Abstract
Developmental genes can harbour multiple transcriptional enhancers that act simultaneously or in succession to achieve robust and precise spatiotemporal expression. However, the mechanisms underlying cooperation between cis-acting elements are poorly documented, notably in vertebrates. The mouse gene Krox20 encodes a transcription factor required for the specification of two segments (rhombomeres) of the developing hindbrain. In rhombomere 3, Krox20 is subject to direct positive feedback governed by an autoregulatory enhancer, element A. In contrast, a second enhancer, element C, distant by 70 kb, is active from the initiation of transcription independent of the presence of the KROX20 protein. Here, using both enhancer knock-outs and investigations of chromatin organisation, we show that element C possesses a dual activity: besides its classical enhancer function, it is also permanently required in cis to potentiate the autoregulatory activity of element A, by increasing its chromatin accessibility. This work uncovers a novel, asymmetrical, long-range mode of cooperation between cis-acting elements that might be essential to avoid promiscuous activation of positive autoregulatory elements.
Author summary
The formation of multicellular organisms from the egg to the adult stage is largely under genetic control. The activation of specific genes is governed by regulatory DNA sequences present nearby on the chromosome. Most of these sequences promote activation and are called enhancers. In this paper, we study two enhancers governing the expression of a gene involved in the formation of the posterior brain in vertebrates. One of these enhancers is involved in a positive feedback loop: it is itself activated by the protein product of the gene that it regulates. The other enhancer was thought to be only involved in the initial accumulation of the protein, necessary for the subsequent activation of the feedback loop. Here we show that the second enhancer directly cooperates with the autoregulatory enhancer to increase its accessibility and its activity. Our work uncovers a novel, long-range mode of cooperation between enhancers that restricts the domain of action of autoregulatory enhancers within embryos and might be essential to avoid their inappropriate activation.
Introduction
DNA cis-acting elements play key roles in the regulation and evolution of gene expression by controlling spatiotemporal transcription patterns. A major class of cis-regulatory elements are transcriptional enhancers, which can recruit combinations of transcription factors (TFs) to modulate transcription initiation from (a) cognate gene promoter(s), in general independently of their relative distance and orientation [1–3]. So far, most enhancers have been functionally characterized by assay of their transcriptional activity using transgenic constructs carrying the enhancer and a reporter gene driven by a minimal promoter [4]. Another strategy consists in the random insertion of a transposon that senses enhancer activity within the surrounding genomic region. It is particularly useful to detect multiple cis-regulatory elements with similar activities and long-distance modulation of gene expression [5,6]. Transgenesis using BACs allows the introduction of large DNA fragments containing enhancers in their native context. This approach is helpful in the analysis of multiple enhancers controlling the same gene [7], but can be challenging for the study of mammalian enhancer that are located far away from the promoter that they control. These different approaches provide useful information on spatial and temporal activity of the putative enhancer element, but they usually do not establish whether and how the enhancer actually participates in the control of the expression of its suspected cognate gene in its full normal genomic context. Answer to this latter question requires in vivo analyses involving deletion or mutation of the endogenous enhancer. This issue is particularly important in situations where multiple, overlapping enhancers operate within the same cis-regulatory landscape. In such cases, various types of regulatory crosstalk can occur between the enhancers, resulting in additive, synergistic, competitive or repressive effects [3]. In vertebrates, very few studies have addressed such situations.
Enhancer activity is intimately linked to chromatin organization. Hence, association of pioneer TFs to an enhancer can lead to chromatin decompaction and facilitate the binding of additional TFs and/or recruitment of various epigenetic machineries [8]. In return, chromatin configuration can affect gene expression by modulating long-range interactions between enhancers and promoters [9], that are usually constrained within regions called topologically associated domains (TADs) [10,11]. TADs, which are approximately Mb-sized in mammals, form constitutive “regulatory neighbourhoods” that provide specificity to enhancer-promoter interactions by reducing aberrant contacts between cis-regulatory elements located in distinct TADs [6,10].
To provide insights into the mechanisms involved in the regulation of a vertebrate gene by multiple enhancers during development, we investigated the case of the mouse Krox20/Egr2 gene [12] for which several hindbrain-specific enhancers have been identified [13]. The hindbrain is an attractive model to investigate the genetic control of morphogenesis in vertebrates, as it is subject to a transient segmentation process leading to the formation of 7–8 segments called rhombomeres (r) [14,15]. The formation and specification of segments r3 and r5 are governed by the transcription factor KROX20/EGR2 [15–17]. So far three evolutionarily conserved sequences exhibiting enhancer activity in the hindbrain have been identified within the Krox20 locus and are termed A, B and C [13]. Element A, located 217 kb upstream of the promoter in the mouse, is active in both r3 and r5. This element carries several KROX20 binding sites and requires direct binding of the protein for its activity, suggesting that it acts as an autoregulatory element [13]. Indeed, upon deletion of element A, Krox20 expression is normally initiated in r3 and r5, but is not amplified nor maintained at later stages [18]. Additional studies have indicated that element A underlies a positive feedback loop that acts as a binary switch for specification of odd- versus even–numbered rhombomere identity [18]. Element B, located 164 kb upstream of the promoter, drives the expression of reporter constructs specifically in r5 [13,19,20]. Finally, element C, located 144 kb upstream of the promoter, is active in the r3-r5 region [13,19–21]. Several observations suggest that elements B and C, in contrast to the autoregulatory element A, are involved in the initial steps of Krox20 expression in r3 or r5 (initiator elements): i) they are transcriptionally active at the early stages of Krox20 hindbrain expression [13]; ii) they are activated by transcription factors known to act upstream of Krox20 [19–21]; iii) they do not require the presence of the KROX20 protein for their activity [13].
In the present study, we have investigated the contribution of element C to Krox20 expression, as it was the only characterized initiator element with an activity in r3. Using a conditional knock-out mutation of element C, we show that, unexpectedly, this element is not necessary for Krox20 initial expression in r3. In contrast, it appears absolutely required for the maintenance of Krox20 expression in this rhombomere. This activity involves a cooperation in cis with element A, element C potentiating its activity and increasing its accessibility. These observations reveal that a cis-acting element can cooperate with other enhancers within the same locus according to different modalities and suggest a scheme for protecting autoregulatory elements from inappropriate activation.
Results
In r3, element C is required for late, but not initial Krox20 expression
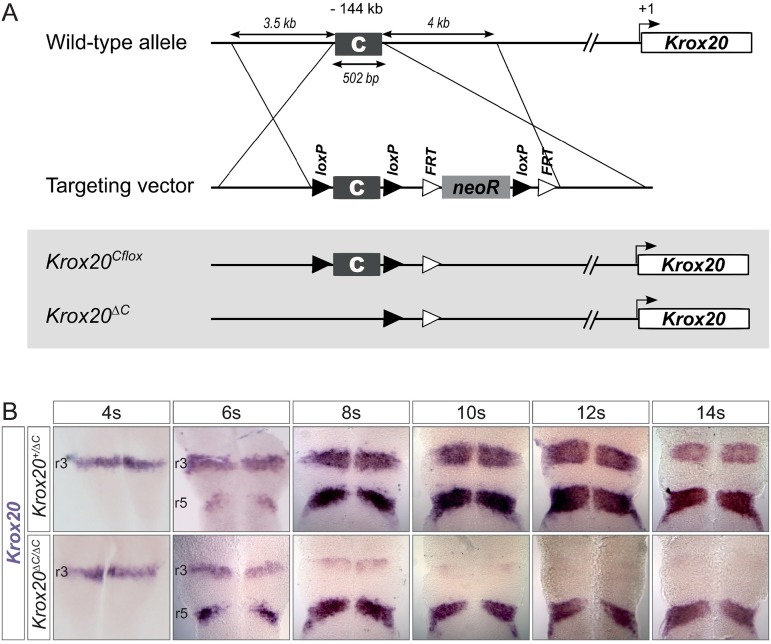
To assess the contribution of element C to the regulation of Krox20 expression, we generated a mouse line carrying a deletion of this element. The details of the knock-out strategy are presented in Fig 1A. Two alleles were generated: Krox20Cflox, in which element C is present, but flanked by two loxP sites, and Krox20ΔC, in which element C is deleted. The impact of element C deletion on Krox20 transcription was analysed by mRNA in situ hybridization on 4 to 14 somite stage (s) embryos, comparing homozygous (Krox20ΔC/ΔC) with heterozygous mutants (Krox20+/ΔC), the knock-out of one allele of Krox20 having no phenotype [15,16,22]. Unexpectedly, up until 6s the expression of Krox20 in r3 and r5 is similar in Krox20ΔC/ΔC embryos and control littermates (Fig 1B). However, at 8s, Krox20 expression is severely reduced in r3 from Krox20ΔC/ΔC embryos as compared to controls and, at later stages, it is completely lost (Fig 1B). During the considered period, although Krox20 expression does not appear to be dramatically affected in r5, in contrast to r3, the width of the corresponding domain of expression appears to be slightly reduced after 10s (Fig 1B).
Fig 1. Genetic analysis of element C function.
(A) Strategy for the construction of conditional and null alleles of element C. The targeting vector was introduced into the locus in ES cells by homologous recombination and one of the ES clones subsequently allowed germ line transmission in the mouse. The floxed allele, Krox20Cflox, was obtained by crossing the founder mouse line with a Flp (targeting FRT sites) deletor line. The null allele, Krox20ΔC, was obtained by crossing the Krox20Cflox line with a Cre (targeting loxP sites) deletor line, PGK-Cre. (B) In situ hybridization for Krox20 mRNA performed on Krox20+/ΔC and Krox20ΔC/ΔC embryos at the indicated somite stages. Embryos were flat-mounted with anterior toward the top. Rhombomere positions are indicated on the left.
To investigate longer-term consequences of element C deletion on cell specification, we analysed the expression of a KROX20 target gene, EphA4 [22], which is known to persist beyond the period of Krox20 expression in r3 and r5 [18]. In control embryos (Krox20+/ΔC), at 10s and 25s, EphA4 is expressed at high levels in r3 and r5 and at a lower level in r2 (S1 Fig). At both stages, the r3 domain, as demarcated by EphA4 expression, is reduced in Krox20ΔC/ΔC embryos as compared to controls, whereas the r5 domain is similar in both genotypes (S1 Fig). This is consistent with the premature loss of Krox20 expression in r3, known to reduce the extension of this rhombomere [18,22]. The limited variation of Krox20 expression in r5 in Krox20ΔC/ΔC embryos after 10s does not appear to perturb the size of this rhombomere at later stages, consistent with the fact that Krox20 expression is not required for the maintenance of EphA4 expression in r3 and r5 [18].
In conclusion, these data indicate that i) element C is dispensable for the initiation of Krox20 expression in r3 or r5, suggesting the existence of other elements in charge of these functions; ii) in r3, in contrast, element C is absolutely required for expression beyond 6s, leading to a reduction in size of this rhombomere at later stages. Notably, the phenotype observed in r3 in Krox20ΔC/ΔC embryos is very similar to what was previously described in Krox20ΔA/ΔA embryos (Figs 1B and 2A, and S1 Fig) [18]; iii) in r5, the contribution of element C to Krox20 expression is rather limited, without significant effect on the size of this rhombomere.
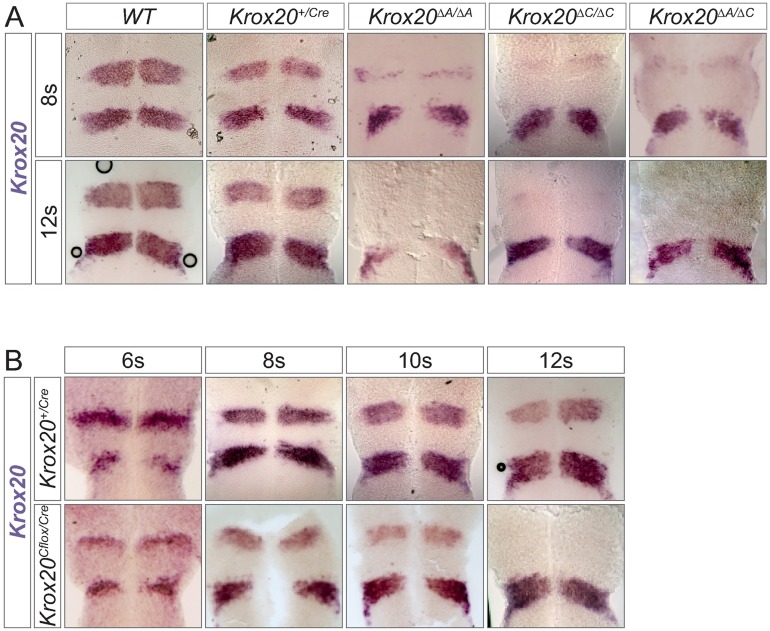
Fig 2.
Cooperation in cis between elements A and C. (A) In situ hybridization for Krox20 mRNA was performed on wild type (WT), Krox20+/Cre, Krox20ΔA/ΔA, Krox20ΔC/ΔC and composite heterozygous Krox20ΔA/ΔC embryos at the indicated somite stages. (B) In situ hybridization for Krox20 mRNA was performed on Krox20+/Cre and Krox20Cflox/Cre embryos at the indicated somite stages. In (A) and (B) embryos were flat-mounted with anterior toward the top.
Elements A and C cooperate in cis for the establishment of the autoregulatory loop
The similarity of the phenotypes observed in r3 upon deletion of elements A or C led us to investigate the possibility of an involvement of element C in Krox20 autoregulation, together with element A. For this purpose, we first analysed the expression of Krox20 in composite heterozygous embryos, Krox20ΔA/ΔC, carrying deletions of element A on one allele and of element C on the other (Fig 2A). Although this combination does not affect Krox20 expression in r3 at early stages, at 8s Krox20 mRNA level is severely reduced and, at 12s, it is completely lost, mimicking the phenotype observed in Krox20ΔA/ΔA or Krox20ΔC/ΔC embryos at both stages (Fig 2A). In r5, Krox20 expression is only slightly affected in Krox20ΔA/ΔC embryos, similarly to Krox20ΔC/ΔC embryos (Fig 2A). This defect in the maintenance of Krox20 expression in r3, combined with apparently normal expression at early stages, contrasts with the fact that a single wild type Krox20 allele is sufficient to activate and maintain the autoregulatory loop (Fig 2A, Krox20+/Cre) [12]. This suggests that in Krox20ΔA/ΔC embryos the level of expression of Krox20 is not a limiting factor for the activation of the only wild type allele of element A. Therefore, the most likely explanation for the defect in Krox20 maintenance is that, in r3, the deletion of element C impairs the activity of element A located on the same chromosome and that the two elements synergistically cooperate, in cis, for the establishment and/or maintenance of the autoregulatory loop. A more conventional, partial redundancy between elements A and C appears much less likely.
This cooperation does not preclude an early involvement of element C, for instance to poise element A for the subsequent autoregulation phase. To investigate whether element C has a function only at the early phase of Krox20 activation, or whether it is required during the autoregulation phase as well, we generated a genetic condition in which element C is initially active, but is deleted at a later stage. This was achieved by combining the Krox20Cflox allele (Fig 1A) with a knock-in allele, Krox20Cre, in which the Krox20 coding sequence has been replaced by the coding sequence of the Cre recombinase [23]. In such embryos, Krox20 and Cre are expected to be synthetized at early somitic stages. Subsequently, the recombinase leads to deletion of element C in r3 and r5. In Krox20Cflox/Cre embryos, Krox20 expression is progressively reduced in r3 from 6s to 10s, as compared to Krox20+/Cre controls (Fig 2B), although less abruptly than in Krox20ΔC/ΔC mutants (Figs 1B and 2A). At 12s, Krox20 expression is completely abolished in r3 in Krox20Cflox/Cre embryos (Fig 2B). These data indicate that the presence of element C only during the early phase of Krox20 expression is not sufficient to establish and/or maintain the autoregulatory loop. The higher level of Krox20 in r3 in Krox20Cflox/Cre as compared to Krox20ΔC/ΔC embryos is likely to originate from transient activation of the loop, followed by termination of its activity, due to Cre excision of element C.
In conclusion, these results indicate that elements A and C synergistically cooperate in cis for establishing and/or maintaining this loop in r3. More precisely, they show that element C is permanently required for activity of the Krox20 feedback loop.
Physical interactions between Krox20 cis-elements
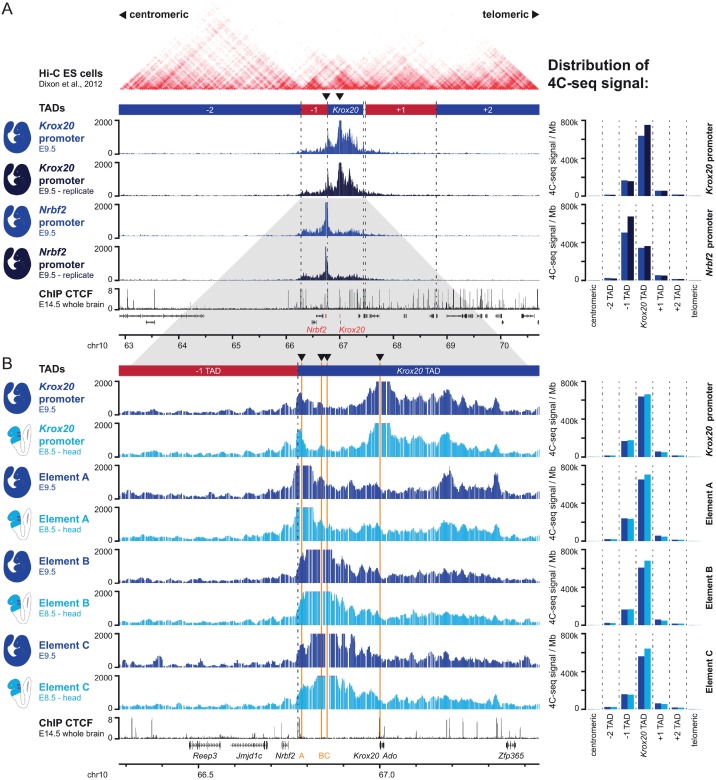
The existence of a cooperation in cis between elements A and C led us to investigate the existence of possible physical 3D interactions between the different Krox20 cis-elements, which are separated by large distances on the mouse chromosome. A previous Hi-C analysis [11] in embryonic stem cells identified a TAD that includes the gene and its cis-regulatory elements (Fig 3A). The left boundary of the TAD spreads out over a relatively large and undefined transition zone (S2A and S2B Fig). To better characterize the Krox20 regulatory neighborhood, we used circular chromosome conformation capture (4C-seq) on multiple viewpoints in the locus [24]. In samples prepared from total embryos at embryonic day (E) 9.5, when Krox20 is no more transcribed [25], the Krox20 gene and its distant regulatory element A (separated by over 200 kb) show highly similar distributions of 4C-seq signal (Fig 3A and S2B and S2C Fig) preferentially located in the Krox20 TAD. In contrast, the distribution of the Nrbf2 gene, which is located in the TAD transition zone and is separated from element A by a 35 kb genomic interval (including a cluster of CTCF binding sites) spreads its interactions about equally over the two neighboring TADs (S2C Fig). Repositioning of the TAD boundary to the cluster of CTCF binding sites results in strongly increased separation of signal between the Nrbf2 gene on one hand and the Krox20 gene and its regulatory elements on the other hand, indicating they are located in different regulatory neighborhood (Fig 3A and S2C Fig).
Fig 3. Physical interactions within the Krox20 locus.
(A) Alignment of data in the Krox20 and adjacent loci from Hi-C in ES cells [11], 4C-seq in E9.5 whole mouse embryos, using the Krox20 and Nrbf2 promoters as viewpoints (this work, 2 biological replicates) and CTCF ChIP-seq in E14.5 mouse brain (ENCODE, [58]). (B) Zoom in on the Krox20 locus, showing 4C-seq data from the Krox20 promoter, element A, element B and element C as viewpoints. CTCF ChIP-seq data in E14.5 mouse brain (ENCODE) are indicated below. Signals from simultaneously processed E9.5 whole embryo (dark blue) and E8.5 embryo head (light blue) samples are shown. On the right, normalized distributions of the 4C-seq signals in different genomic regions are indicated. TADs as defined in [7] or by our additional analysis (S2 Fig) are indicated above, with dashed lines in the graphs demarcating TAD boundaries. Genes (black/red), cis-regulatory elements (orange) and genomic coordinates are indicated below each set of data. Arrowheads above each 4C track pinpoint viewpoints.
To determine if 3D chromatin interactions in the Krox20 regulatory neighborhood were dynamic at these early stages of embryogenesis, and possibly linked to the autoregulatory loop, we compared our E9.5 samples to micro-dissected embryonic heads at E8.5, when the autoregulatory loop is active in a subset of cells [18]. For all viewpoints, very similar patterns were obtained between E8.5 heads and E9.5 (Fig 3B). At both stages, the Krox20 promoter forms long-range interactions within the Krox20 TAD that cover elements A and B (Fig 3B). In addition, bi-directional interactions are formed between elements A and B and, to a lesser extent, between elements A and C (Fig 3B).
In conclusion, these data reveal that the Krox20 regulatory neighbourhood adopts a higher-order configuration that incorporates long-range interactions between the various cis-regulatory elements and is mostly invariant at different positions in the early embryo.
Chromatin accessibility of Krox20 enhancers correlates with transcriptional activity
To investigate the correlation between the activity of the Krox20 cis-regulatory elements and their chromatin modifications and conformation, we first performed ChIP-seq experiments [26] to analyse two histone modifications: H3K4me1 (broad peaks covering active enhancers) and H3K27ac (punctuated peaks covering active enhancers and promoters) [27]. In E8.5 wild type embryo heads, a number of H3K4me1 peaks were observed, including those that expectedly correspond to the A, B and C elements and to a previously known neural crest element (NCE; Fig 4A) [28]. The signals observed for the H3K27ac mark were low across the Krox20 locus except for the promoter (S3A Fig). We can observe the same pattern of H3K27ac at the EphA4 locus with low signal at the enhancer driving its expression in r3 and r5 [29] and higher signal at the promoter (S3B Fig). In contrast, a gene widely expressed at E8.5 in the whole embryo head, like Sox2 [30], displays a high H3K27ac enrichment (S3C Fig). The low signals observed for the Krox20 and EphA4 genes are most likely due to the limited number of Krox20-expressing cells in the sample.
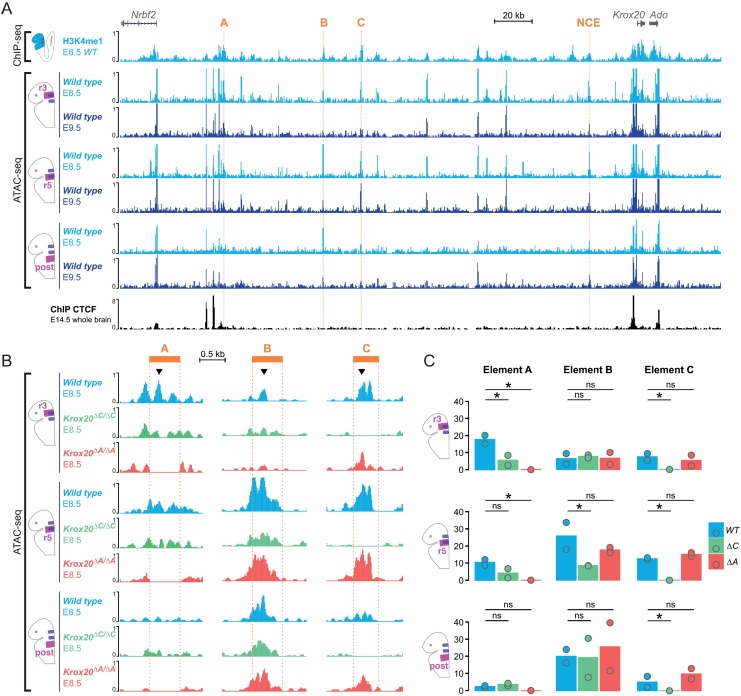
Fig 4. Chromatin modifications and accessibility within the Krox20 locus.
(A) ChIP-seq was performed for the H3K4me1 mark on wild type E8.5 embryo heads (light blue) using biological duplicates. Only one set of data is shown. ATAC-seq was performed on dissected regions (r3, r5 and a more posterior region (“post”; see text) from wild type embryos at E8.5 (light blue) and E9.5 (dark blue) using biological duplicates. Only one set of data is shown. CTCF ChIP-seq data from E14.5 mouse brain (ENCODE) are indicated below (see Fig 3A). Genes, cis-regulatory elements (orange) and a genomic scale are indicated at the top. (B) ATAC-seq was performed on dissected parts from wild type (light blue), and Krox20ΔC/ΔC (green) and Krox20ΔA/ΔA (red) E8.5 embryos using biological duplicates. Only one set of data is shown. Cis-regulatory elements (orange) and a genomic scale are indicated at the top. Arrowheads indicate the summits (defined by macs2 after peak calling, see Material and methods) used for quantifications in (C). (C) Barplots showing signal intensity of ATAC-seq (normalized fragment counts) at the summit of each element (arrowheads in panel B) for wild type (WT, blue) and Krox20ΔC/ΔC (ΔC, green) embryos at E8.5 for each dissected part. The statistical significance was calculated using a negative binomial Wald Test (R package DESeq2) on the 2 replicates, which are represented by dots. Star indicates p-value < 0.05. ns: non-significant.
To overcome this limitation, we performed micro-dissections and assessed chromatin structure by ATAC-seq [31], a technique that requires much lower cell numbers (a few thousand). Enhancer activity is usually associated with increased local chromatin accessibility. E8.5 (8s-10s stage) or E9.5 embryos were dissected to isolate samples derived from three regions: an anterior region, obtained by cutting within r2 and r4 (r3 sample); an intermediate region, for which cutting was performed within r4 and r6 (r5 sample), and a posterior region, for which cutting was performed within r6 and r7-8 (posterior sample). We observed peaks of accessibility at the level of the promoter at both stages and in all of the regional samples (Fig 4A). At both stages, element A was specifically accessible in the r3 and r5 samples, but not in the posterior sample (Fig 4A and 4B), in accordance with its activity restricted to r3 and r5 [13]. At E8.5, element B was compacted in the r3 sample, but highly accessible in the r5 and posterior samples (Fig 4A and 4B). This accessibility largely decreased at E9.5 (Fig 4A). The limited accessibility of element B in r3 is in agreement with its lack of activity in this rhombomere [13]. Finally, element C was particularly accessible in the r3 and r5 samples at E8.5 (Fig 4A and 4B), consistent with its activity that spans the r3-r5 region [13]. This accessibility was only maintained in the r5 sample at E9.5 (Fig 4A).
The pattern of chromatin accessibility observed in our ATAC-seq experiments revealed additional potential enhancers involved in the regulation of the Krox20 gene in the hindbrain. Indeed, we have identified an element located 107 kb downstream to Krox20 with high accessibility at E8.5 (S3A Fig). We have tested the transcriptional activity of this new element (NE) by transgenesis in the zebrafish by cloning it upstream of a GFP reporter gene. In a transgenic line, this element drives specific expression in r3 at the time of the initiation of Krox20 expression in this rhombomere (S4 Fig). These data raise the possibility that element NE might be the missing element involved in the initiation of Krox20 expression in r3, although its activity still needs to be verified in the mouse.
In conclusion, this analysis reveals that the patterns of accessibility of the different known elements largely correlate with their enhancer activities and helped us to identify a novel candidate element for the regulation of Krox20 expression in r3.
Element C modulates the accessibility of enhancer A
A final step was to investigate the effects of enhancer deletions on the accessibility of the other elements. Deletion of element A did not significantly affect the accessibility of elements B or C in any samples (Fig 4B and 4C and S3A Fig). In contrast, deletion of element C significantly reduced the accessibilities of element A in r3 and of element B in r5 (Fig 4B and 4C and S3A Fig). These data establish that element C has the capacity to specifically modulate the accessibility of elements A and B and therefore probably their activities. They may provide a mechanism for the involvement of the late activity of element C in the control of Krox20 autoregulation governed by element A. Furthermore, this analysis establishes the existence of an asymmetry in the relationship between elements A and C: whereas element C affects A accessibility and presumably potentiate its activity, the reverse is not true.
Discussion
In the present study, we have made a further step in the understanding of the molecular mechanisms governing the expression of a master developmental regulator, using both enhancer knock-outs and investigations of chromatin structure. This analysis reveals that Krox20 regulation relies on a complex crosstalk between several cis-acting elements that interact simultaneously according to multiple modes (redundant/additive/synergistic, symmetric/asymmetric) to shape the pattern of expression of the gene. Among these enhancers, element C performs a dual function, as a classical enhancer and as a potentiator in cis of element A. We propose that this latter role may constitute a general means to prevent promiscuous activities of autoregulatory elements.
Control of the early phase of Krox20 expression in r3
Previous analyses had suggested a rather straightforward mode of regulation of the Krox20 gene. Element C was responsible for the initiation of its expression in r3, whereas element B, possibly together with element C, was in charge of the initiation in r5. Subsequently, element A governed the maintenance of the expression through a positive feedback loop towards a definitive engagement into an odd-numbered rhombomere fate [13,18,19]. Knock-out analysis of element C now leads to major revisions of this scenario. Despite the early r3 enhancer activity of element C, as demonstrated by transgenic experiments in mouse and fish [13,32], the deletion of mouse element C does not affect early Krox20 expression in r3 (Fig 1B). This suggests that another cis-acting element contributes to this expression. This is not element B, which is only active in r5, as revealed in transgenic experiments [13], nor element A, which is absolutely dependent on the presence of the KROX20 protein [13,18]. Therefore, another enhancer, active in r3 and not dependent on KROX20, must be involved. Indeed, the identification of the NE element, fully accessible at early time in the hindbrain and specifically active in r3, makes it an attractive candidate for being involved in the initiation of Krox20 expression in this rhombomere (S3A Fig and S4 Fig).
The absence of phenotype in Krox20ΔC/ΔC embryos during the early phase of Krox20 expression does not preclude a role for element C during this phase. In support of this idea, enhancer activity of element C in r3 is dependent on the binding of Meis and Hox/Pbx factors [21], as well as of the Sp5 factor mediating FGF signalling [19,20], factors that are precisely known to act upstream of Krox20 in r3 [33–39]. It is therefore possible that elements C and NE cooperate in a redundant manner (S4 Fig) and further analyses will be required to determine whether this is indeed the case. Several examples of redundancy have been reported for enhancers governing the expression of developmental genes [3,40–42]. Redundant enhancers, or shadow enhancers, often share the same regulatory logic, since their activities have to be, at least in part, concomitant [43]. The analysis of the characteristics of the NE enhancer should reveal whether it depends on the same TFs as element C. In a few cases of redundant cis-acting elements that have been investigated in detail so far, it has been shown that redundancy provides robustness to the system and that, in specific genetic or environmental conditions, phenotypes can be revealed in absence of one of the elements [44].
Dual function of element C
Our study also revealed an unexpected function of element C: it is required for autoregulation, which was thought to be only dependent on element A. Using genetic approaches, we showed that an interaction must occur in cis between the two elements and that it is permanently required during the autoregulatory phase. ATAC-seq experiments indicated that element C is likely to act by modulating the accessibility of element A. Therefore, simultaneous to its classical enhancer function, element C performs another type of activity, which we propose to name enhancer potentiator. Potentiator characteristics (asymmetrical interaction, permanent requirement, long-range) clearly distinguishes this function from classical enhancer cooperative activities (additive, synergistic) and possibly from other hierarchical logic modes of interactions [3,45].
At this point, it is not known whether this additional enhancer potentiator function of element C, which is functionally distinct from its classical enhancer activity, is dependent on enhancer activity. Several recent studies have shown that enhancers can be transcribed and that the products of this transcription can act locally in cis to promote the expression of the target gene [46]. It is possible that such a mechanism could be involved in the potentiator activity of element C. It is interesting to note that Nlz factors, which are likely to repress Krox20 expression by reducing the accessibility of the KROX20 protein to element A [18], are also involved in repressing element C [32]. It is therefore possible that Nlz factors only indirectly affect accessibility of KROX20 on element A, by modulating the potentiator activity of element C.
In r5, Krox20 is almost normally expressed in the absence of element C, suggesting that cooperation between elements A and C is not essential in this rhombomere to support element A activity. It is possible that element B, which is likely to constitute the major initiator element in r5 and physically interacts with element A (Fig 3B) performs a dual function similar to element C and potentiates the activity of element A in this rhombomere, in addition to its classical enhancer activity.
A security lock on autoregulatory elements
Analyses by Hi-C in embryonic stem cells [11] and by 4C-seq in various embryonic samples (this report) revealed the existence of a regulatory neighbourhood that contains interactions between the Krox20 promoter and element A, irrespective of the considered stages or regions of the embryo (Fig 3). This chromatin configuration might therefore constitute a permissive environment for the activation of the autoregulatory loop. Such a situation, in which an autoregulatory element might be only dependent on the presence of its cognate TF for its activity, would be rather dangerous for an organism, as any transcription of the TF gene, even illegitimate, could end up activating the feedback loop and lead to high-level and long-term expression of the gene. This danger would be increased by the stochastic nature of the activation of the autoregulatory loop [19]. Furthermore, developmental genes may possess several positive autoregulatory enhancers that have to function at specific stages or in different tissues. This is exemplified by the Krox20 gene, that has at least three of such elements and that are differentially active in r3/r5, the neural crest and developing bones [13,28]. Therefore, mechanisms must exist as well to prevent the inappropriate activation of these elements in the other embryonic tissues. Indeed, we have shown that ectopic expression of exogenous Krox20 in the entire zebrafish embryo only leads to activation of the autoregulatory loop in the r2-r6 region of the hindbrain [18].
The introduction of an enhancer potentiator within a positive feedback loop may constitute an efficient prevention (safety lock) against inappropriate activation of autoregulatory elements. According to our model (Fig 5A), in the ground state, the autoregulatory element (element A in the case of Krox20) is locked in a configuration that is not accessible to its cognate TF and therefore unable to activate transcription, despite its possible interaction with the promoter. This lock will be released when the potentiator function is provided by a second cis-acting element (element C). It is interesting to note that in transgenic constructs, element A is able to activate a promoter in the absence of element C in cis. This difference in behaviour might be explained by two types of reasons: in transgenic constructs, element A is very close to the promoter, in contrast to the endogenous enhancer, located far upstream to the promoter; the chromatin context of a transgene is likely to be different, possibly more permissive than that of a highly regulated endogenous locus.
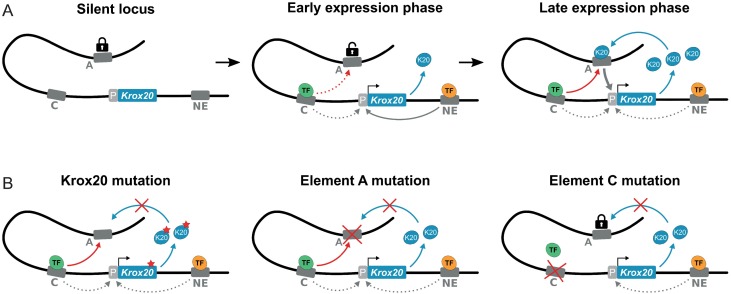
Fig 5. A model for Krox20 regulation and the dual function of element C.
(A) Schematic representation of the regulation of Krox20 in r3. Three situations are envisaged in wild type embryos. Left: silent locus. If both element C and the new enhancer (NE) are inactive, no expression occurs. Middle: early expression phase. At this stage, elements C and NE have been bound by their respective transcription factors and have initiated the expression of Krox20 via their classical enhancer functions. Nevertheless, element C has not yet been unlocked (decompacted) element A and/or the concentration of the KROX20 protein has not reached high enough levels to allow the establishment of a stable feedback loop with a significant probability. Right: late expression phase. Via its potentiator function, element C has unlocked element A, which can bind the KROX20 protein, which has now accumulated at a high enough concentration. Activation of enhancer A establishes the autoregulatory loop. (B) Three mutations that disrupt the positive feedback loop are presented at late expression phase. Left: mutation of the KROX20 protein preventing binding to element A. Middle: mutation of element A, preventing the binding of the KROX20 protein. Right: mutation of element C, preventing unlocking of element A.
In the endogenous locus, if the unlocker element is also responsible for the early accumulation of the cognate TF, through its classical enhancer activity, the autoregulatory element will be placed under the control of the upstream factors regulating the initial expression of the TF. In this way, the asymmetrical cooperation between the two cis-acting elements becomes essential for establishing the appropriate specificity of the positive feedback loop. As indicated in the model, such a feedback loop can be broken by mutation of either the TF or of any of the two cis-acting elements (Fig 5B).
Materials and methods
Ethics statement
All animal experiments were performed in accordance with the guidelines of the council of European Union directive n°2010/63/UE and were approved by the "Comité d'éthique pour l'expérimentation animale Charles Darwin" (Project Number: CE5/2012/120).
Mouse lines and situ hybridization
All mouse lines were maintained in a mixed C57BL6/DBA2 background. We used the following alleles: Krox20Cre [23] and Krox20ΔA [18]; the mouse Krox20Cflox line was generated at the Institut Clinique de la Souris (Illkirch, France) by homologous recombination in ES cells; the Krox20ΔC allele was obtained as described in Fig 1, using the maternally expressed PGK-Cre transgene as deletor [47]. In situ hybridizations were performed on whole embryos as previously described [48], with the following digoxigenin-labelled riboprobe: Krox20 [49] and EphA4 [50].
ChIP-seq
ChIP-seq experiments were performed as previously described [51]. Briefly, 10 embryos at E9.5 or 20 embryos at E8.5 were dissected in cold PBS. Cell suspensions were obtained by passing them through a 21G needle fitted onto a 5ml syringe. The cells were cross-linked with 1% formaldehyde for 10 min and washed twice in PBS, 1 mM PMSF, 1 X PIC (Protease Inhibitor Cocktail). Sonication was performed on a Covaris S220 using the following programme: duty factor = 10/5, peak incident power = 140, cycles per burst = 200 during 600/480 seconds. 5–10 μg of chromatin was used for each IP using 3 μg of the following antibodies: anti H3K4me1 (C15410037, Diagenode) and anti H3K27ac (ab4729, Abcam) in RIPA buffer. The libraries were prepared with the MicroPlex Library Preparation kit (Diagenode, E8.5 embryos) and with the NEXTflex ChIP-Seq Kit (Bioo Scientific, E9.5 embryos). ChIP Seq experiments involved biological duplicates.
Sequencing was performed on multiplexed samples using 50 bp single-end reads on an Illumina HiSeq system (E9.5 embryos) or using 42 bp paired-end reads on an Illumina NextSeq (E8.5 embryos) according to the manufacturer’s specifications. Chip-seq data were analysed as follows, using Eoulsan [52] with extended support for ChIP-seq workflows (https://github.com/GenomicParisCentre/eoulsan/tree/branch-chip-seq). First, reads were filtered out when they would not pass Illumina filters (module filterreads with option illuminaid). Files corresponding to technical replicates were merged (module technicalreplicatemerger, with option format = fastq), followed by trimming of the reads using Trim Galore! (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/; version 0.4.1 in module trimadapt, with cutadapt v1.8.1 and options: length = 41, quality = 20, error = 0.1, stringency = 8, and is.paired = yes for E8.5 data and is.paired = no for E9.5 data). Mapping was performed using STAR [53] (version 2.4.0k in module mapreads with mapper.arguments = “—outFilterMultimapNmax 1—outFilterMismatchNmax 999—outFilterMismatchNoverLmax 0.06—alignIntronMax 1—alignEndsType EndToEnd—alignMatesGapMax 2000—outSAMunmapped Within”). Further filters were applied to the mapped reads before conversion into BAM (module filtersam with removeunmapped = true; module sortsam; module rmdupgalax with is_sort = true; module sam2bam with compression.level = 5). BIGWIG files were created from the resulting BAM files using deepTools’ bamCoverage [54] (version 1.6.0, with options:—binSize 1—normalizeTo1x 200000000—fragmentLength 200—outFileFormat bigwig).
4C-seq
4C-seq libraries were constructed as previously described [55] with small adjustments. 35 E9.5 embryos (for each biological duplicate) or 250 E8.5 embryos were dissected in cold PBS, followed by dissociation in collagenase type I (Gibco). DpnII (New England Biolabs, Ipswich, MA) was used as the primary restriction enzyme and NlaIII (New England Biolabs) was used as the secondary restriction enzyme. For each viewpoint, up to 600 ng of each 4C-seq library were amplified using 16 individual PCR reactions with inverse primers including Illumina adapter sequences (S1 Table). Illumina sequencing was performed on multiplexed samples, containing PCR amplified material of up to 7 viewpoints, using 100 bp single-end reads on the Illumina HiSeq system according to the manufacturer's specifications at the iGE3 Genomics Platform of the University of Geneva (Switzerland). Reads were sorted, aligned, translated to restriction fragments and smoothed (11 fragments running mean) using the 4C-seq pipeline of the BBCF HTSstation [56] according to ENSEMBL Mouse assembly NCBIM37 (mm9). For the calculation of the 4C-seq signal distribution, reads were normalized to the entire chromosome 10, based on an approach adapted from [http://www.ncbi.nlm.nih.gov/pubmed/25959774]. For visualizations, smoothed 4C-seq reads were normalized to the 5 TADs surrounding the Krox20 locus (chr10:62,880,000–70,720,000).
Position of mouse TADs in ES cells and associated 40 kb normalized Hi-C matrices [11] were obtained from http://promoter.bx.psu.edu/hi-c/download.html. Directionality indexes were calculated as described using tools described previously [7]. Interaction matrices are visualized using standard cut-offs.
ATAC-seq
ATAC experiments were performed according to Buenrostros and colleagues [31], using a homemade transposome [57]. 7–8 embryos at 10-12s were dissected in cold PBS for each genotype and cells were mechanically dissociated. Biological duplicates were performed for ATAC experiments. Cells were lysed before transposition using 1 μl of transposome and purified using a Qiagen MinElute Kit with 10 μl of Elution Buffer. Transposed DNA was amplified by PCR as previously described [57] and quantified by qPCR using 5 μl of PCR products. The number of additional cycles was determined by plotting linear Rn versus cycle and corresponded to one third of the maximum fluorescence intensity. The remaining PCR products (45 μl) were treated with the additional number of cycles. The final product was purified with Qiagen PCR Cleanup Kit and eluted in 20 μl Elution Buffer. Sequencing was performed on multiplexed samples using 42 bp paired-end reads on an Illumina NextSeq according to the manufacturer’s specifications. For computational analysis, paired-end reads were mapped onto the mouse genome assembly mm9, using STAR (outFilterMultimapNmax 1; outFilterMismatchNmax 999; outFilterMismatchNoverLmax 0.06; alignIntronMax 1; alignEndsType EndToEnd; alignMatesGapMax 2000). Duplicates reads were removed using Picard (http://picard.sourceforge.net) (MarkDuplicates, REMOVE_DUPLICATES = true). To consider only fragments coming from transcription factors protected DNA (and not from nucleosomes), only fragment with size lower than 100 bp were kept. Bigwig tracks were obtained using DeepTools BamCoverage (1.5.9.1). Peak calling was performed using MACS2 (2.1.0.20140616), using a q-value< = 0.01 threshold (other parameters as default). For quantification, we first defined a set of non-redundant enriched regions for all samples by taking the union of all peak summits from both replicates of all samples, grouped together all summits distant from less than 50 bp, and for each group kept only the summit with the lowest q-value (calculated by MACS2). We then quantified the signal at all summits in each sample by counting the number of fragments (using the R bioconductor package csaw, v. 1.0.7). Normalisation and statistical analysis were performed using the bioconductor DESeq2 package (1.6.3). Library size factors were calculated on fragment counts in genomic bins of 10 kb. Comparison between wild type, Krox20ΔC/ΔC and Krox20ΔA/ΔA embryos was performed using negative binomial Wald Test (DESeq2).
Accession codes
The data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE94716 and is available at the following address: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE94716
Supporting information
(A) Sequence of the deleted element C and associated genomic coordinates (mouse genome assembly mm9). This sequence corresponds to the cloned sequence of the mouse element C described in [13]. (B) In situ hybridization for EphA4 mRNA performed on control (Krox20+/ΔC) and homozygous (Krox20ΔC/ΔC) mouse embryos at the indicated somite stages. Note the reduction of the size of r3 in the homozygous mutant. Rhombomeres positions are indicated on the left of each embryo.
(TIF)
(A) The left boundary of the Krox20 TAD, as previously determined from Hi-C in ES cells [11], spreads out over a transition zone with a low Directionality Index of Hi-C interactions in both ES cells and adult cortex ([11]). Hi-C in ES cells and TADs as defined in [11] or by our additional analysis (‘with CTCF’, this figure) are indicated above. Dashed lines in the graphs demarcate the boundaries between the -2, -1, Krox20 and +1 TADs. The transition zone between the -1 and Krox20 TADs is highlighted in grey. CTCF ChIP-seq data in E14.5 mouse brain (ENCODE, [58]) and genes are indicated below. Orange bars and gene names above pinpoint 4C-seq viewpoints. (B) Coordinates of the different regions used for the analysis of the distribution of 4C-seq signal. (C) Normalized distribution of 4C-seq signal for the Nrbf2, Element A and Krox20 viewpoints using previously determined TAD boundaries ([11], left) or after the repositioning of the TAD boundary to the cluster of CTCF binding sites between the Nrbf2 gene and Element A (right). The ratio of signal between the -1 and Krox20 TADs (average of 2 replicates) is shown above for each viewpoint. When the previously determined TAD boundary is used (left), the 4C-seq signal of the Nrbf2 viewpoint is almost equally distributed over the -1 and Krox20 TADs, whereas a much more discrete distribution is observed when the cluster of CTCF sites is used (right). In contrast, the nearby Element A viewpoint always restricts its strongest signal to the Krox 20 TAD, similar to the associated but much more distant Krox20 gene.
(TIF)
(A-C) ChIP-seq was performed for the H3K27ac mark on wild type E8.5 embryo heads (light blue) in duplicates and only one replicate is shown. ATAC-seq was performed on dissected regions (r3, r5 and a more posterior region (“post”; see text) from wild type (light blue), Krox20ΔC/ΔC (green) and Krox20ΔA/ΔA (red) E8.5 embryos. Genes, cis-regulatory elements (orange) and a genomic scale are indicated at the top. CTCF ChIP-seq data in E14.5 mouse brain (ENCODE) are indicated below each panel.
(TIF)
A zebrafish transgenic line Tg(NE:gfp), carrying a construct in which NE is linked to the gfp gene driven by a minimal promoter, was analysed by double ISH with krox20 (orange) and gfp (purple) probes at 3s and 10s stages.
(TIF)
(PDF)
(PDF)
Acknowledgments
We are grateful to Benoît Noël for help with the UCSC trackhubs, to Melody Matelot for help with the 4C-experiments and to Michel Wassef for critical comments on the manuscript and we thank the genomic platform of the IBENS. 4C-seq data analysis was performed at the Vital-IT (http://www.vital-it.ch) computing center of the Swiss Institute of Bioinformatics using tools developed by the BBCF (http://bbcf.epfl.ch).
Data Availability
The data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE94716.
Funding Statement
The P.C. laboratory was financed by the Institut National de la Recherche Médicale, the Centre National de la Recherche Scientifique, the Ministère de la Recherche et Technologie and the Fondation pour la Recherche Médicale (FRM). It has received support under the PIA launched by the French Government and implemented by the ANR, with the references: ANR-10-LABX-54 MEMOLIFE and ANR-11-IDEX-0001-02 PSL* Research University. The genomic platform of the IBENS is supported by France Génomique national infrastructure and funded as part of the Programme "Investissements d'Avenir" (PIA) managed by the Agence Nationale de la Recherche (ANR, contract ANR-10-INBS-0009). ET and JLM were supported by doctoral grants from Sorbonne Universités and by the FRM. PT is supported by doctoral grant from PSL Research University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27(2 Pt 1):299–308. [DOI] [PubMed] [Google Scholar]
- 2.Maniatis T, Goodbourn S, Fischer J a. Regulation of inducible and tissue-specific gene expression. Science. 1987. June 5;236(4806):1237–45. [DOI] [PubMed] [Google Scholar]
- 3.Long HK, Prescott SL, Wysocka J. Review Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell. 2016;167(5):1170–87. doi: 10.1016/j.cell.2016.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kvon EZ. Using transgenic reporter assays to functionally characterize enhancers in animals. Genomics. 2015;106(3):185–92. doi: 10.1016/j.ygeno.2015.06.007 [DOI] [PubMed] [Google Scholar]
- 5.Ruf S, Symmons O, Uslu VV, Dolle D, Hot C, Ettwiller L, et al. Large-scale analysis of the regulatory architecture of the mouse genome with a transposon-associated sensor. Nat Genet. 2011. April;43(4):379–86. doi: 10.1038/ng.790 [DOI] [PubMed] [Google Scholar]
- 6.Symmons O, Uslu VV, Tsujimura T, Ruf S, Nassari S, Schwarzer W, et al. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014;24(3):390–400. doi: 10.1101/gr.163519.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunipace L, Saunders A, Ashe H, Stathopoulos A. Autoregulatory feedback controls sequential action of cis-regulatory modules at the brinker locus. Dev Cell. 2013;26(5):536–43. doi: 10.1016/j.devcel.2013.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaret KS, Carroll JS. Pioneer transcription factors: Establishing competence for gene expression. Genes Dev. 2011;25(21):2227–41. doi: 10.1101/gad.176826.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013. March 7;49(5):773–82. doi: 10.1016/j.molcel.2013.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012. May 17;485(7398):381–5. doi: 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012. May 17;485(7398):376–80. doi: 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chavrier P, Zerial M, Lemaire P, Almendral J, Bravo R, Charnay P. A gene encoding a protein with zinc fingers is activated during G0/G1 transition in cultured cells. EMBO J. 1988;7(1):29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chomette D, Frain M, Cereghini S, Charnay P, Ghislain J. Krox20 hindbrain cis-regulatory landscape: interplay between multiple long-range initiation and autoregulatory elements. Development. 2006. April;133(7):1253–62. doi: 10.1242/dev.02289 [DOI] [PubMed] [Google Scholar]
- 14.Lumsden A, Krumlauf R. Patterning the vertebrate neuraxis. Science. 1996. November 15;274(5290):1109–15. [DOI] [PubMed] [Google Scholar]
- 15.Schneider-Maunoury S, Topilko P, Seitandou T, Levi G, Cohen-Tannoudji M, Pournin S, et al. Disruption of Krox-20 results in alteration of rhombomeres 3 and 5 in the developing hindbrain. Cell. 1993. December 17;75(6):1199–214. [DOI] [PubMed] [Google Scholar]
- 16.Schneider-Maunoury S, Seitanidou T, Charnay P, Lumsden a. Segmental and neuronal architecture of the hindbrain of Krox-20 mouse mutants. Development. 1997. March;124(6):1215–26. [DOI] [PubMed] [Google Scholar]
- 17.Voiculescu O, Taillebourg E, Pujades C, Kress C, Buart S, Charnay P, et al. Hindbrain patterning: Krox20 couples segmentation and specification of regional identity. Development. 2001. December;128(24):4967–78. [DOI] [PubMed] [Google Scholar]
- 18.Bouchoucha YX, Reingruber J, Labalette C, Wassef M a, Thierion E, Desmarquet-Trin Dinh C, et al. Dissection of a Krox20 positive feedback loop driving cell fate choices in hindbrain patterning. Mol Syst Biol. 2013. September 24;9(690). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labalette C, Bouchoucha YX, Wassef MA, Gongal PA, Le Men J, Becker T, et al. Hindbrain patterning requires fine-tuning of early krox20 transcription by Sprouty 4. Development. 2011. January;138(2):317–26. doi: 10.1242/dev.057299 [DOI] [PubMed] [Google Scholar]
- 20.Labalette C, Wassef MA, Desmarquet-Trin Dinh C, Bouchoucha YX, Le Men J, Charnay P, et al. Molecular dissection of segment formation in the developing hindbrain. Development. 2015. January 1;142(1):185–95. doi: 10.1242/dev.109652 [DOI] [PubMed] [Google Scholar]
- 21.Wassef M a, Chomette D, Pouilhe M, Stedman A, Havis E, Desmarquet-Trin Dinh C, et al. Rostral hindbrain patterning involves the direct activation of a Krox20 transcriptional enhancer by Hox/Pbx and Meis factors. Development. 2008. October;135(20):3369–78. doi: 10.1242/dev.023614 [DOI] [PubMed] [Google Scholar]
- 22.Seitanidou T, Schneider-Maunoury S, Desmarquet C, Wilkinson DG, Charnay P. Krox-20 is a key regulator of rhombomere-specific gene expression in the developing hindbrain. Mech Dev. 1997. July;65(1–2):31–42. [DOI] [PubMed] [Google Scholar]
- 23.Voiculescu O, Charnay P, Schneider-Maunoury S. Expression pattern of a Krox-20/Cre knock-in allele in the developing hindbrain, bones, and peripheral nervous system. Genesis. 2000. February;26(2):123–6. [DOI] [PubMed] [Google Scholar]
- 24.van de Werken HJG, Landan G, Holwerda SJB, Hoichman M, Klous P, Chachik R, et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat Methods. 2012;9(10):969–72. doi: 10.1038/nmeth.2173 [DOI] [PubMed] [Google Scholar]
- 25.Irving C, Nieto M a, DasGupta R, Charnay P, Wilkinson DG. Progressive spatial restriction of Sek-1 and Krox-20 gene expression during hindbrain segmentation. Dev Biol. 1996;173(1):26–38. doi: 10.1006/dbio.1996.0004 [DOI] [PubMed] [Google Scholar]
- 26.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell. 2007;129(4):823–37. doi: 10.1016/j.cell.2007.05.009 [DOI] [PubMed] [Google Scholar]
- 27.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011. January;12(1):7–18. doi: 10.1038/nrg2905 [DOI] [PubMed] [Google Scholar]
- 28.Ghislain J, Desmarquet-Trin-Dinh C, Gilardi-Hebenstreit P, Charnay P, Frain M. Neural crest patterning: autoregulatory and crest-specific elements co-operate for Krox20 transcriptional control. Development. 2003;130(5):941–53. [DOI] [PubMed] [Google Scholar]
- 29.Theil T, Frain M, Gilardi-Hebenstreit P, Flenniken A, Charnay P, Wilkinson DG. Segmental expression of the EphA4 (Sek-1) receptor tyrosine kinase in the hindbrain is under direct transcriptional control of Krox-20. Development. 1998. February;125(3):443–52. [DOI] [PubMed] [Google Scholar]
- 30.Wood HB, Episkopou V. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech Dev. 1999;86(1–2):197–201. [DOI] [PubMed] [Google Scholar]
- 31.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213–8. doi: 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labalette C, Wassef MA, Desmarquet-Trin Dinh C, Bouchoucha YX, Le Men J, Charnay P, et al. Molecular dissection of segment formation in the developing hindbrain. Development. 2015;142(1):185–95. doi: 10.1242/dev.109652 [DOI] [PubMed] [Google Scholar]
- 33.Wiellette EL, Sive H. vhnf1 and Fgf signals synergize to specify rhombomere identity in the zebrafish hindbrain. Development. 2003;130(16):3821–9. [DOI] [PubMed] [Google Scholar]
- 34.Cordes SP, Barsh GS. The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell. 1994. December 16;79(6):1025–34. [DOI] [PubMed] [Google Scholar]
- 35.Waskiewicz AJ, Rikhof H a, Moens CB. Reveals a Hindbrain Ground State. Dev Cell. 2002;3:723–33. [DOI] [PubMed] [Google Scholar]
- 36.Choe S-K, Vlachakis N, Sagerström CG. Meis family proteins are required for hindbrain development in the zebrafish. Development. 2002;129(3):585–95. [DOI] [PubMed] [Google Scholar]
- 37.McNulty CL, Peres JN, Bardine N, van den Akker WMR, Durston AJ. Knockdown of the complete Hox paralogous group 1 leads to dramatic hindbrain and neural crest defects. Development. 2005;132(12):2861–71. doi: 10.1242/dev.01872 [DOI] [PubMed] [Google Scholar]
- 38.Walshe J, Maroon H, McGonnell IM, Dickson C, Mason I. Establishment of hindbrain segmental identity requires signaling by FGF3 and FGF8. Curr Biol. 2002;12(13):1117–23. [DOI] [PubMed] [Google Scholar]
- 39.Aragon F, Pujades C. FGF signaling controls caudal hindbrain specification through Ras-ERK1/2 pathway. BMC Dev Biol. 2009;9:61 doi: 10.1186/1471-213X-9-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong J-W, Hendrix D a, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321(5894):1314 doi: 10.1126/science.1160631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurokawa D, Kiyonari H, Nakayama R, Kimura-Yoshida C, Matsuo I, Aizawa S. Regulation of Otx2 expression and its functions in mouse forebrain and midbrain. Development. 2004. July;131(14):3319–31. doi: 10.1242/dev.01220 [DOI] [PubMed] [Google Scholar]
- 42.Lam DD, de Souza FSJ, Nasif S, Yamashita M, López-Leal R, Otero-Corchon V, et al. Partially Redundant Enhancers Cooperatively Maintain Mammalian Pomc Expression Above a Critical Functional Threshold. PLOS Genet. 2015;11(2):e1004935 doi: 10.1371/journal.pgen.1004935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lagha M, Bothma JP, Levine M. Mechanisms of transcriptional precision in animal development. Trends Genet. 2012. August;28(8):409–16. doi: 10.1016/j.tig.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ludwig MZ, Manu, Kittler R, White KP, Kreitman M. Consequences of eukaryotic enhancer architecture for gene expression dynamics, development, and fitness. PLoS Genet. 2011;7(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leddin M, Perrod C, Hoogenkamp M, Ghani S, Assi S, Heinz S, et al. Two distinct auto-regulatory loops operate at the PU.1 locus in B cells and myeloid cells. Blood. 2011;117(10):2827–38. doi: 10.1182/blood-2010-08-302976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016;17(4):207–23. doi: 10.1038/nrg.2016.4 [DOI] [PubMed] [Google Scholar]
- 47.Lallemand Y, Luria V, Haffner-Krausz R, Lonai P. Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site-specific recombinase. Transgenic Res. 1998;7(2):105–12. [DOI] [PubMed] [Google Scholar]
- 48.Giudicelli F, Taillebourg E, Charnay P, Gilardi-Hebenstreit P. Krox-20 patterns the hindbrain through both cell-autonomous and non cell-autonomous mechanisms. Genes Dev. 2001. March 1;15(5):567–80. doi: 10.1101/gad.189801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilkinson DG, Bhatt S, Chavrier P, Bravo R, Charnay P. Segment-specific expression of a zinc-finger gene in the developing nervous system of the mouse. Nature. 1989. February 2;337(6206):461–4. doi: 10.1038/337461a0 [DOI] [PubMed] [Google Scholar]
- 50.Gilardi-Hebenstreit P, Nieto MA, Frain M, Mattéi MG, Chestier A, Wilkinson DG, et al. An Eph-related receptor protein tyrosine kinase gene segmentally expressed in the developing mouse hindbrain. Oncogene. 1992. December;7(12):2499–506. [PubMed] [Google Scholar]
- 51.Vitobello A, Ferretti E, Lampe X, Vilain N, Ducret S, Ori M, et al. Hox and Pbx Factors Control Retinoic Acid Synthesis during Hindbrain Segmentation. Dev Cell. 2011;20(4):469–82. doi: 10.1016/j.devcel.2011.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jourdren L, Bernard M, Dillies MA, Le Crom S. Eoulsan: A cloud computing-based framework facilitating high throughput sequencing analyses. Bioinformatics. 2012;28(11):1542–3. doi: 10.1093/bioinformatics/bts165 [DOI] [PubMed] [Google Scholar]
- 53.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. DeepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42(W1):187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matelot M, Noordermeer D. Determination of High-Resolution 3D Chromatin Organization Using Circular Chromosome Conformation Capture (4C-seq). 2016;223–41. [DOI] [PubMed]
- 56.David FPA, Delafontaine J, Carat S, Ross FJ, Lefebvre G, Jarosz Y, et al. HTSstation: A web application and open-access libraries for high-throughput sequencing data analysis. PLoS One. 2014;9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Picelli S, Björklund AK, Reinius B, Sagasser S, Winberg G, Sandberg R. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 2014;24(12):2033–40. doi: 10.1101/gr.177881.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488(7409):116–20. doi: 10.1038/nature11243 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Sequence of the deleted element C and associated genomic coordinates (mouse genome assembly mm9). This sequence corresponds to the cloned sequence of the mouse element C described in [13]. (B) In situ hybridization for EphA4 mRNA performed on control (Krox20+/ΔC) and homozygous (Krox20ΔC/ΔC) mouse embryos at the indicated somite stages. Note the reduction of the size of r3 in the homozygous mutant. Rhombomeres positions are indicated on the left of each embryo.
(TIF)
(A) The left boundary of the Krox20 TAD, as previously determined from Hi-C in ES cells [11], spreads out over a transition zone with a low Directionality Index of Hi-C interactions in both ES cells and adult cortex ([11]). Hi-C in ES cells and TADs as defined in [11] or by our additional analysis (‘with CTCF’, this figure) are indicated above. Dashed lines in the graphs demarcate the boundaries between the -2, -1, Krox20 and +1 TADs. The transition zone between the -1 and Krox20 TADs is highlighted in grey. CTCF ChIP-seq data in E14.5 mouse brain (ENCODE, [58]) and genes are indicated below. Orange bars and gene names above pinpoint 4C-seq viewpoints. (B) Coordinates of the different regions used for the analysis of the distribution of 4C-seq signal. (C) Normalized distribution of 4C-seq signal for the Nrbf2, Element A and Krox20 viewpoints using previously determined TAD boundaries ([11], left) or after the repositioning of the TAD boundary to the cluster of CTCF binding sites between the Nrbf2 gene and Element A (right). The ratio of signal between the -1 and Krox20 TADs (average of 2 replicates) is shown above for each viewpoint. When the previously determined TAD boundary is used (left), the 4C-seq signal of the Nrbf2 viewpoint is almost equally distributed over the -1 and Krox20 TADs, whereas a much more discrete distribution is observed when the cluster of CTCF sites is used (right). In contrast, the nearby Element A viewpoint always restricts its strongest signal to the Krox 20 TAD, similar to the associated but much more distant Krox20 gene.
(TIF)
(A-C) ChIP-seq was performed for the H3K27ac mark on wild type E8.5 embryo heads (light blue) in duplicates and only one replicate is shown. ATAC-seq was performed on dissected regions (r3, r5 and a more posterior region (“post”; see text) from wild type (light blue), Krox20ΔC/ΔC (green) and Krox20ΔA/ΔA (red) E8.5 embryos. Genes, cis-regulatory elements (orange) and a genomic scale are indicated at the top. CTCF ChIP-seq data in E14.5 mouse brain (ENCODE) are indicated below each panel.
(TIF)
A zebrafish transgenic line Tg(NE:gfp), carrying a construct in which NE is linked to the gfp gene driven by a minimal promoter, was analysed by double ISH with krox20 (orange) and gfp (purple) probes at 3s and 10s stages.
(TIF)
(PDF)
(PDF)
Data Availability Statement
The data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE94716.