown and overexpression
Specific siRNAs targeting human Reg4 (siReg4-1 and siReg4-2) and non-targeting control siRNA (siCtrl) were designed and provided by Shanghai GenePharma Co., Ltd, China. The siReg4-1 sequences were 5′-AAGUGGCUAUCAGAGAAGCTT-3′ and 5′-GCUUCUCUGAUAGCCACUUTT-3′. The siReg4-2 sequences were 5′-GACAGAAGGAAGAAACUCATT-3′ and 5′-UGAGUU UCUUCCUUCUGUCTT-3′. The siCtrl sequences were 5′-UUCU CCGAACGUGUCACGUTT-3′ and 5′-ACGUGACACGUUCGGAG AATT-3′. For overexpression of Reg4 gene, its cDNA was amplified using polymerase chain reaction (PCR) and subcloned into the p3 × FLAG-CMV-10 vector (Sigma, St. Louis, MO, USA). The p3 × FLAG vector was used as control. All transfections were performed using Lipofectamine3000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. After transfection for 24 hours, the alterations of Reg4 expression were evaluated by Western blot and RT-qPCR.
Treatment of 5-FU and cell viability assay
The 5-FU was purchased from Sigma Chemical Co. (St. Louis, MO, USA). Stock solutions of 5-FU were prepared in sterile DMSO and stored at 4°C prior to use.
Twenty-four hours post transfection, cells were seeded at a density of 3,000 cells per well in 96-well plates. The next day, different concentrations of 5-FU were added into the plates and after 48 hours, cell viability was determined using the 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide (MTT, Sigma) assay.
Two proper concentrations of 5-FU for each cell line were chosen from the results of cell viability assay. Transfected cells were treated with 5-FU in these two concentrations and then harvested for flow cytometry measurement, Western bolt and RT-qPCR analyses.
Flow cytometry analysis of apoptosis cells
Those 5-FU treated transfected cells were harvested by Accutase-Enyme Cell Detachment Medium (eBiosciences, San Diego, CA, USA) and stained with the PE Annexin V Apoptosis Detection kit (BD Biosicences, San Diego, CA, USA) according to the manufacturer’s instructions. Measurements were performed on a FACS Calibur flow cytometer and results were analyzed by CellQuest software (BD Biosicences, San Diego, CA, USA).
Statistical analysis
SPSS 21.0 (IBM) was used for data analysis. Data from a minimum of three independent experiments were expressed as means ± standard deviations (SD). Comparisons between groups were carried out using the one-way ANOVA with Bonferroni post hoc test or Student’s t-test, and a p value of <0.05 was considered statistically significant.
Results
Reg4 knockdown and overexpression
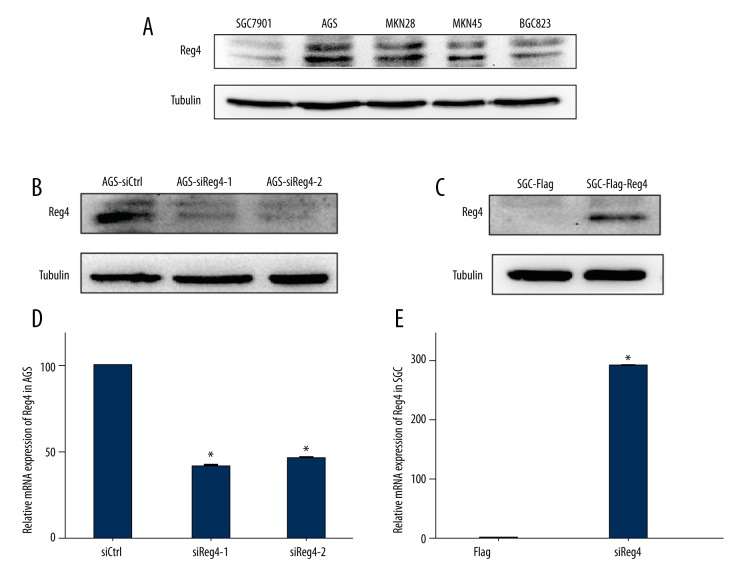
First, we evaluated the expression of Reg4 in five GC cell lines. As shown in Figure 1A, the protein expression of Reg4 was high in AGS, MKN28, and MKN45, but low in BGC-823, and null in SGC7901. Hence AGS and SGC7901 cells were selected to do knockdown and overexpression of Reg4, respectively. As illustrated in Figure 1B–1E, both on protein and mRNA levels, AGS-siReg4-1 and AGS-siReg4-2 expressed less Reg4 than AGS-siCtrl, and SGC-Flag-Reg4 cells expressed more Reg4 than SGC-Flag cells.
Figure 1.
Expression of Reg4 in gastric cancer cell lines. (A) Lysates prepared from SGC7901, AGS, MKN28, MKN45, and BGC823 cells were subjected to Western blot analysis with anti-Reg4 antibody. Tubulin served as a loading control. (B) Lysates prepared from AGS cells transfected with siRNA targeting Reg4 were subjected to Western blot analysis with anti-Reg4 antibody. Tubulin served as a loading control. (C) Lysates prepared from SGC cells transfected with Flag carrying Reg4 were subjected to Western blot analysis with anti-Reg4 antibody. Tubulin served as a loading control. (D) RT-qPCR analysis of Reg4 was performed using total RNA from AGS cells transfected with siRNA against Reg4. GAPDH served as an internal control. Values represent the mean ±SEM, (n=3). (E) RT-qPCR analysis of Reg4 was performed using total RNA from SGC cells transfected with Flag Reg4. GAPDH served as an internal control. Values represent the mean ±SEM, (n=3), * p<0.05.
Reg4 mediated 5-FU resistance of GC cells
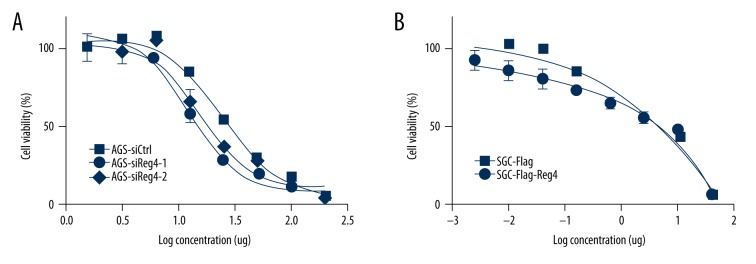
The cell viability assay showed that after 5-FU treatment, AGS-siCtrl cells had better viability than AGS-siReg4-1 and AGS-siReg4-2 cells (Figure 2A) and similarly, SGC-Flag-Reg4 cells had higher cell viability than SGC-Flag cells (Figure 2B). The concentrations of 5-FU for next assays were set as 3 ug/mL and 30 ug/mL for AGS, 0.5 ug/mL and 5 ug/mL for SGC7901.
Figure 2.
Forced Reg4 expression enhanced 5-FU resistance in GC cells. Cell viability was assessed by MTT assay at 48 hours after 5-FU treatment. Bars and error bars represent mean and SD, respectively, from three different experiments. Values represent the mean ±SEM, (n=3).
Expression of the enzymes associated with 5-FU metabolism
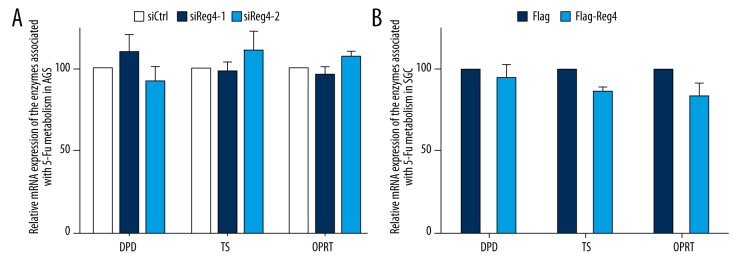
Results from RT-qPCR indicated that there was no obvious difference in expression of enzymes associated with 5-FU metabolism (TS, OPRT, and DPD) between control cells and AGS knockdown or overexpression cells (Figure 3).
Figure 3.
Expression of the enzymes associated with 5-fluorouracil metabolism. (A) RT-qPCR analysis of the mRNA expression of DPD, TS, and OPRT in AGS-siCtrl, AGS-siReg4-1, and AGS-siReg4-2. Values represent the mean ±SD, (n=3), p>0.05. (B) RT-qPCR analysis of the mRNA expression of DPD, TS, and OPRT in SGC-Flag and SGC-Flag-Reg4 cells. Values represent the mean ±SD, (n=3), p>0.05.
Reg4 inhibited apoptosis of GC cells
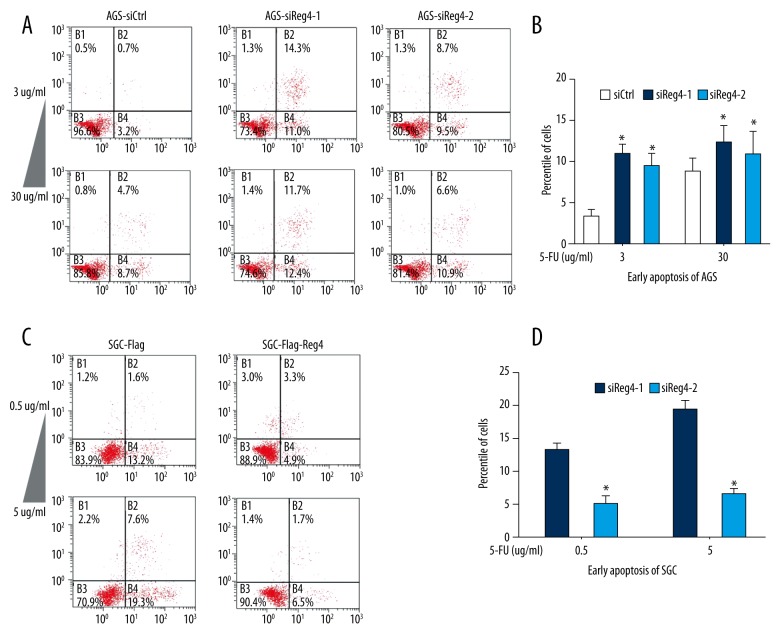
As displayed in Figure 4, after treated with 5-FU, in both of the two concentrations, AGS-siReg4-1 and AGS siReg4-2 cells had significantly higher percentages of apoptosis cells than AGS-siCtrl cells, and SGC-Flag-Reg4 cells had lower apoptosis cells content than SGC-Flag-Ctrl cells.
Figure 4.
High expression of Reg4 decreases apoptosis induced by 5-FU. (A, B) Flow cytometry assays were performed using AGS cells that had been transfected with Reg4 or control siRNA and treated with 3 ug/mL and 30 ug/mL 5-FU for 24 hours. Values represent the mean ±SD, (n=3), * p<0.05. (C, D) Flow cytometry assays were performed using SGC-Flag and SGC-Flag-Reg4 treated with 0.5 ug/mL and 5 ug/mL 5-FU for 24 hours. Values represent the mean ±SD, (n=3), * p<0.05.
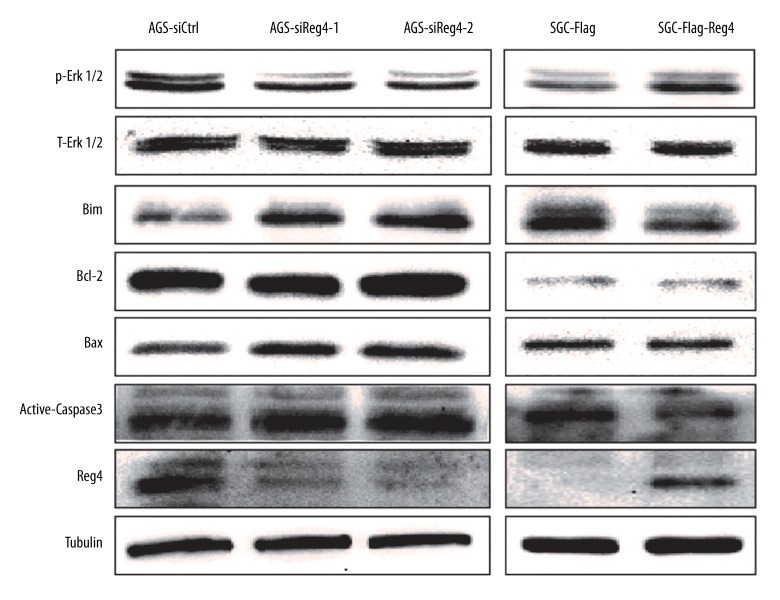
Reg4 regulated the MAPK/Erk/Bim signaling pathway
The Western blot showed that compared with AGS-siCtrl cells, AGS-siReg4-1 and AGS-siReg4-2 cells expressed less phosphorylated Erk1/2 (p-Erk1/2), more apoptosis proteins including Bim, Bax, as well as active-caspase3, and comparable total-Erk1/2 (T-Erk1/2) and Bcl-2 (Figure 5). Consistent trend was detected in SGC-Flag-Reg4 and SGC-Flag cells (Figure 5).
Figure 5.
High expression of Reg4 enhances p-Erk1/2 and some apoptotic proteins’ expression. Lysates prepared from AGS-siCtrl, AGS-siReg4-1, AGS-siReg4-2, SGC-Flag, and SGC-Flag-Reg4 cells were subjected to Western blot analysis with p-Erk1/2, T-Erk1/2, Bim, Bcl-2, Bax, and active-caspase3 antibody. Tubulin served as a loading control.
Discussion
The present study showed that Reg4 increased the 5-FU-tolerability of GC cells in vitro, which was consistent with some previous findings that GC patients with high-expression Reg4 had poor response to 5-FU treatment in comparison to those with low- expression of Reg4 [21].
As has been reported in pancreatic cancer studies, that Reg4 has anti-apoptotic properties when exposed to chemotherapy [22], our results from flow cytometry assay indicated Reg4 decreased 5-FU inducing apoptosis in GC cells. Moreover, our study found that the MAPK/Erk pathway was activated when Reg4 exerted anti-apoptotic effects. It is well acknowledged that the MAPK/Erk pathway plays a crucial role in multiple cellular processes such as cell proliferation, differentiation, adhesion, and migration [23]. Furthermore, it has been reported that Reg4 could modulate receptor tyrosine kinases (RTKs) in prostate cancer and colon cancer [24]. With the addition of acknowledgement that the MAPK pathway is one of the signaling transduction downstream of many RTKs [25], we may suppose that Reg4 may activate the MAPK pathway through influencing RTKs. The mechanism needs to be further explored.
As for apoptotic cascade, MAPK/Erk signaling could repress pro-apoptotic proteins such as Bad and Bim to achieve cell survival [26,27]. In this study, Bim was found to be reduced when Reg4 was overexpressed. Bim, a BH3-domain-only molecule belonging to the Bcl-2 family, is responsible for mitochondrial stress-induced apoptosis [28,29]. As suggested before, Bim can directly bind to and activate Bax; its binding to pro-survival Bcl-2 homologs can indirectly promote the activation of Bax [29]. Thereupon the primed Bax enables the caspase pathway which ultimately leads to cell death [28]. Our results from Western blot analysis (Figure 5) were consistent with the aforementioned understanding, and therefore, we speculated that Reg4 enhanced 5-FU-resistance through affecting the MAPK/Erk/Bim pathway.
However, the present study did not find any alteration of enzymes involved in 5-FU metabolism (DPD, TS, and OPRT) which are known to be important in the formation of 5-FU-resistance [21]. In addition, several studies concentrating on colorectal cancer found that Reg4 could inhibit the expression of Bcl-2 [30,31] while in this study there was no difference in Bcl-2 expression among cells with diverse levels of Reg4. We considered that these disparities were partly caused by the different cell lines used in experiments.
Conclusions
The present study demonstrated that Reg4 inhibited apoptosis through MAPK/Erk/Bim pathway and thereby enhanced the resistance of GC cells to 5-FU. These observations provide clues for further understanding of 5-FU-resistance in GC and other cancers.
Footnotes
Source of support: This study was supported by grants from the Natural Science Foundation of Zhejiang Province (No. LQ13H160017), Zhejiang Provincial Medicine and Health Science Fund (No. 2017PY009), Science Foundation of Zhejiang Chinese Medical University (No. 2014ZY09), and the 1022 program of Zhejiang Cancer Hospita
Compliance with ethical standards
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
No potential conflicts of interest were disclosed.
Reference
- 1.Chen WQ, Zheng RS, Zhang SW. Report of cancer incidence and mortality in China, 2012. China Cancer. 2016;2(7):61. doi: 10.3978/j.issn.2305-5839.2014.04.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uedo NIH, Tatsuta M, Ishihara R, et al. Longterm outcomes after endoscopic mucosal resection for early gastric cancer. Gastric Cancer. 2006;9:88–92. doi: 10.1007/s10120-005-0357-0. [DOI] [PubMed] [Google Scholar]
- 3.Chan BA, JR, Wong RK, Swallow CJ, et al. Improving outcomes in resectable gastric cancer: A review of current and future strategies. Oncology (Williston Park) 2016;30:635–45. [PubMed] [Google Scholar]
- 4.Wang J, Yu JC, Kang WM, Ma ZQ. Treatment strategy for early gastric cancer. Surg Oncol. 2012;21(2):119–23. doi: 10.1016/j.suronc.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Jain VK, Cunningham D, Chau I. Preoperative and postoperative chemotherapy for gastric cancer. Sur Oncol Clin N Am. 2012;21(1):99–112. doi: 10.1016/j.soc.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Wagner AD, Grothe W, Haerting J, et al. Chemotherapy in advanced gastric cancer: A systematic review and meta-analysis based on aggregate data. J Clin Oncol. 2006;24:2903–9. doi: 10.1200/JCO.2005.05.0245. [DOI] [PubMed] [Google Scholar]
- 7.Mackenzie M, Spithoff K, Jonker D. Systemic therapy for advanced gastric cancer: A clinical practice guideline. Curr Oncol (Toronto, Ont) 2011;18(4):e202–9. doi: 10.3747/co.v18i4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters GJ, Backus HH, Freemantle S, et al. Induction of thymidylate synthase as a 5-fluorouracil resistance mechanism. Biochim Biophys Acta. 2002;1587(2–3):194–205. doi: 10.1016/s0925-4439(02)00082-0. [DOI] [PubMed] [Google Scholar]
- 9.Wilson TR, JPaLD Anti-apoptotic mechanisms of drug resistance in cancer. Curr Cancer Drug Targets. 2009;9:307–19. doi: 10.2174/156800909788166547. [DOI] [PubMed] [Google Scholar]
- 10.Wilsker D, Bunz F. Loss of ataxia telangiectasia mutated- and Rad3-related function potentiates the effects of chemotherapeutic drugs on cancer cell survival. Mol Cancer Ther. 2007;6:1406–13. doi: 10.1158/1535-7163.MCT-06-0679. [DOI] [PubMed] [Google Scholar]
- 11.Wyatt MD, Wilson DM., 3rd Participation of DNA repair in the response to 5-fluorouracil. Cell Mol Life Sci. 2009;66:788–99. doi: 10.1007/s00018-008-8557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ying LS, Yu JL, Lu XX, Ling ZQ. Enhanced RegIV expression predicts the intrinsic 5-fluorouracil (5-FU) resistance in advanced gastric cancer. Dig Dis Sci. 2013;58(2):414–22. doi: 10.1007/s10620-012-2381-3. [DOI] [PubMed] [Google Scholar]
- 13.Hartupee JC, Zhang H, Bonaldo MF, et al. Isolation and characterization of a cDNA encoding a novel member of the human regenerating protein family: Reg IV. Biochim Biophys Acta. 2001;1518(3):287–93. doi: 10.1016/s0167-4781(00)00284-0. [DOI] [PubMed] [Google Scholar]
- 14.Oue N, Kuniyasu H, Noguchi T, et al. Serum concentration of Reg IV in patients with colorectal cancer: Overexpression and high serum levels of Reg IV are associated with liver metastasis. Oncology. 2007;72(5–6):371–80. doi: 10.1159/000113147. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Lai M, Lv B, et al. Overexpression of Reg IV in colorectal adenoma. Cancer Lett. 2003;200(1):69–76. doi: 10.1016/s0304-3835(03)00460-9. [DOI] [PubMed] [Google Scholar]
- 16.Oue N, Hamai Y, Mitani Y, et al. Gene expression profile of gastric carcinoma: identification of genes and tags potentially involved in invasion, metastasis, and carcinogenesis by serial analysis of gene expression. Cancer Res. 2004;64(7):2397–405. doi: 10.1158/0008-5472.can-03-3514. [DOI] [PubMed] [Google Scholar]
- 17.Kadowaki Y, Ishihara S, Miyaoka Y, et al. Reg protein is overexpressed in gastric cancer cells, where it activates a signal transduction pathway that converges on ERK1/2 to stimulate growth. FEBS Letters. 2002;530(1–3):59–64. doi: 10.1016/s0014-5793(02)03398-7. [DOI] [PubMed] [Google Scholar]
- 18.Guo Y, Xu J, Li N, et al. RegIV potentiates colorectal carcinoma cell migration and invasion via its CRD domain. Cancer Genet Cytogenet. 2010;199(1):38–44. doi: 10.1016/j.cancergencyto.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 19.Bishnupuri KS, Luo Q, Korzenik JR, et al. Dysregulation of Reg gene expression occurs early in gastrointestinal tumorigenesis and regulates anti-apoptotic genes. Cancer Biol Ther. 2006;5(12):1714–20. doi: 10.4161/cbt.5.12.3469. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Mitani Y, Oue N, Matsumura S, et al. Reg IV is a serum biomarker for gastric cancer patients and predicts response to 5-fluorouracil-based chemotherapy. Oncogene. 2007;26(30):4383–93. doi: 10.1038/sj.onc.1210215. [DOI] [PubMed] [Google Scholar]
- 22.Eguchi HIO, Ohigashi H, Takahashi H, et al. Serum REG4 level is a predictive biomarker for the response to preoperative chemoradiotherapy in patients with pancreatic cancer. Pancreas. 2009;38:791–98. doi: 10.1097/MPA.0b013e3181ac5337. [DOI] [PubMed] [Google Scholar]
- 23.Busca R, Pouyssegur J, Lenormand P. ERK1 and ERK2 map kinases: Specific roles or functional redundancy? Front Cell Dev Biol. 2016;4:53. doi: 10.3389/fcell.2016.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vanderlaag K, Wang W, Fayadat-Dilman L, et al. Regenerating islet-derived family member, 4 modulates multiple receptor tyrosine kinases and mediators of drug resistance in cancer. Int J Cancer. 2012;130(6):1251–63. doi: 10.1002/ijc.26089. [DOI] [PubMed] [Google Scholar]
- 25.Rattanasinchai C, Gallo KA. MLK3 signaling in cancer invasion. Cancers (Basel) 2016;8(5) doi: 10.3390/cancers8050051. pii: E51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cagnol S, Chambard JC. ERK and cell death: mechanisms of ERK-induced cell death – apoptosis, autophagy and senescence. FEBS J. 2010;277(1):2–21. doi: 10.1111/j.1742-4658.2009.07366.x. [DOI] [PubMed] [Google Scholar]
- 27.Balmanno K, Cook SJ. Tumour cell survival signalling by the ERK1/2 pathway. Cell Death Differ. 2009;16(3):368–77. doi: 10.1038/cdd.2008.148. [DOI] [PubMed] [Google Scholar]
- 28.Gross A, McDonnell JM, Korsmeyer SJ. BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 1999;13(15):1899–911. doi: 10.1101/gad.13.15.1899. [DOI] [PubMed] [Google Scholar]
- 29.Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26(9):1324–37. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu YPC, Hu J, Zhang S. The role of Reg IV in colorectal cancer, as a potential therapeutic target. Contemp Oncol (Pozn) 2015;19:261–64. doi: 10.5114/wo.2015.54385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bishnupuri KS, Luo Q, Sainathan SK, et al. Reg IV regulates normal intestinal and colorectal cancer cell susceptibility to radiation-induced apoptosis. Gastroenterology. 2010;138:616–26. doi: 10.1053/j.gastro.2009.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]