Abstract
Clouds are key components in Earth’s functioning. In addition of acting as obstacles to light radiations and chemical reactors, they are possible atmospheric oases for airborne microorganisms, providing water, nutrients and paths to the ground. Microbial activity was previously detected in clouds, but the microbial community that is active in situ remains unknown. Here, microbial communities in cloud water collected at puy de Dôme Mountain’s meteorological station (1465 m altitude, France) were fixed upon sampling and examined by high-throughput sequencing from DNA and RNA extracts, so as to identify active species among community members. Communities consisted of ~103−104 bacteria and archaea mL-1 and ~102−103 eukaryote cells mL-1. They appeared extremely rich, with more than 28 000 distinct species detected in bacteria and 2 600 in eukaryotes. Proteobacteria and Bacteroidetes largely dominated in bacteria, while eukaryotes were essentially distributed among Fungi, Stramenopiles and Alveolata. Within these complex communities, the active members of cloud microbiota were identified as Alpha- (Sphingomonadales, Rhodospirillales and Rhizobiales), Beta- (Burkholderiales) and Gamma-Proteobacteria (Pseudomonadales). These groups of bacteria usually classified as epiphytic are probably the best candidates for interfering with abiotic chemical processes in clouds, and the most prone to successful aerial dispersion.
1. Introduction
The atmospheric envelope is a fundamental component of Earth’s functioning. Apart from holding huge energy exchanges, it transports, transforms and redistributes material at a large scale; it also participates to the spreading of microorganisms over the globe (e.g., [1–5]). Outdoor, the air is dotted with microorganisms (virus, bacteria, archaea, and eukaryotes) originating from surface habitats like vegetation, soil, water, or Humans/animals among natural sources [6–8], at concentrations varying from ~102 to ~106 cells m-3 (e.g., [9,10]. Some of them are regarded with attention for potential health hazards to Humans, animals and plants [11]. Surface ecosystems, also, are exposed to the continuous flow of diverse microbial incomers deposited from the atmosphere, bringing competitors, genetic material, and early colonizers in emerging habitats (e.g., [3,12]. Yet, environmental fitness tends to decrease with increasing distance from the source as habitats diverge [13], while, in addition, atmospheric transport exposes cells to harsh environmental conditions [14,15]. Hence, for microorganisms unable to produce resistance forms (spores), maintaining metabolic activity appears decisive for survival and possibility of successful establishment in the receptacle environment (e.g. [16].
Within the atmospheric system, clouds are genuine atmospheric interfaces with the ground: they physically connect high altitudes with the surface by being to a large extent at the origin of wet deposition of aerosols, including microorganisms [1,17,18]. Cloud water is a complex mixture of soluble gas and particles dissolved into millions of micron-sized water droplets, and forming very reactive and dynamic systems (e.g., [19]. As non-soluble biological particles, some microorganisms can physically impact clouds by acting as embryos for the formation of water droplets and ice crystals [20,21], with subsequent impacts on hydrological cycles [22–26]. Observations of microbiological features in fog and clouds raised the possibility that these also represent habitats for microorganisms [27–29], where they would actively take part in the chemical reactivity through metabolic activity and nutrient utilization [30–34]. So far these active « inhabitants » of clouds remain largely unknown. Microbiological studies in the atmosphere, including precipitation and deposition dust, essentially focused on the biodiversity, pathogens, emission sources and environmental drivers [3,6,7,10,35]. A predominance of Gram-negative bacteria (Alpha-, Beta- and Gamma-Proteobacteria, Bacteroidetes) is often observed, and attributed to inputs from soil and plants, with high temporal and spatial variability [3,6]. Current knowledge about the microorganisms living in clouds is essentially based on cultures approaches, so limited to a small fraction (< 1%) of the whole community. These indicated the presence bacteria and fungi, with prevailing groups, in Proteobacteria notably, and others appearing only once in a while [8,36–38]. Interrogations concerning the actual in situ functioning of microbial communities in clouds remain, starting with the identification of active members. Yet, these are probably better equipped than others (or better fitted) for surviving in the atmosphere and clouds [14,15,39], interfering with abiotic atmospheric processes, and they likely represent the potential successful colonizers of distant habitats. Here, using molecular methods, we investigate the structure of cloud water microbial communities and clarify our current vision of clouds as habitats by identifying active members. This consortium of active microorganisms finally revealed provides crucial information for further research on the interactions existing between microbial communities and abiotic processes in clouds, as well as important insights into the aerial dispersion of microorganisms.
2. Material and methods
2.1. Sample collection
Three cloud water samples were collected during the fall 2013 from the atmospheric station at the summit of puy de Dôme Mountain (1465 m a.s.l., 45.772° N, 2.9655° E, France). Specific permission was not required since the station is operated by OPGC (Observatory of the Globe of Clermont-ferrand), who collaborated this study. The field study carried out did not involve any endangered of protected species. Samples were collected at an air flow rate of 108 m3 h-1 using a cloud droplet impactor similar as in [24,30,40]. It has been slightly modified for allowing immediate fixation of the biological content (DNA and RNA) upon collection using a fixative agent: the water collected was transferred continuously, by gravity through autoclaved silicone tubing, to a sterile glass bottle containing 200 mL of a saturated ammonium sulphate solution used as surrogate for commercial fixative agent (i.e. RNA Later). This later was prepared under sterile conditions from fresh powders dissolved into sterile deionized water, then stored in sterile bottles. When samples froze upon impaction in the sampler, the ice collected was immediately melted into 200 mL of cold fixative solution. Before each sampling occasion, the presence of contaminants along the sampling apparatus and in the fixative solution was controlled by pooring 200 mL of sterile water into the sampler. The resulting 400 mL control sample was then processed and analyzed in parallel. Samples and controls mixed with the fixative solution were processed immediately after sampling using the microbiology facility of the puy de Dôme’s atmospheric observatory. These were filtered on 0.22 μm porosity filters (MoBio 14880-50-WF), within a vertical laminar flow hood previously exposed to UV light for 15 minutes, the filters cut in halves with a sterile scalpel, and each half was finally transferred into bead-beating tubes of the MoBio Power Water kits for DNA or RNA extraction, and stored at -80°C until being further processed, within a week. Samples for routine analyses (cell counts and chemical analyses, see below) were collected during the course of sampling by temporarily exchanging the collection bottle containing the fixative solution for an empty, sterile, glass bottle, until enough volume (~10–15 mL) was collected (~30 min).
2.2. Cell counts and chemical analyses
Cloud water samples collected in the absence of fixative solution were used for chemical and microbiological characterization. Ion analysis was realized within a month by ion chromatography on Dionex DX320 for anions (column AS11, eluant KOH) and Dionex ICS1500 for cations (column CS16, eluant hydroxymethanesulfonate) on samples kept at -25°C, similarly as in [41]. Cell counts were performed by flow cytometry (BD FacsCalibur, Becton Dickinson, Franklin Lakes, NJ) on 450 μL triplicates added with 50 μL 5% glutaraldehyde (0.5% final concentration; Sigma-Aldrich G7651) stored for < 1week at 4°C. For analysis, samples were mixed with 1 vol. of 0.02 μm filtered Tris-EDTA pH 8.0 (40 mM Tris-Base, 1 mM EDTA, acetic acid to pH 8.0) and stained with SYBRGreen I (Molecular Probes Inc., Eugene, OR) from a 100X solution. Counts were performed for 3 minutes or 100,000 events at a flow rate of ~80 μL min-1 (precisely further determined by weighting). Prokaryotes and eukaryotes were distinguished from background particles based on fluorescence and side scattering light intensities (λexc = 488nm; λem = 530nm).
2.3. Meteorological data and backward trajectory plots
Meteorological variables were monitored continuously by the atmospheric observatory of the puy de Dôme’s summit at 5 min intervals. Details on the instrumentation can be found at http://wwwobs.univ-bpclermont.fr/SO/mesures/instru.php. Twenty-four hours backward trajectory plots of the air masses sampled were computed for the puy de Dôme’s site (45.772 N, 2.9655 E; 1465 m above sea level) using the NOAA HYSPLIT trajectory model (HYbrid Single-Particle Lagrangian Integrated Trajectory; [42] using GDAS (1degree) meteorological data archive and default parameters for this site.
2.4. Nucleic acids extraction, amplification and sequencing
DNA and RNA were extracted separately from dedicated filter halves using MoBio PowerWater isolation kits for DNA and for RNA, respectively, following manufacturer’s recommendations and including a DNase treatment step on RNA extracts. The absence (RNA fractions) or presence (DNA fractions) of DNA in the extracts was verified by PCR targeting the 16S rRNA gene of bacteria using the universal primers 1492r and 27f and similar PCR conditions as in [40]. From RNA extracts, cDNA were obtained using Superscript VILO cDNA synthesis kit (Invitrogen). Ribosomal RNA and RNA genes were then amplified and barcoded by PCR from DNA extracts and cDNA products using primer couples targeting either the V4 region of the 16S subunit of prokaryotes (primers 515F and 806R [43], or the V7 region of the 18S subunit of eukaryotes (primers 960F-1200R [44]; S1 Table). The “Marine” cloud was not processed for RNA due to issues related to storage of the corresponding extract. PCR were performed in total volumes of 30 μL, containing 3 μL of 10X NH4 reaction buffer, and final concentrations of 2 mM MgCl2, 0.75 units of of Eurobio Taq II DNA polymerase (Eurobio, 5U/μL), 0.2 mM each dNTP, 0.5 mg mL-1 BSA, and 0.2 μM of each primer. The amplification conditions consisted of an initial denaturation at 94°C for 5 min followed by 30 cycles of 1 min. at 94°C, 45 s. at 58°C (16S) or at 55°C (18S) and 45 s. at 72°C, ended by a final elongation step of 7 min. at 72°C. Amplicons length was verified by agarose gel electrophoresis then purified using MinElute Gel Extraction kits (Qiagen) before quantification by fluorescence using Quant-it PicoGreen (Molecular Probes Inc., Eugene, OR). Finally, an equimolar pool of 14 PCR products was prepared (total amount of 510 ng of DNA (17 ng DNA μL-1 in 30 μL molecular biology grade H2O). Further sample processing and sequencing was realized by Genoscreen (Lille, France). Briefly, DNA libraries were generated by adaptator ligation (section “Perform End Repair and Size Selection”, Illumina reagent kit V3), and controlled on Agilent High Sensitivity microarray. Sequencing (2×300 bp paired-end on Illumina MiSeq platform) yielded a total of 43 763 524 reads (13 129 Mb), 75.7% of which had a quality score Q30.
2.5. Sequence processing
A total of ~11.7 million reads were obtained from MiSeq sequencing. Prokaryotes, including Bacteria and Archaea, contributed ~6.6 millions reads in DNA and ~1.5 million reads in RNA (abbreviated into 16SDNA and 16SRNA, respectively), and eukaryotes ~2.5 millions reads in DNA and ~1.1 million reads in RNA (18SDNA and 18SRNA, respectively). These were assembled with the vsearch tool (https://github.com/torognes/vsearch) and cleaning procedures consisted in the elimination of sequences < 200 bp, presenting a mismatch in the forward or reverse primer, having ambiguous bases “N”, PHRED quality score < 25. The putative chimaeras were detected by vsearch. The remaining rRNA 16S (prokaryotes) and 18S (eukaryotes) sequences were clustered into “molecular species” (Operational Taxonomy Units, OTUs) at a 97% and 95% similarity threshold (OTU0.03 and OTU0.05, respectively), according to [45] and [46] with vsearch (option cluster_small sorted by length). The representative sequence for each OTU was inserted into phylogenetic trees for taxonomic annotation. The seed OTUs were finally affiliated by similarity and phylogeny from reference sequences extracted from the SSURef SILVA database [47], according to the following criteria: length > 1 200 bp, quality score >75% and a pintail value > 50. After comparing the OTUs with the reference sequences using a similarity approach (vsearch tool), trees including OTUs with their closest references were built with FastTree [48]. The different taxonomic affiliations obtained were checked for inconsistency. This process was implemented using the pipeline PANAM (Phylogenetic Analysis of Next-generation AMplicons https://github.com/panammeb/) and is described in more detail in [49,50].
The resulting OTUs were subjected to additional conservative filtering intended to remove potential sequencing artefacts (OTUs represented by less than 3 reads), contaminants (OTUs detected in the control samples) and phantom OTUs (OTUs detected in RNA and not in the DNA fraction of the corresponding sample), totals of 761 729 and 140 645 reads and 48 202 and 37 504 reads remained in the DNA and RNA fractions for prokaryotes and eukaryotes, respectively. The corresponding sequence files were deposited to NCBI’s Sequence Read Archive (SRA BioProject ID PRJNA380262). Data were and normalized (proportions) rather than rarefied to prevent loss of information and possible resulting biases [51]. Results obtained on datasets rarefied at different depth are summarized in S8 Fig for allowing comparison with other studies.
2.6. Data analyses
Data analyses were performed using the R environment version 3.2.2 [52], implemented with the Phyloseq package (version 1.18.1; [53]) for calculating Shannon-Wiener indexes and Abundance-based Coverage Estimators (ACE); Phyloseq was also employed for rarefying the datasets to depths similar as data found in the literature for richness comparison. Gini’s coefficient was calculated using the ineq package (version 0.2.13; [54]). Rarefaction curves were plotted using the ggplot2 package (version 2.2.0; [55]) from community analyses made with the vegan package (version 2.4.1; [56]); Venn diagrams were made using Venny 2.1.0 [57].
3. Results
3.1. Samples characteristics
The basic biological, chemical and meteorological features of the cloud water samples investigated are shown in Table 1; these were usual for clouds collected at the Puy de Dôme Mountain’s atmospheric observatory [40,58]. Based on geographical origin (S1 Fig), pH, and major ions composition, when available, these were classified into “Polluted”, “Continental” or “Marine” type events (S2 Fig)[58]. These categories comprise 9%, 26% and 52% of the clouds observed at puy de Dôme, respectively [58]. Total cell concentration was within the range typically observed in cloud water at this sampling site, with (2.05 to 9.49) ×103 Bacteria and Archaea mL-1 and (0.4 to 8.7) ×102 eukaryotic cells mL-1, equivalent to (0.4 to 2.5) ×103 and 8 to cells 270 cells m-3 of cloudy air, respectively, depending on the sample (Table 1). Bacteria largely dominated the community, both in abundance (cell counts and read number) and richness (OTUs number): they represented ~90% of the DNA reads while eukaryotes contributed ~8%.and Archaea ~2%.
Table 1. Main characteristics of the samples.
| Biological data | Meteorological data | Chemical data | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample ID | Local sampling starting date and time | Local sampling ending date and time | Cloud type* | Sample volume filtered and used for sequencing (mL) [equivalent air volume (m3)] | Total prokaryotic cells concentration (mL-1) | Total eukaryotic cells concentration (mL-1) | T (°C) | Wind speed (m s-1) | Wind direction (°/N) | LWCb (g m-3) | Dissolved ions concentrations (μM) | ||||||||||||
| Mean | SEa | Mean | SEa | Mean | SEc | Mean | SEc | Mean | SEc | Mean | SEc | pH | Na+ | SEa | Cl- | SEa | NO3- | SEa | |||||
| CLOUD 1 | 10/11/13 10:36 AM | 10/11/13 1:25 PM | Polluted | 94 [304 m3 of cloudy air] | 8.23×103 | ± 1.25×103 | 8.67×102 | ± 3.15×102 | -1.2 | ± 0.2 | ND* | - | ND* | - | 0.16 | ± 0.06 | 4.2 | ND | - | ND | - | ND | - |
| CLOUD 2 | 10/14/13 10:00 PM | 10/15/13 11:10 AM | Continental | 350 [1422 m3 of cloudy air] | 9.49×103 | ± 2.87×103 | 2.83×102 | ± 1.05×102 | 6.5 | ± 0.3 | 12.1 | ± 1.5 | 218 | ± 9 | 0.31 | ± 0.05 | 4.7 | 79.0 | ± 6.5 | 15.5 | ± 2.8 | 38.5 | ± 0.8 |
| CLOUD 3 | 11/5/13 2:45 PM | 11/6/13 1:30 PM | Marine | 420 [2457 m3 of cloudy air] | 2.05×103 | ± 1.39×103 | 4.42×101 | ± 0.21×102 | 6.8 | ± 0.8 | 18.3 | ± 3.4 | 337 | ± 4 | 0.40 | ± 0.07 | 5.6 | 188.5 | ± 12.6 | 0.2 | ± 0.1 | 1.13 | ± 0.7 |
ND: Not determined.
*According to the classification of clouds sampled at the puy de Dôme as established by [58] (see S2 Fig), based on pH, ion content and backtrajectory plots (shown in S1 Fig).
aStandard error of 3 replicate measurements.
bLiquid water content.
cStandard error of averages of meteorological variables collected every 5 minutes during sampling.
3.2. The cloud water microbiota: An extremely rich and imbalanced community
A total of 28,143 OTUs were detected in prokaryotes (28,058 in Bacteria and 85 in Archaea) and 2,612 OTUs in eukaryotes. Each sample contained a fraction of the total richness, with ca. 7,800 to 20,500 OTUs0.03 in prokaryotes and ca. 1,900 to 2,100 OTUs0.03 in eukaryotes (Table 2). Inflexions in the rarefaction plots of the different sets of sequences (S3 Fig) indicated that the actual microbial communities targeted were well captured (coverage of 72% to 98%). The corresponding abundance-based coverage estimators (ACE) specified the presence of between ~10,800 and ~21,000 prokaryotic OTUs0.03 per sample and ~2,400 eukaryotic OTUs0.05 (Table 2). The results concerning specifically the composition of prokaryote, eukaryote, then active communities are presented below.
Table 2. Prokaryotic and eukaryotic communities’ richness and distribution.
| Polluted type cloud | Continental type cloud | Marine type cloud | |||
|---|---|---|---|---|---|
| DNA | RNA | DNA | RNA | DNA | |
| Bacteria and Archaea | |||||
| Processed read number | 496,197 | 59,449 | 115,515 | 81,196 | 150,017 |
| Total species richness* | |||||
| Observed | 20,432 | 3,428 | 7,793 | 5,622 | 8,970 |
| Estimated (ACE) | 20,954 | 4,710 | 10,802 | 6,468 | 11,148 |
| Community distribution | |||||
| Shannon's H | 9.1 | 6.3 | 7.2 | 7.3 | 7.4 |
| Gini coefficient | 0.74 | 0.98 | 0.95 | 0.95 | 0.94 |
| Eukaryotes | |||||
| Processed read number | 18,562 | 12,831 | 13,978 | 24,673 | 15,662 |
| Total species richness** | |||||
| Observed | 2,061 | 1,527 | 1,901 | 1,692 | 1,877 |
| Estimated (ACE) | 2,461 | 1,845 | 2,400 | 1,834 | 2,439 |
| Community distribution | |||||
| Shannon's H | 6.58 | 6.28 | 6.48 | 6.32 | 6.26 |
| Gini coefficient | 0.75 | 0.81 | 0.77 | 0.81 | 0.79 |
*OTUs clustered at 97% sequence similarity.
**OTUs clustered at 95% sequence similarity.
3.2.1 Prokaryote community
In total, 30 different phyla were detected (of which 1 in Archaea), distributed over 60 classes, 113 orders, 190 families and 286 genera. A complete list of the abundance and taxonomic affiliation of prokaryotic OTUs is presented in S1 File. In all samples, the community was largely dominated by 4 bacterial phyla, which represented 75% to 79% of the reads in DNA datasets and 85 to 88% in RNA (Fig 1A): Proteobacteria (28 to 51% of the DNA reads, in particular the Gamma- (6–21%), Alpha- (2–21%) and Beta- (3–12%) classes; Fig 1B), Bacteroidetes (5–30%), Firmicutes (5–18%) and Actinobacteria (6–13%). These phyla are almost systematically reported dominant in outdoor airborne bacterial communities studies (e.g. [3,6,8,59–62]).
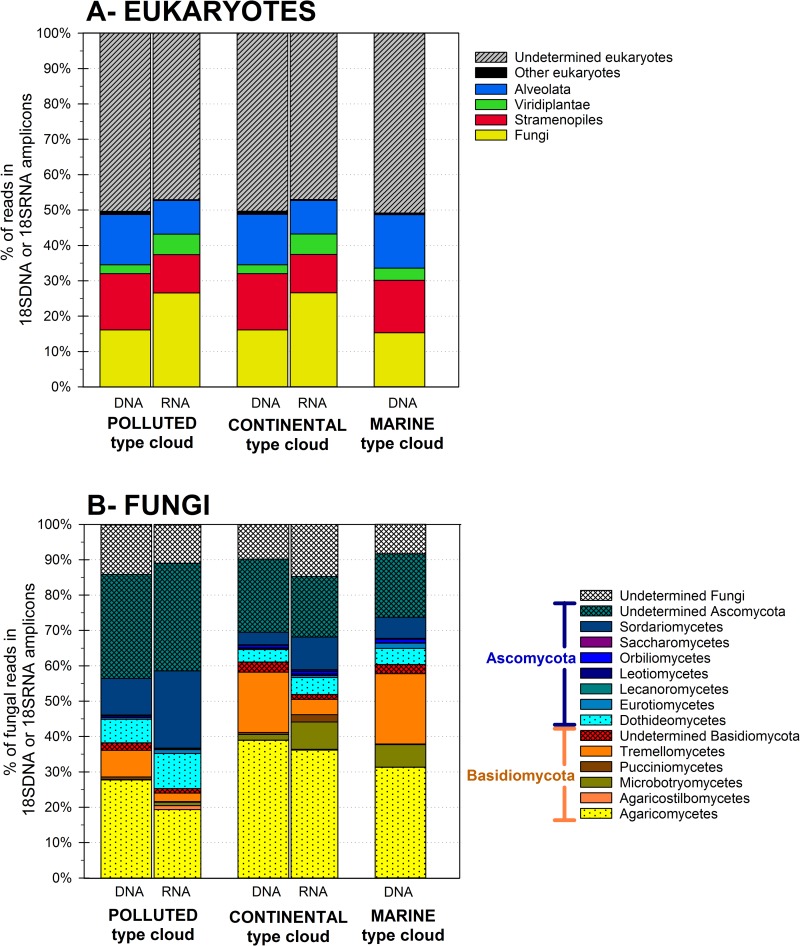
Fig 1.
Prokaryotic total (DNA fraction) and active (RNA fraction) community composition in the cloud water samples at the phylum level (A),and relative distributions of Proteobacteria orders (B).
A total of 1,593 OTUs distributed over 103 genera were common to all samples (S4A Fig). These represented 64% to 96% of the reads identified down to this taxonomic level, and 15%-31% of the total 16SDNA reads in each sample. Their relative contribution to the whole community structure in the different samples is shown as a heat-map in S5A Fig. Dominant genera comprised notably: Rickettsia, Sphingomonas, Methylobacterium and Acidiphilium in Alpha-Proteobacteria; Pseudomonas, Enhydrobacter, Moraxella and Psychrobacter in Gamma-Proteobacteria; Capnocytophaga in Bacteroidetes; Corynebacterium, Arthrobacter and Streptomyces in Actinobacteria; Dolosigranulum, Clostridium in Firmicutes.
The samples had clear distinct patterns: ~70% of the total OTU richness observed in prokaryotes was contributed by the “Polluted” type sample, with most of them being characteristic, i.e. exclusive of this sample (15,152 OTUs representing 72% of the reads of this sample). Comparatively, other samples had 2,600 to 3,000 characteristic OTUs which represented 8–12% of the reads. The “Polluted” type cloud was characterized by relatively high abundance of Dolosigranulum, Corynebacterium, Moraxella and Campylobacter bacteria. The “Marine” type cloud was dominated by Proteobacteria affiliated with Bdellovibrio, Pseudomonas, Methylobacterium, Sphingomonas and Rickettsia; these were also well represented in the “Continental” type cloud, along with some Firmicutes and Actinobacteria (Clostridium, Streptococcus and Corynebacterium).
3.2.2 Eukaryote community
Eukaryotic OTUs were distributed over 12 phyla, with 66 orders identified. A complete list of the abundance and taxonomic affiliation of eukaryotic OTUs can be found in S2 File. A large proportion of reads (~50%) remained unaffiliated at the phylum level, both in DNA and RNA. The reads taxonomically identified in the DNA fraction were evenly distributed among Fungi, Stramenopiles and Alveolata (12% to 18%), while Viridiplantae represented ~3% (Fig 2A). Basidiomycota and Ascomycota largely dominated in Fungi (Fig 2B). By far, most identified Basidiomycota were members of the classes Agaromycetes (52% to 73%, with Polyporales and Agaricales the dominant families), Tremellomycetes (20% to 33%) and Microbotryomycetes (0.2% to 11%). In the phylum Ascomycota, among those identified to the class level, Sordariomycetes (12% to 22%) and Dothideomycetes (12% to 15%) dominated; other classes (Eurotiomycetes, Lecanoromycetes, Leotiomycetes, Orbiliomycetes and Saccharomycetes) represented < 5% of the reads.
Fig 2.
Eukaryotic total (DNA fraction) and active (RNA fraction) community composition in the cloud water samples at the kingdom level (A), and relative distributions of Fungal classes (B).
A total of 1,209 eukaryotic OTUs were shared between the samples (S4B Fig). These were distributed over 39 orders, gathering 15.2% to 16.6% of the total reads of the samples and 91% to 97% of those identified at this taxonomic depth. Their relative abundance in the eukaryotic communities of each sample is shown as a heat-map in S5B Fig. Dominant shared orders in all samples included notably Bicoseocida, Polyporales, Charales and Euplotida.
3.3. Active community
The active community, as detected in RNA extracts, was a fraction of the total community. This included 26.4% of the total richness observed in prokaryotes (7,438 OTUs0.03) and 82% (2,612 OTUs0.05) in eukaryotes. The samples were globally less distinct in their RNA fraction than they were in their DNA fraction (S6 Fig). A total of 1,612 prokaryotic OTUs were shared between the RNA fractions of 2 samples analyzed. These were distributed over 97 identified genera gathering in total 32% to 34% of the 16SRNA reads, of which a few dominant genera contributed each around 1%: Rickettsia, Spirosoma, Enhydrobacter, Corynebacterium, Acidiphilium, Sphingomonas, Pseudomonas and Methylobacterium. In eukaryotes, most RNA reads (18% to 27%) were attributed to Fungi, whereas Stramenopiles and Alveolata each were represented by ~10%, and Viridiplantae by ~6%. A the order level, dominant Fungi included Magnaporthales and Pleosporales in Ascomycota, Polyporales and Sporidiobolales in Basidiomycota), SAR (Bicosoecida) and others such as Syndiniales, a group of dinoflagellates.
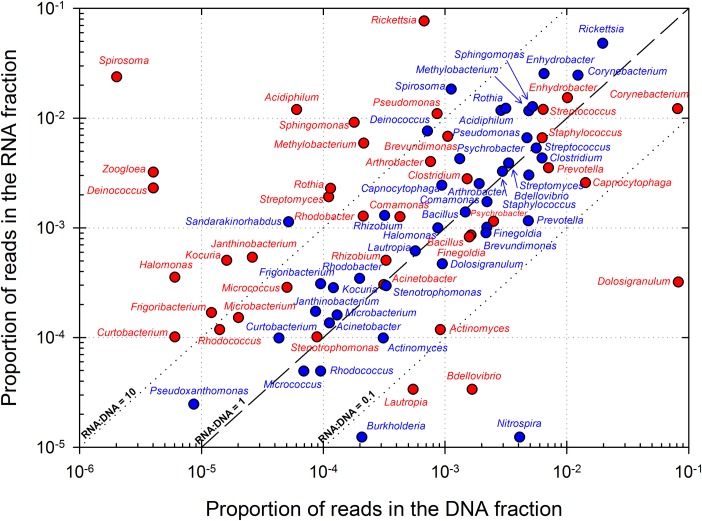
Figs 3 and 4 compile overall most represented bacterial genera and eukaryotic orders, respectively, in corresponding DNA and RNA datasets. The relative abundance of RNA, respect to DNA, in an OTU (abbreviated into RNA:DNA ratio for clarity) is often used for estimating its relative level of metabolic activity, with higher ratios linked with potentially higher metabolic activity [63,64]. RNA:DNA ratio ranged between 0 and 210 in eukaryotes, and from 0 to, exceptionally, 11,760 in prokaryotes in an OTU affiliated to Spirosoma (Bacteroidetes). Low abundance groups tended to exhibit high ratios, in prokaryotes and in a lesser extent also in eukaryotes (see S7 Fig), as observed by others in atmospheric samples [65], but by far, most RNA:DNA ratios were between 0.1 and 10. Alpha- and Gamma-Proteobacteria clearly dominated in bacterial taxas with ratio > 1 (i.e. potentially metabolically active taxa). Notably Rickettsia, Sphingomonas, Methylobacterium, Enhydrobacter, Pseudomonas, and Acidiphilium genera were highly represented and were probably the most active taxas. In bacteria, these included notably Spirosoma, Deinococcus (Deinococcus-Thermus), Janthinobacterium (Beta-Proteobacteria), Frigoribacterium and Curtobacterium (Actinobacteria). Conversely, some bacteria were found abundant but exhibited very low or no activity based on RNA:DNA ratio. These comprised essentially Gram-positive bacteria: Actinobacteria (Corynebacterium, Actinomyces) and Firmicutes (Dolosigranulum, Staphylococcus), and also members of Proteobacteria (Bdellovibrio, Burkholderia), Bacteroidetes (Capnocytophaga) and others like Nitrospira. In eukaryotes the orders Magnaporthales, Syndiniales, Pleosporales, Polyporales, Bicosoecida and Sporidiobolales in particular were markedly abundant in both the DNA and RNA datasets.
Fig 3. Representation of the major prokaryotic genera identified in DNA and RNA datasets.
Dashed and dotted lines depict RNA:DNA ratios of 0.1, 1 and 10. The top 20 genera based on their average position rank over the 3 cloud samples are shown, as well as some selected for high representation in RNA datasets (43 distinct genera in total). POLL: “Polluted” type cloud; CONT: “Continental” type cloud.
Fig 4. Representation of the major eukaryotic orders identified in DNA and RNA datasets.
Dashed and dotted lines depict RNA:DNA ratios of 0.1, 1 and 10. The top 20 genera based on their average position rank over the 3 cloud samples are shown, as well as some selected for high representation in RNA datasets (24 distinct orders in total). POLL: “Polluted” type cloud; CONT: “Continental” type cloud.
4. Discussion
4.1. Clouds are extremely rich and diverse mosaics of multiple sources ecosystems
In this work, we aimed at drawing a picture of the structure of cloudborne microbial communities, including active groups and rare taxa. The detection of eventual environmental drivers such as meteorological variables to the microbial communities observed was beyond the scope of this study. Thus, we chose to orient our investigations toward large sample volumes, associated with deep sequencing. Consistently, species richness reaches here an unprecedented value in atmospheric samples, with ~11,000 to ~21,000 distinct OTUs estimated in prokaryotes and ~2,400 in eukaryotes. Such high richness are uncommon and in general rather reported in soils (e.g., [66,67]. In the atmosphere, although much less is known, it is often described as a highly diverse environment (e.g., [7,8,60,68–70]. The high richness observed in our samples can be related to the large sample volumes considered. It is clear that scale problems arise when estimating community richness in open ecological systems [71,72], especially in dynamic environments like the atmosphere where the biomass is low. Comparatively, DNA analyses were carried out in reference studies from 2.7 to 144 m3 of air at mid-altitude sites (ca. 1,500 to 3,000 m asl.; [35,65], and ~6 m3 of air in the free troposphere (i.e. ~3×104 bacterial cells per sample; [10]). Volumes similar as in our study were notably used for assessing fungal [73] and prokaryotic diversity [4] in aerosols at global scale: up to 1,100 species of fungi and 2,900 species of prokaryotes per sample were observed. Recently [62] detected up to 1,910 species in cloud water volumes of 100 mL, on data rarefied to 9,100 sequences. A richness equivalent was observed in our study when rarefying data to a comparable depth (S8 Fig). Finally, bacterial species richness similar as our findings was reported from large rain samples (7–22 liters), with 13,083 OTUs0.03 [74], in [61].
The structure of the communities was investigated through ecological indexes (Table 2). Shannon’s H indexes ranging from 6.3 to 9.1 demonstrated extreme biodiversity, in a large part contributed to by the numerous rare species. Indeed, the communities, in particular prokaryotes, were highly uneven with a low proportion of abundant species and many rare, as shown by OTU rank-abundance plots (S3C and S3D Fig), Lorenz curves (S9 Fig) and corresponding Gini’s coefficients close to 1 (0 being a perfect equality in OTU abundance distribution and 1 being perfect inequality, i.e. a very contrasted abundance distribution between OTUs). Illustrating this, despite conservative sequence filtering, the 1% most abundant OTUs represented around ~20% of the reads in eukaryotes and ~35% in prokaryotes, respectively, and the top half OTUs more than 90%. This high unevenness suggests that the global functioning of the community is fragile (not robust), sensitive to stress [75], and so, likely to be variable in space and time. If an abundant group was to be lost from the community, i.e. a group that is likely to contribute significantly to the structure and global functioning of the system, there would be a high probability to lose or reduce also the functions associated with it. This ecological theory, that functional stability implies even structure, derives from established ecosystems and it is applied here for apprehending the functioning of cloud’s microbial communities in the frame of clouds as microbial habitats hypothesis; it is possible though that this is not applicable to environments acting mainly as transport areas, where microbial establishment is by essence not possible, like clouds.
Any microbe inhabiting a surface environment has a probability to get aerosolized, though more or less promptly depending on its physiological characteristics (e.g.,[76] and other environmental variables linked with its habitat, like exposure to mechanical disturbances by wind or rainfall for instance [77–79]. The community observed likely resulted from the mixing of microbial inputs from myriads of different sources, albeit not confidently quantifiable here. The high unevenness observed could suggest by itself that there is a marked influence of some specific environments over others; this assumes temporal stability on sources and equivalent strengths among sources and microorganisms, and this is probably not the case (e.g., [67,80,81]. Rather, the variability between the samples indicated that the sources themselves are large and rich, so a variety of possible children communities can emerge from it [82].
As needles in a haystack of complex communities, the presence of more or less specific tracers can inform about emission sources. It is widely observed, and our samples are no exception, that airborne microorganisms outdoors most likely originate from soil, vegetation, surface waters and animals among natural sources; Humans and activities such as composting can also create bioaerosols in high number (e.g., [35,76,79,83,84]). Proteobacteria and Bacteroidetes dominated the communities observed, with also a good representation of Actinobacteria and Firmicutes, as usually in airborne microbial communities [6,8,10,59,60,85]. Prokaryotic endosymbionts or parasites of eukaryotes (Rickettsia, Wolbachia)[86,87] were present in high proportion. To our knowledge, this is the first report of such abundance of these organisms in atmospheric samples. Their presence is not surprising as they probably originated from the numerous plant, insect, arthropod and other animal fragments contained among aerosols [88]. Rickettsia are ultra-small bacteria thought to be at the origin of mitochondria in eukaryotes (endosymbiotic theory) [89]. The abundance of Pseudomonads and Sphingomonads attested of important inputs from vegetation in all samples (e.g., [90,91]), whereas others like Streptomyces rather refer to soils. These apart, Bdellovibrio, a genus of Delta-Proteobacteria found in water environments, dominated in the “Marine” type cloud was, and taxa generally affiliated with soils, decomposing organic material, animals, and humans, like Dolosigranulum, Corynebacterium, Moraxella, Campylobacter and Capnocytophaga (e.g., [92]) were abundant in the “Polluted” type cloud. Wei et al. studied polluted and non-polluted fog events in China and also observed a prevalence of potential Human pathogens in the polluted air masses [61].
In eukaryotes, Basidiomycota tended to dominate over Ascomycota, as a result from continental inputs [11,73,93]. The relative dominance of Basidiomycota over Ascomycota in the air was revealed recently by culture-independent methods [11,93]. More precisely, Basidiomycota tend to dominate in continental air masses, whereas Ascomycota prevail in marine air masses [73].
Despite sampling site’s remoteness from ocean, samples kept a marine biological signature detectable on the taxonomic affiliation of some abundant groups of microorganisms, as also observed for chemical composition: marine or water-related taxa, in prokaryotes (Bdellovibrio, Delta-Proteobacteria) and in eukaryotes [green, brown and red algae (Charales, Ectocarpales, Hildenbrandiales, Fucales, Cladophorales), diatoms (Fragilariales, Thalassiosirales, Hemiaulales), fungi and protozoans affiliated with water environments (Syndiniales, Saprolegniales, Conioscyphales)]. Saprophytic fungi affiliated with vegetation, soils and decomposing litters were also particularly abundant: Polyporales, Sporidiobolales, Agaricales, Pleosporales, and others.
Although it is not statistically verifiable, we observed that the prokaryotic community, and in a lesser extent the eukaryotic community, were richer (ACE estimator), more diverse (Shannon’s index), and less uneven (Gini’s coefficient) in the “Polluted” type cloud than in non-polluted “Continental” or “Marine” type clouds (Table 2). A relationship between Human activities and microbial communities structure in clouds was reported in China [61], with higher diversity in non-polluted clouds. Another study rather pointed out an impact of day and night on the composition of bacterial communities in clouds; a higher representation of Alpha-Proteobacteria during the night, notably, was reported [62], but the reasons for such trend are not clear.
4.2. Clouds are environments open to all, but where only some can thrive: active groups
Among the high diversity of cloud microbial communities, some were capable of maintaining metabolic activity in cloud despite probable stressful conditions. According to criteria of abundance in both DNA and RNA fractions, RNA:DNA ratio (Fig 3), frequency of recovery in cultures in earlier studies [40], and other hints from previous reports [10,65], and at the exception of eukaryotic endosymbionts (Rickettsia), these probable main bacterial “inhabitants” of clouds can be named: Alpha- and Gamma-Proteobacteria, in particular Sphingomonas (order Sphingomonadales), Methylobacterium (Rhizobiales), Acidiphilium (Rhodospirillales), Pseudomonas (Pseudomonadales), Comamonas (Burkholderiales) and, to a lesser extent, Enhydrobacter and Psychrobacter (Pseudomonadales). Among more discrete genera, Curtobacterium, Deinococcus, Spirosoma, Rhizobium and Janthinobacterium notably can also be cited here, along with, in other phyla, Arthrobacter, Staphylococcus. All these have physiological properties compatible with their maintenance in the high atmosphere and clouds, and they probably interact with their cloud water environment with potential impacts on chemistry. Many of these are epiphytic taxa commonly recovered viable from air and clouds [8,9,17,29,84,94–96]. On the other hand, tracers of polluted air masses could reside amongst the most abundant species in DNA, like Dolosigranulum or Capnocytophaga.
Many of the microorganisms identified relate to vegetation: epiphytic, parasitic and endosymbiontes. Plant leaves, like clouds, are subjected to frequent temperature and humidity shifts, high levels of UV light, etc. It is possible that these bacteria acquired physiological traits compatible with survival in clouds from this lifestyle. Pseudomonas and Sphingomonas species are versatile bacteria abundant in the environment, particularly on vegetation (e.g., [80]). Pseudomonas are among the bacteria the most frequently recovered by culture (i.e. viable) from clouds and atmospheric samples [17,27,40], where their presence is particularly interesting for many reasons: plant pathogenicity and epidemiology, degradation of organic compounds in clouds [97,98]; production of siderophores and interactions with iron and radical chemistry [99]; production and release of surfactants, which could facilitate the formation of cloud water droplets [21,68]; ice nucleation, which in clouds can trigger precipitation (hypothesized as “bioprecipitation”) (e.g.[96]). Sphingomonas are pigmented oligotrophic bacteria, frequently described as psychrotolerant bacteria recovered from polar environments and air samples [36,100,101]. Many of them are studied for their intrinsic resistance to numerous antibiotics [102], and for their capacity to degrade xenobiotics [103], alike Comamonas [104]. Methylobacterium are methylotrophic bacteria, i.e. they can develop on one-carbon compounds such as methanol, formaldehyde or formate [105,106], which are abundant in the atmosphere and in cloud water [41,98]. Some species can use compounds shown to be responsible for ozone depletion in the stratosphere, such as chloromethane [107]. The presence and potential activity of methanotrophic bacteria in the air was shown previously [32]. Acidiphilium have high capacities of interaction with iron [108,109] and so is a good candidate for interfering with cloud water oxidant capacity [33]; it remains yet rarely isolated by culture. Enhydrobacter have gas-vacuole helping floatation in aquatic environments [110], and it is possible that this favored its aerosolization from waters bodies (e.g., 104). Finally, Deinococcus and Spirosoma are known for their high resistance to DNA-damages such as those caused by UV light [111,112], so their presence among the common core of the community is no surprise. Spirosoma species have been described from Arctic and Mountain regions [112,113].
In eukaryotes, endosymbiontes and parasites flagellate protists (Syndiniales and Bicosoecida) dominated in DNA, but active groups included mainly plant pathogens and saprophytic fungi from terrestrial or aquatic origins known for aerial dispersion [114–116] in Ascomycota (Pleosporales, Magnaporthales, Xylariales and Conioscyphales), and Basidiomycota (Pucciniales, Hymenochaeales and Sporodiobolales) (Fig 4). Ascomycota were previously reported dominant in the active fraction of airborne fungi [93], and in Basidiomycota, Sporodiobolales includes yeasts frequently isolated from cloud water samples at the same site, Rhodotorula and Sporobolomyces [40], However, if RNA:DNA ratio gives hints about potentially active eukaryotes, ribosome gene number is intrinsically more variable than it is in prokaryotes [117]; so estimating their actual relative activity in cloud water will necessitate more investigations.
Our investigations revealed an incredible richness in the atmosphere, originating from a variety of different sources and meeting in clouds. High inequities suggested high sensibility to perturbations, including potentially stress caused by Human activities. Frequent species probably composed most of the biomass, but the vast majority of the diversity was contributed by rare species. This feature, common in the environment (e.g., [118], funded the “everything is everywhere” concept (e.g., [119]. There is no “global atmosphere” with a specific community structure and functioning, but rather a multitude of different regional to local atmospheres distributed over the globe, as moving airborne imprints of surface ecosystems. On top of this, some atmospheric corridors connecting distant regions together and defining some extent of bio-geographical distribution of microorganisms on the planet have been identified [5,73,118].
Airborne communities are sorts of blurred airborne imprints of surface ecosystems gathering and overlapping with each other in clouds A set of microorganisms able to maintain metabolic activity in clouds was identified among complex communities. In previous studies, many of these active taxa were frequently recovered by culture from cloud water samples [29,40]. These represent the microorganisms the most prone to interfere with their cloud chemical environment. They are also potential competitors brought to surface receptacle ecosystems by atmospheric deposition, and the early colonizers of emerging environments. Their identification certainly helps understanding the atmosphere as a habitat; it will also allow focusing researches for evaluating microbial impact on cloud physical and chemical processes, but their actual functioning, the “what do they do?” question remains to be answered.
Supporting information
(XLSX)
(XLSX)
(DOCX)
(TIF)
(TIF)
Rarefaction curves (A and B) and rank-abundance plots (C and D) of the different set of amplicons.
(TIF)
Venn diagrams depicting similarities and singularities of the 3 samples at the OTU0.03 level for prokaryotes (A) and OTU0.05 for eukaryotes (B).
(TIF)
Relative abundance of shared prokaryotic genera (A) and eukaryotic orders (B).
(TIF)
Bray-Curtis similarity matrices between the different sets of sequence of prokaryotes (A) and eukaryotes (B).
(TIF)
(TIF)
(TIF)
(TIF)
Acknowledgments
We warmly thank the numerous people who helped us concretizing this study: J. Colombet, L. Nauton, J.M. Pichon, I. Mary, F. Enault, J.C. Charvy, A. Mahul and the Mesocentre Calcul Center, and G. Lefebvre for language corrections in the manuscript.
Data Availability
All sequence files are available from the NCBI Sequence Read Archive SRA database (BioProject ID PRJNA380262).
Funding Statement
Funding provided by CNRS MicrobiEN program (http://www.insu.cnrs.fr/ec2co), "FONCOMIC" project, attributed to PA. MJ and LB were hired on the projects ANR-15-CE01-0002 ("INHALE") and ANR-DFG-14-CE35-005-02 ("CHLOROFILTER"), respectively.
References
- 1.Burrows SM, Butler T, Jöckel P, Tost H, Kerkweg A, Pöschl U, et al. Bacteria in the global atmosphere–Part 2: Modeling of emissions and transport between different ecosystems. Atmos Chem Phys. 2009;9: 9281–9297. doi: 10.5194/acp-9-9281-2009 [Google Scholar]
- 2.Griffin DW, Garrison VH, Herman JR, Shinn EA. African desert dust in the Caribbean atmosphere: Microbiology and public health. Aerobiologia. 2001;17: 203–213. doi: 10.1023/A:1011868218901 [Google Scholar]
- 3.Barberán A, Henley J, Fierer N, Casamayor EO. Structure, inter-annual recurrence, and global-scale connectivity of airborne microbial communities. Sci Total Environ. 2014;487: 187–195. doi: 10.1016/j.scitotenv.2014.04.030 [DOI] [PubMed] [Google Scholar]
- 4.Smith DJ, Timonen HJ, Jaffe DA, Griffin DW, Birmele MN, Perry KD, et al. Intercontinental dispersal of bacteria and archaea by transpacific winds. Appl Environ Microbiol. 2013;79: 1134–1139. doi: 10.1128/AEM.03029-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Womack AM, Bohannan BJM, Green JL. Biodiversity and biogeography of the atmosphere. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2010;365: 3645–3653. doi: 10.1098/rstb.2010.0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowers RM, Clements N, Emerson JB, Wiedinmyer C, Hannigan MP, Fierer N. Seasonal Variability in Bacterial and Fungal Diversity of the Near-Surface Atmosphere. Environ Sci Technol. 2013;47: 12097–12106. doi: 10.1021/es402970s [DOI] [PubMed] [Google Scholar]
- 7.Brodie EL, DeSantis TZ, Parker JPM, Zubietta IX, Piceno YM, Andersen GL. Urban aerosols harbor diverse and dynamic bacterial populations. PNAS. 2007;104: 299–304. doi: 10.1073/pnas.0608255104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Šantl-Temkiv T, Finster K, Hansen BM, Nielsen NW, Karlson UG. The microbial diversity of a storm cloud as assessed by hailstones. FEMS microbiology ecology. 2012;81: 684–695. doi: 10.1111/j.1574-6941.2012.01402.x [DOI] [PubMed] [Google Scholar]
- 9.Bowers RM, Lauber CL, Wiedinmyer C, Hamady M, Hallar AG, Fall R, et al. Characterization of Airborne Microbial Communities at a High-Elevation Site and Their Potential To Act as Atmospheric Ice Nuclei. Appl Environ Microbiol. 2009;75: 5121–5130. doi: 10.1128/AEM.00447-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLeon-Rodriguez N, Lathem TL, Rodriguez-R LM, Barazesh JM, Anderson BE, Beyersdorf AJ, et al. Microbiome of the upper troposphere: Species composition and prevalence, effects of tropical storms, and atmospheric implications. PNAS. 2013;110: 2575–2580. doi: 10.1073/pnas.1212089110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fröhlich-Nowoisky J, Pickersgill DA, Després VR, Pöschl U. High diversity of fungi in air particulate matter. PNAS. 2009;106: 12814–12819. doi: 10.1073/pnas.0811003106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris CE, Sands DC. Impacts of microbial aerosols on natural and agro-ecosystems: immigration, invasions and their consequences. Microbiology of aerosols. John Wiley and Sons Inc. Hoboken, New Jersey, USA: Delort, A.M. and Amato, P.; 2017. [Google Scholar]
- 13.Nekola JC, White PS. The distance decay of similarity in biogeography and ecology. Journal of Biogeography. 1999;26: 867–878. doi: 10.1046/j.1365-2699.1999.00305.x [Google Scholar]
- 14.Joly M, Amato P, Sancelme M, Vinatier V, Abrantes M, Deguillaume L, et al. Survival of microbial isolates from clouds toward simulated atmospheric stress factors. Atmospheric Environment. 2015;117: 92–98. doi: 10.1016/j.atmosenv.2015.07.009 [Google Scholar]
- 15.Smith DJ, Griffin DW, McPeters RD, Ward PD, Schuerger AC. Microbial survival in the stratosphere and implications for global dispersal. Aerobiologia. 2011;27: 319–332. doi: 10.1007/s10453-011-9203-5 [Google Scholar]
- 16.Price PB, Sowers T. Temperature dependence of metabolic rates for microbial growth, maintenance, and survival. Proc Natl Acad Sci U S A. 2004;101: 4631–4636. doi: 10.1073/pnas.0400522101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monteil CL, Bardin M, Morris CE. Features of air masses associated with the deposition of Pseudomonas syringae and Botrytis cinerea by rain and snowfall. ISME J. 2014;8: 2290–2304. doi: 10.1038/ismej.2014.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao TL, Gong SL, Zhang XY, McKendry IG. Modeled size-segregated wet and dry deposition budgets of soil dust aerosol during ACE-Asia 2001: Implications for trans-Pacific transport. J Geophys Res. 2003;108: 8665 doi: 10.1029/2002JD003363 [Google Scholar]
- 19.Herrmann H, Schaefer T, Tilgner A, Styler SA, Weller C, Teich M, et al. Tropospheric Aqueous-Phase Chemistry: Kinetics, Mechanisms, and Its Coupling to a Changing Gas Phase. Chemical Reviews. 2015;115: 4259–4334. doi: 10.1021/cr500447k [DOI] [PubMed] [Google Scholar]
- 20.Möhler O, DeMott PJ, Vali G, Levin Z. Microbiology and atmospheric processes: the role of biological particles in cloud physics. Biogeosciences. 2007;4: 1059–1071. doi: 10.5194/bg-4-1059-2007 [Google Scholar]
- 21.Renard P, Canet I, Sancelme M, Wirgot N, Deguillaume L, Delort A-M. Screening of cloud microorganisms isolated at the Puy de Dôme (France) station for the production of biosurfactants. Atmos Chem Phys. 2016;16: 12347–12358. doi: 10.5194/acp-16-12347-2016 [Google Scholar]
- 22.Bigg EK, Soubeyrand S, Morris CE. Persistent after-effects of heavy rain on concentrations of ice nuclei and rainfall suggest a biological cause. Atmos Chem Phys. 2015;15: 2313–2326. doi: 10.5194/acp-15-2313-2015 [Google Scholar]
- 23.Creamean JM, Suski KJ, Rosenfeld D, Cazorla A, DeMott PJ, Sullivan RC, et al. Dust and Biological Aerosols from the Sahara and Asia Influence Precipitation in the Western U.S. Science. 2013;339: 1572–1578. doi: 10.1126/science.1227279 [DOI] [PubMed] [Google Scholar]
- 24.Joly M, Amato P, Deguillaume L, Monier M, Hoose C, Delort A-M. Quantification of ice nuclei active at near 0°C temperatures in low-altitude clouds at the Puy de Dôme atmospheric station. Atmos Chem Phys. 2014;14: 8185–8195. doi: 10.5194/acp-14-8185-2014 [Google Scholar]
- 25.Morris CE, Sands DC, Bardin M, Jaenicke R, Vogel B, Leyronas C, et al. Microbiology and atmospheric processes: research challenges concerning the impact of airborne micro-organisms on the atmosphere and climate. Biogeosciences. 2011;8: 17–25. [Google Scholar]
- 26.Stopelli E, Conen F, Morris CE, Herrmann E, Bukowiecki N, Alewell C. Ice nucleation active particles are efficiently removed by precipitating clouds. Scientific Reports. 2015;5: 16433 doi: 10.1038/srep16433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuzzi S, Mandrioli P, Perfetto A. Fog droplets—an atmospheric source of secondary biological aerosol particles. Atmospheric Environment. 1997;31: 287–290. doi: 10.1016/1352-2310(96)00160-4 [Google Scholar]
- 28.Sattler B, Puxbaum H, Psenner R. Bacterial growth in supercooled cloud droplets. Geophysical Research Letters. 2001;28: 239–242. [Google Scholar]
- 29.Amato P, Parazols M, Sancelme M, Laj P, Mailhot G, Delort A-M. Microorganisms isolated from the water phase of tropospheric clouds at the Puy de Dôme: major groups and growth abilities at low temperatures. FEMS Microbiology Ecology. 2007;59: 242–254. doi: 10.1111/j.1574-6941.2006.00199.x [DOI] [PubMed] [Google Scholar]
- 30.Amato P, Ménager M, Sancelme M, Laj P, Mailhot G, Delort A-M. Microbial population in cloud water at the Puy de Dôme: Implications for the chemistry of clouds. Atmospheric Environment. 2005;39: 4143–4153. doi: 10.1016/j.atmosenv.2005.04.002 [Google Scholar]
- 31.Hill KA, Shepson PB, Galbavy ES, Anastasio C, Kourtev PS, Konopka A, et al. Processing of atmospheric nitrogen by clouds above a forest environment. J Geophys Res. 2007;112: D11301 doi: 10.1029/2006JD008002 [Google Scholar]
- 32.Šantl-Temkiv T, Finster K, Hansen BM, Pašić L, Karlson UG. Viable methanotrophic bacteria enriched from air and rain can oxidize methane at cloud-like conditions. Aerobiologia. 2013; 1–12. [Google Scholar]
- 33.Vaïtilingom M, Deguillaume L, Vinatier V, Sancelme M, Amato P, Chaumerliac N, et al. Potential impact of microbial activity on the oxidant capacity and organic carbon budget in clouds. PNAS. 2013;110: 559–564. doi: 10.1073/pnas.1205743110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krumins V, Mainelis G, Kerkhof LJ, Fennell DE. Substrate-Dependent rRNA Production in an Airborne Bacterium. Environ Sci Technol Lett. 2014;1: 376–381. doi: 10.1021/ez500245y [Google Scholar]
- 35.Bowers RM, McLetchie S, Knight R, Fierer N. Spatial variability in airborne bacterial communities across land-use types and their relationship to the bacterial communities of potential source environments. ISME J. 2011;5: 601–612. doi: 10.1038/ismej.2010.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fahlgren C, Hagström Å, Nilsson D, Zweifel UL. Annual Variations in the Diversity, Viability, and Origin of Airborne Bacteria. Appl Environ Microbiol. 2010;76: 3015–3025. doi: 10.1128/AEM.02092-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amato P, Parazols M, Sancelme M, Mailhot G, Laj P, Delort A-M. An important oceanic source of micro-organisms for cloud water at the Puy de Dôme (France). Atmospheric Environment. 2007;41: 8253–8263. doi: 10.1016/j.atmosenv.2007.06.022 [Google Scholar]
- 38.Dueker ME, O’Mullan GD, Weathers KC, Juhl AR, Uriarte M. Coupling of fog and marine microbial content in the near-shore coastal environment. Biogeosciences. 2012;9: 803–813. doi: 10.5194/bg-9-803-2012 [Google Scholar]
- 39.Amato P, Joly M, Schaupp C, Attard E, Möhler O, Morris CE, et al. Survival and ice nucleation activity of bacteria as aerosols in a cloud simulation chamber. Atmos Chem Phys. 2015;15: 6455–6465. doi: 10.5194/acp-15-6455-2015 [Google Scholar]
- 40.Vaïtilingom M, Attard E, Gaiani N, Sancelme M, Deguillaume L, Flossmann AI, et al. Long-term features of cloud microbiology at the puy de Dôme (France). Atmospheric Environment. 2012;56: 88–100. doi: 10.1016/j.atmosenv.2012.03.072 [Google Scholar]
- 41.Marinoni A, Laj P, Sellegri K, Mailhot G. Cloud chemistry at the Puy de Dôme: variability and relationships with environmental factors. Atmos Chem Phys. 2004;4: 715–728. doi: 10.5194/acp-4-715-2004 [Google Scholar]
- 42.Draxler RR, Rolph GD. HYSPLIT (HYbrid Single-Particle Lagrangian Integrated Trajectory) Model access via NOAA ARL READY Website (http://ready.arl.noaa.gov/HYSPLIT.php). NOAA Air Resources Laboratory, Silver Spring, MD: 2013; [Google Scholar]
- 43.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6: 1621–1624. doi: 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gast RK, Mitchell BW, Holt PS. Detection of airborne Salmonella enteritidis in the environment of experimentally infected laying hens by an electrostatic sampling device. Avian Dis. 2004;48: 148–154. doi: 10.1637/7086 [DOI] [PubMed] [Google Scholar]
- 45.Kim M, Morrison M, Yu Z. Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes. J Microbiol Methods. 2011;84: 81–87. doi: 10.1016/j.mimet.2010.10.020 [DOI] [PubMed] [Google Scholar]
- 46.Mangot J-F, Domaizon I, Taib N, Marouni N, Duffaud E, Bronner G, et al. Short-term dynamics of diversity patterns: evidence of continual reassembly within lacustrine small eukaryotes. Environ Microbiol. 2013;15: 1745–1758. doi: 10.1111/1462-2920.12065 [DOI] [PubMed] [Google Scholar]
- 47.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35: 7188–7196. doi: 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Price MN, Dehal PS, Arkin AP. FastTree 2 –Approximately Maximum-Likelihood Trees for Large Alignments. PLOS ONE. 2010;5: e9490 doi: 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taib N, Mangot J-F, Domaizon I, Bronner G, Debroas D. Phylogenetic affiliation of SSU rRNA genes generated by massively parallel sequencing: new insights into the freshwater protist diversity. PLoS ONE. 2013;8: e58950 doi: 10.1371/journal.pone.0058950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taib N, Bronner G, Charvy JC, Mephu Nguifo E, Breton V, Debroas D. ePANAM: a web server dedicated to the analysis of next generation amplicons. Copenhaguen, Denmark; 2012.
- 51.McMurdie PJ, Holmes S. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. PLOS Computational Biology. 2014;10: e1003531 doi: 10.1371/journal.pcbi.1003531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria: ). Available at: http://wwwR-project.org. 2011; [Google Scholar]
- 53.McMurdie PJ, Holmes S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLOS ONE. 2013;8: e61217 doi: 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeileis A, Kleiber C. ineq: Measuring Inequality, Concentration, and Poverty [Internet]. 2014. Available: https://cran.r-project.org/web/packages/ineq/index.html
- 55.Wickham H. Ggplot2: Elegant Graphics for Data Analysis. 2nd ed. Springer Publishing Company, Incorporated; 2009. [Google Scholar]
- 56.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. vegan: Community Ecology Package [Internet]. 2017. Available: https://cran.r-project.org/web/packages/vegan/index.html
- 57.Oliveros JC. Venny 2.1.0 [Internet]. 2015 2007 [cited 13 Mar 2017]. Available: http://bioinfogp.cnb.csic.es/tools/venny/
- 58.Deguillaume L, Charbouillot T, Joly M, Vaïtilingom M, Parazols M, Marinoni A, et al. Classification of clouds sampled at the puy de Dôme (France) based on 10 yr of monitoring of their physicochemical properties. Atmos Chem Phys. 2014;14: 1485–1506. doi: 10.5194/acp-14-1485-2014 [Google Scholar]
- 59.Fahlgren C, Bratbak G, Sandaa R-A, Thyrhaug R, Zweifel UL. Diversity of airborne bacteria in samples collected using different devices for aerosol collection. Aerobiologia. 2010;27: 107–120. doi: 10.1007/s10453-010-9181-z [Google Scholar]
- 60.Maron P-A, Lejon DPH, Carvalho E, Bizet K, Lemanceau P, Ranjard L, et al. Assessing genetic structure and diversity of airborne bacterial communities by DNA fingerprinting and 16S rDNA clone library. Atmospheric Environment. 2005;39: 3687–3695. doi: 10.1016/j.atmosenv.2005.03.002 [Google Scholar]
- 61.Wei M, Xu C, Chen J, Zhu C, Li J, Lv G. Characteristics of bacterial community in fog water at Mt. Tai: similarity and disparity under polluted and non-polluted fog episodes. Atmospheric Chemistry and Physics Discussions. 2016; 1–30. doi: 10.5194/acp-2016-776 [Google Scholar]
- 62.Xu C, Wei M, Chen J, Sui X, Zhu C, Li J, et al. Investigation of diverse bacteria in cloud water at Mt. Tai, China. Science of The Total Environment. 2017;580: 258–265. doi: 10.1016/j.scitotenv.2016.12.081 [DOI] [PubMed] [Google Scholar]
- 63.Baldrian P, Kolařík M, Stursová M, Kopecký J, Valášková V, Větrovský T, et al. Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J. 2012;6: 248–258. doi: 10.1038/ismej.2011.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y, Zhao Z, Dai M, Jiao N, Herndl GJ. Drivers shaping the diversity and biogeography of total and active bacterial communities in the South China Sea. Mol Ecol. 2014;23: 2260–2274. doi: 10.1111/mec.12739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klein AM, Bohannan BJM, Jaffe DA, Levin DA, Green JL. Molecular Evidence for Metabolically Active Bacteria in the Atmosphere. Front Microbiol. 2016; 772 doi: 10.3389/fmicb.2016.00772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Youssef NH, Elshahed MS. Species richness in soil bacterial communities: A proposed approach to overcome sample size bias. Journal of Microbiological Methods. 2008;75: 86–91. doi: 10.1016/j.mimet.2008.05.009 [DOI] [PubMed] [Google Scholar]
- 67.Kivlin SN, Hawkes CV. Temporal and Spatial Variation of Soil Bacteria Richness, Composition, and Function in a Neotropical Rainforest. PLOS ONE. 2016;11: e0159131 doi: 10.1371/journal.pone.0159131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ahern HE, Walsh KA, Hill TCJ, Moffett BF. Fluorescent pseudomonads isolated from Hebridean cloud and rain water produce biosurfactants but do not cause ice nucleation. Biogeosciences. 2007;4: 115–124. [Google Scholar]
- 69.Zweifel UL, Hagström Å, Holmfeldt K, Thyrhaug R, Geels C, Frohn LM, et al. High bacterial 16S rRNA gene diversity above the atmospheric boundary layer. Aerobiologia. 2012;28: 481–498. doi: 10.1007/s10453-012-9250-6 [Google Scholar]
- 70.Kourtev PS, Hill KA, Shepson PB, Konopka A. Atmospheric cloud water contains a diverse bacterial community. Atmospheric Environment. 2011;45: 5399–5405. doi: 10.1016/j.atmosenv.2011.06.041 [Google Scholar]
- 71.Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJM. Counting the Uncountable: Statistical Approaches to Estimating Microbial Diversity. Appl Environ Microbiol. 2001;67: 4399–4406. doi: 10.1128/AEM.67.10.4399-4406.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.O’Brien SL, Gibbons SM, Owens SM, Hampton-Marcell J, Johnston ER, Jastrow JD, et al. Spatial scale drives patterns in soil bacterial diversity. Environ Microbiol. 2016;18: 2039–2051. doi: 10.1111/1462-2920.13231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fröhlich-Nowoisky J, Burrows SM, Xie Z, Engling G, Solomon PA, Fraser MP, et al. Biogeography in the air: fungal diversity over land and oceans. Biogeosciences. 2012;9: 1125–1136. doi: 10.5194/bg-9-1125-2012 [Google Scholar]
- 74.Cho BC, Jang GI. Active and diverse rainwater bacteria collected at an inland site in spring and summer 2011. Atmospheric Environment. 2014;94: 409–416. doi: 10.1016/j.atmosenv.2014.05.048 [Google Scholar]
- 75.Wittebolle L, Marzorati M, Clement L, Balloi A, Daffonchio D, Heylen K, et al. Initial community evenness favours functionality under selective stress. Nature. 2009;458: 623–626. doi: 10.1038/nature07840 [DOI] [PubMed] [Google Scholar]
- 76.Aller JY, Kuznetsova MR, Jahns CJ, Kemp PF. The sea surface microlayer as a source of viral and bacterial enrichment in marine aerosols. Journal of Aerosol Science. 2005;36: 801–812. doi: 10.1016/j.jaerosci.2004.10.012 [Google Scholar]
- 77.Burrows SM, Elbert W, Lawrence MG, Pöschl U. Bacteria in the global atmosphere–Part 1: Review and synthesis of literature data for different ecosystems. Atmos Chem Phys. 2009;9: 9263–9280. doi: 10.5194/acp-9-9263-2009 [Google Scholar]
- 78.Huffman JA, Pöhlker C, Prenni AJ, DeMott PJ, Mason RH, Robinson NH, et al. High concentrations of biological aerosol particles and ice nuclei during and after rain. Atmospheric Chemistry and Physics Discussions. 2013;13: 1767–1793. doi: 10.5194/acpd-13-1767-2013 [Google Scholar]
- 79.Joung YS, Ge Z, Buie CR. Bioaerosol generation by raindrops on soil. Nature Communications. 2017;8: 14668 doi: 10.1038/ncomms14668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Copeland JK, Yuan L, Layeghifard M, Wang PW, Guttman DS. Seasonal Community Succession of the Phyllosphere Microbiome. MPMI. 2015;28: 274–285. doi: 10.1094/MPMI-10-14-0331-FI [DOI] [PubMed] [Google Scholar]
- 81.Blanchard DC, Syzdek LD. Water-to-Air Transfer and Enrichment of Bacteria in Drops from Bursting Bubbles. Appl Environ Microbiol. 1982;43: 1001–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Curtis TP, Sloan WT. Prokaryotic diversity and its limits: microbial community structure in nature and implications for microbial ecology. Current Opinion in Microbiology. 2004;7: 221–226. doi: 10.1016/j.mib.2004.04.010 [DOI] [PubMed] [Google Scholar]
- 83.Galès A, Bru-Adan V, Godon J-J, Delabre K, Catala P, Ponthieux A, et al. Predominance of single bacterial cells in composting bioaerosols. Atmospheric Environment. 2015;107: 225–232. doi: 10.1016/j.atmosenv.2015.02.035 [Google Scholar]
- 84.Lindemann J, Constantinidou HA, Barchet WR, Upper CD. Plants as sources of airborne bacteria, including ice nucleation-active bacteria. Applied and Environmental Microbiology. 1982;44: 1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maki T, Hara K, Iwata A, Lee KC, Kawai K, Kai K, et al. Variations in airborne bacterial communities at high altitudes over the Noto Peninsula (Japan) in response to Asian dust events. Atmos Chem Phys Discuss. 2017;2017: 1–32. doi: 10.5194/acp-2016-1095 [Google Scholar]
- 86.Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Micro. 2008;6: 741–751. doi: 10.1038/nrmicro1969 [DOI] [PubMed] [Google Scholar]
- 87.Perlman SJ, Hunter MS, Zchori-Fein E. The emerging diversity of Rickettsia. Proceedings of the Royal Society of London B: Biological Sciences. 2006;273: 2097–2106. doi: 10.1098/rspb.2006.3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wittmaack K, Wehnes H, Heinzmann U, Agerer R. An overview on bioaerosols viewed by scanning electron microscopy. Sci Total Environ. 2005;346: 244–255. doi: 10.1016/j.scitotenv.2004.11.009 [DOI] [PubMed] [Google Scholar]
- 89.Emelyanov VV. Evolutionary relationship of Rickettsiae and mitochondria. FEBS Letters. 2001;501: 11–18. doi: 10.1016/S0014-5793(01)02618-7 [DOI] [PubMed] [Google Scholar]
- 90.Gnanamanickam SS. Plant-Associated Bacteria. Springer Science & Business Media; 2007. [Google Scholar]
- 91.Lindow SE, Brandl MT. Microbiology of the phyllosphere. Applied and environmental microbiology. 2003;69: 1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Camarinha-Silva A, Jáuregui R, Chaves-Moreno D, Oxley APA, Schaumburg F, Becker K, et al. Comparing the anterior nare bacterial community of two discrete human populations using Illumina amplicon sequencing. Environ Microbiol. 2014;16: 2939–2952. doi: 10.1111/1462-2920.12362 [DOI] [PubMed] [Google Scholar]
- 93.Womack AM, Artaxo PE, Ishida FY, Mueller RC, Saleska SR, Wiedemann KT, et al. Characterization of active and total fungal communities in the atmosphere over the Amazon rainforest. Biogeosciences. 2015;12: 6337–6349. doi: 10.5194/bg-12-6337-2015 [Google Scholar]
- 94.Constantinidou HA. Atmospheric Dispersal of Ice Nucleation-Active Bacteria: The Role of Rain. Phytopathology. 1990;80: 934 doi: 10.1094/Phyto-80-934 [Google Scholar]
- 95.Després VR, Hufffman JA, Burrows SM, Hoose C, Safatov AS, Buryak G, et al. Primary biological aerosol particles in the atmosphere: a review. Tellus B. 2012;64 doi: 10.3402/tellusb.v64i0.15598 [Google Scholar]
- 96.Morris CE, Sands DC, Vinatzer BA, Glaux C, Guilbaud C, Buffière A, et al. The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J. 2008;2: 321–334. doi: 10.1038/ismej.2007.113 [DOI] [PubMed] [Google Scholar]
- 97.Amato P, Demeer F, Melaouhi A, Fontanella S, Martin-Biesse AS, Sancelme M, et al. A fate for organic acids, formaldehyde and methanol in cloud water: their biotransformation by micro-organisms. Atmos Chem Phys. 2007;7: 4159–4169. [Google Scholar]
- 98.Vaïtilingom M, Amato P, Sancelme M, Laj P, Leriche M, Delort A-M. Contribution of Microbial Activity to Carbon Chemistry in Clouds. Appl Environ Microbiol. 2010;76: 23–29. doi: 10.1128/AEM.01127-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vinatier V, Wirgot N, Joly M, Sancelme M, Abrantes M, Deguillaume L, et al. Siderophores in Cloud Waters and Potential Impact on Atmospheric Chemistry: Production by Microorganisms Isolated at the Puy de Dôme Station. Environ Sci Technol. 2016;50: 9315–9323. doi: 10.1021/acs.est.6b02335 [DOI] [PubMed] [Google Scholar]
- 100.Busse H-J, Denner EBM, Buczolits S, Salkinoja-Salonen M, Bennasar A, Kämpfer P. Sphingomonas aurantiaca sp. nov., Sphingomonas aerolata sp. nov. and Sphingomonas faeni sp. nov., air- and dustborne and Antarctic, orange-pigmented, psychrotolerant bacteria, and emended description of the genus Sphingomonas. Int J Syst Evol Microbiol. 2003;53: 1253–1260. doi: 10.1099/ijs.0.02461-0 [DOI] [PubMed] [Google Scholar]
- 101.Foght J, Aislabie J, Turner S, Brown CE, Ryburn J, Saul DJ, et al. Culturable bacteria in subglacial sediments and ice from two Southern Hemisphere glaciers. Microb Ecol. 2004;47: 329–340. doi: 10.1007/s00248-003-1036-5 [DOI] [PubMed] [Google Scholar]
- 102.Vaz-Moreira I, Nunes OC, Manaia CM. Diversity and antibiotic resistance patterns of Sphingomonadaceae isolates from drinking water. Appl Environ Microbiol. 2011;77: 5697–5706. doi: 10.1128/AEM.00579-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Neef A, Witzenberger R, Kämpfer P. Detection of sphingomonads and in situ identification in activated sludge using 16S rRNA-targeted oligonucleotide probes. J Ind Microbiol Biotechnol. 1999;23: 261–267. doi: 10.1038/sj/jim/2900768 [DOI] [PubMed] [Google Scholar]
- 104.Haritash AK, Kaushik CP. Biodegradation aspects of Polycyclic Aromatic Hydrocarbons (PAHs): A review. Journal of Hazardous Materials. 2009;169: 1–15. doi: 10.1016/j.jhazmat.2009.03.137 [DOI] [PubMed] [Google Scholar]
- 105.Corpe WA, Rheem S. Ecology of the methylotrophic bacteria on living leaf surfaces. FEMS Microbiol Ecol. 1989;5: 243–249. doi: 10.1111/j.1574-6968.1989.tb03698.x [Google Scholar]
- 106.Haber CL, Allen LN, Zhao S, Hanson RS. Methylotrophic Bacteria: Biochemical Diversity and Genetics. Science. 1983;221: 1147–1153. doi: 10.1126/science.221.4616.1147 [DOI] [PubMed] [Google Scholar]
- 107.Nadalig T, Greule M, Bringel F, Keppler F, Vuilleumier S. Probing the diversity of chloromethane-degrading bacteria by comparative genomics and isotopic fractionation. Front Microbiol. 2014;5 doi: 10.3389/fmicb.2014.00523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bridge TAM, Johnson DB. Reductive Dissolution of Ferric Iron Minerals by Acidiphilium SJH. Geomicrobiology Journal. 2000;17: 193–206. doi: 10.1080/01490450050121161 [Google Scholar]
- 109.Wakao N, Yokoi N, Isoyama N, Hiraishi A, Shimada K, Kobayashi M, et al. Discovery of Natural Photosynthesis using Zn-Containing Bacteriochlorophyll in an Aerobic Bacterium Acidiphilium rubrum. Plant Cell Physiol. 1996;37: 889–893. doi: 10.1093/oxfordjournals.pcp.a029029 [Google Scholar]
- 110.Staley JT, Irgens RL, Brenner DJ. Enhydrobacter aerosaccus gen. nov., sp. nov., a Gas-Vacuolated, Facultatively Anaerobic, Heterotrophic Rod. International Journal of Systematic and Evolutionary Microbiology. 1987;37: 289–291. doi: 10.1099/00207713-37-3-289 [Google Scholar]
- 111.Battista JR. Against all odds: the survival strategies of Deinococcus radiodurans. Annu Rev Microbiol. 1997;51: 203–224. doi: 10.1146/annurev.micro.51.1.203 [DOI] [PubMed] [Google Scholar]
- 112.Lee J-J, Kang M-S, Joo ES, Kim MK, Im W-T, Jung H-Y, et al. Spirosoma montaniterrae sp. nov., an ultraviolet and gamma radiation-resistant bacterium isolated from mountain soil. J Microbiol. 2015;53: 429–434. doi: 10.1007/s12275-015-5008-5 [DOI] [PubMed] [Google Scholar]
- 113.Finster KW, Herbert RA, Lomstein BA. Spirosoma spitsbergense sp. nov. and Spirosoma luteum sp. nov., isolated from a high Arctic permafrost soil, and emended description of the genus Spirosoma. Int J Syst Evol Microbiol. 2009;59: 839–844. doi: 10.1099/ijs.0.002725-0 [DOI] [PubMed] [Google Scholar]
- 114.Zhang Y, Schoch CL, Fournier J, Crous PW, de Gruyter J, Woudenberg JHC, et al. Multi-locus phylogeny of Pleosporales: a taxonomic, ecological and evolutionary re-evaluation. Studies in Mycology. 2009;64: 85–102. doi: 10.3114/sim.2009.64.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Luo J, Qiu H, Cai G, Wagner NE, Bhattacharya D, Zhang N. Phylogenomic analysis uncovers the evolutionary history of nutrition and infection mode in rice blast fungus and other Magnaporthales. Scientific Reports. 2015;5: 9448 doi: 10.1038/srep09448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stadler M, Hellwig V. Chemotaxonomy of the Xylariaceae and remarkable bioactive compounds from Xylariales and their associated asexual stages [Internet]. 2005. Available: http://cat.inist.fr/?aModele=afficheN&cpsidt=17833910
- 117.Stoddard SF, Smith BJ, Hein R, Roller BRK, Schmidt TM. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 2015;43: D593–598. doi: 10.1093/nar/gku1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Griffin DW. Atmospheric movement of microorganisms in clouds of desert dust and implications for human health. Clin Microbiol Rev. 2007;20: 459–477, table of contents. doi: 10.1128/CMR.00039-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.De Wit R, Bouvier T. ‘Everything is everywhere, but, the environment selects’; what did Baas Becking and Beijerinck really say? Environmental Microbiology. 2006;8: 755–758. doi: 10.1111/j.1462-2920.2006.01017.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(DOCX)
(TIF)
(TIF)
Rarefaction curves (A and B) and rank-abundance plots (C and D) of the different set of amplicons.
(TIF)
Venn diagrams depicting similarities and singularities of the 3 samples at the OTU0.03 level for prokaryotes (A) and OTU0.05 for eukaryotes (B).
(TIF)
Relative abundance of shared prokaryotic genera (A) and eukaryotic orders (B).
(TIF)
Bray-Curtis similarity matrices between the different sets of sequence of prokaryotes (A) and eukaryotes (B).
(TIF)
(TIF)
(TIF)
(TIF)
Data Availability Statement
All sequence files are available from the NCBI Sequence Read Archive SRA database (BioProject ID PRJNA380262).