Abstract
Aim:
Assess patient-level utility of suggested pretreatment biomarkers of sunitinib in advanced renal cell carcinoma.
Patients & methods:
Kaplan–Meier analysis of data from a randomized, Phase II study (n = 292) suggested baseline predictive value for circulating soluble Ang-2 and MMP-2 and HIF-1α percentage of tumor expression. Using this dataset, the sensitivity, specificity and area under the curve (AUC) were calculated, using receiver operating characteristic (ROC) curves.
Results:
Based on a ROC (sensitivity vs 1 - specificity) threshold AUC value of >0.8, neither Ang-2 (0.67) nor MMP-2 (0.65), nor HIF-1α percentage of tumor expression (0.65), performed appropriately from a patient-selection standpoint.
Conclusion:
To properly assess potential biomarkers, sensitivity and specificity characteristics should be obtained by ROC analysis.
KEYWORDS : biomarkers, renal cell carcinoma, ROC curves, serum soluble protein, sunitinib
Intensive investigations are ongoing to identify and validate predictive molecular markers for antiangiogenic agents for solid tumors, including advanced renal cell carcinoma (RCC). These efforts typically generate lists of potential markers in small sample sets that are suggested as worthy candidates for further research, based on p-values < 0.05 from comparison of survival curves using the log rank test (Kaplan–Meier method) or the likelihood ratio test (Cox proportional hazards model). In subsequent studies, many of these candidate biomarkers are not validated, or, at best, the p-value in the larger dataset is reduced and too often the perspective of clinical utility is either not examined or not properly addressed.
There are multiple examples of promising biomarkers associated with a p-value of 0.05 or less generated by Kaplan–Meier analysis from studies involving the use of sunitinib in the context of advanced or metastatic RCC therapy [1–3]. For example, the following biomarkers were identified as part of the sunitinib Renal EFFECT Trial [4], and have been previously presented: circulating Ang-2 and MMP-2, identified via two independent multiplex platforms, and HIF-1α percentage of tumor expression, as assessed by immunohistochemistry (IHC) (details in Table 1) [3].
Table 1. . Biomarkers associated with favorable outcome for sunitinib-treated advanced renal cell carcinoma patients in the Renal EFFECT trial.
| Biomarker associated with favorable outcome | p-value |
|---|---|
| Lower serum Ang-2† |
0.0197‡ |
| Higher serum MMP-2† |
0.0327‡ |
| Lower tumor HIF-1α§ | 0.0341 |
There were no statistically significant associations with efficacy for germline VEGF-A or VEGFR3 single nucleotide polymorphisms assayed or VHL mutation alone, deletion alone, methylation alone or aberrancies combined.
†Serum soluble proteins identified by both SOMAscan and SearchLight platforms using Response Evaluation Criteria in Solid Tumors-defined response.
‡SOMAscan platform.
§Assessed by immunohistochemistry.
Data taken with permission from [3].
It is proposed that the methodological approach for identifying ‘promising’ patient selection biomarkers should be reconsidered for targeted therapies, in particular for antiangiogenic agents. Rather than rely upon Kaplan–Meier-derived statistical significance (p-values < 0.05), a more stringent performance assessment that incorporates sensitivity and specificity characteristics should be considered. To test this hypothesis, sensitivity and specificity characteristics of the three statistically significant biomarkers reported in the biomarker analysis from the Renal EFFECT Trial (circulating Ang-2 and MMP-2, and tumor HIF-1α) were evaluated with a receiver operating characteristics (ROC) model in order to determine whether these biomarkers have any clinical utility as potential patient selection markers. In general, the development of any clinical grade laboratory test [5] requires such an approach. Note that for the continuous data set of a range of tumor protein expression data or soluble protein concentrations assessed in this study, an ROC approach is appropriate. For other biomarker data type where the outputs are dichotomous (yes or no), assessment of clinical utility would be simpler with outputs of only sensitivity, specificity, as well as positive and negative predictive value.
Patients & methods
• Study design & treatment
Data for this analysis were obtained from a multicenter, randomized Phase II study (n = 292) in which adult patients with histologically confirmed advanced RCC were randomly assigned 1:1 to receive either sunitinib 50 mg/day on Schedule 4/2 (4 weeks on treatment and 2 weeks off; n = 146) or 37.5 mg/day on a continuous daily dosing (CDD) schedule (n = 146) as first-line therapy [4]. Randomization was stratified by risk factors based on published Memorial Sloan–Kettering Cancer Center (MSKCC) data [6]. Patients continued treatment up to 2 years or until disease progression, significant toxicity, or consent withdrawal. The primary end point was comparison of time to tumor progression between patients on the two schedules.
The study was run in accordance with the International Conference on Harmonization Good Clinical Practice guidelines and applicable local regulatory requirements and laws, and was approved by the institutional review board or independent ethics committee of each center (ClinicalTrials.gov: NCT00267748). All patients gave written informed consent.
• ROC analysis
An ROC (curve) analysis is an approach that assesses both the sensitivity and specificity performance of a biomarker test signal according to the percentage of true positives identified by the test (sensitivity) and percentage of true negatives identified by the test (specificity). The ROC approach can help to identify optimal ‘cut-points’ for a given analyte for prospective follow-up for diagnostic use [7]. A test with no false positives and no false negatives would have a perfect performance.
Sensitivity (also called the true positive rate) measures the proportion of actual positives which are correctly identified as such (e.g., the percentage of cancer patients with progressive disease who are correctly identified as having progressive disease). Specificity measures the proportion of negatives which are correctly identified as such (e.g., the percentage of cancer patients who are correctly identified as not having progressive disease, sometimes called the true negative rate). In an ROC curve, the true positive rate (sensitivity) is plotted as a function of the false-positive rate (1 - specificity) for different cut-off points of a parameter, such as numeric assessments of proteins in tumor tissue or blood from cancer patients. Each point on the ROC curve represents a sensitivity/specificity pair corresponding to a particular decision threshold. ROC analysis generates an area under the curve (AUC), which is a measure of how well a parameter can distinguish between two diagnostic groups (those with progressive disease/those without progressive disease), where a value of 1 shows perfect performance (sensitivity rises from 0 to 1.0 at 1 - specificity of 0, and specificity rises from 0 to 1.0 at sensitivity of 1.0 on a ROC plot), and where an AUC of 0.5 has the performance equivalent of a coin toss (represented by a 45º line on a ROC plot). Although extensive literature is absent on acceptable cut-offs for companion diagnostics for patient use, an AUC value <0.8 is viewed by the authors not to have predictive value. However, around or above 0.8, there is no literature agreement as to what an acceptable value would be for patient selection utility and must be put into context of diagnostic [5,7] or companion diagnostic use [8]. Therefore, the authors acknowledge that refinement of the AUC value from the 0.8 value is probable following validation of patient selection markers for RCC patients receiving targeted therapies.
ROC curves with censored data have an additional complication. If one waits until the end of the study, then every subject has a binary outcome, and one ROC curve and AUC is produced. However, if one wishes to evaluate the outcome at another (earlier) time point, a time point is selected and subjects are classified as having the event or are censored. The choice of time point is important, as it can influence the conclusions. When a specific time point is not clear, one may choose several time points, present the results, and select the curve with the largest AUC [9].
Using the previously derived biomarker datasets from the randomized, Phase II sunitinib study (Renal EFFECT Trial) [3], circulating Ang-2 and MMP-2, and HIF-1α percent of tumor expression were assessed for sensitivity and specificity characteristics using the ROC method. The serum soluble protein dataset comprised 74 baseline biospecimens (and 26 end-of-treatment/withdrawal biospecimens with associated paired baseline values) randomly selected from patients on schedule 4/2 only (the approved regimen for advanced RCC). Ang-2 and MMP-2 were identified by two distinct analysis platforms from a common list of 37 analytes. The IHC dataset comprised tumor biospecimens from 149 evaluable patients in both schedules combined. Clinical outcomes in patients comprising these biospecimen sets were representative of those from the entire study.
Results
In the previously reported biomarker analysis for the Renal EFFECT Trial [3], which utilized a Kaplan–Meier analysis or Wilcoxon rank-sum test, three biomarkers emerged with p-values less than 0.05: baseline concentrations of circulating Ang-2 and MMP-2, and HIF-1α percent of tumor expression. Statistical analysis for the circulating soluble proteins in patients in the Schedule 4/2 arm was conducted using Wilcoxon rank-sum test to compare serum concentrations in responders versus nonresponders, as defined by Response Evaluation Criteria in Solid Tumors (RECIST), version 1.0 [10]. Lower baseline Ang-2 and higher baseline MMP-2 levels were identified by both platforms as significantly associated with RECIST-defined tumor response.
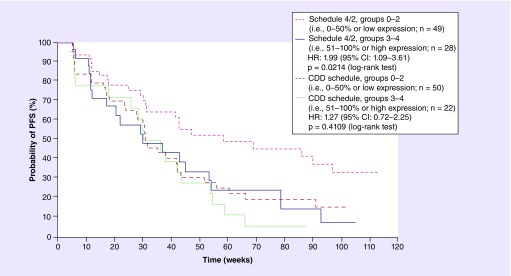
Statistical analysis for HIF-1α percent of tumor expression was conducted using Kaplan–Meier analysis. Figure 1, obtained from the original analysis, shows a Kaplan–Meier analysis of progression-free survival (PFS) after stratification into groups based on levels of HIF-1α percentage of tumor expression. In the schedule 4/2 arm, which is the approved dosing schedule in RCC and thus directly relates to assessment of biomarker clinical utility, improved PFS was observed for patients with less than 50% expression of tumor HIF-1α compared with greater than 50% expression of tumor HIF-1α. This improvement was a statistically significant observation based upon a hazard ratio of 1.99 and a p-value of 0.02. There was no statistically significant association in the CDD arm.
Figure 1. . Kaplan–Meier curves of progression-free survival after stratification by level of HIF-1α percentage of tumor expression according to sunitinib dosing schedule.
HIF-1α percentage of tumor expression (individual variable score) was stratified in groups (0, 1, 2, 3, 4) and the cut-off point was set at 3. Groups 0–2 correspond to 0–50% (i.e., low HIF-1α expression) and groups 3–4 correspond to 51–100% (i.e., high HIF-1α expression). An HR greater than 1 indicates a reduction in hazard rate in favor of groups 0–2, whereas an HR less than 1 indicates a reduction in hazard rate in favor of groups 3–4.
CDD: Continuous daily dosing; HR: Hazard ratio; PFS: Progression-free survival; Schedule 4/2: 4 weeks on treatment and 2 weeks off treatment.
• ROC analyses
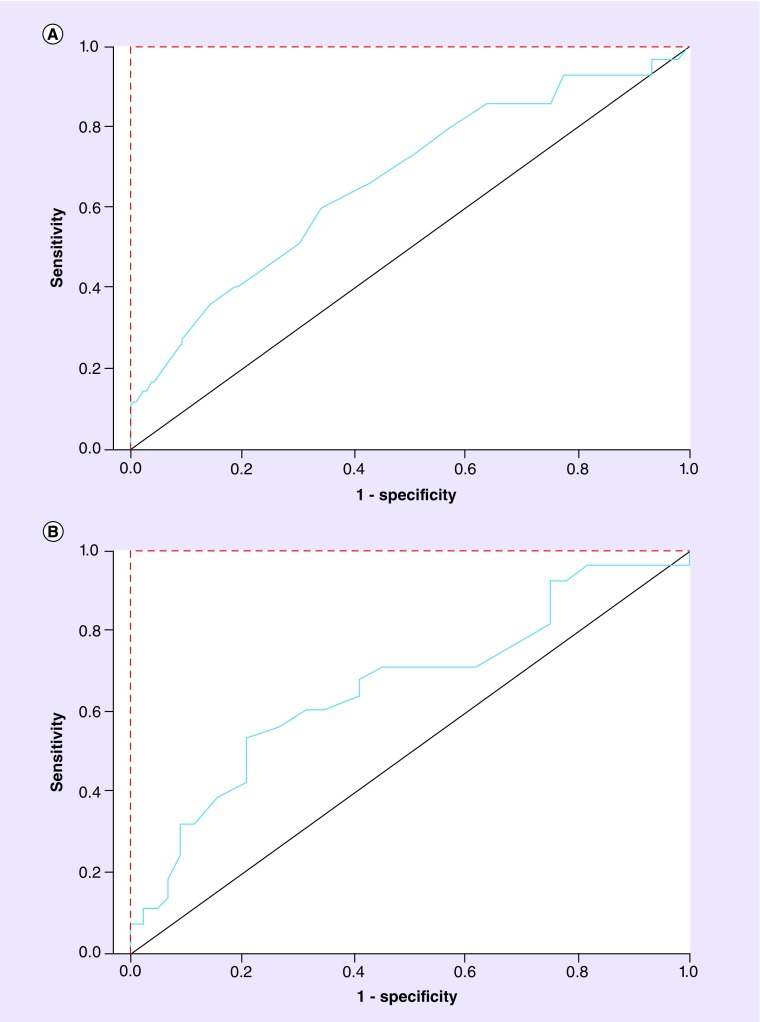
As shown in Figure 2, the blue plot lines, representing the sensitivity and specificity performances of Ang-2 (Figure 2A) and MMP-2 (Figure 2B) to predict RECIST-defined tumor response, are closer to the black diagonal lines, the equivalent of a series of coin tosses, than to the theoretical perfect performance. Although the software macro generating the output shown in Figure 2 produce a smoothened line, a tabular output is also generated from this continuous data set, which could conceivably provide sensitivity and specificity values at any point along the blue line, corresponding to a specific circulating concentration value. In both cases, the AUC for the circulating soluble proteins was less than 0.7.
Figure 2. . Receiver operating characteristics curves of (A) plasma Ang-2 concentrations (area under the curve = 0.67) and (B) plasma MMP-2 concentrations (area under the curve = 0.65) in RECIST-defined responders versus non-responders.
Red dotted line: theoretical perfect performance of an efficacy biomarker. Black solid line: theoretical performance of an efficacy biomarker equivalent to a coin toss. Blue line: actual performance of the results.
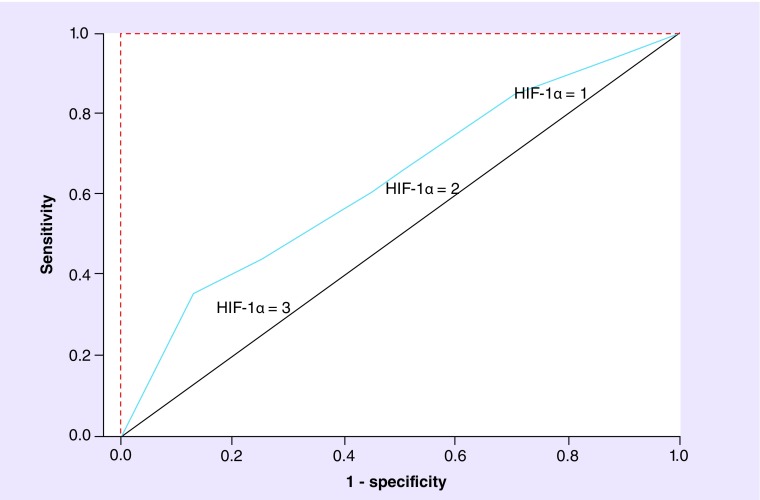
The results of the performance analysis of HIF-1α percentage of tumor expression to detect PFS at 38 weeks, the median PFS for both arms combined, are shown in Figure 3. The blue plot line, representing the actual sensitivity and specificity results across various prespecified cut-points (25, 50 and 75%), is again closer to the black diagonal line, equivalent to the performance of a coin toss, rather than the shape of the theoretical perfect performance. In addition, the AUC was 0.65. Note that the prespecified cut-points for analysis of HIF-1α tumor expression were analyzed by the CLIA-certified commercial laboratory conducting the IHC analysis, before the availability of any clinical efficacy data.
Figure 3. . Receiver operating characteristics curve of HIF-1α percentage of tumor expression (area under the curve = 0.65) in patients with versus without RECIST-defined progression-free survival at 38 weeks after initial sunitinib dose.
HIF-1α cut-points = 1, 2 and 3 represent percent of tumor expression = 25, 50 and 75%, respectively. Red dotted line: theoretical perfect performance of an efficacy biomarker. Black solid line: theoretical performance of an efficacy biomarker equivalent to a coin toss. Blue line: actual performance of the HIF-1α percentage of tumor expression result at the prespecified cut-points.
The tabular output generated by the software macro for the ROC plot of HIF-1α percentage of tumor expression in Figure 3 is shown in Table 2. As with the figure, the HIF-1α cut-points represent percent expression values; therefore, 25, 50 and 75% are reported with the corresponding sensitivities and specificities. Where the sensitivity identified by the test (percentage of true positives) is high, the specificity (percentage of true negatives) is low, and vice versa. At the 50% cut-point, used to classify observations into greater than or less than or equal to the median values in Figure 3, sensitivity was 45% and specificity was 75% (Table 2). This would clearly not be useful to select patients as only 45% of the patients benefiting from sunitinib treatment would be identified. Furthermore, only 75% of the patients who did not derive clinical benefit from sunitinib at 38 weeks would have been excluded from treatment.
Table 2. . Tumor immunohistochemistry results: predicting progression-free survival at 38 weeks (median progression-free survival) from baseline HIF-1α.
| HIF-1α cut-point (% stained cells)† | Sensitivity | Specificity | 1 - specificity |
|---|---|---|---|
| 0 |
0.842 |
0.305 |
0.695 |
| 25 |
0.614 |
0.551 |
0.449 |
| 50 |
0.447 |
0.746 |
0.254 |
| 75 |
0.350 |
0.876 |
0.124 |
| 100 | 0 | 1.00 | 0 |
†The 25, 50 and 75% stain cells (percentage of tumor expression) represent the HIF-1α cut-points = 1, 2 and 3 in Figure 3, respectively. Data were reported as 0, 1, 2, 3, 4.
The data in Table 2 provide a more detailed output than the plot in Figure 3, indicating low sensitivity and specificity characteristics across the prespecified cut-points, which is consistent with the low AUC of 0.65. Ultimately, the performance of this biomarker is therefore not promising for the individual patient, when assessed using the ROC methodology.
Discussion
In summary, based on data from the Renal EFFECT Trial, HIF-1α percentage of tumor expression, and baseline circulating soluble Ang-2 and MMP-2, were initially identified for future research as promising patient selection biomarkers for sunitinib [3]. However, based on a threshold AUC of 0.8 by ROC analysis, neither HIF-1α percent of tumor expression (AUC = 0.65) nor baseline circulating Ang-2 (0.67) and MMP-2 (0.65) have true potential as patient selection markers.
Additional ad-hoc ROC analyses of other examples of potential biomarkers were conducted for further assessment of previously generated data. The circulating soluble protein VEGFR-3 and VEGF-C data from a previous publication [1] showing ‘promising’ statistically significant associations with efficacy (p < 0.01) also generated AUC values (0.44 and 0.75, respectively) below the minimal 0.8 threshold (data not shown).
Taken together, these results are consistent with our assertion that p-values < 0.05 that separate Kaplan–Meier curves do not necessarily translate into sensitivity and specificity characteristics needed for a clinically useful patient selection diagnostic. It is commonly observed by the authors that traditional group-versus-group comparison approaches are most commonly driven by small numbers of subjects that influence the overall group performance. Therefore, Kaplan–Meier analyses and other similar approaches using arbitrarily predefined p-values of 0.10, 0.05, or even 0.01, in isolation, are unable to provide insight into false-positive and false-negative rates, which contribute to the overall key features of a patient selection biomarker.
Resistance to antiangiogenic agents can be innate and/or acquired [11]. It is important to recognize that angiogenesis biology is complex and that multiple indirect mechanisms of resistance contribute to the challenge in identifying and validating a single baseline predictive test for patient selection for antiangiogenic tyrosine kinase inhibitors in advanced RCC [12].
Upregulation of these mechanisms can be assessed in patients receiving treatment, including enumeration and characterization of infiltrating myeloid suppressor cells, or measurement of tumor or circulating proangiogenic proteins (e.g., FGF) or upregulation of invasion/resistance proteins such as hepatocyte growth factor receptor. However, such complexity does not mean that predictive biomarkers do not exist. Larger hypothesis-generating studies, powered to adjust for multiple comparisons, may be helpful in this regard. Of note, the ROC model can be applied equally well for a single candidate biomarker or for a composite biomarker, obtained from a multivariate analysis [13].
Although we strongly support the inclusion of sensitivity and specificity assessments, including ROC, for potential biomarkers of antiangiogenic agents, there are limitations to the outputs described in this manuscript. These include nonmandatory biospecimen collection, small sample size and absence of prospective testing. In future clinical trials, routine biospecimens collection and analysis, as well as statistical analysis, will help address clinical utility of biomarkers simultaneously with the clinical results.
Conclusion
In conclusion, an ROC analysis should be the next step in evaluating the clinical utility of ‘promising’ patient selection biomarkers rather than increasing sample size and repeating studies or meta-analysis of multiple studies. The later approaches may demonstrate a lower p-value, but the ROC characteristics of the biomarkers under evaluation are unlikely to change. This sequential approach can be expected to help the clinician separate truly promising markers from those that, realistically, have no chance of helping patient selection for a given therapeutic agent in RCC or any other indication. While there is no consensus on a specific AUC value for good predictive purposes, there have been literature recommendations of an AUC >0.8 for a predictive marker with potential clinical utility, which we support. Finally, for identification and validation of baseline predictive markers for antiangiogenic agents, the complexity of angiogenesis biology and existence of multiple mechanisms of resistance should also be taken into account.
Future perspective
Promising biomarkers for use in advanced RCC and other tumor types, in particular for antiangiogenic agents, will continue to be preliminarily identified via Kaplan–Meier analysis. However, in the future, ROC analysis will increasingly be used for continuous biomarker data in a step-wise fashion to distinguish those biomarkers for antiangiogenic agents, immune checkpoint inhibitors and other targeted agents with true clinical utility for patient selection, rather than validating results with larger studies or meta-analysis of multiple studies.
EXECUTIVE SUMMARY.
Kaplan–Meier analysis of data from a randomized, Phase II study of first-line sunitinib in patients with advanced renal cell carcinoma (RCC; n = 292) identified circulating soluble Ang-2 and MMP-2 and HIF-1α percentage of tumor expression as potential pretreatment biomarkers of sunitinib.
The methodological approach for identifying ‘promising’ patient selection biomarkers should be reconsidered for targeted therapies, in particular for antiangiogenic agents. Rather than solely relying upon Kaplan–Meier derived statistical significance (e.g., p-values < 0.05), a more stringent performance assessment, such as receiver operator characteristics (ROC) for continuous data, that incorporates sensitivity and specificity characteristics should be considered.
Patients & methods
Using the same dataset, sensitivity and specificity characteristics of the three previously identified statistically significant biomarkers were evaluated with an ROC model in order to determine whether these biomarkers have any clinical utility as potential patient selection markers.
Results
Based on a ROC (sensitivity vs 1 - specificity) threshold AUC value of >0.8, neither Ang-2 (0.67) nor MMP-2 (0.65), nor HIF-1α percentage of tumor expression (0.65), performed appropriately from a patient-selection standpoint.
Discussion
None of the three previously identified biomarkers have true potential as patient selection markers for sunitinib in advanced RCC, supporting the hypothesis that p-values < 0.05 that separate Kaplan–Meier curves do not necessarily translate into sensitivity and specificity characteristics needed for a clinically useful patient selection diagnostic.
To properly assess potential biomarkers, sensitivity and specificity characteristics for continuous biomarker data should be obtained by ROC analysis.
Acknowledgements
The authors would like to thank all of the participating patients and their families, as well as the network of investigators, research nurses, study coordinators, and operations staff.
Footnotes
Financial & competing interests disclosure
This study was sponsored by Pfizer Inc. PA English, JF Martini, O Valota and RE Buller are full-time employees of Pfizer with stock ownership. RJ Motzer has served in an advisory role with Pfizer, received research funding from Pfizer, and has been compensated for expert testimony by Pfizer. JA Williams is a former employee of Pfizer. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Medical writing support was provided by Andy Gannon at ACUMED©, an Ashfield company, part of UDG Healthcare plc (NY, USA), with funding from Pfizer Inc.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Open access
This work is licensed under the Creative Commons Attribution-NonCommercial 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
Papers of special note have been highlighted as: • of interest
- 1.Rini BI, Michaelson MD, Rosenberg JE, et al. Antitumor activity and biomarker analysis of sunitinib in patients with bevacizumab – refractory metastatic renal cell carcinoma. J. Clin. Oncol. 2008;26(22):3743–3748. doi: 10.1200/JCO.2007.15.5416. [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Donas J, Esteban E, Leandro-García LJ, et al. Single nucleotide polymorphism associations with response and toxic effects in patients with advanced renal cell carcinoma treated with first-line sunitinib: a multicentre, observational, prospective study. Lancet Oncol. 2011;12(12):1143–1150. doi: 10.1016/S1470-2045(11)70266-2. [DOI] [PubMed] [Google Scholar]
- 3.Motzer RJ, Hutson TE, Hudes GR, et al. Investigation of novel circulating proteins, germ line single-nucleotide polymorphisms, and molecular tumor markers as potential efficacy biomarkers of first-line sunitinib therapy for advanced renal cell carcinoma. Cancer Chemother. Pharmacol. 2014;74(4):739–750. doi: 10.1007/s00280-014-2539-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Previous study in which the biomarkers analyzed for utility using ROC curves were identified via Kaplan–Meier analysis.
- 4.Motzer RJ, Hutson TE, Olsen MR, et al. Randomized Phase II trial of sunitinib on an intermittent versus continuous dosing schedule as first-line therapy for advanced renal cell carcinoma. J. Clin. Oncol. 2012;30(12):1371–1377. doi: 10.1200/JCO.2011.36.4133. [DOI] [PubMed] [Google Scholar]; • The multicenter, randomized Phase II study (Renal EFFECT trial) from which the dataset used for this analysis was obtained.
- 5.Geomini P, Kruitwagen R, Bremer GL, Cnossen J, Mol BW. The accuracy of risk scores in predicting ovarian malignancy. Obstet. Gynecol. 2009;113(2 Pt 1):384–394. doi: 10.1097/AOG.0b013e318195ad17. [DOI] [PubMed] [Google Scholar]
- 6.Motzer RJ, Bacik J, Murphy BA, Russo P, Mazumdar M. Interferon-alfa as a comparative treatment for clinical trials of new therapies against advanced renal cell carcinoma. J. Clin. Oncol. 2002;20(1):289–296. doi: 10.1200/JCO.2002.20.1.289. [DOI] [PubMed] [Google Scholar]
- 7.Marinovich ML, Houssami N, Macaskill P, et al. Meta-analysis of magnetic resonance imaging in detecting residual breast cancer after neoadjuvant therapy. J. Natl Cancer Inst. 2013;105(5):321–333. doi: 10.1093/jnci/djs528. [DOI] [PubMed] [Google Scholar]
- 8.Frati A, Chereau E, Coutant C, et al. Comparison of two nomograms to predict pathologic complete responses to neoadjuvant chemotherapy for breast cancer: evidence that HER2-positive tumors need specific predictors. Breast Cancer Res. Treat. 2012;132(2):601–607. doi: 10.1007/s10549-011-1897-0. [DOI] [PubMed] [Google Scholar]
- 9.Gönen M. Analyzing Receiver Operating Characteric Curves with SAS®. SAS Institute Inc.; Cary, NC, USA: 2007. [Google Scholar]
- 10.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors: European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J. Natl Cancer Inst. 2000;92(3):205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 11.Bergers G, Hanahan D. Modes of resistance to anti-angiogenic therapy. Nat. Rev. Cancer. 2008;8(8):592–603. doi: 10.1038/nrc2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sonpavde G, Choueiri TK, Escudier B, et al. Sequencing of agents for metastatic renal cell carcinoma: can we customize therapy? Eur. Urol. 2012;61(2):307–316. doi: 10.1016/j.eururo.2011.10.032. [DOI] [PubMed] [Google Scholar]
- 13.Shultz EK. MuItivariate receiver-operating characteristic curve analysis: prostate cancer screening as an example. Clin. Chem. 1995;41(8 Pt 2):1248–1255. [PubMed] [Google Scholar]