Abstract
Fibroproliferative diseases are common complex traits featuring scarring and overgrowth of connective tissue which vary widely in presentation because they affect many organ systems. Most fibroproliferative diseases are more prevalent in African-derived populations than in European populations, leading to pronounced health disparities. It is hypothesized that the increased prevalence of these diseases in African-derived populations is due to selection for pro-fibrotic alleles that are protective against helminth infections. We constructed a genetic risk score (GRS) of fibroproliferative disease risk-increasing alleles using 147 linkage disequilibrium-pruned variants identified through genome-wide association studies of seven fibroproliferative diseases with large African-European prevalence disparities. A comparison of the fibroproliferative disease GRS between 1000 Genomes Phase 3 populations detected a higher mean GRS in AFR (mean = 148 risk alleles) than EUR (mean = 136 risk alleles; T-test p-value = 1.75x10-123). To test whether differences in GRS burden are systematic and may be due to selection, we employed the quantitative trait loci (QTL) sign test. The QTL sign test result indicates that population differences in risk-increasing allele burdens at these fibroproliferative disease variants are systematic and support a model featuring selective pressure (p-value = 0.011). These observations were replicated in an independent sample and were more statistically significant (T-test p-value = 7.26x10-237, sign test p-value = 0.015). This evidence supports the role of selective pressure acting to increase frequency of fibroproliferative alleles in populations of African relative to European ancestry populations.
Introduction
Fibroproliferative diseases are a consequence of dysregulated scarring and connective tissue overgrowth, affect many organ systems, vary widely in presentation, and are very common in humans[1, 2]. Uterine fibroids, keloid scars, pulmonary fibrosis, cirrhosis, Crohn’s disease, and atherosclerosis are examples of diseases with fibroproliferative features. Many fibroproliferative diseases are more prevalent in recently African-derived populations than in European populations[3], collectively contributing to pronounced overall health disparity (Table 1). For example, keloids are more common in those with darker pigmentation[4], and systemic scleroderma[5, 6], nephrosclerosis[7], and sarcoidosis[8] are more prevalent in African American individuals. However, this is not the case for all fibroproliferative diseases. Dupuytren contracture is a disease predominantly affecting European American individuals[9], as are pulmonary fibrosis[10] and multiple sclerosis[11], though they are not uncommon in those of recent African ancestry.
Table 1. Fibroproliferative diseases with increased prevalence in African-derived populations.
| Disease | Prevalence Ratio (AA:EA) | Number of SNPs included in GRS1 |
|---|---|---|
| Nephrosclerosis | 3–20[35–41] | 69 |
| Keloids | 20[42] | 3 |
| Sarcoidosis | 3–17[43–51] | 1 |
| Hypertension2 | 1.4–7[52–57] | 42 |
| Glaucoma | 4–5[58, 59] | 21 |
| Scleroderma | 3[43, 60–62] | 8 |
| Uterine Fibroids | 1.5–3[63–66] | 3 |
Table modified from Russell et al, 2015[3]. AA-African American; EA-European American
1Number of loci reaching genome-wide significance in the NHGRI/EBI GWAS Catalog (www.ebi.ac.uk/gwas). Complete list of SNPs is contained in S1 Table
2Hypertension is associated with a systemic inflammatory state which can lead to target organ fibrosis[67–70], and also occurs in the presence of atherosclerosis which is itself fibroproliferative[71–73]. There are shared characteristics between hypertension and other fibroproliferative diseases[54, 74–78], and fibrotic growth factors are often upregulated in hypertension as well[53, 55, 79, 80]. Hypertension has been presented as a fibroproliferative condition in other publications[1, 3, 81], which discuss the evidence for this in more detail.
Beyond the large prevalence disparities across continental ancestral groups, there is observational association study evidence to suggest that these conditions are heritable[12–17]. Genome-wide association studies (GWAS) have identified many common susceptibility variants for several fibroproliferative diseases[18, 19]. However, like most phenotypes, these studies have been performed predominantly in European American populations.
It has been suggested that fibroproliferative diseases may share a common genetic background[20]. There is also evidence for pathological similarities across fibroproliferative phenotypes[1, 2]. A recent review by Russell et al presents the hypothesis and presents evidence for the increased prevalence of fibroproliferative diseases in African American populations as a result of selection for anti-helminthic, pro-fibrotic alleles in response to helminth infections on the African continent[3]. Similar scenarios of diseases arising due to selective response to pathogens in African populations have been seen in sickle cell disease conferring resistance to malaria[21, 22], and the increased frequency of chronic kidney disease in carriers of Apolipoprotein L1 (APOL1) variants, which offer enhanced ability to resist trypanosome infections that cause African sleeping sickness[23].
When a trait is under adaptive selective pressure, allele frequencies at all loci with an influence on that trait will change over generations. In contrast, under neutrality allele frequencies will drift randomly. Thereby, if individuals with higher values for the trait enjoy higher relative fitness, then trait-increasing alleles will tend to become more common over generations, and the effect of selection on allele frequency will be proportional to the effect of the allele on the trait under selection. For a complex trait under selection with many genetic determinants with subtle effects, small changes in allele frequency will occur with relatively undetectable differences in linkage disequilibrium (LD) and haplotype diversity, as has been seen in human height[24]. Across trait loci, systematic differences in trait- or risk-increasing allele frequencies that are consistent with the disparity between two populations are detectable, given a sufficient number of known causal loci and precision to estimate allele frequencies[25, 26]. In this study, we were able to assess systematic frequency differences of known fibroproliferative risk-increasing alleles across populations; however, we could not assume or assess proportionality of effect sizes to allele frequency differences between African and other continental populations. The available effect size estimates for these alleles are not for their putative protective effects on helminth infections, but for their consequences across organ systems in various fibroproliferative traits.
Results and discussion
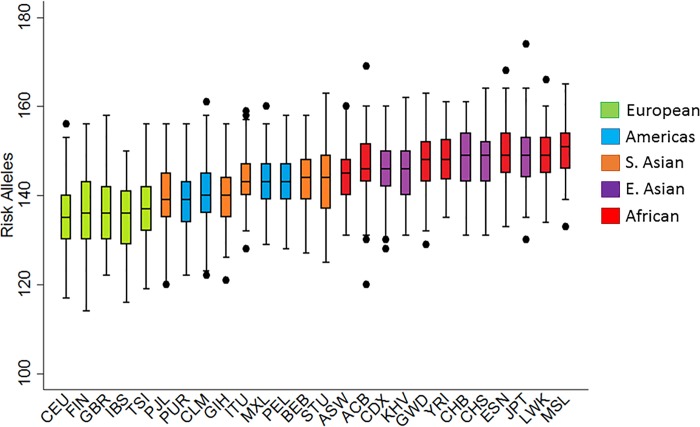
This study sought to determine whether allele frequency differences at known fibroproliferative risk loci are consistent with evidence for selective pressure and may explain racial disparities associated with fibroproliferative diseases (and related quantitative phenotypes) through evaluating loci implicated by genome wide association studies (GWAS). To do this, a genetic risk score (GRS) utilizing single nucleotide polymorphisms (SNPs) from the GWAS catalog[19] was constructed for seven fibroproliferative diseases with increased prevalence (>2 fold on average) in African ancestry populations compared to European ancestry populations (Table 1). The total number of LD-pruned (r2<0.2) independent SNPs included in the GRS (excluding the HLA region) was 147 (S1 Table). This unweighted GRS was calculated in all 1000 Genomes[27] (a sample without phenotypic selection bias) samples as the number of risk-increasing alleles in each individual. The individual-level GRS burdens ranged from 114 to 174 risk alleles (Table 2, Fig 1, S2 Table). Overall, the burden of risk alleles was consistently higher in African-derived (AFR) populations (mean GRS = 148.07) than European-derived (EUR) populations (mean GRS = 136.03), which have the lowest overall GRS burden (Fig 1). The difference in the mean GRS values between the EUR and AFR populations was 12.04 risk alleles (t-test p-value = 1.75x10−123). This result became marginally more significant when limiting the AFR population to only continental African populations (difference in means = 12.69 risk alleles, p-value = 1.37x10-125).
Table 2. Summary statistics for fibroproliferative GRS among AFR and EUR populations from 1000 Genomes, and among BioVU samples.
| Population | N | Mean GRS |
Minimum GRS |
Maximum GRS |
|---|---|---|---|---|
| AFR | 661 | 148.07 | ||
| ACB | 96 | 146.57 | 120 | 169 |
| ASW | 61 | 145.00 | 131 | 160 |
| ESN | 99 | 149.90 | 133 | 168 |
| GWD | 113 | 147.61 | 129 | 163 |
| LWK | 99 | 148.71 | 134 | 166 |
| MSL | 85 | 149.69 | 133 | 165 |
| YRI | 108 | 148.06 | 135 | 161 |
| BioVU AA (combined) | 1382 | 145.11 | ||
| BioVU AA cases | 578 | 145.20 | 126.44 | 167.47 |
| BioVU AA controls | 804 | 145.05 | 117.41 | 164.25 |
| EUR | 503 | 136.03 | ||
| CEU | 99 | 135.34 | 117 | 156 |
| FIN | 99 | 136.07 | 114 | 156 |
| GBR | 91 | 136.67 | 122 | 156 |
| IBS | 107 | 135.01 | 116 | 150 |
| TSI | 107 | 137.10 | 119 | 156 |
| BioVU EA (combined) | 2359 | 136.34 | ||
| BioVU EA cases | 1195 | 136.13 | 113.20 | 162.18 |
| BioVU EA controls | 1164 | 136.55 | 113.94 | 161.10 |
GRS: Genetic risk score; AA: African American; EA: European American; CEU: Utah Residents (CEPH) with Northern and Western Ancestry; TSI: Toscani in Italia; FIN: Finnish in Finland; GBR: British in England and Scotland; IBS: Iberian Population in Spain; YRI: Yoruba in Ibadan, Nigeria; LWK: Luhya in Webuye, Kenya; GWD: Gambian in Western Divisions in the Gambia; MSL: Mende in Sierra Leone; ESN: Esan in Nigeria; ASW: Americans of African Ancestry in SW USA; ACB: African Caribbeans in Barbados
Fig 1. Distribution of fibroproliferative disease GRS in populations from 1000 Genomes.
Results are sorted by median risk allele burden. Bars represent the 25th and 75th percentiles and are color coded by super-population (Green = EUR, Blue = AMR, Orange = SAS, Purple = EAS, Red = AFR).
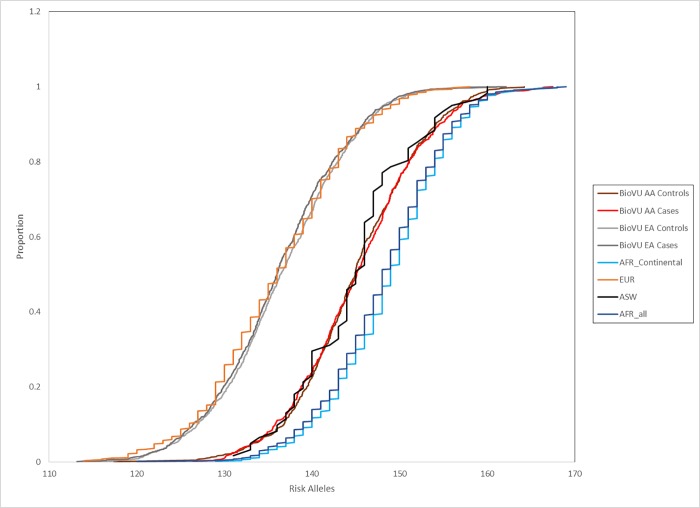
This was replicated in a larger independent population consisting of uterine fibroids cases and controls from BioVU, an electronic health record-linked DNA biorepository (Table 2, Fig 2), ascertained as part of another study[28]. In BioVU, the mean GRS in the African American samples was 145.11 (N = 1382; range: 117.41–167.47), while the mean in European American samples was 136.34 (N = 2359; range: 113.20–162.18). Cases and controls had similar means within racial groups, which were not significantly different from each other (African American controls mean = 145.05, African American cases mean = 145.20, t-test p-value = 0.69; European American controls mean = 136.55, European American cases mean = 136.13, t-test p-value = 0.16). The difference between African American and European American in this set was also highly significant (t-test p-value = 7.26x10-237), and remained so when evaluating controls only, as cases may be enriched for fibroproliferative alleles above those in the general population (t-test p-value = 6.38x10-127).
Fig 2. Cumulative distribution of GRS in 1000 Genomes AFR, EUR, ASW, and BioVU populations.
*AFR includes only the continental African populations.
We further evaluated whether there are systematic differences in allele frequency at fibroproliferative disease risk alleles in the same direction as the prevalence disparity between African ancestry and European ancestry populations. Many approaches to evaluate selective pressure at multiple loci with small effects rely upon the proportionality assumption between disease effect sizes and allele frequency differences[26, 29]. However, those tests require that the disease of interest is itself under selective pressure, which is inconsistent with the hypothesis of selection in fibroproliferative diseases.
Therefore, evidence for selection was examined using the quantitative trait loci (QTL) sign test, which evaluates whether the number of variants at higher frequency in one population compared to the other deviates from neutrality under a binomial distribution[30]. This approach does not utilize phenotypic effect sizes, which is ideal for this study sample in which phenotypes are not observed and in this scenario where the effects of alleles on helminth infections have not been estimated. In this analysis, 85 of the 147 SNPs had risk alleles at higher frequency in African-derived populations than in European-derived populations (S3 Table), which is significant from the QTL sign test with a p-value of 0.011. Differences in risk allele frequency ranged from 0.006 to 0.73, with greater than 35% of the variants with an African-European frequency difference of 0.2 or larger. Thirty-three of the 53 variants (62%) with the largest (>0.2) differences were more common in African-derived populations (p-value = 0.022). The number of SNPs at higher frequency (and therefore the p-value for the sign test) did not change when limiting to samples from continental Africa. The sign test analysis also replicated in BioVU, with 84 of the 147 SNPs available in this dataset being more common in the African American samples compared to European American samples (p-value = 0.015).
Conclusion
Overall, this analysis supports the possibility that selective pressure on an as-yet undetermined phenotype may have impacted genetic variants predisposing to seven fibroproliferative diseases. The observation made in the present study of higher genetic burden of fibroproliferative alleles was observed in both a population-based sample as well as within an independent sample ascertained from a hospital-based biobank. It is of note that nearly all of the genetic variants identified though GWAS (and thus included in the GRS) were implicated through studies of European or East Asian populations. This supports our conclusion, and identification of additional fibroproliferative risk variants that are more common in African populations will likely make the trends observed between these two population groups even more apparent.
Methods
Samples and genotypes
The discovery phase of these analyses utilized publically available genotype and population data from phase 3 of the 1000 Genomes Project was downloaded from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/[31]. Individual population and super-population assignments were the only non-genetic information evaluated.
For replication individuals with imaging-confirmed uterine fibroids and genome-wide genotype data were included from BioVU, Vanderbilt University Medical Center’s de-identified biorepository linked to electronic health records. The phenotyping algorithms used to identify case and control subjects have been previously published [28]. Briefly, this algorithm used a combination of demographic inclusion and exclusion criteria, International Classification of Diseases 9th edition (ICD-9) diagnostic codes, Current Procedural Terminology (CPT) codes, and keywords exclusions from specific notes and reports of a participant in order to identify cases and controls. Cases required evidence of a fibroid diagnosis defined by either an ICD-9 code indicating the presence of fibroids or ICD and CPT codes indicating a history of fibroid treatment procedures (e.g. myomectomy or uterine artery embolization). An individual was included as a control if they had two imaging events on separate dates and did not have a fibroid diagnosis or history of fibroid treatment procedures. Excluded from controls were women without an intact uterus (e.g. having had a prior hysterectomy) based on CPT procedural codes and text mentions of hysterectomy. This study was reviewed and approved by the Vanderbilt University Institutional Review Board (#161378).
BioVU subjects were genotyped using both the Affymetrix BioBank array (European American and African American subjects) and the Axiom World array 2 (Affymetrix Inc., Santa Clara, CA) was additionally genotyped in the African Americans in order to attain better coverage for African-derived variants. Genotype quality control was performed separately for European American and African American datasets, including a 95% SNP and individual call rate threshold, removal of related individuals, gender checks, alignment of alleles to the genomic ‘+’ strand, and visualization of ancestry by principal components analysis using Eigenstrat software [32]. The genotype data were imputed to the 1000 Genomes phase 3 reference panel using SHAPEIT2 [33] for haplotype phasing and IMPUTE2 [34] for genotype imputation, with phasing and imputation performed separately for each dataset.
Genetic risk score construction
A genetic risk score (GRS) utilizing single nucleotide polymorphisms (SNPs) from the GWAS catalog[19] was constructed for seven fibroproliferative diseases with increased prevalence (>2 fold on average) in African ancestry populations compared to European ancestry populations (Table 1). The total number of LD-pruned (r2<0.2) independent SNPs included in the GRS (excluding the HLA region) was 147 (S1 Table). This unweighted GRS was calculated in all 1000 Genomes[27] as the number of risk-increasing alleles in each individual. The GRS was also computed across the imputed BioVU data utilizing the dosage data to account for the number of risk-increasing alleles. Of the 147 variants evaluated, 70 were directly genotyped in the African American set, and 63 were genotyped in the European American set. Mean info scores among imputed SNPs were 0.979 and 0.971 in African American and European American sets, respectively.
Supporting information
(PDF)
(PDF)
AFR (cont.) represents continental AFR samples only (i.e. not including ACB and ASW populations).
(PDF)
Data Availability
1000 Genomes data is publically available and may be downloaded via FTP (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/), Aspera, or Globus GridFTP (http://www.internationalgenome.org/data#download). Genotype data from BioVU used in this study is publically available, and can be found in dbGaP, accession number phs001409.v1.p1. Phenotype data from BioVU is available to researchers, who meet the criteria for access to confidential data, upon request and clearance from Vanderbilt University Medical Center Institutional Review Board and BioVU. Interested and eligible researchers may contact the BioVU data access team at biovu@vanderbilt.edu for more detailed information regarding access to phenotype data from BioVU.
Funding Statement
This project was supported by National Institutes of Health grants R01HD074711 (DRVE), R03HD078567 (DRVE), R21AR067938 (DRVE), and R21HL121429 (TLE and DRVE), as well as the Vanderbilt Molecular and Genetic Epidemiology of Cancer (MAGEC) training program, which was funded by the US National Cancer Institute grant number R25 CA160056 (PI: X.-O. Shu). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wynn TA. Common and unique mechanisms regulate fibrosis in various fibroproliferative diseases. J Clin Invest. 2007;117(3):524–9. Epub 2007/03/03. doi: 10.1172/JCI31487 ; PubMed Central PMCID: PMCPMC1804380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C, Ogawa R. Fibroproliferative disorders and their mechanobiology. Connective tissue research. 2012;53(3):187–96. Epub 2012/02/15. doi: 10.3109/03008207.2011.642035 . [DOI] [PubMed] [Google Scholar]
- 3.Russell SB, Smith JC, Huang M, Trupin JS, Williams SM. Pleiotropic Effects of Immune Responses Explain Variation in the Prevalence of Fibroproliferative Diseases. PLoS Genet. 2015;11(11):e1005568 Epub 2015/11/06. doi: 10.1371/journal.pgen.1005568 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ud-Din S, Bayat A. Strategic management of keloid disease in ethnic skin: a structured approach supported by the emerging literature. The British journal of dermatology. 2013;169 Suppl 3:71–81. Epub 2013/10/23. doi: 10.1111/bjd.12588 . [DOI] [PubMed] [Google Scholar]
- 5.Gelber AC, Manno RL, Shah AA, Woods A, Le EN, Boin F, et al. Race and association with disease manifestations and mortality in scleroderma: a 20-year experience at the Johns Hopkins Scleroderma Center and review of the literature. Medicine. 2013;92(4):191–205. Epub 2013/06/25. doi: 10.1097/MD.0b013e31829be125 ; PubMed Central PMCID: PMCPMC4553970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayes MD, Lacey JV Jr., Beebe-Dimmer J, Gillespie BW, Cooper B, Laing TJ, et al. Prevalence, incidence, survival, and disease characteristics of systemic sclerosis in a large US population. Arthritis and rheumatism. 2003;48(8):2246–55. Epub 2003/08/09. doi: 10.1002/art.11073 . [DOI] [PubMed] [Google Scholar]
- 7.Meyrier A. Nephrosclerosis: a term in quest of a disease. Nephron. 2015;129(4):276–82. Epub 2015/04/15. doi: 10.1159/000381195 . [DOI] [PubMed] [Google Scholar]
- 8.Mirsaeidi M, Machado RF, Schraufnagel D, Sweiss NJ, Baughman RP. Racial difference in sarcoidosis mortality in the United States. Chest. 2015;147(2):438–49. Epub 2014/09/05. doi: 10.1378/chest.14-1120 ; PubMed Central PMCID: PMCPMC4314818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saboeiro AP, Porkorny JJ, Shehadi SI, Virgo KS, Johnson FE. Racial distribution of Dupuytren's disease in Department of Veterans Affairs patients. Plastic and reconstructive surgery. 2000;106(1):71–5. Epub 2000/07/07. . [DOI] [PubMed] [Google Scholar]
- 10.Swigris JJ, Olson AL, Huie TJ, Fernandez-Perez ER, Solomon J, Sprunger D, et al. Ethnic and racial differences in the presence of idiopathic pulmonary fibrosis at death. Respiratory medicine. 2012;106(4):588–93. Epub 2012/02/03. doi: 10.1016/j.rmed.2012.01.002 ; PubMed Central PMCID: PMCPMC3294009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cree BA, Khan O, Bourdette D, Goodin DS, Cohen JA, Marrie RA, et al. Clinical characteristics of African Americans vs Caucasian Americans with multiple sclerosis. Neurology. 2004;63(11):2039–45. Epub 2004/12/15. . [DOI] [PubMed] [Google Scholar]
- 12.Luoto R, Kaprio J, Rutanen EM, Taipale P, Perola M, Koskenvuo M. Heritability and risk factors of uterine fibroids—the Finnish Twin Cohort study. Maturitas. 2000;37(1):15–26. Epub 2000/12/02. . [DOI] [PubMed] [Google Scholar]
- 13.Marneros AG, Norris JE, Olsen BR, Reichenberger E. Clinical genetics of familial keloids. Archives of dermatology. 2001;137(11):1429–34. Epub 2001/11/16. . [DOI] [PubMed] [Google Scholar]
- 14.Satko SG, Sedor JR, Iyengar SK, Freedman BI. Familial clustering of chronic kidney disease. Seminars in dialysis. 2007;20(3):229–36. Epub 2007/06/09. doi: 10.1111/j.1525-139X.2007.00282.x . [DOI] [PubMed] [Google Scholar]
- 15.Sverrild A, Backer V, Kyvik KO, Kaprio J, Milman N, Svendsen CB, et al. Heredity in sarcoidosis: a registry-based twin study. Thorax. 2008;63(10):894–6. Epub 2008/06/07. doi: 10.1136/thx.2007.094060 . [DOI] [PubMed] [Google Scholar]
- 16.Allanore Y, Wipff J, Kahan A, Boileau C. Genetic basis for systemic sclerosis. Joint, bone, spine: revue du rhumatisme. 2007;74(6):577–83. Epub 2007/09/15. doi: 10.1016/j.jbspin.2007.04.005 . [DOI] [PubMed] [Google Scholar]
- 17.Teikari JM. Genetic factors in open-angle (simple and capsular) glaucoma. A population-based twin study. Acta ophthalmologica. 1987;65(6):715–20. Epub 1987/12/01. . [DOI] [PubMed] [Google Scholar]
- 18.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42(Database issue):D1001–6. Epub 2013/12/10. doi: 10.1093/nar/gkt1229 ; PubMed Central PMCID: PMCPMC3965119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The NHGRI-EBI Catalog of published genome-wide association studies. [Internet]. [cited 02/08/2016].
- 20.Wenzke KE, Cantemir-Stone C, Zhang J, Marsh CB, Huang K. Identifying common genes and networks in multi-organ fibrosis. AMIA Joint Summits on Translational Science proceedings AMIA Summit on Translational Science. 2012;2012:106–15. Epub 2012/07/11. ; PubMed Central PMCID: PMCPMC3392050. [PMC free article] [PubMed] [Google Scholar]
- 21.Elguero E, Delicat-Loembet LM, Rougeron V, Arnathau C, Roche B, Becquart P, et al. Malaria continues to select for sickle cell trait in Central Africa. Proc Natl Acad Sci U S A. 2015;112(22):7051–4. Epub 2015/05/06. doi: 10.1073/pnas.1505665112 ; PubMed Central PMCID: PMCPMC4460506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams TN, Mwangi TW, Roberts DJ, Alexander ND, Weatherall DJ, Wambua S, et al. An immune basis for malaria protection by the sickle cell trait. PLoS Med. 2005;2(5):e128 Epub 2005/05/27. doi: 10.1371/journal.pmed.0020128 ; PubMed Central PMCID: PMCPMC1140945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science (New York, NY). 2010;329(5993):841–5. doi: 10.1126/science.1193032 ; PubMed Central PMCID: PMCPMC2980843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lohmueller KE. The impact of population demography and selection on the genetic architecture of complex traits. PLoS Genet. 2014;10(5):e1004379 Epub 2014/05/31. doi: 10.1371/journal.pgen.1004379 ; PubMed Central PMCID: PMCPMC4038606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joshi PK, Esko T, Mattsson H, Eklund N, Gandin I, Nutile T, et al. Directional dominance on stature and cognition in diverse human populations. Nature. 2015;523(7561):459–62. Epub 2015/07/02. doi: 10.1038/nature14618 ; PubMed Central PMCID: PMCPMC4516141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berg JJ, Coop G. A population genetic signal of polygenic adaptation. PLoS Genet. 2014;10(8):e1004412 Epub 2014/08/08. doi: 10.1371/journal.pgen.1004412 ; PubMed Central PMCID: PMCPMC4125079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632 ; PubMed Central PMCID: PMCPMC3498066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feingold-Link L, Edwards TL, Jones S, Hartmann KE, Velez Edwards DR. Enhancing uterine fibroid research through utilization of biorepositories linked to electronic medical record data. Journal of women's health (2002). 2014;23(12):1027–32. doi: 10.1089/jwh.2014.4978 ; PubMed Central PMCID: PMCPMC4267124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turchin MC, Chiang CW, Palmer CD, Sankararaman S, Reich D, Hirschhorn JN. Evidence of widespread selection on standing variation in Europe at height-associated SNPs. Nat Genet. 2012;44(9):1015–9. Epub 2012/08/21. doi: 10.1038/ng.2368 ; PubMed Central PMCID: PMCPMC3480734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orr HA. Testing natural selection vs. genetic drift in phenotypic evolution using quantitative trait locus data. Genetics. 1998;149(4):2099–104. Epub 1998/08/05. ; PubMed Central PMCID: PMCPMC1460271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. Epub 2015/10/04. doi: 10.1038/nature15393 ; PubMed Central PMCID: PMCPMC4750478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–9. Epub 2006/07/25. doi: 10.1038/ng1847 . [DOI] [PubMed] [Google Scholar]
- 33.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9(2):179–81. Epub 2011/12/06. doi: 10.1038/nmeth.1785 . [DOI] [PubMed] [Google Scholar]
- 34.Duan Q, Liu EY, Auer PL, Zhang G, Lange EM, Jun G, et al. Imputation of coding variants in African Americans: better performance using data from the exome sequencing project. Bioinformatics. 2013;29(21):2744–9. doi: 10.1093/bioinformatics/btt477 ; PubMed Central PMCID: PMCPMC3799474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.August P, Suthanthiran M. Transforming growth factor beta and progression of renal disease. Kidney international Supplement. 2003;(87):S99–104. Epub 2003/10/09. . [DOI] [PubMed] [Google Scholar]
- 36.Ferguson R, Grim CE, Opgenorth TJ. The epidemiology of end-state renal disease: the six-year South-Central Los Angeles experience, 1980–85. American journal of public health. 1987;77(7):864–5. Epub 1987/07/01. ; PubMed Central PMCID: PMCPMC1647229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tracy RE, Bhandaru SY, Oalmann MC, Guzman MA, Newmann WP, 3rd. Blood pressure and nephrosclerosis in black and white men and women aged 25 to 54. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 1991;4(5):602–9. Epub 1991/09/01. . [PubMed] [Google Scholar]
- 38.Freedman BI, Bowden DW, Rich SS, Valis CJ, Sale MM, Hicks PJ, et al. A genome scan for all-cause end-stage renal disease in African Americans. Nephrol Dial Transplant. 2005;20(4):712–8. Epub 2005/02/11. doi: 10.1093/ndt/gfh704 . [DOI] [PubMed] [Google Scholar]
- 39.URD S. USRDS 2003 Annual Data Report: Atlas of End-Stage Renal Disease in the United States. Bethesda, MD: National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, 2003.
- 40.URDS U. 1998 Annual Data Report. Bethesda, MD: The National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, 1998.
- 41.Toto RB. Hypertensive nephrosclerosis in African Americans. Kidney Int. 2003;64(6):2331–41. Epub 2003/11/25. doi: 10.1046/j.1523-1755.2003.00333.x . [DOI] [PubMed] [Google Scholar]
- 42.Barrett J. Keloid In: Bergsma D, editor. Birth Defect Compendium. Baltimore: Williams and Wilkins Company; 1973. p. 553. [Google Scholar]
- 43.Polednak AP. Connective tissue responses in blacks in relation to disease: further observations. American journal of physical anthropology. 1987;74(3):357–71. Epub 1987/11/01. doi: 10.1002/ajpa.1330740308 . [DOI] [PubMed] [Google Scholar]
- 44.Rybicki BA, Maliarik MJ, Major M, Popovich J Jr., Iannuzzi MC. Epidemiology, demographics, and genetics of sarcoidosis. Seminars in respiratory infections. 1998;13(3):166–73. Epub 1998/10/09. . [PubMed] [Google Scholar]
- 45.Terris M, Chaves AD. An epidemiologic study of sarcoidosis. The American review of respiratory disease. 1966;94(1):50–5. Epub 1966/07/01. doi: 10.1164/arrd.1966.94.1.50 . [DOI] [PubMed] [Google Scholar]
- 46.Keller AZ. Hospital, age, racial, occupational, geographical, clinical and survivorship characteristics in the epidemiology of sarcoidosis. Am J Epidemiol. 1971;94(3):222–30. Epub 1971/09/01. . [DOI] [PubMed] [Google Scholar]
- 47.Sartwell PE, Edwards LB. Epidemiology of sarcoidosis in the U.S. Navy. Am J Epidemiol. 1974;99(4):250–7. Epub 1974/04/01. . [DOI] [PubMed] [Google Scholar]
- 48.Rybicki BA, Major M, Popovich J Jr., Maliarik MJ, Iannuzzi MC. Racial differences in sarcoidosis incidence: a 5-year study in a health maintenance organization. Am J Epidemiol. 1997;145(3):234–41. Epub 1997/02/01. . [DOI] [PubMed] [Google Scholar]
- 49.Rybicki BA, Harrington D, Major M, Simoff M, Popovich J Jr., Maliarik M, et al. Heterogeneity of familial risk in sarcoidosis. Genet Epidemiol. 1996;13(1):23–33. Epub 1996/01/01. doi: 10.1002/(SICI)1098-2272(1996)13:1<23::AID-GEPI3>3.0.CO;2-7 . [DOI] [PubMed] [Google Scholar]
- 50.Kucera GP, Rybicki BA, Kirkey KL, Coon SW, Major ML, Maliarik MJ, et al. Occupational risk factors for sarcoidosis in African-American siblings. Chest. 2003;123(5):1527–35. Epub 2003/05/13. . [DOI] [PubMed] [Google Scholar]
- 51.Iannuzzi MC, Rybicki BA, Teirstein AS. Sarcoidosis. N Engl J Med. 2007;357(21):2153–65. Epub 2007/11/23. doi: 10.1056/NEJMra071714 . [DOI] [PubMed] [Google Scholar]
- 52.Polednak AP. Various chronic disorders and other conditions Racial and Ethnic Differences in Disease. New York: Oxford University Press; 1989. p. 228–9,46–47. [Google Scholar]
- 53.Suthanthiran M, Li B, Song JO, Ding R, Sharma VK, Schwartz JE, et al. Transforming growth factor-beta 1 hyperexpression in African-American hypertensives: A novel mediator of hypertension and/or target organ damage. Proc Natl Acad Sci U S A. 2000;97(7):3479–84. Epub 2000/03/22. doi: 10.1073/pnas.050420897 ; PubMed Central PMCID: PMCPMC16265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dustan HP. Does keloid pathogenesis hold the key to understanding black/white differences in hypertension severity? Hypertension. 1995;26(6 Pt 1):858–62. Epub 1995/12/01. . [DOI] [PubMed] [Google Scholar]
- 55.August P, Leventhal B, Suthanthiran M. Hypertension-induced organ damage in African Americans: transforming growth factor-beta(1) excess as a mechanism for increased prevalence. Current hypertension reports. 2000;2(2):184–91. Epub 2000/09/12. . [DOI] [PubMed] [Google Scholar]
- 56.Racial/ethnic disparities in prevalence, treatment, and control of hypertension—United States, 1999–2002. MMWR Morbidity and mortality weekly report. 2005;54(1):7–9. Epub 2005/01/14. . [PubMed] [Google Scholar]
- 57.Lackland DT, Keil JE. Epidemiology of hypertension in African Americans. Seminars in nephrology. 1996;16(2):63–70. Epub 1996/03/01. . [PubMed] [Google Scholar]
- 58.Racette L, Wilson MR, Zangwill LM, Weinreb RN, Sample PA. Primary open-angle glaucoma in blacks: a review. Survey of ophthalmology. 2003;48(3):295–313. Epub 2003/05/15. . [DOI] [PubMed] [Google Scholar]
- 59.Tielsch JM, Sommer A, Katz J, Royall RM, Quigley HA, Javitt J. Racial variations in the prevalence of primary open-angle glaucoma. The Baltimore Eye Survey. Jama. 1991;266(3):369–74. Epub 1991/07/17. . [PubMed] [Google Scholar]
- 60.Mayes MD. Scleroderma epidemiology. Rheumatic diseases clinics of North America. 2003;29(2):239–54. Epub 2003/07/05. . [DOI] [PubMed] [Google Scholar]
- 61.Masi AT, D'Angelo WA. Epidemiology of fatal systemic sclerosis (diffuse scleroderma). A 15-year survey in Baltimore. Annals of internal medicine. 1967;66(5):870–83. Epub 1967/05/01. . [DOI] [PubMed] [Google Scholar]
- 62.Arnett FC, Cho M, Chatterjee S, Aguilar MB, Reveille JD, Mayes MD. Familial occurrence frequencies and relative risks for systemic sclerosis (scleroderma) in three United States cohorts. Arthritis and rheumatism. 2001;44(6):1359–62. Epub 2001/06/16. doi: 10.1002/1529-0131(200106)44:6<1359::AID-ART228>3.0.CO;2-S . [DOI] [PubMed] [Google Scholar]
- 63.Flake GP, Andersen J, Dixon D. Etiology and pathogenesis of uterine leiomyomas: a review. Environmental health perspectives. 2003;111(8):1037–54. Epub 2003/06/27. ; PubMed Central PMCID: PMCPMC1241553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baird DD, Dunson DB, Hill MC, Cousins D, Schectman JM. High cumulative incidence of uterine leiomyoma in black and white women: ultrasound evidence. American journal of obstetrics and gynecology. 2003;188(1):100–7. Epub 2003/01/28. . [DOI] [PubMed] [Google Scholar]
- 65.Christopherson WM, Nealon NA. Uterine cancer: a comparative study of black and white women. Progress in clinical and biological research. 1981;53:185–95. Epub 1981/01/01. . [PubMed] [Google Scholar]
- 66.Marshall LM, Spiegelman D, Barbieri RL, Goldman MB, Manson JE, Colditz GA, et al. Variation in the incidence of uterine leiomyoma among premenopausal women by age and race. Obstetrics and gynecology. 1997;90(6):967–73. Epub 1997/12/16. . [DOI] [PubMed] [Google Scholar]
- 67.Zanchetti A. Impact of hypertension and antihypertensive treatment on organ damage. Am J Cardiol. 1999;84(2a):18k–24k. Epub 1999/08/07. . [DOI] [PubMed] [Google Scholar]
- 68.Muller DN, Kvakan H, Luft FC. Immune-related effects in hypertension and target-organ damage. Current opinion in nephrology and hypertension. 2011;20(2):113–7. Epub 2011/01/20. doi: 10.1097/MNH.0b013e3283436f88 . [DOI] [PubMed] [Google Scholar]
- 69.McMaster WG, Kirabo A, Madhur MS, Harrison DG. Inflammation, immunity, and hypertensive end-organ damage. Circ Res. 2015;116(6):1022–33. Epub 2015/03/15. doi: 10.1161/CIRCRESAHA.116.303697 ; PubMed Central PMCID: PMCPMC4535695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Irigoyen MC, De Angelis K, Dos Santos F, Dartora DR, Rodrigues B, Consolim-Colombo FM. Hypertension, Blood Pressure Variability, and Target Organ Lesion. Current hypertension reports. 2016;18(4):31 Epub 2016/03/24. doi: 10.1007/s11906-016-0642-9 . [DOI] [PubMed] [Google Scholar]
- 71.Ross R. Atherosclerosis is an inflammatory disease. Am Heart J. 1999;138(5 Pt 2):S419–20. Epub 1999/10/28. . [DOI] [PubMed] [Google Scholar]
- 72.Sakakura K, Nakano M, Otsuka F, Ladich E, Kolodgie FD, Virmani R. Pathophysiology of atherosclerosis plaque progression. Heart, lung & circulation. 2013;22(6):399–411. Epub 2013/04/02. doi: 10.1016/j.hlc.2013.03.001 . [DOI] [PubMed] [Google Scholar]
- 73.Lan TH, Huang XQ, Tan HM. Vascular fibrosis in atherosclerosis. Cardiovascular pathology: the official journal of the Society for Cardiovascular Pathology. 2013;22(5):401–7. Epub 2013/02/05. doi: 10.1016/j.carpath.2013.01.003 . [DOI] [PubMed] [Google Scholar]
- 74.Silver MA, Raghuvir R, Fedirko B, Elser D. Systemic hypertension among women with uterine leiomyomata: potential final common pathways of target end-organ remodeling. Journal of clinical hypertension (Greenwich, Conn). 2005;7(11):664–8. Epub 2005/11/10. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boynton-Jarrett R, Rich-Edwards J, Malspeis S, Missmer SA, Wright R. A prospective study of hypertension and risk of uterine leiomyomata. Am J Epidemiol. 2005;161(7):628–38. Epub 2005/03/23. doi: 10.1093/aje/kwi072 ; PubMed Central PMCID: PMCPMC4586055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Okolo S. Incidence, aetiology and epidemiology of uterine fibroids. Best practice & research Clinical obstetrics & gynaecology. 2008;22(4):571–88. Epub 2008/06/07. 10.1016/j.bpobgyn.2008.04.002. doi: 10.1016/j.bpobgyn.2008.04.002 [DOI] [PubMed] [Google Scholar]
- 77.Haan YC, Oudman I, de Lange ME, Timmermans A, Ankum WM, van Montfrans GA, et al. Hypertension risk in Dutch women with symptomatic uterine fibroids. Am J Hypertens. 2015;28(4):487–92. Epub 2014/09/23. doi: 10.1093/ajh/hpu183 . [DOI] [PubMed] [Google Scholar]
- 78.Huang C, Ogawa R. The link between hypertension and pathological scarring: does hypertension cause or promote keloid and hypertrophic scar pathogenesis? Wound repair and regeneration: official publication of the Wound Healing Society [and] the European Tissue Repair Society. 2014;22(4):462–6. Epub 2014/06/06. doi: 10.1111/wrr.12197 . [DOI] [PubMed] [Google Scholar]
- 79.Dustan HP. Growth factors and racial differences in severity of hypertension and renal diseases. Lancet. 1992;339(8805):1339–40. Epub 1992/05/30. . [DOI] [PubMed] [Google Scholar]
- 80.August P, Suthanthiran M. Transforming growth factor beta signaling, vascular remodeling, and hypertension. N Engl J Med. 2006;354(25):2721–3. Epub 2006/06/23. doi: 10.1056/NEJMcibr062143 . [DOI] [PubMed] [Google Scholar]
- 81.Wernig G, Chen SY, Cui L, Van Neste C, Tsai JM, Kambham N, et al. Unifying mechanism for different fibrotic diseases. Proc Natl Acad Sci U S A. 2017;114(18):4757–62. Epub 2017/04/21. doi: 10.1073/pnas.1621375114 ; PubMed Central PMCID: PMCPMC5422830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
AFR (cont.) represents continental AFR samples only (i.e. not including ACB and ASW populations).
(PDF)
Data Availability Statement
1000 Genomes data is publically available and may be downloaded via FTP (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/), Aspera, or Globus GridFTP (http://www.internationalgenome.org/data#download). Genotype data from BioVU used in this study is publically available, and can be found in dbGaP, accession number phs001409.v1.p1. Phenotype data from BioVU is available to researchers, who meet the criteria for access to confidential data, upon request and clearance from Vanderbilt University Medical Center Institutional Review Board and BioVU. Interested and eligible researchers may contact the BioVU data access team at biovu@vanderbilt.edu for more detailed information regarding access to phenotype data from BioVU.