Abstract
The nonstructural protein 5A (NS5A) encoded by the human hepatitis C virus RNA genome is shown here to induce the activation of NF-κB and STAT-3 transcription factors from its cytoplasmic residence via oxidative stress. NS5A causes the disturbance of intracellular calcium. Ca2+ signaling triggers the elevation of reactive oxygen species in mitochondria, leading to the translocation of NF-κB and STAT-3 into the nucleus. Evidence is presented for the constitutive activation of STAT-3 by NS5A. In the presence of antioxidants [pyrrolidine dithiocarbamate (PDTC), N-acetyl l-cysteine (NAC)] or Ca2+ chelators (EGTA-AM, TMB-8), NS5A-induced activation of NF-κB and STAT-3 was eliminated. These results provide an insight into the mechanism by which NS5A can alter intracellular events relevant to liver pathogenesis associated with the viral infection.
Hepatitis C virus (HCV) causes acute/chronic hepatitis with a significant risk of end-stage cirrhosis and hepatocellular carcinoma (1). The single-stranded RNA genome of the human HCV is a 9.6-kb-long positive-sense molecule, which encodes a polyprotein of about 3,000 aa (2, 3). The polyprotein is posttranslationally cleaved by both viral/cellular proteases to produce about 10 polypeptides that include structural (core and E1 and E2) and nonstructural (NS2, NS3-NS5A/B) proteins (2, 3). The single long ORF is preceded by 332 or 342 nt of the 5′ untranslated region, which harbors an internal ribosome entry site capable of initiating translation at an internal site (4–6). The 3′ end of the RNA genome contains a unique sequence of the untranslated region that is necessary for initiating RNA replication. Although the infectious cDNA clones that have been generated could infect chimpanzees, they failed to replicate in vitro in cultured cell lines (7). Lohmann et al. (8) described the generation of efficient replicating HCV RNA subgenomic replicons coexpressing a neomycin-resistance marker. A high level of replication of subgenomic replicons was characterized, resulting from adaptive mutations, which were scattered throughout the HCV genes of the replicon (9, 10).
The HCV NS5A has generated a significant level of interest as several cellular targets have been identified. NS5A is a serine phosphoprotein, which exists as a polypeptide of p56 or p58 with varying degrees of phosphorylation (11, 12). The identity of cellular kinase(s) responsible for NSS5A phosphorylation has not been firmly established. NS4A, an integral membrane protein, has been shown to modulate NS5A phosphorylation at least for some HCV subtypes (13). NS5A is localized to the cytoplasm in the perinuclear reticular network characteristic of the endoplasmic reticulum (ER) membrane (3, 14). NS5A came into prominence because of its suggested role in IFN resistance. It was shown that NS5A directly interacted with double-stranded RNA-dependent kinase (PKR) and inactivated its function, thus modulating the IFN-stimulated antiviral response (15). Neither the sites of hyperphosphorylation nor the region designated ISDR (IFN-sensitive determining region) within NS5A was found to be necessary for the HCV replicative functions (8, 16). Among other cellular targets of NS5A, an interaction with the VAP-30 membrane protein, along with NS5B, has been described (17), implicating the membrane association of the replication complex. Membrane association of HCV replication complex is consistent with other models of RNA viral replication (18). Upon viral infection, cells become programmed to produce large amounts of viral proteins that are processed through ER. This may elicit an ER overload response, as has been documented for a number of viral proteins (19, 20).
It was previously shown that NS5A can function as a transcriptional trans-activator (refs. 21–23 and unpublished results). Although these reports implicate a functional role of NS5A in transcription, the exact nature of its role or the mechanism(s) involved in regulating the cellular transcription has not been investigated. This study was undertaken to investigate the function of NS5A as a transcriptional trans-activator. NS5A is localized to the ER, whereas transcriptional trans-activation traditionally requires the protein to be in the nucleus. We reasoned that NS5A must participate in signal transduction pathways that are initiated in the cytoplasm where it resides.
In this study, we report that NS5A can activate cellular transcription factors by inducing oxidative stress in the cells. We have focused on NF-κB and STAT-3, both of which exist in latent form in the cytoplasm and respond to signals for activation and translocation to the nucleus. These data further demonstrate a potential role of NS5A in altering calcium homeostasis in cells. The evidence in support of these conclusions is based on studies in which antioxidant reagents abrogated NS5A's ability to activate transcriptional factors. Similarly, the reagents that chelate calcium also reduced NS5A's ability to activate transcriptional factors. Direct measurements of reactive oxygen species (ROS) established its ability to induce oxidative stress. These studies further revealed an intriguing property of NS5A in constitutively activating STAT-3, which normally requires cytokine signaling for its activation (24). These studies provide insight into the mechanisms by which NS5A may contribute to liver disease pathogenesis as well as hepatocellular carcinoma associated with the viral infection.
Methods
Plasmids.
Plasmids p3x-kB-Luc (luciferase reporter driven by minimal fos promoter with three upstream NF-κB binding sites from MHC class I) and p3x-mut-Luc (mutated NF-κB site) were a generous gift of J. Martin (University of Colorado, Boulder). The HCV NS5A-coding sequences and the mutated versions of it were generated by PCR amplification of HCV plasmid pCMV729-3010 (14) (a gift of K. Shimotohno, Kyoto Univ., Kyoto, Japan) (13). The plasmid pCMV729-3010 contains coding sequences of all of the nonstructural proteins. A flag tag was added at the N terminus of the NS5A sequences. The primers used to PCR-amplify NS5A sequences contained HindIII and XbaI restriction sites, respectively. The PCR-generated fragments were cloned into the HindIII and XbaI sites of pRc/CMV vector (Invitrogen) to produce pCNS5A, pCNS5A-M1, pCNS5A-M2, pCNS5A-M3, and pCNS5A-M4, respectively. Manganese superoxide dismutase (Mn-SOD) expression vector was a gift from S. Flores (Univ. of Colorado Health Sciences Center, Denver).
Cell Transfections.
About 60–70% confluent cells in 60-mm dishes were transfected with indicated plasmids with Lipofectin reagent (GIBCO). Antioxidants N-acetyl l-cysteine (NAC), pyrrolidine dithiocarbamate (PDTC), and metal ion chelators EGTA-AM, EGTA, TMB-8, and Ruthenium red (RR) were added to the transfected cells for various incubation times before harvesting cells for mobility-shift assays and measuring luciferase activities. RR and TMB-8 were purchased from Aldrich and Sigma, respectively. Luciferase activity was determined by standard procedure.
Electrophoretic Mobility-Shift Assay (EMSA).
EMSA was carried out on nuclear extracts prepared from untransfected or pCNS5A-transfected Huh 7 cells by standard methods. NF-κB, STAT-3, AP-1, and Oct-1 consensus oligonucleotides were radiolabeled with [γ-32P]ATP in the presence of T4 polynucleotide kinase. About 15,000 cpm of each oligonucleotide probe and 5 μg of indicated nuclear extracts were subjected to 5% PAGE. The gel was dried and autoradiographed.
Immunoprecipitation and Western Blot Analysis of STAT-3.
Nuclear lysates were prepared as described above from Huh-7 cells untransfected or transfected with pCNS5A, pCNSM4, and pCMV729-3010 expression vectors. The lysates were immunoprecipitated with anti-STAT-3 serum and fractionated by SDS/PAGE. Gels were electroblotted onto poly(vinylidene difluoride) membrane (Amersham Pharmacia) and probed with monoclonal antiphosphotyrosine antibody and visualized by using the ECL detection system (Amersham Pharmacia).
Detection of Intracellular ROS.
Thirty-six hours posttransfection, cells were treated with dihydroethidium for 45 min. Cells were harvested in 50% FBS/DMEM, washed two times with PBS, and finally suspended in 300 μl of PBS; superoxide levels were measured by using an XL00W42322 flow cytometer with an excitation emission at 605 nm.
Results
NS5A Induces Activation of NF-κB and STAT-3.
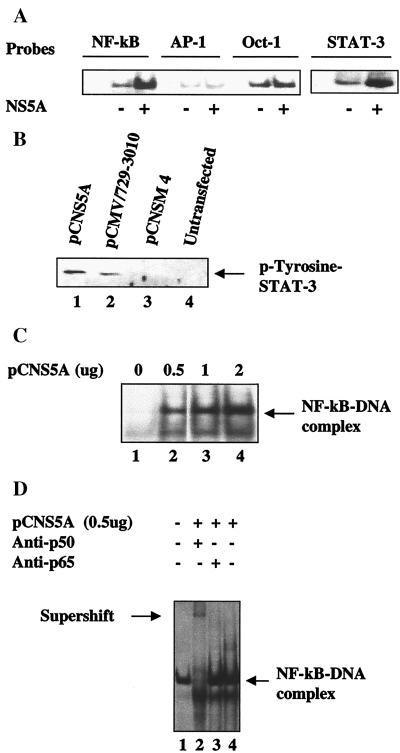
To demonstrate the role of NS5A in regulating cellular transcription, EMSA studies were initiated with four oligonucleotides representing cognate sequences for the transcription factors NF-κB, AP-1, STAT-3, and Oct-1. The EMSA results clearly show the activation of NF-κB and STAT-3 transcription factors in nuclear lysates expressing NS5A (Fig. 1A). Activation of STAT-3 was surprising because it is known to be activated by cytokines such as IL-6 and epidermal growth factor (24). We next examined the status of STAT-3 tyrosine phosphorylation in NS5A-expressing cells. The results of the Western blot analysis are presented in Fig. 1B and demonstrate constitutive induction of tyrosine phosphorylation of STAT-3 by NS5A (Fig. 1B, lane 1). In this analysis, we included two additional controls. An NS5A deletion mutant, pCNSM4 (Fig. 2A), was included, which failed to stimulate STAT-3 activation (Fig. 1B, lane 3). No STAT-3 stimulation was observed in untransfected Huh-7 hepatoma cells (Fig. 1B, lane 4). We further used an expression vector (pCMV 729-3010) (14), which expresses all of the nonstructural proteins (NS2–NS5) and is capable of activating STAT-3 (Fig. 1B, lane 2). The rationale behind using this vector is to examine the ability of NS5A in inducing STAT-3 activation in the context of other viral proteins it is known to associate with (3, 14, 25). These results clearly document the ability of NS5A to constitutively activate STAT-3. HCV nonstructural proteins including NS3, NS4, and NS5A/B are believed to form a ribonucleoprotein complex that is associated with the ER membrane (14, 25).
Figure 1.
HCV NS5A activates NF-κB and STAT-3. (A) Activation of cellular transcription factors by NS5A. EMSA was carried out in the presence of 32P-labeled oligonucleotide probes corresponding to NF-κB, AP-1, Oct-1, or STAT-3 in the presence of equal amounts of nuclear lysates prepared from untransfected and Huh-7 cells transfected with wild-type pCNS5A expression vector. (B) NS5A constitutively activates STAT-3 tyrosine phosphorylation. Western blot analysis of STAT-3 protein in cell extracts transfected with wild-type pCMV 729-3010, pCNS5A, or pCNSM4 expression vectors. Cellular lysates were immunoprecipitated with anti-STAT-3 polyclonal serum and Western-blotted with antiphosphotyrosine mAb. Blot was detected by using an ECL kit (Amersham Pharmacia). Lanes 1–3, lysates from cells transfected with pCNS5A, pCMV 729-3010, or pCNSM4 mutant expression vectors, respectively. Lane 4, untransfected lysates. (C) NS5A induces DNA binding activity of NF-κB. EMSA was carried out with 32P-labeled NF-κB probe and increasing concentrations of NS5A-transfected lysates. Lane 1, probe alone; lanes 2–4, NF-κB probe incubated with 0.5, 1, and 2 μg of NS5A transfected lysates, respectively. (D) Supershift of NF-κB protein–DNA complex. EMSA was carried out as described above. Lanes 1 and 4, NF-κB protein–DNA complex; lanes 2 and 3, complex incubated with anti-p50 and anti-p65, respectively.
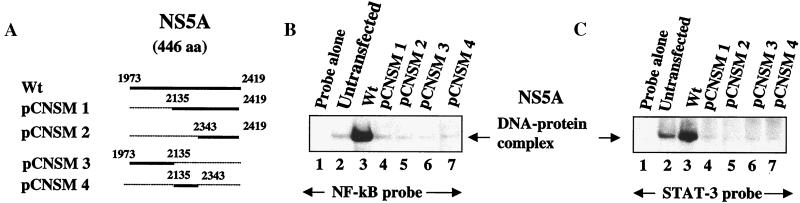
Figure 2.
NS5A deletion mutants do not activate NF-κB and STAT-3. (A) Schematic of NS5A deletion mutants. Dotted lines represent the extent of deletion of NS5A sequences. (B) Wild-type NS5A, but not the deletion mutants, activates NF-κB. EMSA was carried out with lysates transfected with wild-type NS5A (lane 3) and a series of NS5A deletion mutants as indicated in A (lanes 4–7). Lane 1, probe alone; lane 2, untransfected lysates. Lane 3, NS5A-transfected nuclear lysates; lanes 4–7, NS5A-deletion mutants as indicated in A. (C) NS5A activates transcription factor STAT-3. EMSA was carried out as described above in B. Lane 1, probe alone; lane 2, untransfected lysates; lane 3, NS5A-transfected nuclear lysates; lanes 4–7, NS5A-deletion mutants as indicated in A.
In the next series of experiments, we present evidence for the stimulated NF-κB and STAT-3 DNA binding in the lysates prepared from NS5A-transfected Huh-7 cells. A dose-dependent activation of NF-κB as a function of DNA concentration of the NS5A expression vector is shown during EMSA (Fig. 1C). The DNA–protein complex migrated slowly in the presence of antibody to the NF-κB subunit p50, but the p65 antiserum failed to show similar pattern. The failure to produce a supershift of the DNA–protein–antibody complex could be caused by the poor recognition of the DNA–protein complex containing NF-κB for anti-p65 antibody (Fig. 1D). To identify the activation domain of NS5A, deletion mutations were generated representing different regions of the protein (Fig. 2A). All four NS5A mutants express truncated proteins, which was assessed by immunofluorescence (data not shown). EMSA performed using NF-κB and STAT-3 probes and showed activation of those factors by wild-type NS5A, but the four deletion mutants failed to induce that activity (Fig. 2 B and C), suggesting the presence of more than one interactive domain within the NS5A protein.
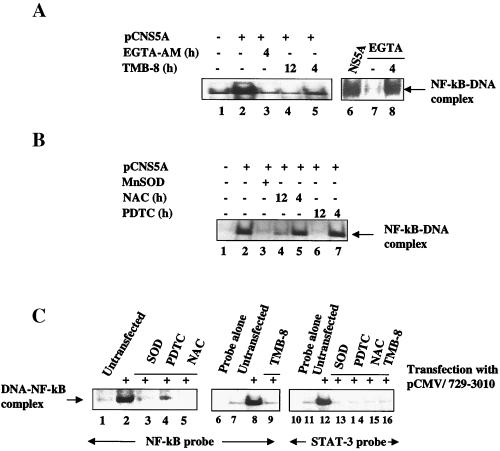
Next, we carried out similar analyses in the presence of chelators of calcium. The rationale behind this line of inquiry is that NS5A expression in the membrane may cause alteration of calcium homeostasis. These changes in intracellular calcium levels have been shown to contribute to the activation of NF-κB (26, 27). During transfection, the cell monolayers were treated with EGTA-AM, EGTA, and TMB-8. EGTA-AM and TMB-8 are chelators of calcium. As can be seen in Fig. 3A, the activation of NF-κB was significantly reduced in the presence of these reagents. EGTA is an extracellular Ca2+ chelator and has little effect on reducing NF-κB activation, whereas the esterified Ca2+ chelator, EGTA-AM, effectively reduced the NF-κB activation, suggesting that the intracellular Ca2+ is responsible for the observed activation of NF-κB. These results suggest the requirement of calcium as messenger for NF-κB activation. The use of these chelators distinguishes the origin of Ca2+ between extracellular versus intracellular stores (26). Alteration of calcium homeostasis within the ER has been shown to trigger production of reactive oxygen intermediates/ROS (19, 27). ROS production can occur in the ER or mitochondria (19). The Ca2+ released from ER is readily taken up by the mitochondria (19, 20). To determine whether reactive oxygen intermediates are involved in NF-κB activation, we used oxidative stress inhibitors NAC and PDTC, which counteract the oxygen free radical effects (26, 27). EMSA was carried out by using the lysates prepared from Huh-7 cells transfected with NS5A expression vector and subsequently incubated with PDTC and NAC for 4 or 12 h, respectively. We also cotransfected Huh-7 cells with both NS5A and Mn-SOD expression vectors. Activation of NF-κB was significantly reduced in the presence of NAC and PDTC after 12 h of exposure (Fig. 3B). Similarly, the overexpression of Mn-SOD led to a dramatic abrogation of NF-κB activation. Overexpression of Mn-SOD has been shown to inhibit activation of NF-κB (28). These data clearly support the view that ROS levels are elevated as a result of NS5A expression and that antioxidants that counteract oxygen free radicals eliminated these effects.
Figure 3.
Calcium chelators and antioxidants inhibit NF-κB and STAT-3. (A) EMSA with NF-κB probe with NS5A-transfected lysates as described in Fig. 1. Lane 1, untransfected; lanes 2–7, NS5A-transfected nuclear lysates treated with EGTA-AM, TMB-8, and EGTA for 4 or 12 h, respectively. The data described in lanes 6 and 7 were from a different experiment. (B) Inhibition of NS5A-induced NF-κB activation by antioxidants. EMSA with NF-κB probe and lysates from untransfected (lane 1), transfected with NS5A vector (lane 2), and Mn-SOD expression vector (lane 3). Lanes 4–7, NS5A-transfected lysates treated with 30 mM NAC (lanes 4 and 5) or 100 μM PDTC (lanes 6 and 7) at 4 or 12 h, respectively, as indicated. (C) HCV nonstructural proteins (NS2–NS5) activate transcription factors NF-κB and STAT-3. EMSA was carried out in the presence of NF-κB probe (lanes 1–9) or STAT-3 probe (lanes 10–16), and nuclear lysates were prepared from cells transfected with pCMV729-3010. Lanes 6 and 10, probes alone; lanes 1, 7, and 11, untransfected lysates. Lanes 3 and 13, cotransfected with Mn-SOD and pCMV729-3010 expression vectors. All other lanes, pCMV729-3010-transfected nuclear lysates treated with antioxidants, 100 μM PDTC, 30 mM NAC, or Ca2+ chelator TMB-8 (100 μM) as indicated.
We next used a vector pCMV 729-3010, which has been previously described to express all of the HCV nonstructural proteins that are appropriately processed during transient transfection (14). EMSA analysis was carried out using lysates from Huh-7 cells transiently transfected with pCMV 729-3010 along with the indicated pharmacological reagents. The results described in Fig. 3C show that expression of all of the nonstructural proteins, including NS5A, produced the same results during EMSA, i.e., NF-κB and STAT-3 activation occurred and was eliminated in the presence of antioxidants NAC and PDTC. NAC and PDTC did produce low levels of inhibition of NF-κB activation in untransfected cells, as has been previously seen in other studies (data not shown; ref. 19). Collectively, these data document the ability of NS5A to induce Ca2+ signaling, leading to the generation of ROS. These cellular events ultimately manifest in the activation of NF-κB and STAT-3.
NS5A Stimulates NF-κB-Dependent Luciferase Activity.
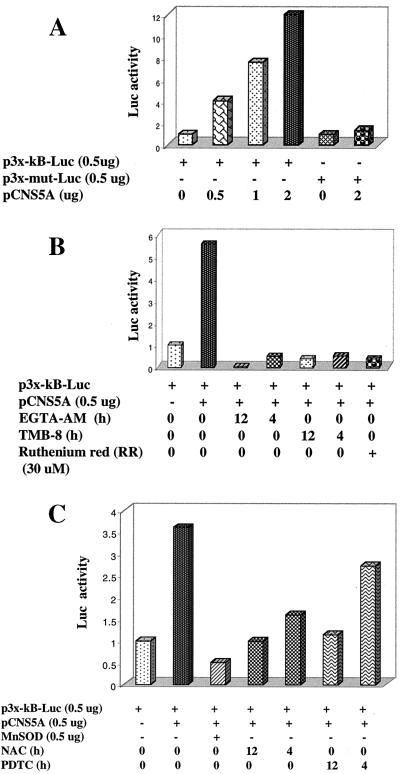
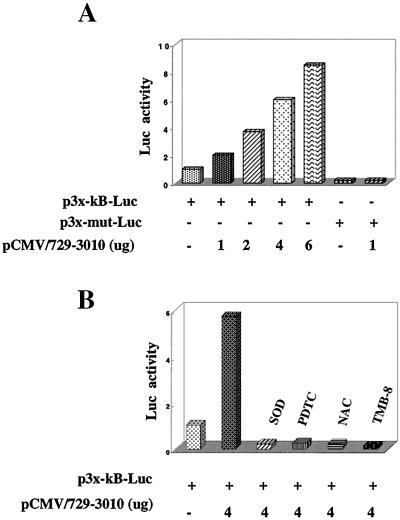
A luciferase expression vector under the transcriptional control of NF-κB motifs (multimers) was used to monitor the trans-activating effect of NS5A. A series of cotransfections was carried out with similar reagents used above to confirm the activation of NF-κB by cell-based assays. The initial analysis of these cotransfections also included a mutant form of the NF-κB luciferase vector as a negative control. Results shown in Fig. 4 demonstrate that in the presence of increasing concentrations of NS5A, luciferase activity was significantly increased, whereas similar levels of NS5A expression did not lead to any luciferase expression under the control of mutant NF-κB–luciferase vector (Fig. 4A). These studies were extended to include the calcium chelators EGTA-AM and TMB-8, all of which abrogated luciferase expression (Fig. 4B). In the presence of RR, the NF-κB-regulated expression was significantly reduced (Fig. 4B). The reduction in luciferase activity in the presence of RR suggests that mitochondrial uptake of intracellular Ca2+ is required for the activation of NF-κB. RR inhibits mitochondrial uptake of Ca2+ (29). The role of antioxidants, NAC, PDTC, and overexpression of Mn-SOD was studied during the expression of NF-κB luciferase, and the results shown in Fig. 4C support the EMSA results shown above. NAC and PDTC reduced NF-κB-controlled luciferase expression. Similar results were obtained during cotransfection with an Mn-SOD expression vector (Fig. 4C). We next confirmed the luciferase activity in the presence of all of the HCV nonstructural proteins (pCMV729-3010). Results described in Fig. 5A show that expression of HCV nonstructural proteins, which includes NS5A, also led to an increase of luciferase activity as a function of HCV protein concentration. However, luciferase activity was dramatically reduced in the presence of NAC, PDTC, and Mn-SOD (Fig. 5B). These results reinforce the issue that NS5A can induce NF-κB in the context of the rest of the nonstructural proteins; however, this property of NS5A is sensitive to chelators of Ca2+ and antioxidants. These results suggest that activation of transcriptional factors requires signaling from both Ca2+ and ROS, whose intracellular levels are affected by NS5A expression.
Figure 4.
Effect of NS5A on the NF-κB controlled luciferase expression. (A) Dose-dependent activation of NF-κB by NS5A. Huh-7 cells were cotransfected with increasing amounts of pCNS5A along with the wild-type or mutant NF-κB luciferase plasmids. (B) Calcium chelators inhibit NS5A-mediated NF-κB-controlled luciferase activity. Cells were transfected as described in A and treated with EGTA-AM (500 μM), TMB-8 (100 μM), and RR (30 μM) for various times of inhibition before luciferase activity. (C) Antioxidants inhibit NS5A-mediated NF-κB-controlled luciferase activity. Transfected cells were treated with NAC (30 mM) and PDTC (100 μM) for 6 h before preparing lysates for luciferase activity. Cells were cotransfected with NS5A and Mn-SOD expression vector along with the reporter plasmids.
Figure 5.
HCV nonstructural protein induced NF-κB-controlled luciferase expression. (A) Dose-dependent activation of NF-κB by HCV nonstructural proteins (pCMV729-3010) as described in Fig. 4. (B) Antioxidants and calcium chelators inhibit the transcriptional activation of NF-κB-controlled luciferase expression by HCV nonstructural proteins. Cells were transfected with 4 μg of pCMV729-3010 vector and Mn-SOD or were treated with NAC (30 mM) and PDTC (100 μM) for 6 h and TMB-8 (100 μM) for 45 min before preparing lysates for luciferase activity, respectively.
NS5A Induces ROS Production.
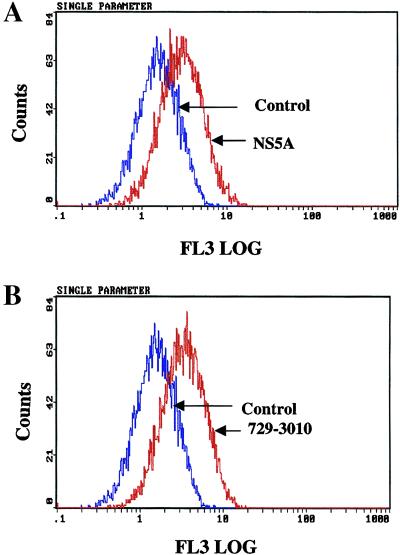
Next, we investigated whether NS5A expression triggers the generation of ROS. NS5A-transfected cells were stained with dihydroethidium and subjected to flow cytometry. Results described in Fig. 6A clearly show the production of ROS as evidenced by an increase in ethidium staining of DNA. Dihydroethidium is oxidized to ethidium by superoxide anions. Once oxidized, ethidium is free to intercalate with DNA in the nucleus, where it emits fluorescence at 605 nm (FL3 log scale) (30). This pattern also was observed in cells expressing all of the nonstructural proteins (Fig. 6B). These data suggest a direct involvement of NS5A in the ROS production.
Figure 6.
HCV NS5A and nonstructural proteins induce the production of intracellular ROS/superoxide anion radicals. (A) Untransfected (control) or NS5A-transfected Huh-7 cells were treated with 4 μM of dihydroethidium for 45 min. Cells were harvested, and superoxide levels were measured by using flow cytometer with excitation emission at 605 nm. ROS/superoxide levels were assessed in untransfected (blue) and cells transfected with pCNS5A (red). (B) ROS/superoxide levels were determined in untransfected (blue) and cells transfected with pCMV729-3010 (red).
In summary, the data described above demonstrates that HCV NS5A expression is capable of an ER–nucleus signal transduction pathway, which involves both Ca2+ and ROS as second messengers. These intracellular events ultimately contribute to the activation of NF-κB and STAT-3.
Discussion
The HCV nonstructural proteins including NS5A are associated with the membrane in the reticular network of the ER in the perinuclear area (2, 3, 14, 17, 25). They form an ribonucleoprotein complex, which is assembled to link translation of the viral genome with the subsequent events of replication, a function needed to perpetuate and establish the infectious process in hepatocytes. NS5A is one of the components of the ribonucleoprotein complex, which also includes the viral RNA genome poised to initiate minus strand synthesis soon after RNA has been translated. RNA viruses replicate principally on membranes including ER, golgi, endosomes, or lysosomes (18). We reasoned that NS5A being associated with the membrane is capable of inducing ER stress. As a consequence of ER stress, Ca2+ is released from the ER, where it is mainly stored and is readily taken up by mitochondria, where it affects the transmembrane potential and induces oxidative stress exhibited by the rising levels of ROS in mitochondria (19, 31, 32). Increased Ca2+ efflux from the ER can therefore directly affect the mitochondrial oxidant production and activation of transcription factors. Studies have shown that calcium release precedes the production of ROS in cells exhibiting this phenomenon (19).
In the present study, we set out to investigate the possible mechanism by which NS5A trans-activates gene expression in the nucleus. Various pharmacological reagents were included in the EMSA and luciferase reporter cell-based assays, which either chelated calcium ions or eliminated oxygen free radicals. NAC and PDTC, which eliminate oxygen free radicals (33), dramatically reduced NF-κB and STAT-3 activation induced by NS5A. Similarly, the chelators of calcium, EGTA-AM, and TMB-8 effectively inhibited NS5A-induced NF-κB and STAT-3 activation, indicating the requirement of calcium signaling for the observed phenomenon. Several deletion mutants of NS5A were included in this analysis, and all of these were unable to induce the activation of these transcription factors. Because NS5A is associated with other viral proteins, we investigated the ability of NS5A to induce these effects in the context of other nonstructural proteins. The results of this analysis clearly support the ability of NS5A to activate NF-κB and STAT-3 in the context of other nonstructural proteins, which requires both Ca2+ and ROS to mediate these effects. It is pertinent to examine the biological and biochemical function(s) of a given HCV protein in the context of other viral proteins because the HCV nonstructural proteins associate with each other, and in doing so, they may fold in conformations different when expressed individually. The data presented here suggest the involvement of ER and mitochondria in the activities of NS5A in inducing the activation of NF-κB and STAT-3. Whether other HCV proteins contribute to oxidative stress or alteration of Ca2+ homeostasis will be investigated in future.
Recent imaging techniques indicate that mitochondria exist as a continuous interconnected mitochondrial reticulum, closely associated with ER (34). Ca2+ ions released from the ER in response to stimuli are transferred to the mitochondrial matrix, causing local induction of oxidative stress leading to the elevation of ROS (27). RR, an inhibitor of electrogenic calcium import in mitochondria (29), inhibited NS5A-induced NF-κB activation, indicating the involvement of mitochondria in the generation of ROS (Fig. 4). Collectively, these studies imply the involvement of both the ER and mitochondria in the activation of NF-κB. STAT-3 activation was also impaired by RR (data not shown).
NF-κB stays latent in the cytoplasm as a complex of proteins consisting of p65–p50–ikB and other proteins (33). The oxidative stress-induced activation of NF-κB has been recently shown to be modulated by tyrosine phosphorylation and the PEST sequences of iKB (35). It was further demonstrated that such iKB molecules are degraded by calpains rather than by the proteasome pathway (35). Future work should focus on whether NS5A induces NF-κB via these motifs of iKB. The direct consequence of activation of these transcriptional factors is the induction of genes whose functions can be protective and/or antiapoptotic (33, 36).
STAT-3 is activated by cytokines such as epidermal growth factor or IL-6 (24). Similar constitutive activation of STAT-3 has been previously reported for vSrc tyrosine kinase (37). NS5A does not possess tyrosine kinase activity, and how it induces tyrosine phosphorylation of STAT-3 is not clear. A complete understanding of tyrosine kinase pathways is presently lacking. There are multiple tyrosine kinase signaling pathways in addition to the JAK family of kinases, which can induce STAT signaling (38). It is likely that NS5A induction of NF-κB and STAT-3 targets to signaling pathways affecting tyrosine phosphorylation.
STAT-3 and NF-κB motifs are found in a wide variety of cellular genes. Several lines of evidence suggest that the NF-κB family of proteins is involved in tumor growth and metastasis, where a high level of NF-κB activity has been observed (33, 36). Similarly, the chronic hepatitis C liver tissues displayed elevated levels of NF-κB compared with the normal liver (39). NF-κB induces expression of antiapoptotic genes, such as IAP protein, or genes that contribute to proliferation such as myc and Bcl-2 (36). It is the persistent expression of NF-κB that ensures the expression of genes providing protection against apoptotic stimuli. Apart from growth stimulatory functions of STAT-3, it is also the major inducer of acute phase of liver disease (27), which may be relevant to the acute hepatitis associated with HCV infection (1).
An overwhelming number of studies support the role of free radicals in the initiation and progression of multistage carcinogenesis (40). Consistent with this idea, free radical scavengers and antioxidant enzymes are down-regulated in tumor cells (41). The production of hepatocellular carcinoma by HCV probably involves a combination of indirect mechanisms. For instance, chronic liver injury that leads to necrosis, inflammation, and liver regeneration over a period of time can contribute to cirrhosis, thus paving the way for events preceding liver neoplasia (42). In keeping with this notion, NS5A has been shown to participate in signal transduction pathways leading to growth promotion (43). In summary, the evidence presented here for the participation of NS5A in the ER-nucleus signal transduction pathway implicates a potential role of this protein in the establishment of chronic liver disease in infected hepatocytes as well as protection from apoptosis.
Abbreviations
- HCV
hepatitis C virus
- ROS
reactive oxygen species
- NS5A
nonstructural protein 5A
- RR
ruthenium red
- ER
endoplasmic reticulum
- Mn-SOD
manganese superoxide dismutase
- EMSA
electrophoretic mobility-shift assay
- PDTC
pyrrolidine dithiocarbamate
- NAC
N-acetyl l-cysteine
References
- 1.Di Bishcegli A M. Hepatology. 1997;26:34S–38S. [Google Scholar]
- 2.Reed K E, Rice C M. Curr Top Microbiol Immunol. 2000;242:55–84. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]
- 3.Bartenschlager R, Lohmann V. J Gen Virol. 2000;81:1631–1648. doi: 10.1099/0022-1317-81-7-1631. [DOI] [PubMed] [Google Scholar]
- 4.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C, Sarnow P, Siddiqui A. J Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rijnbrand R, Lemon S M. Curr Top Microbiol Immunol. 1999;242:85–116. doi: 10.1007/978-3-642-59605-6_5. [DOI] [PubMed] [Google Scholar]
- 7.Kolykhalov A, Agapove E, Blight K, Michalik S, Feinstone S, Rice C M. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 8.Lohmann V, Korner F, Koch J O, Herian U, Theilmann L, Bartenschlager R. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 9.Blight K J, Kolykhalov A A, Rice C M. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 10.Lohmann V, Korner F, Dobierzewska A, Bartenschlager R. J Virol. 2001;75:1437–1449. doi: 10.1128/JVI.75.3.1437-1449.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kankeo T, Tanji Y, Satoh S, Hijikata M, Asabe S, Kimura K, Shimotohno K. Biochem Biohpys Res Commun. 1994;205:320–326. doi: 10.1006/bbrc.1994.2667. [DOI] [PubMed] [Google Scholar]
- 12.Reed K E, Xu J, Rice C M. J Virol. 1997;71:7187–7197. doi: 10.1128/jvi.71.10.7187-7197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koch J O, Bartenschlager R. J Virol. 1999;73:7138–7146. doi: 10.1128/jvi.73.9.7138-7146.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hijikata M, Mizushima H, Tanji Y, Komoda Y, Hirowatari Y, Akagi T, Kato N, Kimura K, Shimotohno K. Proc Natl Acad Sci USA. 1993;90:10773–10777. doi: 10.1073/pnas.90.22.10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gale M J, Blakely S M, Kwieciszewski B, Tan S-L, Dossett M, Tang N M, Korth M J, Polyak S J, Gretch D R, Katze M G. Mol Cell Biol. 1998;18:5208–5218. doi: 10.1128/mcb.18.9.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francois C, Duverlie G, Rebouillat D, Khorsi H, Castelain S, Blum H E, Gatignol A, Wychowski C, Moradpour D, Meurs E F. J Virol. 2000;74:5587–5596. doi: 10.1128/jvi.74.12.5587-5596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tu H, Gao L, Shi T, Taylor D, Yang T, Mircheff A, Wen Y, Gorbalenyz A, Hwang S, Lai M. Virology. 1999;263:30–41. doi: 10.1006/viro.1999.9893. [DOI] [PubMed] [Google Scholar]
- 18.Lai M. Virology. 1998;244:1–12. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- 19.Pahl H L. Physiol Rev. 1999;70:683–701. doi: 10.1152/physrev.1999.79.3.683. [DOI] [PubMed] [Google Scholar]
- 20.Kaufman R K. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 21.Kato N, Keng-Hsin L, Ono-Nita, Shiratori Y, Omato M. J Virol. 1997;71:8856–8859. doi: 10.1128/jvi.71.11.8856-8859.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Majumdar M, Ghosh A, Steele R, Ray R, Ray R B. J Virol. 2001;75:1401. doi: 10.1128/JVI.75.3.1401-1407.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosh A, Steele R, Meyer K, Ray R, Ray R B. J Gen Virol. 1999;80:1179–1183. doi: 10.1099/0022-1317-80-5-1179. [DOI] [PubMed] [Google Scholar]
- 24.Darnell J E., Jr Science. 1997;277:630–635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 25.Moradpour D, Kery P, Rice C M, Blum H. Hepatology. 1998;28:192–201. doi: 10.1002/hep.510280125. [DOI] [PubMed] [Google Scholar]
- 26.Sen C K, Roy S, Packer L. FEBS Lett. 1996;385:58–62. doi: 10.1016/0014-5793(96)00346-8. [DOI] [PubMed] [Google Scholar]
- 27.Pahl H L, Baeuerle P A. FEBS Lett. 1996;392:129–136. doi: 10.1016/0014-5793(96)00800-9. [DOI] [PubMed] [Google Scholar]
- 28.Manna S K, Zhang H J, Tao Y, Oberley L W, Aggarwal B B. J Biol Chem. 1998;273:13245–13254. doi: 10.1074/jbc.273.21.13245. [DOI] [PubMed] [Google Scholar]
- 29.Van der Water B, Zoeteweij J, de Bont H, Mulder G, Nagelkerke J. J Biol Chem. 1994;269:14546–14552. [PubMed] [Google Scholar]
- 30.Costa A, Cotter T. In: Apoptosis: A Practical Approach. Studzinski G, editor. London: Oxford Univ. Press; 1999. pp. 141–156. [Google Scholar]
- 31.Berridge M J, Bootman M D, Lipp P. Nature (London) 1998;395:645–648. doi: 10.1038/27094. [DOI] [PubMed] [Google Scholar]
- 32.Murphy A, Bredesen G, Cortopaasi C, Wang E, Fiskum G. Proc Natl Acad Sci USA. 1996;93:9893–9898. doi: 10.1073/pnas.93.18.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li N, Karin M. FASEB J. 1999;13:1137–1143. [PubMed] [Google Scholar]
- 34.Rutter G, Rizzuto R. Trends Biochem Sci. 2000;25:215–217. doi: 10.1016/s0968-0004(00)01585-1. [DOI] [PubMed] [Google Scholar]
- 35.Schoonbroodt S, Ferreira V, Best-Belpomme M, Boelaert J, Legrand-Poels S, Korner M, Peitte J. J Immunol. 2000;164:4292–4300. doi: 10.4049/jimmunol.164.8.4292. [DOI] [PubMed] [Google Scholar]
- 36.Mercurio F, Manning A. Oncogene. 1999;18:6163–6171. doi: 10.1038/sj.onc.1203174. [DOI] [PubMed] [Google Scholar]
- 37.Zong C, Yan R, August A, Darnell J E, Jr, Hanafusa H. EMBO J. 1996;15:4515–4525. [PMC free article] [PubMed] [Google Scholar]
- 38.Jove R. Oncogene. 2000;19:2466–2467. doi: 10.1038/sj.onc.1203549. [DOI] [PubMed] [Google Scholar]
- 39.Tai D, Tsai S, Chen Y, Chuang Y, Peng C, Sheen I, Yeh C, Chang K, Huang S, Kuo G, Liaw Y. Hepatology. 2000;31:656–664. doi: 10.1002/hep.510310316. [DOI] [PubMed] [Google Scholar]
- 40.Sun Y. Free Radical Biol Med. 1990;8:583–599. doi: 10.1016/0891-5849(90)90156-d. [DOI] [PubMed] [Google Scholar]
- 41.Corrocher R, Csaril M, Belllisola G, Gabrielli G, Guidi G, De Sandre G. Cancer. 1986;58:1658–1662. doi: 10.1002/1097-0142(19861015)58:8<1658::aid-cncr2820580814>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 42.Butel J. Carcinogenesis. 2000;21:405–426. doi: 10.1093/carcin/21.3.405. [DOI] [PubMed] [Google Scholar]
- 43.Tan S-L, Nakao H, Yupeng H, Vijaysri S, Neddermann P, Jacobs B, Mayer B, Katze M. Proc Natl Acad Sci USA. 1999;96:5533–5538. doi: 10.1073/pnas.96.10.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]