Abstract
Current heart valve prostheses are associated with significant complications, including aggressive immune response, limited valve life expectancy, and inability to grow in juvenile patients. Animal derived “tissue” valves undergo glutaraldehyde fixation to mask tissue antigenicity; however, chronic immunological responses and associated calcification still commonly occur. A heart valve formed from an unfixed bovine pericardium (BP) extracellular matrix (ECM) scaffold, in which antigenic burden has been eliminated or significantly reduced, has potential to overcome deficiencies of current bioprostheses. Decellularization and antigen removal methods frequently use sequential solutions extrapolated from analytical chemistry approaches to promote solubility and removal of tissue components from resultant ECM scaffolds. However, the extent to which such prefractionation strategies may inhibit removal of antigenic tissue components has not been explored. We hypothesize that presence of magnesium in prefractionation steps causes DNA precipitation and reduces removal of nuclear-associated antigenic proteins. Keeping all variables consistent bar the addition or absence of magnesium (2 mM magnesium chloride hexahydrate), residual BP ECM scaffold antigenicity and removed antigenicity were assessed, along with residual and removed DNA content, ECM morphology, scaffold composition, and recellularization potential. Furthermore, we used proteomic methods to determine the mechanism by which magnesium presence or absence affects scaffold residual antigenicity. This study demonstrates that absence of magnesium from antigen removal solutions enhances solubility and subsequent removal of antigenic nuclear-associated proteins from BP. We therefore conclude that the primary mechanism of action for magnesium removal during antigen removal processes is avoidance of DNA precipitation, facilitating solubilization and removal of nuclear-associated antigenic proteins. Future studies are necessary to further facilitate solubility and removal of nuclear-associated antigenic proteins from xenogeneic ECM scaffolds, in addition to an in vivo assessing of the material.
Keywords: : antigen removal, xenogeneic scaffold, decellularization, extracellular matrix, heart valve, tissue engineering
Introduction
Approximately 2.5% of the population suffers from valvular heart disease, and 100,000 heart valve replacements are performed annually in the United States.1,2 The National Heart, Lung, and Blood Institute recognizes that current bioprosthetic heart valve replacements are far from ideal due to chronic immune response, limited valve life expectancy, and inability of the prosthesis to grow in juvenile patients.3
Native extracellular matrix (ECM) architecture of xenogeneic tissues, such as bovine pericardium (BP), provides appropriate structure/function properties and an ECM niche environment which is potentially ideal for cell differentiation and proliferation.4–7 However, antigenic components of xenogeneic tissues represent the critical barrier to increasing use of unfixed biomaterials in clinical practice.8–10 A heart valve formed from an unfixed BP ECM scaffold, where antigenic burden has been eliminated or significantly reduced, has potential to overcome deficiencies of current bioprostheses.11–15
Previous attempts at production of unfixed ECM scaffolds largely utilize decellularization approaches focusing on cellular removal.11,16 Decellularization approaches assume that xenoantigens are exclusively cellular in origin, and therefore, absence of cellular elements equates to removal of antigenic components.17, 18 However, assumptions of the decellularization approach have recently been questioned due to identification of noncellular antigenic components and demonstration of persistence of both known and unknown antigenic components in acellular ECM scaffolds.8,11,18 Consequently, several groups have suggested assessment of residual antigenicity as a specific outcome measure in development of unfixed xenogeneic ECM scaffolds.8,11,16,19–22
Decellularization and antigen removal methods frequently extrapolate protein chemistry and analytical proteomic approaches of solubilizing and promoting removal of undesirable tissue components to produce unfixed xenogeneic ECM scaffolds.23 In doing so, reported ECM scaffold generation processes have utilized a wide range of detergents (e.g., sodium dodecyl sulfate [SDS], Triton X-100, sodium deoxycholate, amidosulfobetaine 14 [ASB-14]), enzymes (e.g., trypsin, DNase, and RNase), buffers (e.g., phosphate- or tris-buffered saline), chelating agents (e.g., EDTA), or salts (e.g., NaCl, MgCl2) previously used in analytical applications.12,24–30 The diversity and complexity of antigenic protein solubilization profiles make it unlikely that a single solution is capable of solubilizing and removing all antigens within a tissue. Consequently, stepwise application of solutions designed to maximize solubilization and removal of antigenic proteins based on shared physiochemical properties may be necessary. However, the question remains as to whether all analytical protein chemistry mechanisms have a positive effect on removal of antigenic proteins from intact tissues, or whether some approaches may actually result in detrimental effects on ultimate material antigenicity?
Magnesium presence has been extensively used in analytical chemistry applications as a prefractionation method, designed to reduce sample complexity, avoid nucleic acid contamination, and, thereby, increase protein solubility during protein extraction.31–34 Coprecipitation of proteins during such initial steps of prefractionation may result in inability to achieve protein resolubilization for later analyses.35,36 The potential for magnesium to precipitate and reduce the removal of antigenic proteins in previously published decellularization or antigen removal protocols has not been investigated.8,30,37 The process of coprecipitation is not limited to magnesium, but can also be achieved using other divalent cations, such as calcium, which may be problematic in tissues with large intracellular calcium stores.31,32 In addition to precipitating DNA, magnesium catalyzes the oxidation of thiol residues and, therefore, produces both intra- and intermolecular disulfide bonds.38–42 Consequently, presence of magnesium or other divalent cations during antigen removal processes has potential to induce protein precipitation either directly through thiol residue oxidation or indirectly through coprecipitation with DNA, limiting removal of such antigens in later solubilization steps.
We hypothesize that absence of magnesium during stepwise antigen removal will enhance protein solubility and subsequent removal of antigenic proteins from BP, while maintaining native ECM structural integrity, composition, and recellularization capacity. This study determines: (1) the effect of magnesium presence or absence during hydrophilic solubilization and/or lipophilic solubilization steps on residual antigenicity of BP ECM scaffolds following antigen removal (BP-AR), (2) the mechanism by which presence or absence of magnesium results in differential removal of protein and DNA from BP-AR ECM scaffolds, (3) whether nuclear-associated proteins that precipitate due to magnesium presence during prefractionation steps are available for later removal, and (4) the effect of presence or absence of magnesium on resultant scaffold structure and recellularization capacity.
Materials and Methods
Unless stated otherwise, all chemicals were purchased from Sigma-Aldrich (St. Louis, MO). Tissue harvest and anti-native BP serum production methods are located in the Supplementary data (Supplementary Data are available online at www.liebertpub.com/tea).
Antigen removal
All removal steps were performed in 2 mL of solution at 4°C, 125 g and changed twice daily, unless otherwise stated. Previously frozen BP strips (see Antigen removal section) were thawed and cut into 0.2 g intact pieces (approximately 1.0 × 1.5 cm). Pieces were incubated in hydrophile solubilizing solution (HSS) for 48 h (100 mM DTT, 100 mM KCl, 0.5 mM Pefabloc, 1% (v/v) AAS, 10 mM Tris-HCl (pH 8.0), in the presence or absence of 2 mM MgCl2–6H2O) followed by 48 h of incubation in lipophile solubilizing solution (LSS) (1% (w/v) amidosulfobetaine-14 in HSS in presence or absence of 2 mM MgCl2–6H2O) at RT.
Each piece of BP had an anatomical control that only underwent 1 min of incubation in HSS and LSS. Subsequently, all samples (treatment and anatomical) underwent 24 h of nuclease digestion (nuclease digest) (2.5 Kunitz units/mL DNAse I, 7.5 Kunitz units/mL RNAse A, 1% (v/v) AAS, 150 mM NaCl, 5 mM MgCl2–6H2O, and 10 mM tris-HCl (pH 7.6)) and 48 h of Tris-buffered saline (TBS) washout (0.5 mM Pefabloc and 1% [v/v] AAS and 10% [v/v] tris-buffered saline). All antigen removal supernatants were retained and stored at −80°C. BP-AR scaffolds were stored in DMEM with 15% (v/v) DMSO at −80°C. All antigen removal experiments were conducted with n = 12 experimental replicates per group, with the three groups designated as: AR+/+ (presence of 2 mM MgCl2–6H2O in both HSS and LSS), AR+/− (presence of 2 mM MgCl2–6H2O in only HSS), and AR−/− (absence of 2 mM MgCl2–6H2O from both HSS and LSS).
Protein extraction
BP-AR scaffolds (see Antigen removal section) underwent manual mincing and were incubated in standard extraction solution (10 mM tris–HCl (pH 8.0), 1 mM DTT, 2 mM MgCl2–6H2O, 10 mM KCl, and 0.5 mM Pefabloc) containing 0.1% (w/v) sodium dodecyl sulfate (SDS) (Bio-Rad, Hercules, CA) at 1000 rpm, 4°C for 1 h. Following centrifugation at 17,000 g, 4°C for 30 min supernatant was recovered and designated as Residual Hydrophile Protein Extract. The remaining pellet was washed twice and then incubated in standard extraction solution containing 1% (w/v) SDS at 1400 rpm, 4°C for 1 h. Following centrifugation, supernatant was recovered and designated as Residual Lipophile Protein Extract. All extracts were stored at −80°C.8
One-dimensional electrophoresis and western blot
Equal volumes of residual hydrophile protein extract, residual lipophile protein extract (see DNA content analysis section), HSS, LSS, nuclease digest, and TBS washout (see Protein extraction section) were assessed using one-dimensional electrophoresis and western blot.8 All blots were probed with anti-native BP serum (1:100 dilution) and assessed specifically for IgG positivity using peroxidase-conjugated mouse anti-rabbit secondary antibody (1:5000 dilution; Jackson ImmunoResearch, West Grove, PA).
Residual antigenicity for hydrophile protein extracts (hydrophiles) and lipophile protein extracts (lipophiles) was quantified using band intensity and densitometry. Residual hydrophile or lipophile antigenicity was calculated as percentage of initial antigenicity (anatomic control) remaining in each BP-AR scaffold. Antigen content of HSS, LSS, nuclease digest, and TBS washout solutions (antigens removed from BP-AR scaffolds) was quantified using band intensity and densitometry in Relative fluorescence units (RFU).
DNA content analysis
DNA content of native BP, BP-AR scaffolds, HSS, LSS, nuclease digest, and TBS washout was quantified using Quant-iT PicoGreen Assay Kit (Invitrogen, Paisley, PA). For BP-AR scaffold analysis, Miltex 3 mm biopsy punches (Integra, Plainsboro, NJ) were taken (wet weight ∼5 mg) and digested in papain solution (99.6 mL phosphate buffer (0.032 M monobasic potassium phosphate and 0.071 M dibasic potassium phosphate), 0.46 mL papain (Sigma P-3125, 27.8 mg/mL), and 0.0816 g N-acetyl-L-cysteine, and 0.1901 g EDTA) for 18 h at 60°C. Solutions were cooled for 30 min at RT and 25 μL of sample added with 75 μL tris-EDTA buffer (10 mM Tris-HCl (pH 8.0) and 1 mM EDTA) and 100 μL PicoGreen Reagent (in DMSO) in opaque 96-well plates.
Plates were incubated at RT and 125 rpm for 5 min before fluorescent detection at 480/520 nm. HSS, LSS, nuclease digest, and TBS washout were assessed similarly, but without initial papain digestion (n = 12 for all groups).
Proteomic analysis
Samples (n = 12 per antigen removal step) were pooled in pairs (final n = 6) to generate sufficient volume for analysis. Nuclease digest and TBS washout samples were concentrated for 1 h and 3 h, respectively, at 4°C, 7500 g using Amicon Ultra-4 Centrifugal Filter Devices (Millipore, Billerica, PA). Nuclease digest and TBS washout samples were precipitated with nine volumes of 100% ethanol and incubated for 1 h at −20°C. Samples were centrifuged at 15,000 g for 15 min at 4°C, and pellets were stored at −80°C for 24 h and then resuspended in 6 M urea/50 mM ammonium bicarbonate. Following proteolytic digestion using a 1:25 ratio of Lys-C/Trypsin (Promega, Madison, WI) and protein, samples were diluted in 1 M urea overnight at 37°C.
Before analysis, samples were concentrated and desalted with C18 resin (MacroSpin Column; Nest Group, Southborough, MA) and subject to liquid chromatography–mass spectrometry on a Exactive Plus Orbitrap Mass Spectrometer (Thermo Scientific, Waltham, MA) in conjunction with an EASY-nLC II nano UHPLC and Proxeon nanospray source. Tandem mass spectra were extracted and charge state deconvoluted with Proteome Discoverer 1.4 (Thermo Scientific) and searched using X! Tandem (The GPM, thegpm.org; version CYCLONE 2013.02.01.1), as well as cRAP database of common laboratory contaminants and an equal number of reverse protein sequences.
Samples were uploaded into Scaffold 4 Proteomics Software, and repeating proteins and protein domains were disregarded. Protein identifications were accepted with greater than 99% probability and replicates containing an average of two or more spectral counts. Effect of magnesium presence or absence was assessed under each of following four categories: (1) Nuclear-associated antigens, (2) Histones, (3) Nuclear-associated proteins according to Genotype Ordering from NCBI protein database, and (4) total protein removal. Spectral sampling assessed average fold changes, with relative abundance orders of magnitude of two or greater taken to be statistically significant.43,44
Histology
Three millimeters biopsy punches from native and BP-AR scaffolds (n = 6 per group) were subjected to formalin-fixation and paraffin embedding followed by Hematoxylin and Eosin (H&E) staining for ECM morphology and nuclei visualization. Verhoeff van Gieson staining (VVG) and Picro-Sirius Red (PSR) staining were utilized for assessment of elastin and collagen organization, respectively. Birefringence quantification was performed throughout the full thickness of the tissue (from the serous parietal surface to the fibrous adventitial surface of the pericardium). Birefringence was calculated using limit-to-threshold within ImageJ to determine the percent area of collagen alignment. All images were taken at 200× and 400× magnification using Nikon Eclipse Ni-E microscope (Nikon, Melville, NY).
Recellularization capacity
All hMSC experiments were performed in accordance with guidelines outlined by the Institutional Stem Cell Research Oversight committee.45 Discs of native and BP-AR scaffolds were generated using 5 mm biopsy punches (Acuderm, Inc., Fort Lauderdale, FL). Discs were washed for 24 h at 125 rpm and 4°C in D20 media (500 mL of DMEM with high glucose (Hyclone Laboratories, South Logan, UT), 1% (v/v) L-glutamine, 1% (v/v) penicillin-streptomycin (10,000 U/mL), and 20% (v/v) fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA)). Discs were transferred to 1.7 mL Eppendorf tubes (n = 7 BP-AR and n = 4 native BP), seeded with 1 × 105 enhanced green fluorescent protein human mesenchymal stem cells (eGFP-hMSCs P4-P7), and cultured in 250 μL D20 media at 37°C, 5% CO2 for 24 h.
Scaffolds were transferred to 96-well plates and incubated in 250 μL D20 media at 37°C, 5% CO2. Cell morphology was subjectively tracked throughout passaging, plating, and seeding using fluorescent inverted microscopy (Nikon TS100; Nikon, Melville, NY). Following 3 days of static culture, scaffolds were imaged using fluorescent microscopy (Leica DMI6000 B; Leica Company, Buffalo Grove, IL). Percent recellularization was calculated using ImageJ software and limit-to-threshold to determine the percent area of eGFP labeled cells on each scaffold following cell seeding.
Statistical analysis
All data are expressed as mean ± standard deviation and were analyzed using one-way analysis of variance with Tukey HSD post hoc test and statistical significance defined at p < 0.05.
For proteomic analysis, individual nuclear-associated proteins with a fold change of two or greater histones and nuclear-associated antigens were analyzed using one-way analysis of variance with Tukey HSD post hoc test and statistical significance defined at p < 0.05.
Results
Residual ECM scaffold antigenicity following antigen removal
Residual hydrophile antigenicity
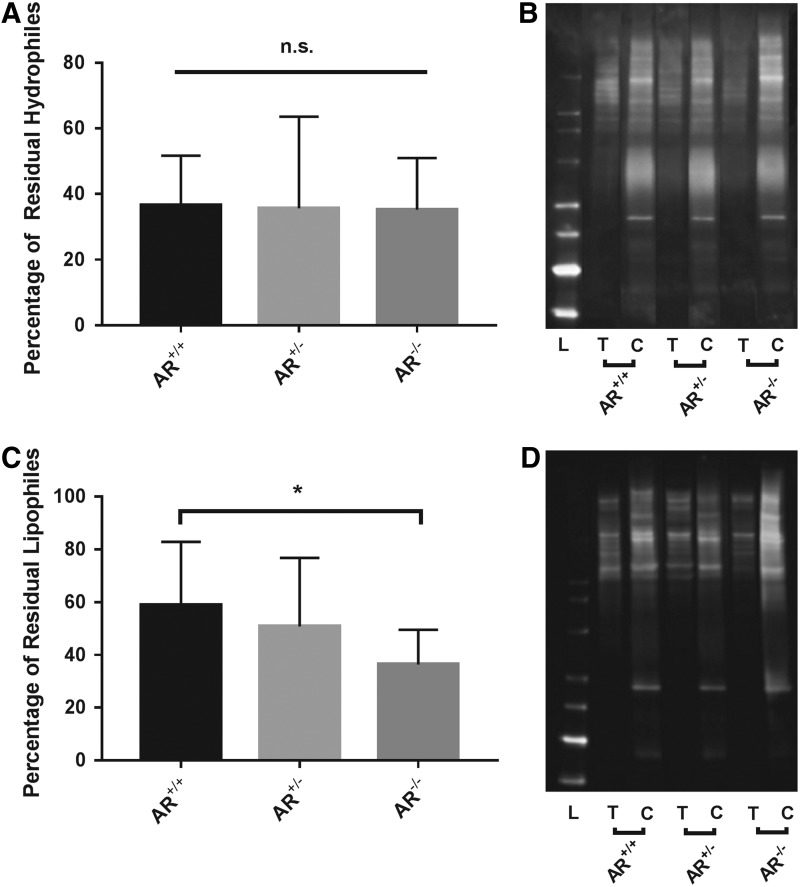
No significant difference in residual hydrophile antigenicity was found among AR−/− (35.22% ± 20.94%), AR+/− (35.60% ± 28.00%), and AR+/+ (36.60% ± 15.09%) scaffolds (p = 0.9877, n = 6 per group) (Fig. 1A, B).
FIG. 1.
Residual hydrophile and lipophile antigenicity of bovine pericardium following antigen removal in the presence or absence of magnesium in solubilizing solutions. Residual hydrophile antigenicity does not change in response to presence or absence of magnesium (A). Representative western blot demonstrating that all treatment groups result in equal percentage reduction of antigenicity compared to their respective controls (B). Absence of magnesium from both hydrophile and lipophile solubilizing solutions results in a statistically significant reduction in residual lipophile antigenicity (C). Representative western blot demonstrating that compared to AR+/+, AR−/− treatment group results in a greater percentage reduction of antigenicity compared to respective control (D). *Signifies p < 0.05, L = Ladder, T = Treated group, and C = Control group (n = 12 per group). n.s. signifies no significant differences between groups.
Residual lipophile antigenicity
Absence of magnesium significantly decreased residual lipophile antigenicity of AR−/− scaffolds (36.35% ± 13.21%) compared to AR+/+ scaffolds (58.86% ± 23.99%, p = 0.04979). Residual lipophile antigenicity of AR+/− scaffolds (50.81% ± 25.96%) did not significantly differ from either AR−/− or AR+/+ (p = 0.2492 and p = 0.6407, respectively, n = 6 per group) (Fig. 1C, D).
Assessment of antigens removed from BP-AR during each antigen removal step
No statistical difference was found in the number of hydrophilic antigens (RFU) removed during the HSS step among AR−/− (10099.51 ± 1172.71), AR+/− (9338.11 ± 1086.35), and AR+/+ (9323.71 ± 1197.47) (p = 0.431, n = 6 per group).
No statistical difference was found in the number of lipophilic antigens removed during the LSS step among AR−/− (13007.90 ± 9439.43), AR+/− (10488.41 ± 8955.90), and AR+/+ (14547.85 ± 10915.70) (p = 0.774, n = 6 per group).
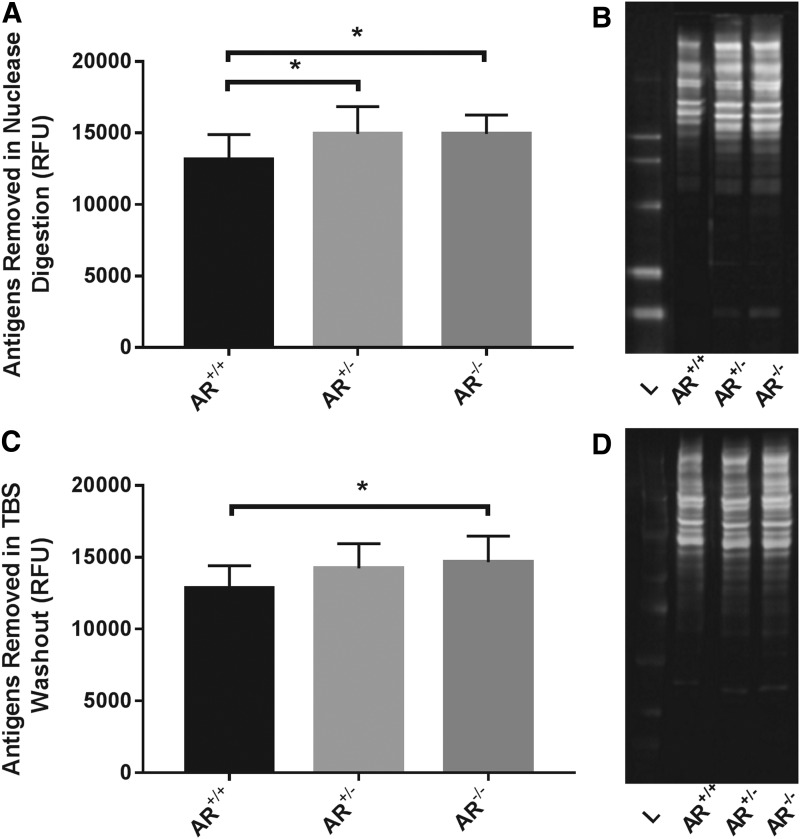
AR−/− (14949.61 ± 1306.02) and AR+/− (14936.83 ± 1907.74) significantly increased antigen removal during the nuclease digestion step compared to AR+/+ (13156.66 ± 1736.36) (p = 0.0335 and p = 0.0350, respectively, n = 12 per group) (Fig. 2A, B).
FIG. 2.
Antigens removed from scaffolds during sequential and differential solubilization steps of antigen removal. Absence of magnesium from hydrophile solubilizing solution alone (AR+/−) and hydrophile and lipophile solubilizing solutions (AR−/−) results in statistically significant increase in removal of antigens during the nuclease digestion step (A). Representative western blot demonstrating increase in antigens removed in absence of magnesium (B). Absence of magnesium from both hydrophile and lipophile solubilizing solutions (AR−/−) results in statistically significant increase in antigens removed in the TBS washout step (C). Representative western blot demonstrating increase in antigens removed in absence of magnesium (D). *Signifies p < 0.05, L = Ladder, T = Treated group, and C = Control group (n = 12 per group). TBS, tris-buffered saline.
AR−/− (14652.51 ± 1823.57) significantly increased antigen removal during the TBS washout step compared to AR+/+ (12820.25 ± 1593.78, p = 0.0340) (Fig. 2C, D). There was no difference in removal of antigens during the TBS washout step between AR+/− (14229.52 ± 1706.44) and AR+/+ (p = 0.1237) or AR−/− (p = 0.8180) (n = 12 per group).
Scaffold DNA content
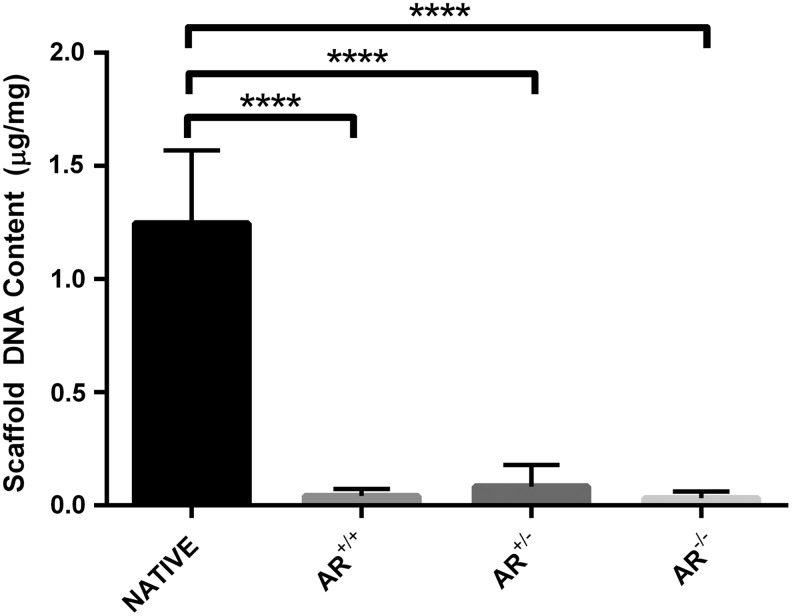
No significant difference in residual DNA was found between BP-AR scaffolds, regardless of presence or absence of magnesium before nuclease digestion (AR−/−, AR+/−, and AR+/+). However, all three groups demonstrated >97% removal of DNA compared to anatomical controls (1.25 ± 0.08 μg/mg DNA, p < 0.0001) (Fig. 3). AR−/− had the greatest reduction in DNA, (0.03 ± 0.03 μg/mg), followed by AR+/+ (0.04 ± 0.03 μg/mg), and finally AR+/− (0.08 ± 0.1 μg/mg) (n = 12 per group) (Fig. 3).
FIG. 3.
Residual DNA content of native bovine pericardium compared to bovine pericardium following antigen removal in the presence or absence of magnesium. Presence or absence of magnesium during antigen removal did not alter scaffold residual DNA content. However, compared to anatomical control, antigen removed scaffolds demonstrated a greater than 97% reduction in residual DNA content. ****Signifies p < 0.0001 (n = 12 per group).
Removed supernatant DNA content
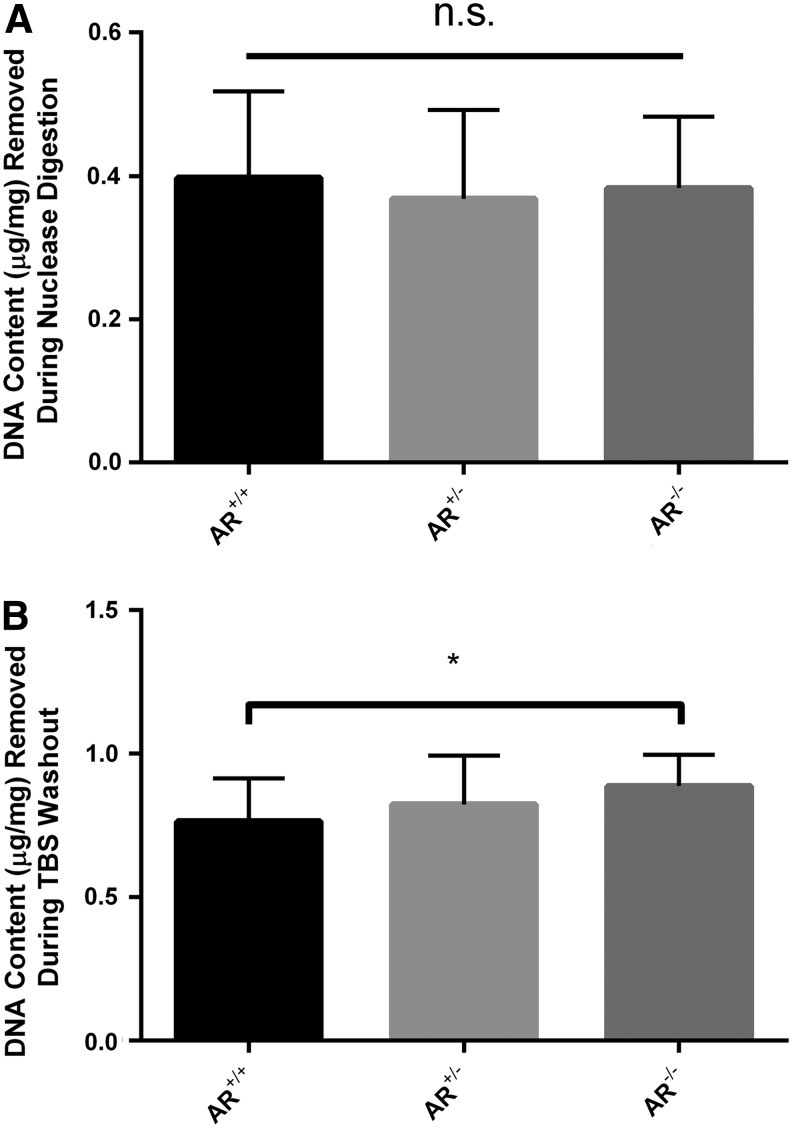
Presence or absence of magnesium in the HSS and LSS did not result in a significant difference in the amount of DNA removed during the nuclease digestion step. AR−/− nuclease digest contained 0.38 ± 0.10 μg of DNA/mg of tissue, whereas AR+/− and AR+/+ contained 0.37 ± 0.12 μg/mg and 0.40 ± 0.12 μg/mg, respectively (p = 0.820, n = 12 per group) (Fig. 4A).
FIG. 4.
DNA content removed from scaffolds during nuclease digestion (NUC) and Tris-buffered saline (TBS) washout steps. DNA removal during nuclease digestion was unaltered by presence or absence of magnesium (A). A trend toward progressively increasing DNA removal during TBS washout was evident in the absence of magnesium compared to its presence (B), although this finding failed to reach statistical significance (n = 12 per group). * signifies p < 0.05. n.s. signifies no significant differences between groups.
No statistically significant difference in DNA was noted in TBS washout when analyzed using a one-way ANOVA. However, a trend was identified which indicated that absence of magnesium in both HSS and LSS results in a greater amount of DNA removed during the TBS washout step, which was significant between AR−/− (0.89 ± 0.11 μg of DNA/mg of tissue) and AR+/+ (0.76 ± 0.15 μg/mg) using a Student t-test (p = 0.0311) (n = 12 per group) (Fig. 4B).
Proteomic analysis
Removal of nuclear-associated antigens glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and elongation factor 1-alpha 1 (EF1α1) during the nuclease digestion step was not significantly affected by presence or absence of magnesium (p = 0.0952 and p = 0.5522, respectively).46–49 Conversely, during TBS washout step both antigens demonstrated statistically greater removal in the absence of magnesium (94.67 ± 9.97 and 35.5 ± 4.42 spectra, respectively) compared to its presence (69.17 ± 17.60 and 15.2 ± 3.90 spectra, respectively) (p = 0.0418 and p = 0.0095, respectively) (n = 6 per group).
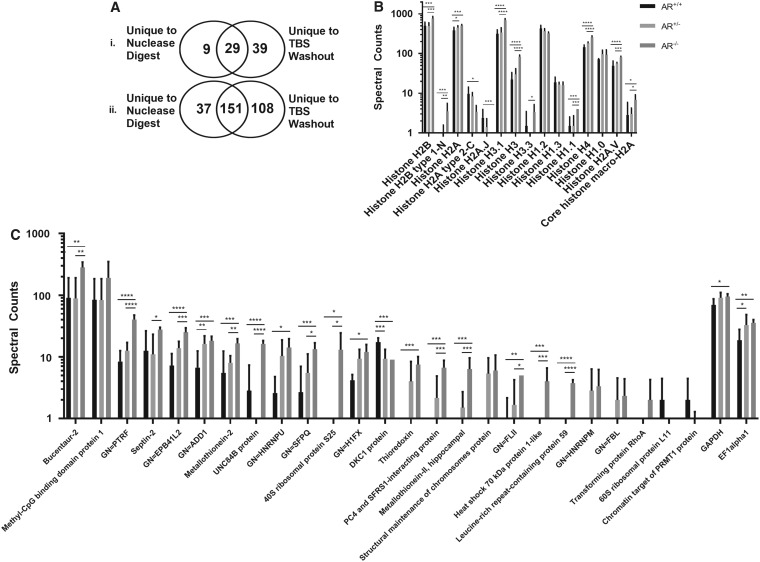
Histones were examined as a depiction of nuclear-associated protein solubilization during antigen removal. Of the 12 histones removed from BP-AR scaffolds during the nuclear digestion step, 9 demonstrated increased removal in AR+/+ although only 1 (Histone H3.3) was statistically significantly increased (p = 0.0498) (Supplementary Fig. S1A). Of the 15 histones removed during TBS washout, 11 had increased removal in AR−/−, 9 of which were statistically significantly increased compared to AR+/+ (p < 0.05) (Fig. 5B).
FIG. 5.
Effect of magnesium presence or absence on protein removal during nuclease digestion and TBS washout steps. Nuclear-associated protein removal is greater during TBS washout than during nuclease digestion (Ai). Total protein removal is greater during TBS washout than during nuclease digestion (Aii). Absence of magnesium increased both histone (B) and nuclear-associated protein (C) removal during TBS washout. Results plotted in Log [10] scale. ****signifies p < 0.0001, ***signifies p < 0.005, **signifies p < 0.015, and *signifies p < 0.05 (n = 6 per group).
Nuclease digest and TBS washout solutions shared 29 nuclear-associated proteins, with 9 unique proteins removed during nuclease digestion step and 39 unique proteins removed during TBS washout (Fig. 5A). Of the nuclear-associated proteins removed during nuclease digest of AR−/−, 11 proteins had increased removal, three of which had >2-fold increase. Conversely, 26 nuclear-associated proteins had increased removal in nuclease digest AR+/+, 7 of which had >2-fold increase in spectral counts, yet only 2 (Major Vault Protein and an uncharacterized protein, p < 0.005 and p < 0.05, respectively) were statistically significant (Supplementary Fig. S1B). Four additional proteins unique to AR+/+ were also present.
Overwhelmingly though, 61 of the nuclear-associated proteins removed in TBS washout step had greater removal in AR−/−, 22 of which had >2-fold increase in spectral counts and 17 of those demonstrated a statistically significant (p < 0.025) increase in spectral counts. In addition to the 22 nuclear-associated proteins with >2-fold increase, 12 proteins were completely unique to AR−/−. Seven nuclear-associated proteins in TBS washout step had a greater number of spectra in AR+/+, three of which had >2-fold increase in spectral counts and one of those (DKC1 protein, p < 0.005) was statistically relevant (Fig. 5C).
When examining the distribution of proteins as a whole from the study, 188 proteins were removed in nuclease digest, 37 of which were completely unique to nuclease alone. Conversely, 259 proteins were removed in TBS washout, 108 of which were unique solely to TBS (Fig. 5A).
Histology
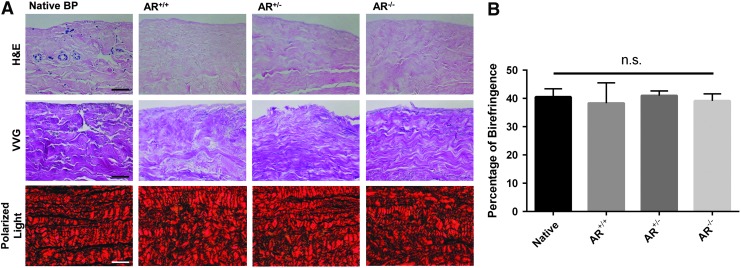
No qualitative differences in ECM morphology were observed between the three groups (AR−/−, AR+/−, AR+/+) and native BP on H&E sections. However, all three groups following antigen removal exhibited almost complete elimination of nuclei or nuclear remnants compared to native BP (Fig. 6A).
FIG. 6.
Gross histological morphology of native and antigen removed bovine pericardium in presence or absence of magnesium. Preservation of histological ECM morphology and marked reduction in residual nuclei are evident in H&E staining compared to native tissue. Verhoeff van Gieson (VVG) staining demonstrates preservation of collagen and elastin structure following antigen removal. Collagen structure was unchanged in PSR stained samples. PSR birefringence indicated no change in type I (thick fibers, yellow-orange) and type III (thin fibers, green) collagen (A). Quantification of birefringence demonstrates no change in collagen alignment between native and antigen removed BP (B). (n = 6 per antigen removed group, n = 5 per native group). Polarized light scale bar represents 100 μm, otherwise scale bar represents 50 μm. BP, bovine pericardium; ECM, extracellular matrix; H&E, hematoxylin and eosin; PSR, Picro-Sirius Red. n.s. signifies no significant differences between groups.
In VVG-stained sections, both collagen and elastin content and organization were grossly maintained between the three antigen removed groups (AR−/−, AR+/−, AR+/+) and native BP (Fig. 6A).
In PSR-stained sections no difference in collagen content or organization was observed between the three antigen removed groups and native BP (Fig. 6A). Collagen alignment was not statistically significantly different between native BP (40.48% ± 2.87%) and the three antigen removed groups (AR+/+ 38.29% ± 7.19%, AR+/− 40.99% ± 1.62%, AR−/− 39.11 ± 2.46; p > 0.05) (Fig. 6B) (n = 6 per group, n = 5 for native BP).
Recellularization capacity
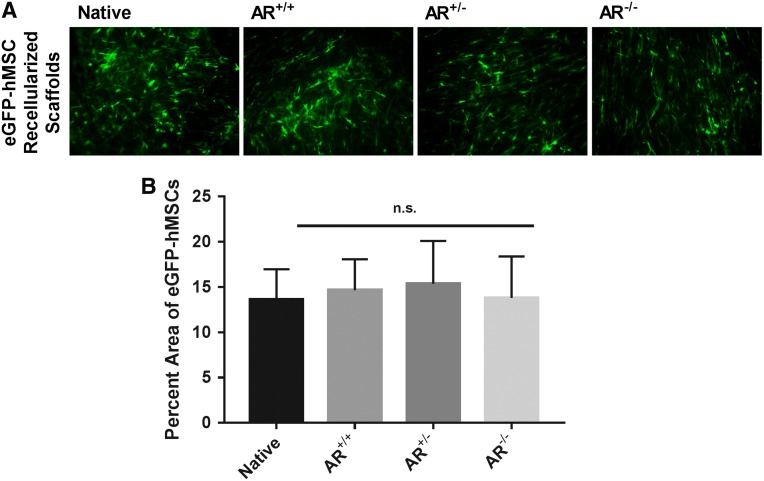
There were no observed changes in hMSC morphology or recellularization capacity (p = 0.9113, (Fig. 7B) between the three groups compared to native BP. All scaffolds exhibited cellular confluency and similar cell morphology (n = 7 for AR−/−, AR+/−, AR+/+ and n = 4 for native BP) (Fig. 7A).
FIG. 7.
Recellularization capacity of native and antigen removed bovine pericardium in the presence or absence of magnesium. hMSC morphology was unchanged following seeding on either native BP or BP-AR scaffolds (A). No significant difference in recellularization capacity, assessed by percent area of scaffolds positive for eGFP labeled hMSCs, was present between groups (B). (n = 7 per antigen removed group, n = 4 per native group). Scale bar represents 100 μm. eGFP, enhanced green fluorescent protein; hMSC, human mesenchymal stem cell. n.s. signifies no significant differences between groups.
Discussion
This study demonstrates that proteomic methods of prefractionation through presence of magnesium in antigen removal solutions reduce efficiency of antigen removal from BP, although mechanism is predominantly due to DNA precipitation (Fig. 8). This study specifically establishes that: (1) absence of magnesium in both hydrophile and lipophile steps of antigen removal significantly reduces residual lipophilic antigen content of BP-AR ECM scaffolds. (2) This reduction in antigenicity is achieved due to increased removal of nuclear-associated antigenic proteins during the nuclease digestion and washout steps. (3) Nuclear-associated proteins that precipitate due to magnesium presence are unavailable for later removal. (4) BP ECM scaffolds generated in absence of magnesium exhibit native ECM structure, collagen organization, and recellularization potential.
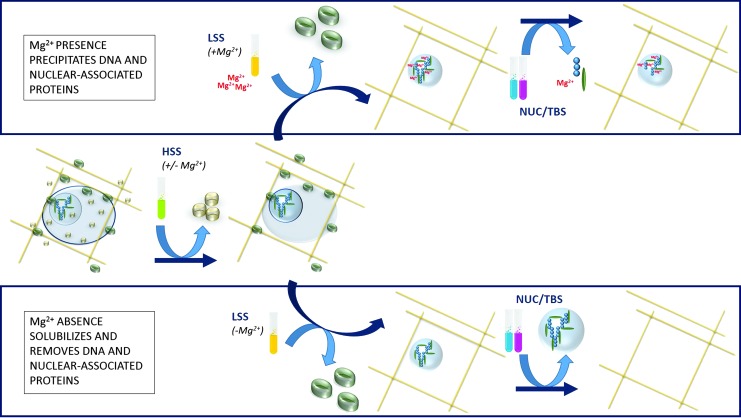
FIG. 8.
Proposed mechanism for effects of magnesium on nuclear-associated protein precipitation and subsequent removal from bovine pericardium. Magnesium presence before nuclease digestion causes greater DNA precipitation, and antigenic nuclear-associated proteins are unavailable for subsequent removal. Magnesium absence before nuclease digestion avoids DNA precipitation and, thereby, promotes antigenic nuclear-associated protein removal.
NHLBI xenotransplantation working group identified material antigenicity as the principle barrier to expanding use of xenogeneic tissues in clinical practice.3 Consequently, reduction or elimination of biomaterial antigenicity is the stated goal in production of xenogeneic ECM scaffolds for heart valve tissue engineering. Both in vitro and in vivo studies have exhibited that assessment of xenogeneic scaffold cellularity is poorly predictive of in vivo recipient host immune response. Indeed, Kasimir et al. indicated that insufficient removal of xenogeneic tissue antigens in decellularized SynerGraft porcine aortic valve precipitated a catastrophic recipient graft-specific immune response.11,20,22 Recent studies have demonstrated that specific assessment of ECM scaffold antigen content more strongly correlates with recipient graft-specific adaptive immune responses.11,20,22,50 In this study we demonstrate that absence of magnesium increased removal of lipophilic antigens by 38.2% compared to presence of magnesium (Fig. 1C, D), while presence or absence of magnesium made no difference in removal of hydrophilic antigens (Fig. 1A, D). To determine at what step the reduction in lipophile antigenicity occurred, supernatants of all four steps in antigen removal were examined. Statistically significantly increased removal of lipophilic antigens was identified in both nuclease digestion and TBS washout steps in absence of magnesium (Fig. 2A–D). Furthermore, proteomic analysis confirmed statistically significantly increased removal of two nuclear-associated antigens (GADPH and EF1α1) during TBS washout (26.9% and 57.2% increase in removal, respectively) (Fig. 5C).46,51 Although both antigens are known to also reside in the cytoplasm, their increased removal following nuclease digestion supports the conclusion that presence or absence of magnesium alters availability of the nuclear-associated fraction of each antigen for later removal.48,52–56 Currently, the threshold below which antigenicity must be reduced to avoid recipient graft-specific immune responses remains unknown. Consequently, future in vivo studies are necessary to ascertain the effects of decreased lipophile antigenicity due to absence of magnesium on recipient graft-specific immune response. In summary, due to its ability to increase removal of lipophilic antigens, a BP-AR scaffold generated in absence of magnesium has potential as a clinically relevant biomaterial.
Maintaining solubility of antigenic components is critical to achieving their removal from xenogeneic tissues during decellularization or antigen removal processes. Two possible mechanisms exist which could explain the effect of magnesium on residual lipophile antigenicity: (1) Magnesium causes coprecipitation of DNA through electrostatic interactions resulting from counterion condensation (for weak affinity), or binding to a specific site and production of a hydrophobic complex (for strong affinity).31 (2) Magnesium causes the catalyst of thiol oxidation and formation of protein precipitation through development of inter- and intramolecular disulfide bonds.40–42 Regardless of underlying mechanism, resolubilization efficiency following protein precipitation is frequently less than 100%.35,36 Consequently, allowing precipitation of antigenic components during decellularization/antigen removal processes would be expected to increase residual ECM scaffold antigenicity. The differential effect of magnesium absence on lipophile residual antigenicity supports magnesium-induced DNA precipitation as the predominant mechanism for this beneficial effect, since thiol oxidation would be expected to affect both hydrophilic and lipophilic proteins equally. Although the remaining DNA content in BP-AR was too small to see a significant difference between groups (Fig. 3), the increased removal of DNA in the absence of magnesium during TBS washout (Fig. 4B) supports avoidance of DNA precipitation, especially given the greater volume of protein removal during TBS washout (Fig. 5A). To further confirm this mechanism, proteins removed during nuclease digestion and TBS washout steps underwent liquid chromatography–mass spectrometry to ascertain specific changes in nuclear-associated protein removal.57 Histone removal as a marker of DNA precipitation was overwhelmingly increased in the absence of magnesium during TBS washout (Fig. 5B), with 60% of identified histones demonstrating statistically increased removal in the absence of magnesium compared to its presence. Other nuclear-associated proteins compiled using NCBI Gene Ontology were also compared and showed remarkably similar data to histones. Absence of magnesium resulted in statistically increased removal of 68% of identified nuclear-associated proteins (Fig. 5C). These proteomic data further support the proposed mechanism of action that magnesium presence results in precipitation of nuclear-associated proteins, which cannot be resolubilized for later removal. More so, precipitation of DNA can occur through a multitude of divalent cations. One such cation is calcium, which has a slightly higher efficiency curve in prefractionation than magnesium.31 This has implications for calcium-rich tissues, like cardiac muscle, that have endogenous calcium stores which may lead to DNA precipitation during ECM matrix scaffold production.31,58,59 Future studies are needed to ascertain whether other divalent cations inhibit antigen removal by a similar mechanism as that of magnesium. Furthermore, the potential additive benefits of heavy metal chelators (e.g., EDTA) on scaffold residual antigenicity also require further investigation in a tissue-specific manner.60,61 The current findings demonstrate that absence of magnesium before nuclear digestion prevents DNA precipitation and, thereby, maintains DNA-associated protein solubility for later removal.
An ideal antigen removal method would decrease scaffold antigenicity, while preserving the structure and recellularization capacity of the resultant ECM scaffold.11,16,19–22 Previously published data indicated no change in the gross morphological appearance of BP-AR scaffolds in the presence of magnesium.8,18 Conversely, Tsuchiya et al. noted detrimental effects of changes in pH during rat lung decellularization, leading to altered ECM structure, morphology, recellularization, and host response toward the ECM scaffold.62 Their findings included a significant decrease in total collagen content at pH 10 and above, as well as significant removal of laminin, fibronectin, and elastin. Similarly, Poornejad et al. noted that depending on the decellularization method, deleterious effects on cell viability and proliferation, collagenous fiber networks, glycosaminoglycans, fibroblast growth factor content, and overall ECM structure could be identified.63 In this study, despite the marked reduction in residual nuclei and DNA content, no change in ECM scaffold structure was found between BP-AR scaffolds and native tissue, regardless of magnesium presence or absence (Fig. 6A). Quantitatively there was no significant change in collagen type I and III (yellow/red and green birefringence, respectively), which is important since these are the two most prominent structural proteins in native BP, comprising approximately 80–90% the biomaterial's dry weight (Fig. 6B).64–66 The immunomodulatory properties of human mesenchymal stem cells (hMSCs) may be beneficial to modulate residual recipient graft-specific immune responses toward xenogeneic ECM scaffolds.67–70 Consequently, we utilized hMSC as a potentially important cell type for scaffold repopulation studies. Previously reported xenogeneic ECM scaffold generation studies have reported variable recellularization efficiency depending on decellularization method used.62,63 In the current study, no change in recellularization capacity of BP-AR scaffolds generated in the absence of magnesium was found compared to native BP (Fig. 7A, B). Although recellularization of native BP is not an essential parameter for a transplant biomaterial, the fact that cellular repopulation and migration is unaltered compared to native BP demonstrates that: (1) the resultant scaffold is nontoxic to repopulating cells, (2) and scaffold structural preservation results in conservancy of hMSC elongated cellular morphology and binding capacity. Despite these promising results, further in vivo studies are necessary to determine the effect of hMSC repopulation on recipient immune and regenerative response toward BP-AR scaffolds. The combination of decreased antigen removal, retained ECM properties, and recellularization capacity suggests that scaffolds made in the absence of magnesium have potential application as a biomaterial for heart valve prostheses.
Conclusions
In this study, we demonstrate that direct application of analytical proteomic approaches cannot be directly translated to ECM scaffold production. Consequently, special consideration of the effect of each component within the solution is required in development of antigen removal strategies derived from proteomics in ECM scaffold generation. Mechanistically, the presence of magnesium, and potentially other cations, in antigen removal solutions results in increased DNA precipitation and, subsequently, increased precipitation of antigenic nuclear-associated proteins. Based on the results of this study, we conclude that absence of magnesium significantly reduced resultant ECM scaffold antigenicity, while maintaining matrix structure and compatibility with cellular repopulation. Therefore BP-AR ECM scaffolds generated in the absence of magnesium may represent a potentially ideal biomaterial for future heart valve prostheses.
Supplementary Material
Acknowledgments
The authors acknowledge funding from the National Institutes of Health (NIH R01HL115205). In addition, the authors thank the laboratory of Dr. Clare E. Yellowley for use of their facilities.
Disclosure Statement
No competing financial interests exist.
References
- 1.Go A.S., Mozaffarian D., Roger V.L., Benjamin E.J., Berry J.D., Borden W.B., Bravata D.M., Dai S., Ford E.S., Fox C.S., Franco S., Fullerton H.J., Gillespie C., Hailpern S.M., Heit J.A., Howard V.J., Huffman M.D., Kissela B.M., Kittner S.J., Lackland D.T., Lichtman J.H., Lisabeth L.D., Magid D., Marcus G.M., Marelli A., Matchar D.B., McGuire D.K., Mohler E.R., Moy C.S., Mussolino M.E., Nichol G., Paynter N.P., Schreiner P.J., Sorlie P.D., Stein J., Turan T.N., Virani S.S., Wong N.D., Woo D., Turner M.B., American Heart Association Statistics, C., and Stroke Statistics S. Heart disease and stroke statistics—2013 update: a report from the American Heart Association. Circulation 127, e6, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roger V.L., Go A.S., Lloyd-Jones D.M., Adams R.J., Berry J.D., Brown T.M., Carnethon M.R., Dai S., de Simone G., Ford E.S., Fox C.S., Fullerton H.J., Gillespie C., Greenlund K.J., Hailpern S.M., Heit J.A., Ho P.M., Howard V.J., Kissela B.M., Kittner S.J., Lackland D.T., Lichtman J.H., Lisabeth L.D., Makuc D.M., Marcus G.M., Marelli A., Matchar D.B., McDermott M.M., Meigs J.B., Moy C.S., Mozaffarian D., Mussolino M.E., Nichol G., Paynter N.P., Rosamond W.D., Sorlie P.D., Stafford R.S., Turan T.N., Turner M.B., Wong N.D., and Wylie-Rosett J. American Heart Association Statistics C and Stroke Statistics S. Heart disease and stroke statistics—2011 update: a report from the American Heart Association. Circulation 123, e18, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumgartner W.A., Burrows S., del Nido P.J., Gardner T.J., Goldberg S., Gorman R.C., Letsou G.V., Mascette A., Michler R.E., Puskas J.D., Rose E.A., Rosengart T.K., Sellke F.W., Shumway S.J., Wilke N., National Heart L and Blood Institute Working Group on Future Direction in Cardiac S. Recommendations of the National Heart, Lung, and Blood Institute Working Group on Future Direction in Cardiac Surgery. Circulation 111, 3007, 2005 [DOI] [PubMed] [Google Scholar]
- 4.Badylak S.F., Freytes D.O., and Gilbert T.W. Extracellular matrix as a biological scaffold material: structure and function. Acta Biomater 5, 1, 2009 [DOI] [PubMed] [Google Scholar]
- 5.Iop L., Renier V., Naso F., Piccoli M., Bonetti A., Gandaglia A., Pozzobon M., Paolin A., Ortolani F., Marchini M., Spina M., De Coppi P., Sartore S., and Gerosa G. The influence of heart valve leaflet matrix characteristics on the interaction between human mesenchymal stem cells and decellularized scaffolds. Biomaterials 30, 4104, 2009 [DOI] [PubMed] [Google Scholar]
- 6.Griffiths L.G., Choe L., Lee K.H., Reardon K.F., and Orton E.C. Protein extraction and 2-DE of water- and lipid-soluble proteins from bovine pericardium, a low-cellularity tissue. Electrophoresis 29, 4508, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt C.E., and Baier J.M. Acellular vascular tissues: natural biomaterials for tissue repair and tissue engineering. Biomaterials 21, 2215, 2000 [DOI] [PubMed] [Google Scholar]
- 8.Wong M.L., Wong J.L., Athanasiou K.A., and Griffiths L.G. Stepwise solubilization-based antigen removal for xenogeneic scaffold generation in tissue engineering. Acta Biomater 9, 6492, 2013 [DOI] [PubMed] [Google Scholar]
- 9.Simionescu D.T., Lovekamp J.J., and Vyavahare N.R. Degeneration of bioprosthetic heart valve cusp and wall tissues is initiated during tissue preparation: an ultrastructural study. J Heart Valve Dis 12, 226, 2003 [PubMed] [Google Scholar]
- 10.Liao J., Joyce E.M., and Sacks M.S. Effects of decellularization on the mechanical and structural properties of the porcine aortic valve leaflet. Biomaterials 29, 1065, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goncalves A.C., Griffiths L.G., Anthony R.V., and Orton E.C. Decellularization of bovine pericardium for tissue-engineering by targeted removal of xenoantigens. J Heart Valve Dis 14, 212, 2005 [PubMed] [Google Scholar]
- 12.Vesely I. Heart valve tissue engineering. Circ Res 97, 743, 2005 [DOI] [PubMed] [Google Scholar]
- 13.Human P., and Zilla P. Inflammatory and immune processes: the neglected villain of bioprosthetic degeneration? J Long Term Eff Med Implants 11, 199, 2001 [PubMed] [Google Scholar]
- 14.Zilla P., Human P., and Bezuidenhout D. Bioprosthetic heart valves: the need for a quantum leap. Biotechnol Appl Biochem 40, 57, 2004 [DOI] [PubMed] [Google Scholar]
- 15.Erdbrugger W., Konertz W., Dohmen P.M., Posner S., Ellerbrok H., Brodde O.E., Robenek H., Modersohn D., Pruss A., Holinski S., Stein-Konertz M., and Pauli G. Decellularized xenogenic heart valves reveal remodeling and growth potential in vivo. Tissue Eng 12, 2059, 2006 [DOI] [PubMed] [Google Scholar]
- 16.Meyer S.R., Chiu B., Churchill T.A., Zhu L., Lakey J.R., and Ross D.B. Comparison of aortic valve allograft decellularization techniques in the rat. J Biomed Mater Res A 79, 254, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Griffiths L.G., Choe L.H., Reardon K.F., Dow S.W., and Christopher Orton E. Immunoproteomic identification of bovine pericardium xenoantigens. Biomaterials 29, 3514, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong M.L., Leach J.K., Athanasiou K.A., and Griffiths L.G. The role of protein solubilization in antigen removal from xenogeneic tissue for heart valve tissue engineering. Biomaterials 32, 8129, 2011 [DOI] [PubMed] [Google Scholar]
- 19.Sandor M., Xu H., Connor J., Lombardi J., Harper J.R., Silverman R.P., and McQuillan D.J. Host response to implanted porcine-derived biologic materials in a primate model of abdominal wall repair. Tissue Eng Part A 14, 2021, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Kasimir M.T., Rieder E., Seebacher G., Nigisch A., Dekan B., Wolner E., Weigel G., and Simon P. Decellularization does not eliminate thrombogenicity and inflammatory stimulation in tissue-engineered porcine heart valves. J Heart Valve Dis 15, 278; discussion 286, 2006 [PubMed] [Google Scholar]
- 21.Bloch O., Golde P., Dohmen P.M., Posner S., Konertz W., and Erdbrugger W. Immune response in patients receiving a bioprosthetic heart valve: lack of response with decellularized valves. Tissue Eng Part A 17, 2399, 2011 [DOI] [PubMed] [Google Scholar]
- 22.Kasimir M.T., Rieder E., Seebacher G., Wolner E., Weigel G., and Simon P. Presence and elimination of the xenoantigen gal (alpha1, 3) gal in tissue-engineered heart valves. Tissue Eng 11, 1274, 2005 [DOI] [PubMed] [Google Scholar]
- 23.Leimgruber R. Extraction and solubilization of Proteins for Proteomic Studies. In: Walker J., ed. The Proteomics Protocols Handbook. Towana, NJ: Humana Press, 2005, pp. 1–18 [Google Scholar]
- 24.Cordwell S.J. Sequential extraction of proteins by chemical reagents. Methods Mol Biol 424, 139, 2008 [DOI] [PubMed] [Google Scholar]
- 25.Lehner I., Niehof M., and Borlak J. An optimized method for the isolation and identification of membrane proteins. Electrophoresis 24, 1795, 2003 [DOI] [PubMed] [Google Scholar]
- 26.Molloy M.P., Herbert B.R., Walsh B.J., Tyler M.I., Traini M., Sanchez J.C., Hochstrasser D.F., Williams K.L., and Gooley A.A. Extraction of membrane proteins by differential solubilization for separation using two-dimensional gel electrophoresis. Electrophoresis 19, 837, 1998 [DOI] [PubMed] [Google Scholar]
- 27.Ramsby M.L., Makowski G.S., and Khairallah E.A. Differential detergent fractionation of isolated hepatocytes: biochemical, immunochemical and two-dimensional gel electrophoresis characterization of cytoskeletal and noncytoskeletal compartments. Electrophoresis 15, 265, 1994 [DOI] [PubMed] [Google Scholar]
- 28.Little M.H. Whole Organ Decellularization. In: Little M.H., ed. Kidney Development, Disease, Repair, and Regeneration. London: Academic Press, 2016, p. 573 [Google Scholar]
- 29.Crapo P.M., Gilbert T.W., and Badylak S.F. An overview of tissue and whole organ decellularization processes. Biomaterials 32, 3233, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Genovese L., Zawada L., Tosoni A., Ferri A., Zerbi P., Allevi R., Nebuloni M., and Alfano M. Cellular localization, invasion, and turnover are differently influenced by healthy and tumor-derived extracellular matrix. Tissue Eng Part A 20, 2005, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Frutos M., Raspaud E., Leforestier A., and Livolant F. Aggregation of nucleosomes by divalent cations. Biophys J 81, 1127, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J., and Russell D.W. Calcium-phosphate-mediated Transfection of Eukaryotic Cells with Plasmid DNAs. CSH Protoc 2006, pii: , 2006 [DOI] [PubMed] [Google Scholar]
- 33.Lin J., Yan Y.Y., Ou T.M., Tan J.H., Huang S.L., Li D., Huang Z.S., and Gu L.Q. Effective detection and separation method for G-quadruplex DNA based on its specific precipitation with Mg(2+). Biomacromolecules 11, 3384, 2010 [DOI] [PubMed] [Google Scholar]
- 34.Skovgaard O. Selective precipitation of RNA with Mg2+ improves the purification of plasmid DNA. Trends Genet 6, 140, 1990 [DOI] [PubMed] [Google Scholar]
- 35.Gorg A., Weiss W., and Dunn M.J. Current two-dimensional electrophoresis technology for proteomics. Proteomics 4, 3665, 2004 [DOI] [PubMed] [Google Scholar]
- 36.Doucette A.A., Vieira D.B., Orton D.J., and Wall M.J. Resolubilization of precipitated intact membrane proteins with cold formic acid for analysis by mass spectrometry. J Proteome Res 13, 6001, 2014 [DOI] [PubMed] [Google Scholar]
- 37.Yang C. Decellularized Omentum Matrix and Uses Thereof. Oct. 4, 2013; US 20140271784 A1
- 38.Rabilloud T. Protein Solubility in Two-Dimensional Electrophoresis. In: Walker J., ed. The Protein Protocols Handbook. Totowa: Humana Press, 2002, pp. 78–79 [Google Scholar]
- 39.Weiss W., and Gorg A. Protein Solubilization. In: Hagen J., ed. Proteomics Sample Preparation. Weinheim: Wiley-VCH, 2008, p. 137 [Google Scholar]
- 40.MacLellan S.R., and Forsberg C.W. Properties of the major non-specific endonuclease from the strict anaerobe Fibrobacter succinogenes and evidence for disulfide bond formation in vivo. Microbiology 147, 315, 2001 [DOI] [PubMed] [Google Scholar]
- 41.Clancey C.J., and Gilbert H.F. Thiol/disulfide exchange in the thioredoxin-catalyzed reductive activation of spinach chloroplast fructose-1,6-bisphosphatase. Kinetics and thermodynamics. J Biol Chem 262, 13545, 1987 [PubMed] [Google Scholar]
- 42.Kolesnik B., Heine C.L., Schmidt R., Schmidt K., Mayer B., and Gorren A.C. Aerobic nitric oxide-induced thiol nitrosation in the presence and absence of magnesium cations. Free Radic Biol Med 76, 286, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu H., Sadygov R.G., and Yates J.R., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 76, 4193, 2004 [DOI] [PubMed] [Google Scholar]
- 44.Dicker L., Lin X., and Ivanov A.R. Increased power for the analysis of label-free LC-MS/MS proteomics data by combining spectral counts and peptide peak attributes. Mol Cell Proteomics 9, 2704, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin X., Robinson M., Petrie T., Spandler V., Boyd W.D., and Sondergaard C.S. Small intestinal submucosa-derived extracellular matrix bioscaffold significantly enhances angiogenic factor secretion from human mesenchymal stromal cells. Stem Cell Res Ther 6, 164, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costa R.M., Nogueira F., de Sousa K.P., Vitorino R., and Silva M.S. Immunoproteomic analysis of Plasmodium falciparum antigens using sera from patients with clinical history of imported malaria. Malar J 12, 100, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krause R.G., Grobler A.F., and Goldring J.P. Comparing Antibody Responses in Chickens Against Plasmodium falciparum Lactate Dehydrogenase and Glyceraldehyde-3-phosphate Dehydrogenase with Freund's and Pheroid(R) Adjuvants. Immunol Invest 44, 627, 2015 [DOI] [PubMed] [Google Scholar]
- 48.Bergquist J., Gobom J., Blomberg A., Roepstorff P., and Ekman R. Identification of nuclei associated proteins by 2D-gel electrophoresis and mass spectrometry. J Neurosci Methods 109, 3, 2001 [DOI] [PubMed] [Google Scholar]
- 49.Doyle A., Crosby S.R., Burton D.R., Lilley F., and Murphy M.F. Actin bundling and polymerisation properties of eukaryotic elongation factor 1 alpha (eEF1A), histone H2A-H2B and lysozyme in vitro. J Struct Biol 176, 370, 2011 [DOI] [PubMed] [Google Scholar]
- 50.Kasimir M.T., Rieder E., Seebacher G., Silberhumer G., Wolner E., Weigel G., and Simon P. Comparison of different decellularization procedures of porcine heart valves. Int J Artif Organs 26, 421, 2003 [DOI] [PubMed] [Google Scholar]
- 51.Fu Q., Wei Z., Liu X., Xiao P., Lu Z., and Chen Y. Glyceraldehyde-3-phosphate dehydrogenase, an immunogenic Streptococcus equi ssp. zooepidemicus adhesion protein and protective antigen. J Microbiol Biotechnol 23, 579, 2013 [DOI] [PubMed] [Google Scholar]
- 52.Gangwani L., Mikrut M., Galcheva-Gargova Z., and Davis R.J. Interaction of ZPR1 with translation elongation factor-1alpha in proliferating cells. J Cell Biol 143, 1471, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanders J., Brandsma M., Janssen G.M., Dijk J., and Moller W. Immunofluorescence studies of human fibroblasts demonstrate the presence of the complex of elongation factor-1 beta gamma delta in the endoplasmic reticulum. J Cell Sci 109(Pt 5), 1113, 1996 [DOI] [PubMed] [Google Scholar]
- 54.Hara M.R., Cascio M.B., and Sawa A. GAPDH as a sensor of NO stress. Biochim Biophys Acta 1762, 502, 2006 [DOI] [PubMed] [Google Scholar]
- 55.Hara M.R., and Snyder S.H. Nitric oxide-GAPDH-Siah: a novel cell death cascade. Cell Mol Neurobiol 26, 527, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kornberg M.D., Sen N., Hara M.R., Juluri K.R., Nguyen J.V., Snowman A.M., Law L., Hester L.D., and Snyder S.H. GAPDH mediates nitrosylation of nuclear proteins. Nat Cell Biol 12, 1094, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pavelka N., Fournier M.L., Swanson S.K., Pelizzola M., Ricciardi-Castagnoli P., Florens L., and Washburn M.P. Statistical similarities between transcriptomics and quantitative shotgun proteomics data. Mol Cell Proteomics 7, 631, 2008 [DOI] [PubMed] [Google Scholar]
- 58.Ye X.F., Wang H.Z., Gong W.H., Li S., Li H.Q., Wang Z., and Zhao Q. Impact of decellularization on porcine myocardium as scaffold for tissue engineered heart tissue. J Mater Sci Mater M 27, 2016 [DOI] [PubMed] [Google Scholar]
- 59.Edwards J.N., and Blatter L.A. Cardiac alternans and intracellular calcium cycling. Clin Exp Pharmacol Physiol 41, 524, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hulsmann J., Grun K., El Amouri S., Barth M., Hornung K., Holzfuss C., Lichtenberg A., and Akhyari P. Transplantation material bovine pericardium: biomechanical and immunogenic characteristics after decellularization vs. glutaraldehyde-fixing. Xenotransplantation 19, 286, 2012 [DOI] [PubMed] [Google Scholar]
- 61.Pagoulatou E., Triantaphyllidou I.E., Vynios D.H., Papachristou D.J., Koletsis E., Deligianni D., and Mavrilas D. Biomechanical and structural changes following the decellularization of bovine pericardial tissues for use as a tissue engineering scaffold. J Mater Sci Mater Med 23, 1387, 2012 [DOI] [PubMed] [Google Scholar]
- 62.Tsuchiya T., Balestrini J.L., Mendez J., Calle E.A., Zhao L., and Niklason L.E. Influence of pH on extracellular matrix preservation during lung decellularization. Tissue Eng Part C Methods 20, 1028, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poornejad N., Schaumann L.B., Buckmiller E.M., Momtahan N., Gassman J.R., Ma H.H., Roeder B.L., Reynolds P.R., and Cook A.D. The impact of decellularization agents on renal tissue extracellular matrix. J Biomater Appl 31, 521, 2016 [DOI] [PubMed] [Google Scholar]
- 64.Schoen F.J., Tsao J.W., and Levy R.J. Calcification of bovine pericardium used in cardiac valve bioprostheses. Implications for the mechanisms of bioprosthetic tissue mineralization. Am J Pathol 123, 134, 1986 [PMC free article] [PubMed] [Google Scholar]
- 65.Cuttle L., Nataatmadja M., Fraser J.F., Kempf M., Kimble R.M., and Hayes M.T. Collagen in the scarless fetal skin wound: detection with picrosirius-polarization. Wound Repair Regen 13, 198, 2005 [DOI] [PubMed] [Google Scholar]
- 66.Wong M.L., Wong J.L., Horn R.M., Sannajust K.C., Rice D.A., and Griffiths L.G. Effect of urea and thiourea on generation of xenogeneic extracellular matrix scaffolds for tissue engineering. Tissue Eng Part C Methods 22, 700, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yi T., and Song S.U. Immunomodulatory properties of mesenchymal stem cells and their therapeutic applications. Arch Pharm Res 35, 213, 2012 [DOI] [PubMed] [Google Scholar]
- 68.Aggarwal S., and Pittenger M.F. Human mesenchymal stem cells modulate allogeneic immune cell responses. Blood 105, 1815, 2005 [DOI] [PubMed] [Google Scholar]
- 69.Yagi H., Soto-Gutierrez A., Parekkadan B., Kitagawa Y., Tompkins R.G., Kobayashi N., and Yarmush M.L. Mesenchymal stem cells: mechanisms of immunomodulation and homing. Cell Transplant 19, 667, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Batten P., Sarathchandra P., Antoniw J.W., Tay S.S., Lowdell M.W., Taylor P.M., and Yacoub M.H. Human mesenchymal stem cells induce T cell anergy and downregulate T cell allo-responses via the TH2 pathway: relevance to tissue engineering human heart valves. Tissue Eng 12, 2263, 2006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.