Abstract
Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 has been applied to edit genomes in a wide variety of model systems. Although this process can be quite efficient, editing at precise locations in the genome remains difficult without a suitable single guide RNA (sgRNA). We have developed a method for screening sgRNA function in vitro, using reagents that most zebrafish laboratories are already using. The results from our in vitro assay correlate with function in vivo in every sgRNA that we have examined so far. When combined with endonucleases with alternative protospacer adjacent motif site specificities and alternative sgRNAs, this method will streamline genome editing at almost any locus.
Keywords: : Cas9, CRISPR, in vitro, validation, zebrafish, knock in
The clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 prokaryotic immune response has recently emerged as a key tool in modifying genomic DNA.1–8 Since its discovery, CRISPR has been adapted to modification of a plethora of model systems, including zebrafish.4,8 Mutagenesis is driven by the Cas9 endonuclease, specifically targeted to a genomic region by a locus-specific single guide RNA (sgRNA), adjacent to a protospacer adjacent motif (PAM), such as NGG.9 Although this process can be quite efficient,2 editing at precise locations in the genome remains difficult without an effective sgRNA,10 especially in the rapidly developing zebrafish. Validation of sgRNAs is a time-consuming and labor-intensive process, where success can only be determined by sacrificing a portion of the F0 generation, and waiting to screen for germline transmission. We demonstrate in this study, a method for cost-effective production of Cas9 protein and a method for validating sgRNAs in vitro, before F0 in vivo confirmation assays. Prediction algorithms can be inaccurate, and we show here that sgRNAs can be prescreened for activity before F0 validation using our in vitro assay. When used in combination with alternative PAM site endonucleases and their cognate sgRNAs, this method will streamline genome editing at almost any locus.
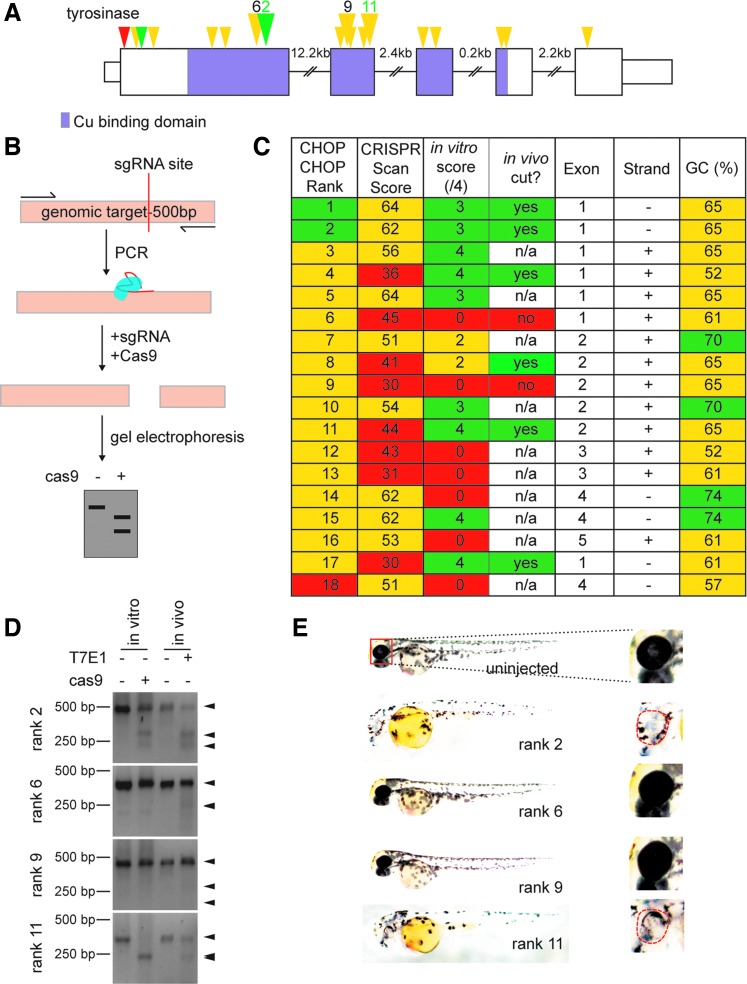
Our strategy for enriching untagged Cas9 suitable for in vitro testing of sgRNAs is detailed in the Supplementary Data (Supplementary Data are available online at www.liebertpub.com/zeb). Briefly, Cas9 protein produced in HEK293T cells can be crudely extracted and combined with a candidate sgRNA and a polymerase chain reaction (PCR) product from the genomic region of interest. The reaction products are resolved on an agarose gel to determine if the PCR product was cleaved. To validate our Cas9-containing crude protein (hereafter called Cas9 for simplicity), we began by using a sgRNA previously shown to function in vivo in zebrafish, and we found that when combined with our Cas9 protein, it could also cleave target sequences in vitro (Supplementary Fig. S4, lanes 1 and 2). As a proof of concept, we expanded our study to include the top 18 sgRNAs with 5′GG sequences predicted by CHOPCHOP to target tyrosinase (Fig. 1A), a gene encoding an enzyme essential for proper melanin production and pigment formation.11 Presence of the sgRNA target site and PAM in our wild-type strain was confirmed by sequencing. We tested each of these sgRNAs in vitro, and found that 11/18 of the sgRNAs were able to cleave target sequences in vitro (Fig. 1B–D). These included sgRNAs predicted both as strong and weak candidates by CHOPCHOP. Comparison of our in vitro test results against prediction scores of these sgRNAs from another algorithm, CRISPRScan,12 also demonstrated that sgRNA function was not accurately predicted by this algorithm. Finally, we did not find a trend between GC content or strand direction and sgRNA success (Fig. 1C). Taken together, these results indicated that prediction algorithms alone do not predict sgRNA success in all circumstances. By using a combination of prediction algorithms and our sgRNA prescreening method, users can streamline their mutagenesis approach. This is of vital importance when designing genome editing strategies.
FIG. 1.
Guide RNA screening of the tyrosinase locus. (A) Schematic of tyrosinase gene structure. The tyrosinase gene is encoded by five exons. The sequence encoding the enzymatic Cu-binding domain is highlighted in purple. The CHOPCHOP algorithm was used to predict the top 18 sgRNA binding sites that began with GG, the positions of which are indicated with arrowheads. The CHOPCHOP confidence scores are indicated as high (green), medium (yellow), or low (red). The positions of sgRNAs validated in vivo are represented with larger triangles and their CHOPCHOP rank number. (B) Schematic of assay to detect CRISPR/Cas9-mediated cleavage in vitro. Sequences surrounding the target site were PCR amplified from genomic DNA and combined with Cas9 proteins and sgRNA; reactions were allowed to proceed and run out on a gel to detect cleavage. (C) Summary of CHOPCHOP rank (1–18; 1 is best), CRISPRscan score (1–100; 100 is best), in vitro assay results, in vivo T7EI assay results, exon, strand, and GC content (confidence scores are indicated as high (green), medium (yellow), or low (red)). Note that there was no significant correlation with reactivity in vitro and CHOPCHOP rank, CRISPRscan score, strand, or GC content. n/a sgRNAs were not tested in vivo. (D) Sample from chosen sgRNAs with Cas9 assay run next to in vivo T7E1 assay. (E). Zebrafish at 48 h postfertilization after injection with chosen guides. Note the loss of melanocytes compared with the uninjected control in rank 2 and 11. There was no change in melanocyte distribution and coloration in rank 6 and rank 9 injected fish. CRISPR, clustered regularly interspaced short palindromic repeats; PCR, polymerase chain reaction; sgRNA, single guide RNA.
To determine the extent of correlation between in vitro and in vivo activity, we selected seven sgRNAs to compare the rank 1 control sgRNA, five of which we found to be effective in vitro (CHOPCHOP ranks 2, 4, 8, 11, and 17) and two of which we found to be ineffective in vitro (CHOPCHOP ranks 6 and 9). Injecting each of these sgRNAs into zygotes confirmed that in vitro testing does predict in vivo function, as measured by both assessment of indel formation by T7 endonuclease assay (Fig. 1C, D, and Supplementary Fig. S5) and loss of pigmentation (Fig. 1E). Furthermore, we have applied this strategy to test 50 other sgRNAs in the laboratory so far, including guides predicted by CHOPCHOP and CRISPRScan,12 neither of which showed a significant correlation to the in vitro test (two-tailed Pearson correlation test CHOPCHOP to in vitro test R2 = 0.006399; CRISPRScan to in vitro R2 = 0.05282) (Supplementary Table S3). We further tested these for somatic mutation in F0 fish and found that all 16 sgRNAs that failed the in vitro test also failed in vivo, whereas all 34 that had at least some cleavage in vitro also produced somatic mutations in vivo (Supplementary Table S3). Finally, sgRNAs that pass the in vitro test have resulted in germline transmission upon first attempt in 11 of 13 cases examined so far (Supplementary Table S4). Altogether, these results indicate that sgRNAs can be screened in vitro to predict in vivo success.
CRISPR/Cas9 has revolutionized our ability to manipulate genomes; however, finding a sgRNA suitable for cleavage at the desired location remains an obstacle to ubiquitous precise genome editing. It has now been shown that slight modifications to sgRNAs can be tolerated,12 and also that Cas9 can be modified to allow for alternative PAM sequences,13 expanding our repertoire of editable sequences. The direct method that we demonstrate here allows for screening of sgRNAs in vitro, before F0 validation. In particular, generation of specific targeted knock-ins to introduce, for example, fluorescent labels, peptide tags, or nucleotide substitutions, is dependent on having a robust, functional sgRNA site in close proximity to the region of intended knock-in.14 Many sgRNA prediction algorithms have been developed to focus on predicting robust sgRNAs with limited off-target effects, largely based on a limited pool of experimental results.12,15–17 Although success rates based on high confidence predicted that sgRNAs is improving, even those in the highest tier of prediction algorithms are not always successful.12 Pairing computer algorithms to identify potential sgRNAs and our in vitro screen to select the most active sgRNAs will streamline the process of precise genome editing at virtually any locus.
Supplementary Material
Acknowledgments
The authors are indebted to R. Rainville and K. Ong for fish husbandry. They extend their gratitude to J. Richter, C. Pouget, B. Weijts, P. Sahai, B. Goertner, S. Espanola, and X. Zeng for providing validation of the protocol and reading of the article. They thank P. Mali for thoughtful discussion of the article.
Authors' Contributions
S.G. designed and conducted experiments, and performed experimental analysis. B.L., C.H.O., and N.N. performed experiments. S.G. wrote the article and made the figures. K.W. and D.T. provided experimental guidance and edited the article.
Disclosure Statement
No competing financial interests exist.
References
- 1.Xiao A, et al. . Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res 2013;41:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jao L-E, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A 2013;110:13904–13909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cong L, et al. . Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013;339:819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hwang WY, et al. . Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 2013;31:227–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hruscha A, et al. . Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development 2013;140:4982–4987 [DOI] [PubMed] [Google Scholar]
- 6.Chen B, et al. . Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 2013;155:1479–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang H, et al. . One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 2013;154:1370–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hwang WY, et al. . Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS One 2013;8:e68708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, La Russa M, Qi LS. CRISPR/Cas9 in Genome Editing and Beyond. Annu Rev Biochem 2016;85:227–264 [DOI] [PubMed] [Google Scholar]
- 10.Gagnon JA, et al. . Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One 2014;9:e98186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ota S, Hisano Y, Ikawa Y, Kawahara A. Multiple genome modifications by the CRISPR/Cas9 system in zebrafish. Genes Cells 2014;19:555–564 [DOI] [PubMed] [Google Scholar]
- 12.Moreno-Mateos MA, et al. . CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods 2015;12:982–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleinstiver BP, et al. . Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015;523:481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auer TO, Duroure K, Cian AD, Concordet J-P, Bene FD. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res 2014;24:142–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 2014;42:W401–W407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu PD, et al. . DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 2013;31:827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim D, et al. . Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods 2015;12:237–243, 1 p following 243 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.