Summary
Image-based assays, such as alkaline phosphatase staining or immunocytochemistry for pluripotent markers, are common methods used in the stem cell field to assess pluripotency. Although an increased number of image-analysis approaches have been described, there is still a lack of software availability to automatically quantify pluripotency in large images after pluripotency staining. To address this need, we developed a robust and rapid image processing software, Pluri-IQ, which allows the automatic evaluation of pluripotency in large low-magnification images. Using mouse embryonic stem cells (mESC) as a model, we combined an automated segmentation algorithm with a supervised machine-learning platform to classify colonies as pluripotent, mixed, or differentiated. In addition, Pluri-IQ allows the automatic comparison between different culture conditions. This efficient user-friendly open-source software can be easily implemented in images derived from pluripotent cells or cells that express pluripotent markers (e.g., OCT4-GFP) and can be routinely used, decreasing image assessment bias.
Keywords: ESC, pluripotency, automated image analysis, alkaline phosphatase, pluripotency quantification, Pluri-IQ
Graphical Abstract
Highlights
-
•
Open-source software to evaluate pluripotency in low-magnification images
-
•
Automatic colony detection and segmentation
-
•
Supervised machine-learning platform with high characterization accuracy
-
•
Software tools for easy data validation, visualization, and data analysis comparison
Perestrelo et al. show an image-analysis methodology coupled with a machine-learning platform to analyze and quantify pluripotency with high accuracy in different contexts. Requiring low user input, this software allows the pluripotency evaluation of large low-magnification images, with colony visualization and automated data analysis comparison among experiments.
Introduction
Embryonic stem cells (ESCs) are characterized by their self-renewal and pluripotent capacities. Due to their properties, ESCs serve as an important research model to study key factors that maintain pluripotency, as well as factors that trigger differentiation. ESCs are morphologically distinct from differentiated cells, featuring a high nuclear-to-cytoplasmic ratio and growth as 3D colonies. In the undifferentiated state, ESCs are characterized by high levels of pluripotency-related transcription factors, such as OCT 3/4, NANOG, and SOX2 (Avilion et al., 2003, Chambers et al., 2003, Nichols et al., 1998, Hart et al., 2004, Hay et al., 2004). In addition, the expression of enzyme alkaline phosphatase (AP) is another hallmark of pluripotency (Palmqvist et al., 2005). AP is an enzyme that catalyzes the hydrolysis of phosphate esters (Stefková et al., 2015). ESCs have high levels of AP, which decreases upon ESC differentiation (Stefková et al., 2015, Palmqvist et al., 2005). The expression specificity of AP and the transcription factors described above makes them crucial proteins to evaluate ESC pluripotency in vitro.
Image-based assays are common methods used in stem cell research to evaluate maintenance and loss of pluripotency. AP staining is commonly used to assess maintenance/loss of pluripotency after different stimuli, such as drug treatments, gene silencing, or overexpression (Martello and Smith, 2014). Immunocytochemistry with antibodies specific to pluripotent markers is another image-based method used to analyze stem cell fate (Martello and Smith, 2014). Immunocytochemistry and AP assays are fast and easy to perform. Contrary to methods that require suspension cells, such as flow cytometry, these image-based assays allow colony morphological analysis as they maintain the spatial information of each cell in the colony. Therefore, the increase of ESC-image acquisitions creates a demand for image-analysis programs suitable for ESC-image quantification.
Considering that ESC cultures are usually heterogeneous, with varying degrees of pluripotency and irregular colony sizes, several imaging quantification programs have been developed specifically for ESC. Using phase-contrast images, different imaging analysis pipelines have been reported to segment ESC colonies and automatically track their growth and morphology over time (Jaccard et al., 2014a, Jaccard et al., 2014b, Narkilahti et al., 2007, Scherf et al., 2012). Despite their high ability to segment ESC in culture and monitor ESC growth and confluency, these pipelines do not provide any other pluripotency measurements. More recently, phase-contrast images have been used to evaluate induced pluripotent stem cells (iPSCs) quality (Tokunaga et al., 2014, Joutsijoki et al., 2016, Maddah et al., 2014). However, in all these studies, morphology is the key feature, where iPSCs are classified as good- or bad-quality colonies, but no other pluripotency measurement is obtained. In addition to phase-contrast images, other studies have been reported to segment ESC colonies using immunofluorescence images (Chalfoun et al., 2015, Gorman et al., 2014, Lou et al., 2014, Paduano et al., 2013). Although these pipelines allow the location analysis of labeled cells, which makes them suitable for pluripotency quantification, the pipelines developed by some groups rely in high-magnification images to evaluate pluripotency marker expression (Gorman et al., 2014, Lou et al., 2014), whereas others, despite analyzing labeled cells in low magnification, they only determine the signal location of different markers in a colony, constraining the automatic and global pluripotency determination (Paduano et al., 2013).
Therefore, none of the current image-analysis pipelines allow an easy and robust automatic quantification of ESC pluripotency in an environment where different degrees of pluripotency occur, i.e., the presence of pluripotent colonies, mixed colonies, and differentiated cells. For instance, to even answer the simple question of whether or not a treatment induces loss of pluripotency, stem cell biologists rely on manual scoring, or culture observations after AP staining. This manual quantification not only limits reproducibility and objectivity, but it is also time consuming.
Here, using mouse ESC (mESC) as a model we present an accurate, open-source, and user-friendly software, Pluri-IQ, which can automatically quantify the percentage of pluripotent, mixed, or differentiated cells through culture images. Pluri-IQ is able to analyze different low-magnification image sizes, and through core cascade modules (segmentation, machine learning, validation, and automatic scoring) accurately quantifies pluripotency through the analysis of the pluripotency markers present in the image. Segmentation provides the outline of each colony and is performed automatically by Pluri-IQ. Classifiers are built through a machine-learning interactive process. Manual validation provides the user with the classifier accuracy, while automatically updating the classifier, which guarantees high accuracy of automatic score classification of each colony as pluripotent, mixed, or differentiated, with low user input required.
Results
Image-Based Analysis Outline
To evaluate the performance of Pluri-IQ, we tested its classification precision over a set of mESCs images. These mESCs were cultured in different medium conditions that promote maintenance of pluripotency or induce mESC differentiation. Cells were stained with AP. Subsequently, low-magnification phase-contrast images and a fluorescence channel for AP staining were acquired.
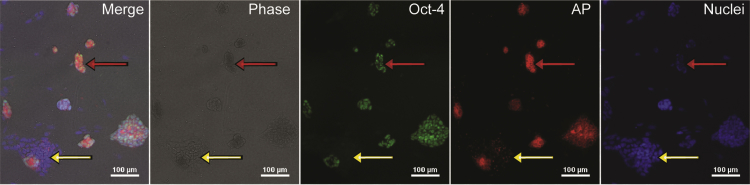
AP staining was selected as a method for pluripotency assessment since it is a fast and common protocol to evaluate pluripotency (Martello and Smith, 2014). To confirm that the AP assay utilized only stained pluripotent cells, we performed the AP assay followed by immunostaining against the pluripotent marker OCT-4 (Figure 1). Phase-contrast images, as well as nuclear staining, were used to provide a general outlook of all colonies. Then, OCT-4 expression was compared with AP staining, and colonies positive for OCT-4 were also positive for AP (Figure 1, red arrow), while the portions of colonies that fail to express OCT-4 do not stain for AP (Figure 1, yellow arrow). These results show that the AP assay utilized in these experiments is specific for pluripotent cells.
Figure 1.
AP Co-localizes with OCT-4 Expression
Cropped image of mESCs cultured for 4 days in pluripotent conditions and stained for AP assay and the pluripotent marker OCT-4. The yellow arrow shows a portion of a colony differentiated, where no OCT-4 or AP staining is visible; the red arrow shows a pluripotent colony positive to OCT-4 and AP staining. Scale bar, 100 μm. AP, alkaline phosphatase.
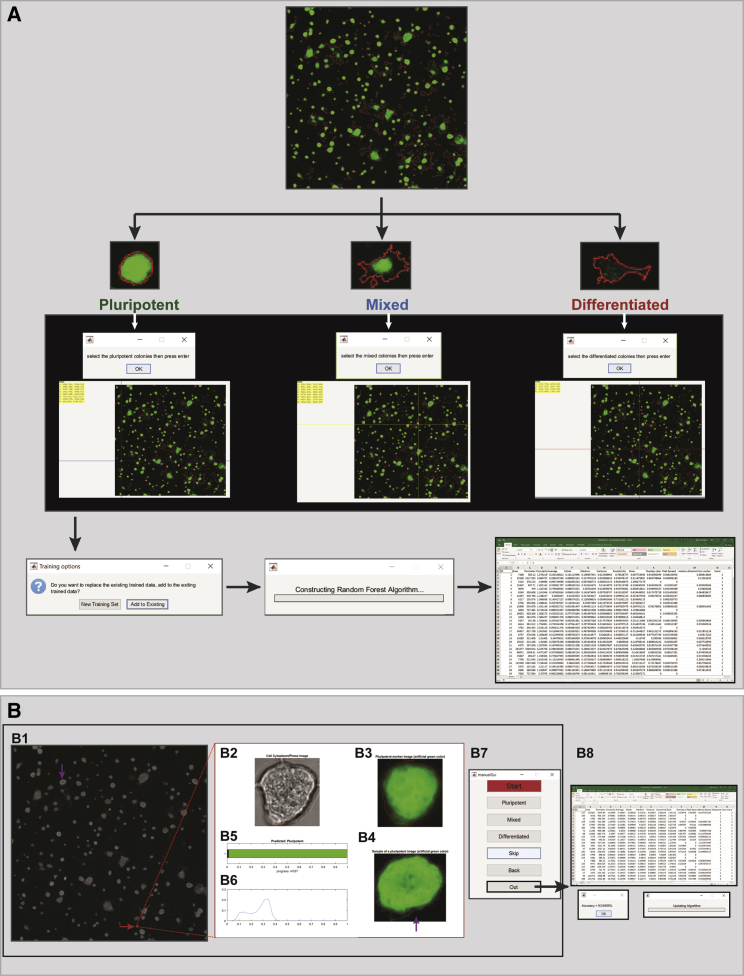
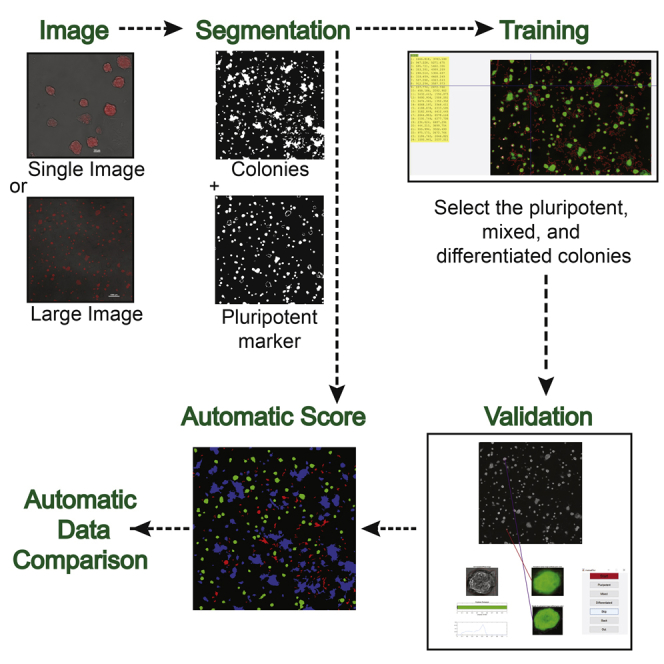
One of the challenges in characterizing pluripotency automatically is that the majority of image-based assays, including AP staining, are specific for pluripotent cells, which gives a positive signal when a colony is pluripotent. However, differentiated colonies are only defined as such when the positive signal is non-detectable (Figure 1). In addition, pluripotent colonies acquire different morphologies upon different treatments, and usually, during ESC differentiation, pluripotent staining is firstly reduced before it is completely lost. Therefore, mixed colonies, which we consider a mixture of pluripotent cells and differentiated cells, are prevalent in most cultures and difficult to impartially classify. Taking all these factors into consideration, we developed an image-analysis approach that incorporates three major steps in order to automatically quantify pluripotency: a fast segmentation algorithm capable of identifying different colony types in both large phase-contrast and fluorescent images, an interactive machine-learning algorithm to classify colonies as pluripotent, mixed, or differentiated, and a validation algorithm responsible for the measurement of classification accuracy to increase the trust on the automatic pluripotency quantification and the comparison of different conditions (Figure 2).
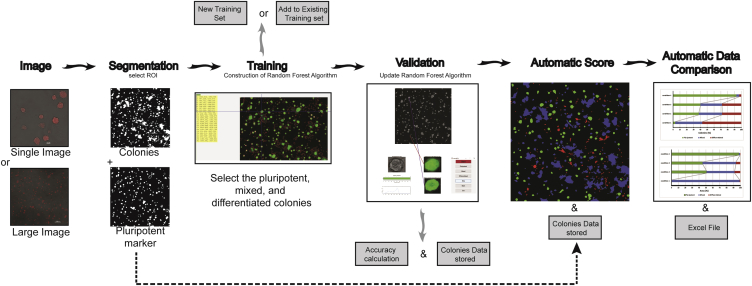
Figure 2.
Pluri-IQ Approach to Quantify ESCs Pluripotency
Users provide a single image or a large image input, as shown. Upon selection of the ROI, Pluri-IQ segments both phase-contrast image channel to get all colonies, and the pluripotent marker channel. After segmentation, training is required, through selection of what is considered pluripotent, mixed, and differentiated colonies, in order to construct a random forest algorithm, which can be added to an existing training system or to a new training system. Validation is performed by the user, and the accuracy of the Pluri-IQ is calculated. The random forest algorithm is updated and colonies features are stored in both Excel and MATLAB files. After all these steps are completed, and segmentation performed to all images, the automatic score is then available. Finally, automatic data comparison is available and allows the comparison of different conditions automatically, providing figures with the percentage of colonies or area pluripotent, mixed and differentiated, in addition to storage as Excel file the data comparisons. Scale bars, 100 μm (single image); 1,000 μm (large image). Segmentation, training, validation, and automatic score example derived from the raw large image.
Image Properties and Colony Segmentation
A reliable pluripotency evaluation requires an overview of the entire well plate. However, automatic image acquisition often leads to colonies on the border of the image and, consequently, loss of partial colony information. Such colonies are thus, not suitable for quantification. Therefore, we designed a pipeline that allows the analysis of single or multiple TIFF images combined (stitched together during or after single image acquisition) in order to fulfill the criteria described above (Figure 1).
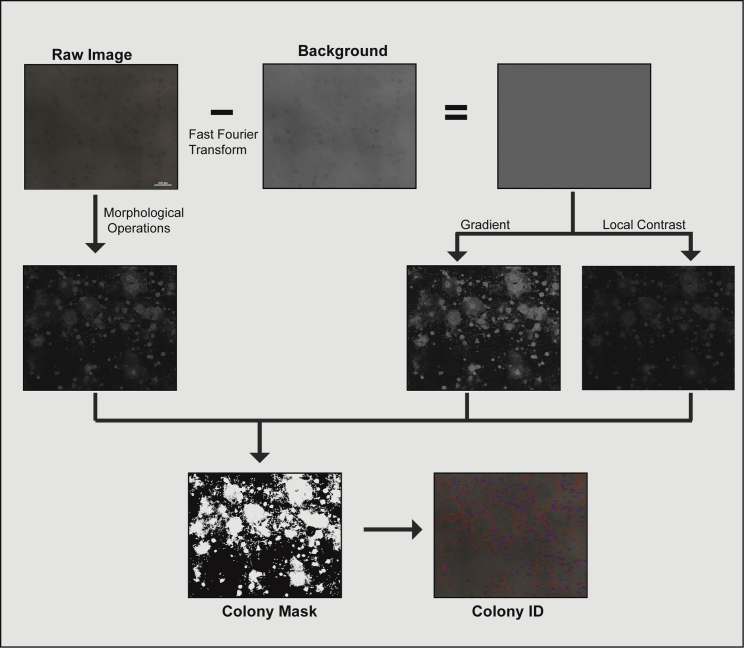
To detect and segment the colonies, we developed a custom script written in MATLAB (MathWorks), FACT (fast and accurate colony tracing), which does not require user input (Figure 3). Colony detection and segmentation is primarily performed in the phase-contrast image. The image background is first calculated: fast Fourier transform is applied to the raw image; the transformed image obtained is filtered with a cutoff frequency of 0.2 in the Fourier domain, followed by an inverse fast Fourier transform, resulting in a reconstructed image with only low frequency features. The background is removed from the raw image through subtraction of the low frequency image from the raw image, which isolates the high-frequency information, cells, and colonies (Figure 3). Then, the local contrast is highlighted using a 3 × 3 high-frequency filter and binarized using Otsu thresholding, which separates the foreground from the background. A sobel filter is applied to the subtracted background image to obtain the image gradient. Raw images are also processed by morphological operations erosion and dilation, and the difference between these two processes is used to obtain a binary mask. The binary mask from the local contrast, gradient, and morphological operations applied to the raw image is added together, and the final mask is produced when the value of the cumulative mask is greater than 1 (Figure 3). Given that ESCs grow in colonies, we added two particular restrictions in the algorithm: (1) the segmentation algorithm was adapted to ignore single cells (however, single cells can be included by manually adapting this segmentation algorithm parameter); and (2) differentiated and mixed colonies tend to occupy a large area and, by eliminating all colonies on the periphery of the region of interest (ROI), biased results were obtained. Therefore, colonies in the periphery are only discarded if their pixel size is below 7 × 104 pixels. After colony segmentation is completed, all colonies detected are assigned with a specific number (ID), with the colony ID derived from the phase-contrast image applied to the pluripotent marker image (Figure 3). After segmentation is performed, the results are saved as new TIFF images, which allows for segmentation inspection. A similar procedure is applied for fluorescence images, with a small nuance: if there are saturated pixel in the image, the software will automatically fill them in as a positive hint for a colony.
Figure 3.
Colony Detection and Segmentation Pipeline
The raw image is processed by the difference of erosion and dilation followed by Otsu thresholding to create the first mask. The raw image is also processed by removing the background level through fast Fourier transform, creating a background subtracted image, from which we extract the gradient and local contrast and produce two more masks through Otsu thresholding. The three masks are added together and if the pixel value is greater than 1, the object is incorporated into the final mask. The outline and label of the final mask is added to the raw image to show the outcome of the segmentation. Scale bar, 500 μm.
Segmentation Accuracy
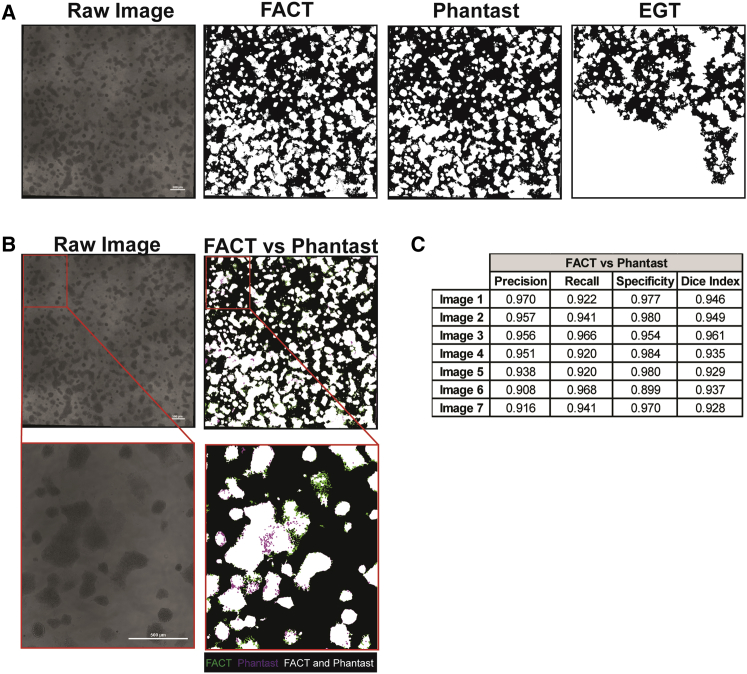
To evaluate segmentation accuracy, large phase-contrast images (10,078 × 10,054 pixels), with different degrees of pluripotency, morphology, and confluency were analyzed by FACT and compared with previously published segmentation algorithms: Phantast (Jaccard et al.,. 2014b), and empirical gradient threshold (EGT) (Chalfoun et al., 2015) (Figures 4A and S1). We selected Phantast and EGT as they have both been shown to successfully segment ESCs images. While Phantast requires user input for accurate segmentation, EGT does not. Our results show that both Phantast and FACT are able to segment large images, whereas EGT fails to segment some image fractions (Figures 4A and S1). We then decided to directly compare FACT segmentation with Phantast segmentation, and calculate FACT precision, recall, specificity, and dice index (Figures 4B and 4C). Quantitatively, we achieved an average precision of 94.24 ± 0.02; an average recall of 93.95 ± 0.02; an average specificity of 96.35 ± 0.03, and an average dice index of 94.06 ± 0.01. These results show that our segmentation method (FACT) can provide accurate image segmentation on a wide range of cell colony morphologies, from a large sheet of cells to individualized colonies, without any additional user input. Moreover, these results demonstrate that our segmentation algorithm, without tuning requirements, is as robust and accurate as Phantast.
Figure 4.
Comparison between FACT Algorithm Segmentation and Phantast and EGT Segmentation Algorithms
(A) FACT, Phantast, and EGT segmentation results. Binary images, where white is representative of presence of a colony. Scale bar, 500 μm.
(B) Direct comparison between FACT and Phantast segmentation. White regions represent segmentation overlap between Phantast and FACT; purple regions represent pixel only positively segmented by Phantast; and green regions represent pixel only positively segmented by FACT. Scale bar, 500 μm.
(C) Precision, recall, specificity, and dice index of FACT segmentation was compared with Phantast segmentation. EGT, empirical gradient threshold; FACT, fast and accurate colony tracing.
See also Figure S1.
Machine-Learning Classification
Once segmentation is performed, automatic pluripotency classification poses a further challenge. Pluripotent cells in culture conditions have different colony height, area, and circularity. To classify a colony as pluripotent, the intensity of the pluripotent marker must be high, whereas a differentiated colony has low or undetectable pluripotent signal. Mixed colonies are described to (1) have a combination of high and low intensity pluripotency markers, or (2) have a dim positive signal, above the positive staining threshold. However, intensity signal is dependent on the colony morphology, which makes automatic pluripotency classification through image-based analysis a challenge. To tackle this issue, we used a supervised colony classification approach to differentiate between the three colony types. We selected the random forest classifier (Breiman, 2001), due to its robustness and computational simplicity. We designed an interactive classification approach, where users can build a classifier by interactively training and validating their own image sets (Figure 5A). During this cycle, users are expected to after selecting the training set images, to pick the best subset of colonies that fit the pluripotent, mixed, and differentiated standard parameters from each image. After selecting the colonies, the classifier is built taking into account different colony features from both phase-contrast and pluripotent marker images (Table S1), and an Excel file is created with each colony feature (Figure 5A). Thus, when a new colony is presented to the program, the software evaluates all the features of the new colony and classifies this colony according to the classifier pool where its features best fit. Since fluorescence intensity is one of the features to characterize pluripotency, and intensity is dependent on microscopy settings upon acquisition, different training sets can be created and uploaded to attenuate the misclassification due to the difference between independent experiments.
Figure 5.
–Machine-Learning and Manual Validation Overview
(A) Machine-learning overview. Upon selection of the images to use as training set, the user interactively selects first the pluripotent colonies, then the mixed colonies and, finally, the differentiated colonies presented in the image. After this procedure is complete for all the images in the training set, the classifier is built, with the possibility to add the training set to a previous classifier, or create a new training set. An Excel file with all the colony features as well as the pluripotency score is created, where score 1 is differentiated, 2 mixed, and 3 pluripotent colony. Green, pluripotent marker AP; red, colony border obtained by the phase-contrast segmentation image.
(B) Manual validation overview: (B1) image overview. Purple arrow shows an example of a pluripotent colony. Red arrow shows the colony picked to validate the classifier prediction. (B2) Phase-contrast image and (B3) pluripotent marker image of the colony picked to validate the classifier prediction. (B4) Example of a pluripotent colony. (B5) Classifier prediction and progress bar with the total number of colonies present in the image and the number of colonies already validated. (B6) Normalized number of pixel versus pluripotent marker intensity. (B7) Graphical user interface used to validate each colony as pluripotent, mixed, or differentiated. (B8) After manual validation completed, the manual accuracy is automatic shown, and the random forest algorithm is updated. Segmentation information summary is saved as an Excel file.
See Table S1.
Manual Validation Algorithm
To evaluate the accuracy of colony classification, we developed an interactive validation algorithm that allows the user to validate the classification of the selected training set (Figure 5B). After the selection of different images, a phase-contrast and fluorescent marker image derived from a random selected colony is shown in addition to its intensity pluripotent marker plot (Figure 5B1–5B6). Manual validation is allowed through the selection of the pluripotent, mixed, or differentiated button (Figure 5B7), with the opportunity to go back to a previous colony and overwrite the previous classification (back button), to skip a colony (skip button), or to finalize the manual validation process without the need to validate the entire image (out button) (Figure 5B7). The classification results are exported with each colony ID to Excel and MATLAB files. These files provide a detailed summary of each colony feature, as well as the classifier prediction and user manual validation (Figure 5B8). In addition, the accuracy score is shown at the end of each large image-analysis. Finally, in order to increase precision, every time that manual validation is completed, the classifier is updated.
Performance Evaluation
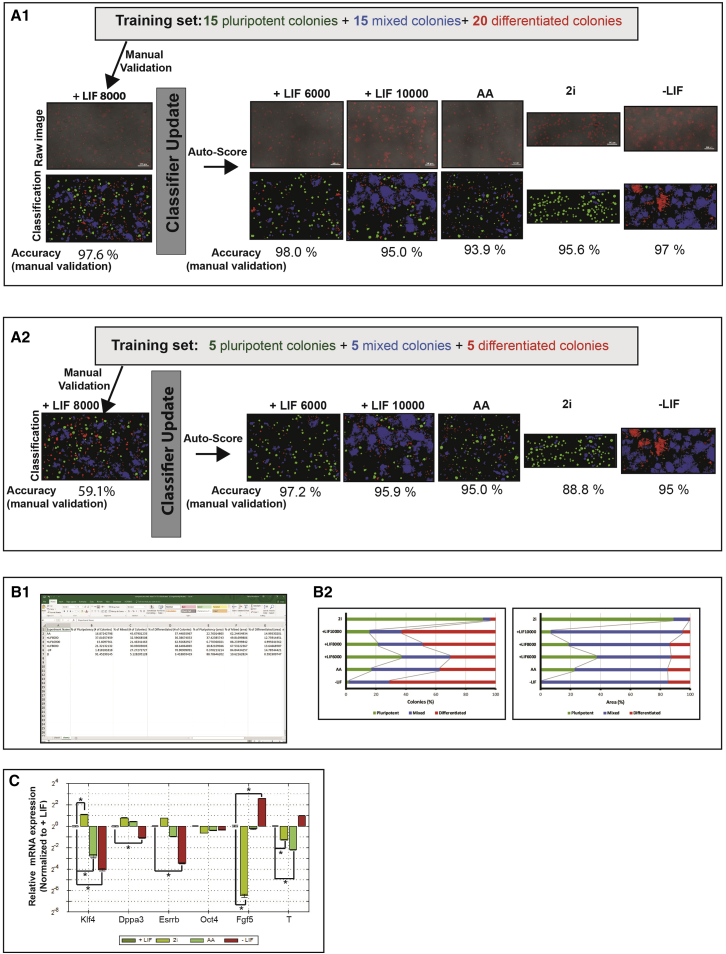
To evaluate our colony classification approach, we used six large images derived from different mESCs cultures, with a wide range of pluripotent percentages and colony shapes. We started by creating a training set selecting 15 pluripotent colonies, 15 mixed colonies, and 20 differentiated colonies from one of the large images (Figure 6A1). Then, we ran the manual validation on the same image, evaluating all the colonies, and we achieved an accuracy of 97.6%. Afterward, we ran the automatic score on the other 5 large images and manually evaluated 100 colonies from each image to obtain the pluripotency classification accuracy (Figure 6A1). All images had an accuracy classification above 90%. To test the number of colonies that should be selected to train the classifier, we went back and selected only five pluripotent, five mixed, and five differentiated colonies, and created a new classification set (Figure 6A2). The accuracy decreased to 59% when we manually validated 150 colonies from the same image. After the manual validation classifier update, we ran the automatic score on the five large images that we had previously ran, and manually validated the results. All images had similar accuracy values to the previous classifier set (Figure 6A2). These results demonstrate that our manual validation is important and capable of maintaining high accuracy values, without the need to tune the training system multiple times, since manual validation updates the classifier.
Figure 6.
Pluri-IQ Performance Evaluation in Different mESC Culture Conditions
(A1) The training set was built by selecting 15 pluripotent, 15 mixed, and 20 differentiated colonies. Manual validation showed that accuracy of the classifier is ∼ 97%. Classifier was updated after manual validation and automatic score was run in five different large images. Manual validation performed afterward showed accuracy values all above 90%. Scale bar, 500 μm (in raw images).
(A2) After selection of a reduced number of colonies to train the classifier, the manual accuracy decreased to ∼59%. However, the update of the classifier after the manual training leads to an increase of accuracy classification.
(B1) Excel sheet output derived from the automatic data comparison.
(B2) Percentage of pluripotent, mixed, and differentiated colonies (left) and area (right) of each condition. A minimum of 100 colonies was analyzed per condition. Color code: green, pluripotent colonies; blue, mixed colonies; red, differentiated colonies.
(C) Normalized mRNA expression fold change (determined by RT-qPCR) of mESC cultured for 4 days in the presence of 2i medium, LIF, 50 nM AA or in the absence of LIF. All samples were analyzed in two technical replicates. All data are presented as means ± SEM. ∗p < 0.01. AA, Antimycin A; LIF, leukemia inhibitory factor; 2i, serum-free medium with CHIR99021, PD0325901, and LIF.
To validate Pluri-IQ pipeline, mESCs were cultured in medium with serum and leukemia inhibitory factor (LIF), and seeded with different densities (6,000, 8,000, or 10,000 cells/cm2), in medium with serum but in absence of LIF (referred as −LIF), in medium with serum and Antimycin A (referred as AA), and in serum-free medium with CHIR99021 and PD0325901 (referred as 2i). After validation and automatic score, automatic comparison between different conditions was performed, with an Excel file as output (Figure 6B1). Our results show that the highest number of pluripotent colonies is obtained when mESCs are cultured in 2i medium, whereas the absence of LIF in medium with serum induces the highest differentiation. In addition, the increase in mESC density induces a decrease in the percentage of pluripotent colonies and an increase in differentiated colonies. Finally, when mESCs are cultured in the absence of LIF but in the presence of a complex III mitochondria inhibitor, AA, mESC differentiation is decreased (Figure 6B2). To confirm the cell fate in different conditions, we performed qRT-PCR analysis in mESC cultured in the presence of LIF (6,000 cells/cm2), 2i, AA, and in the absence of LIF, conditions known to affect pluripotency (Figure 6C). To evaluate mESC fate, we selected four pluripotent genes (Klf4, Dppa3, Esrrb, and Oct4) and two early differentiation genes (Fgf5 and T) and compared the relative mRNA expression of the different conditions with +LIF condition. Cells cultured in the absence of LIF had a low expression of pluripotent markers (Klf4, Dppa3, and Esrrb) and an increased expression of differentiated markers (Fgf5 and T) (Figure 6C), which suggests that this cell culture condition had a high percentage of differentiated colonies and low percentage of pluripotent colonies, in agreement with the results obtained by Pluri-IQ software (Figure 6B2). Cells cultured in the presence of 2i had a significant increase of the pluripotent marker Klf4, and low expression of the differentiated markers Fgf5 and T (Figure 6C). This result suggests that these cells have a higher percentage of pluripotent colonies than mESC cultured in the presence of LIF, which is in accordance with the results obtained by Pluri-IQ (Figure 6B2). Finally, when cells were cultured in the presence of AA they had low expression of Klf4, when compared with cells cultured in the presence of LIF (Figure 6C), which suggests that AA culture conditions promotes a decrease of pluripotent colonies when compared with mESC cultured in the presence of LIF. Thus, through qRT-PCR, we also verify that mESC cultured in the presence of 2i medium have the highest pluripotency levels, whereas mESC cultured in the absence of LIF promote colony differentiation. These results are also in agreement with the literature (Ying et al., 2008, Pereira et al., 2013, Palmqvist et al., 2005), which demonstrates that our pipeline is able to accurately classify colony pluripotency even in the presence of different colony densities and morphologies.
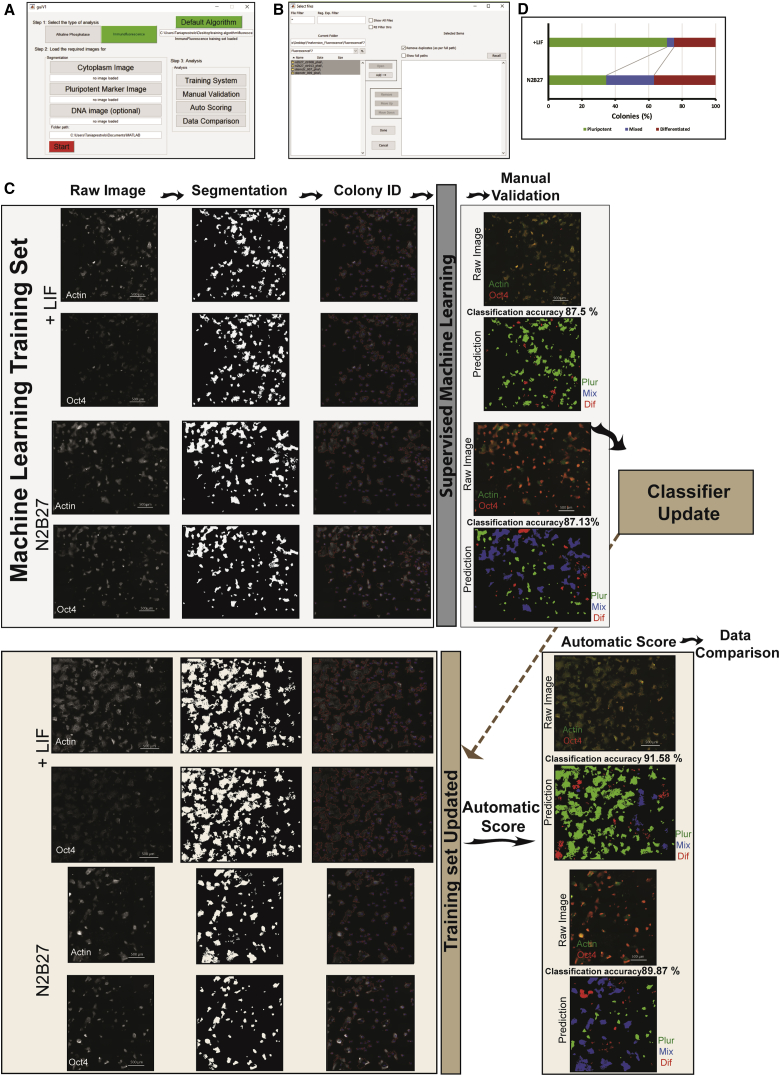
We decided to use the same rationale, and evaluate Pluri-IQ accuracy in fluorescence images (Figure 7). We utilized images from mESC cultured in serum with LIF (pluripotency medium) or in a neuronal differentiation medium (referred as N2B27). Cells were stained for the pluripotent marker OCT-4. After uploading the images and their segmentation performed, the classifier was created utilizing 16 pluripotent colonies, 14 mixed colonies, and 10 differentiated colonies selected from two large images (Figure 7C, upper panel). Manual validation was performed on the same images, and an accuracy of 87% was achieved. We then used the training set to automatic score two new images (Figure 7C, bottom panel). The mESC classification accuracy was approximately 90%. After comparing both conditions, we saw that, in agreement with the literature, mESCs cultured in the presence of neuronal differentiation medium have more differentiated and mixed colonies than cells cultured in the presence of LIF (Figure 7D). In addition, when we measured colony parameters such as nuclear cytoplasmic ratio, the results obtained were in agreement with previous studies: nuclear/cytoplasm ratio decreased with colony differentiation (Figure S2). These results demonstrate that our pipeline also accurately classifies pluripotency in fluorescence images.
Figure 7.
Pluri-IQ Application Pipeline and Its Performance Evaluation in Immunofluorescence Images
(A) The main graphical user interface (GUI) of Pluri-IQ.
(B) GUI used to select different folders containing the images to perform manual validation, autoscoring or data comparison.
(C) Pluri-IQ pipeline: two different images with different degrees of pluripotency were used to create the machine-learning training set (upper panel). After each channel segmentation and colony identification, a fluorescence training set was created and manually validated. After the classifier automatic update, two new images were scored automatically by Pluri-IQ and classification accuracy was evaluated (bottom panel). Scale bar, 500 μm (in raw images). Color code on the raw images: green, actin; red, OCT4. Color code on the images prediction: green, pluripotent colonies (Plur); blue, mixed colonies (Mix); red, differentiated colonies (Dif).
(D) Percentage of pluripotent, mixed, and differentiated colonies in the two different experimental conditions. Data derived from the automatic data comparison in Pluri-IQ. Results derived from two replicates. LIF, pluripotency medium; N2B27, neuronal differentiation medium.
See also Figure S2.
Graphical User Interface
We created a simple and straightforward graphical user interface, which confers an easy comprehension of the processing pipeline (Figures 7A and 7B). Users are first required to select their type of image staining, AP or immunofluorescence, and upload two images: a single channel image of phase-contrast (or fluorescence cytoplasmic) image, and a single channel pluripotent marker image. In addition to these two images, nuclear staining can also be uploaded in order to calculate the nucleus to cytoplasmic ratio. After successfully uploading the images and selecting the ROI, segmentation is performed and the results are saved as new TIFF images, which allows for segmentation inspection (Figure 7C). To proceed to the automatic pluripotency quantification, the interface requires the uploading of a training classifier set and selection of each condition folder (Figure 7B). The colony quantification results are exported as color-coded images and Excel files. Finally, data comparison interface automatically quantifies the colony pluripotency percentage, pluripotency colony area percentage, pluripotency mean area and circularity, and nuclear/cytoplasm ratio, exporting these results to an Excel file.
Discussion
Considering the widespread practice in stem cell laboratories to quantify pluripotency through image-based assays, we sought to develop a software that allows the automatic quantification of pluripotency, with a low requirement of user input. Here, we report the development of an efficient, accurate, open-source, and user-friendly pipeline for pluripotency quantification of low-magnification images, Pluri-IQ. This software segments colonies from large images with high precision, without the requirement of user input. Subsequently, through a machine-learning process, it automatically and accurately classifies pluripotent, mixed, and differentiated colonies. In parallel, we implemented a manual validation algorithm, which allows for the validation of the program by the user, through visualization of each colony and its corresponding pluripotent marker expression. The storage of each colony features, as well as its pluripotency score in Excel file, enables post-data treatment result analysis.
Importantly, we developed a user-friendly software that is accurate and efficient, with low user input requirements. Pluri-IQ uses as input large images, and presents relevant advantages compared with others since it does not require segmentation parameters to be refined in order to discharge background or detect and segment colonies, as in ilastik (http://ilastik.org/), and does not require users to create a specific pipeline of analysis as in CelProfiler (http://cellprofiler.org/). Although training data is required, the software interactivity allows the user to quickly select the best colonies of each classification, without requiring the selection of the best features to tune machine learning, as in ilastik. After machine learning is complete, manual validation promotes the evaluation of the classifier accuracy at the same time that the classifier is updated. Finally, after high accuracy is achieved, it is possible to run a fast automatic score, followed by data comparison. Pluri-IQ was already tested in two independent laboratories, and high precision classification of mESC colonies was achieved. Although mESC were used as a model in this study, the procedure should be easily extendable to other types of pluripotent cells (hESC, iPSCs, etc). Nevertheless, it is important to take into account that Pluri-IQ relies on image quality and user experience of pluripotency classification. Moreover, for certain experimental protocols that involve pluripotency assessment it may be deemed necessary to extend image analysis to include also qRT-PCR or western blot.
Different approaches to characterize cell fate have been developed in order to increase data consistency and reproducibility (French et al., 2015). Although Pluri-IQ is specifically designed to measure pluripotency percentage in images, image-based assays to evaluate pluripotency are routinely performed. The necessity to infer if a different medium, small hairpin RNA, or other stimuli maintain pluripotency or induces differentiation makes quantification of pluripotency a common and required practice in stem cell laboratories (French et al., 2015). Usually, fast and easy assays, such as image-based-assays, are performed to quantify pluripotency; however, this quantification is still evaluated manually. We have presented a robust method to evaluate the pluripotency of colonies through a fast Fourier transform-based segmentation, which works in both phase-contrast as well as in fluorescence images. Using the information produced by the segmentation, we can very efficiently classify the colonies as pluripotent, mixed or differentiated without using time-consuming methodologies that are currently employed. Based on our case study, we can conclude that Pluri-IQ is applicable in both large phase-contrast and fluorescence low-magnification images. Moreover, Pluri-IQ is able to analyze conditions that promote or destabilize pluripotency, allowing result collection in a faster and more impartial manner, thus increasing unbiased reproducibility.
Experimental Procedures
Recall, Precision, Specificity, and Dice Index Calculations
To compare FACT and Phantast segmentation we calculated recall, precision, specificity, and dice index values defined in (Equation 1), (Equation 2), (Equation 3), (Equation 4), as described previously (Intawong et al., 2013, Zou et al., 2004). In brief, TP (true positives) represents the number of segmented colonies in FACT that were also segmented in Phantast; FN (false negatives) represent the number of colonies not segmented in FACT but segmented in Phantast; FP (false positives) represent the number of colonies segmented in FACT and not segmented in Phantast; TN (true negatives) represent the number of pixel that are not considered a colony in both FACT and Phantast; X represents FACT segmentation regions and Y represents the Phantast segmentation regions.
| (Equation 1) |
| (Equation 2) |
| (Equation 3) |
| (Equation 4) |
Classification Accuracy
Classification accuracy was measured by comparing the colony prediction of the classifier with the classification given by the user through manual validation. The number of positive hits was then divided by the total number of colonies evaluated in order to obtain the accuracy percentage.
Data Comparison
The percentage of pluripotent, mixed, and differentiated colonies, as well as the percentage of pluripotent, mixed, and differentiated colony area are calculated for each large image. In addition, mean area, circularity, and nuclear/cytoplasm ratio, as well as SEM, are provided. The percentage of pluripotent, mixed, and differentiated colonies in an image is measured by calculating the number of colonies in each classification and dividing it by the total number of colonies present in the image. The pluripotent, mixed, and differentiated area of each image is measured by the summation of each classifier area, followed by ratio between each sum of classifier area and the total area occupied by colonies. Mean area and mean circularity are calculated by averaging the pluripotent, mixed, and differentiated area and circularity in each image, respectively. Nuclear/cytoplasm ratio of each large image is calculated using colony average area derived from DNA staining image, and colony average area derived from cytoplasm image.
Software Availability
Pluri-IQ was implemented using MATLAB on a 64-bit Windows OS laptop with intel i7 processor with 8 GB of RAM memory. The software will be hosted at the CNC website (http://www.cnbc.pt/equipment/microscopyUnit.asp#divImageAnalysis) both as a compiled MATLAB standalone application (requires installation of 64 bit MATLAB runtime, available for free at www.mathworks.com/products/compiler/mcr.html) and MATLAB.m files. In our application, we made use of Custom GINPUT by Jiro Doke, Nov 07, 2012 (https://www.mathworks.com/matlabcentral/fileexchange/38703-custom-ginput/content/ginputc.m, retrieved June 2016) and uipickfiles: uigetfile on steroids by Douglas Schwarz, Apr 25, 2006 (https://www.mathworks.com/matlabcentral/fileexchange/10867-uipickfiles--uigetfile-on-steroids, retrieved May 2016).
Cell Culture
Mouse embryonic cell line (E14Tg2a.4, derived from 129P2/OlaHsd, RRID:MMRRC_015890-UCD) was cultured at 37°C, 5% CO2 and two different culture media were used for ESC maintenance and propagation: (1) medium with serum (KODMEM), consisting in KnockOut-DMEM (Thermo Fisher Scientific) supplemented with 15% fetal bovine serum, ESC-qualified, USDA-approved regions (Thermo Fisher Scientific), 2 mM L-glutamine (Thermo Fisher Scientific), 1% non-essential amino acids (Sigma-Aldrich), 100 U/mL penicillin/streptomycin (Thermo Fisher Scientific), 0.1 mM 2-mercapthoethanol (Thermo Fisher Scientific) and 1,000 U/mL of ESGRO LIF (Merck Millipore); and (2) serum-free medium (2i), consisting of 1:1 mix of DMEM/F12 (Thermo Fisher Scientific) and Neurobasal medium (Thermo Fisher Scientific), N2 (Clontech), and B27 (Thermo Fisher Scientific) supplements, 100 U/mL penicillin/streptomycin, 0.1 mM 2-mercapthoethanol, 2 mM L-glutamine, and 1,000× dilution of the supplements LIF and MEK/GSK3 inhibitors (Merck Millipore). To induce spontaneous differentiation, mESCs were maintained in KODMEM in the absence of LIF for 4 days. AA (50 nM, Sigma-Aldrich) was used to block complex III mitochondria respiratory chain. AA was added to cells cultured in KODMEM medium in the absence of LIF.
To induce neuronal differentiation, 10,000 cells/cm2 mESC were seeded on 0.1% gelatin-coated plates and cultured in N2B27 serum-free medium for 5 days. Medium was replaced every 2 days. N2B27 is a 1:1 mix of Neurobasal medium (Thermo Fisher Scientific), 1:2 mix of minimum essential medium (Thermo Fisher Scientific) and 1:2 mix of Ham's F12 Nutrient Mix (Thermo Fisher Scientific), supplemented with 1 mM L-glutamine, 100 U/mL penicillin/streptomycin, 0.1 mM 2-mercapthoethanol, 1.5 g/L D-glucose, 1.5 g/L AlbuMAX I Lipid-Rich BSA (Thermo Fisher Scientific), 7.5 mM HEPES (Thermo Fisher Scientific), B27 (Thermo Fisher Scientific), and N2 (Thermo Fisher Scientific).
AP Staining
mESCs were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) at room temperature (RT) and stained with Vector Red AP vector kit (no. SK5100, Vector Laboratories) as described in the manufacturer's instructions. In brief, after mESC were fixed, cells were washed twice with PBS and incubated with substrate working solution (2 drops of reagents 1, 2, and 3 diluted in 5 mL of Tris-HCl 150 mM, pH 8.5, buffer containing 0.1% Tween [Sigma-Aldrich]) for 30 min on the dark. After incubation, cells were washed once with 150 mM Tris-HCl and then PBS was added. Fluorescence as well as phase-contrast images were taken.
Immunocytochemistry
mESCs were fixed with 4% paraformaldehyde for 15 min at RT. Cells were then washed 3 times with PBS, permeabilized with 0.1% Triton X-100 (Sigma-Aldrich) in PBS for 10 min, and blocked for non-specific binding with 1% BSA (Sigma-Aldrich) in PBS for 1 hr at RT, followed by incubation overnight at 4°C primary antibody: rabbit anti-OCT-4 (no. 2840, Cell Signaling Technology) at 1:100 dilution. Cells were then washed with PBS for 3 times and incubated for 1 hr at RT with a solution containing secondary antibody: anti-rabbit Alexa Fluor 488 (Thermo Fisher Scientific) at 1:200 dilution; Hoechst 33342 (Sigma-Aldrich) at 1:50 dilution and phalloidin Alexa Fluor 647 (no. A22287, Thermo Fisher Scientific) at a dilution 1:40 dilution. Cells were then washed 3 times with PBS and images were taken.
Imaging Acquisition
All images were collected with a Nikon DS-QiMc camera installed on a customized Nikon TE300 epifluorescent microscope (Nikon, Melville, NY), equipped with a motorized stage and motorized excitation and emission filters (Prior Scientific, Rockland, MA) controlled by Nikon NIS Elements. Images were acquired with a 10× Plan Fluor lens (N.A. 0.3, Nikon, Melville, NY) and different grid numbers with a 20% overlap were acquired in order to ensure that the entire well was imaged. Image size from the camera was 1,280 × 1,280 pixels, and the pixel size 0.57 μm. For immunofluorescence images, three fluorescence channels for Hoechst 33342, Alexa Fluor 488 and Alexa Fluor 647 were recorded, while for AP staining the fluorescence channel Alexa Fluor 568 and phase-contrast channel were recorded.
RNA Extraction and Purification
RNA was extracted of each condition by incubating cells with trizol (Life Technologies) for 1 min at RT. The solution was then collected to eppendorf tubes and mechanical disrupted by vortex each sample for 30 s. Then, RNA extraction was performed according to manufacturer's instructions (Direct-zol RNA MiniPrep, Zymo).
cDNA Synthesis and Quantitative Real-Time PCR
One microgram of total RNA was used to synthesize first-strand DNA through the iScriptTM cDNA Synthesis Kit (Bio-Rad), according to manufacturer's instructions.
qPCR was performed using mouse-specific primers and iTaq Universal SYBR Green master mix (Bio-Rad). Primer sequences, which were obtained in the PrimerBank database (Spandidos et al., 2008, Spandidos et al., 2010, Wang and Seed, 2003) are described in the Table S2. All samples were analyzed in technical duplicates. The expression of each target mRNA was calculated based on the threshold cycle (Ct) as 2−Δ(ΔCT), where ΔCt = Cttarget – Ct Rplp0 and –Δ(ΔCT) = ΔCttest – ΔCtcontrol. Control condition refers to mESC cultured in the presence of LIF. All data are presented as mean ± SEM. qPCR data analysis was performed using the Bio-Rad CFX manager software 3.1, and gene expression was considered significantly different to the control when regulation threshold was higher than 2.0 and p value threshold was lower than 0.05.
Author Contributions
T.P., J.R.S., and D.W. conceived and designed the experiments; T.P. and C.L. performed the experiments; T.P. and W.C. planned the software; W.C. developed the software; T.P., W.C., M.C., C.L., S.P., A.S.R., and M.I.S. analyzed the data; and T.P., J.R.S., and D.W. wrote the manuscript. All authors reviewed, edited, and approved the manuscript.
Acknowledgments
The authors thank Fundação para a Ciência e Tecnologia (FCT) Portugal, for PhD scholarships attributed to T.P. (SFRH/BD/51684/2011), M.C. (SFRH/BD/51681/2011), and M.I.S. (SFRH/BD/86260/2012) and a postdoc fellowship attributed to S.P. (SFRH/BPD/98995/2013). Center for Neuroscience and Cell Biology (CNC) is also funded by FCT under the strategic project UID/NEU/04539/2013. The authors also give thanks for the grant support by FCT and FEDER/COMPETE 2020 (POCI-01-0145-FEDER-007440) and HealthyAging2020:CENTRO-01-0145-FEDER-000012. Rui Portela (REQUIMTE/FCT/UNL) is thanked for scientific discussions and for reviewing the Manuscript. All members of Wirtz and Ramalho-Santos Labs are acknowledged for helpful discussions.
Published: July 13, 2017
Footnotes
Supplemental Information includes two figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.06.006.
Contributor Information
João Ramalho-Santos, Email: jramalho@uc.pt.
Denis Wirtz, Email: wirtz@jhu.edu.
Supplemental Information
References
- Avilion A.A., Nicolis S.K., Pevny L.H., Perez L., Vivian N., Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Random forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- Chalfoun J., Majurski M., Peskin A., Breen C., Bajcsy P., Brady M. Empirical gradient threshold technique for automated segmentation across image modalities and cell lines. J. Microsc. 2015;260:86–99. doi: 10.1111/jmi.12269. [DOI] [PubMed] [Google Scholar]
- Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- French A., Bravery C., Smith J., Chandra A., Archibald P., Gold J.D., Artzi N., Kim H.W., Barker R.W., Meissner A. Enabling consistency in pluripotent stem cell-derived products for research and development and clinical applications through material standards. Stem Cells Transl. Med. 2015;4:217–223. doi: 10.5966/sctm.2014-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman B.R., Lu J., Baccei A., Lowry N.C., Purvis J.E., Mangoubi R.S., Lerou P.H. Multi-scale imaging and informatics pipeline for in situ pluripotent stem cell analysis. PLoS One. 2014;9:e116037. doi: 10.1371/journal.pone.0116037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart A.H., Hartley L., Ibrahim M., Robb L. Identification, cloning and expression analysis of the pluripotency promoting Nanog genes in mouse and human. Dev. Dyn. 2004;230:187–198. doi: 10.1002/dvdy.20034. [DOI] [PubMed] [Google Scholar]
- Hay D.C., Sutherland L., Clark J., Burdon T. Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells. 2004;22:225–235. doi: 10.1634/stemcells.22-2-225. [DOI] [PubMed] [Google Scholar]
- Intawong K., Scuturici M., Miguet S. A new pixel-based quality measure for segmentation algorithms integrating precision, recall and specificity. In: Wilson R., Hancock E., Bors A., Smith W., editors. Computer Analysis of Images and Patterns: 15th International Conference, CAIP 2013, York, UK, August 27-29, 2013, Proceedings, Part I. Springer; 2013. pp. 188–195. [Google Scholar]
- Jaccard N., Macown R.J., Super A., Griffin L.D., Veraitch F.S., Szita N. Automated and online characterization of adherent cell culture growth in a microfabricated bioreactor. J. Lab. Autom. 2014;19:437–443. doi: 10.1177/2211068214529288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaccard N., Griffin L.D., Keser A., Macown R.J., Super A., Veraitch F.S., Szita N. Automated method for the rapid and precise estimation of adherent cell culture characteristics from phase contrast microscopy images. Biotechnol. Bioeng. 2014;111:504–517. doi: 10.1002/bit.25115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joutsijoki H., Haponen M., Rasku J., Aalto-Setälä K., Juhola M. Machine learning approach to automated quality identification of human induced pluripotent stem cell colony images. Comput. Math. Methods Med. 2016;2016:3091039. doi: 10.1155/2016/3091039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou X., Kang M., Xenopoulos P., Muñoz-Descalzo S., Hadjantonakis A.K. A rapid and efficient 2D/3D nuclear segmentation method for analysis of early mouse embryo and stem cell image data. Stem Cell Rep. 2014;2:382–397. doi: 10.1016/j.stemcr.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddah M., Shoukat-Mumtaz U., Nassirpour S., Loewke K. A system for automated, noninvasive, morphology-based evaluation of induced pluripotent stem cell cultures. J. Lab. Autom. 2014;19:454–460. doi: 10.1177/2211068214537258. [DOI] [PubMed] [Google Scholar]
- Martello G., Smith A. The nature of embryonic stem cells. Annu. Rev. Cell Dev. Biol. 2014;30:647–675. doi: 10.1146/annurev-cellbio-100913-013116. [DOI] [PubMed] [Google Scholar]
- Narkilahti S., Rajala K., Pihlajamäki H., Suuronen R., Hovatta O., Skottman H. Monitoring and analysis of dynamic growth of human embryonic stem cells: comparison of automated instrumentation and conventional culturing methods. Biomed. Eng. Online. 2007;6:11. doi: 10.1186/1475-925X-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Paduano V., Tagliaferri D., Falco G., Ceccarelli M. Automated identification and location analysis of marked stem cells colonies in optical microscopy images. PLoS One. 2013;8:1–11. doi: 10.1371/journal.pone.0080776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmqvist L., Glover C.H., Hsu L., Lu M., Bossen B., Piret J.M., Humphries R.K., Helgason C.D. Correlation of murine embryonic stem cell gene expression profiles with functional measures of pluripotency. Stem Cells. 2005;23:663–680. doi: 10.1634/stemcells.2004-0157. [DOI] [PubMed] [Google Scholar]
- Pereira S.L., Grãos M., Rodrigues A.S., Anjo S.I., Carvalho R.A., Oliveira P.J., Arenas E., Ramalho-Santos J. Inhibition of mitochondrial complex III blocks neuronal differentiation and maintains embryonic stem cell pluripotency. PLoS One. 2013;8:1–16. doi: 10.1371/journal.pone.0082095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf N., Herberg M., Thierbach K., Zerjatke T., Kalkan T., Humphreys P., Smith A., Glauche I., Roeder I. Imaging, quantification and visualization of spatio-temporal patterning in mESC colonies under different culture conditions. Bioinformatics. 2012;28:i556–i561. doi: 10.1093/bioinformatics/bts404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandidos A., Wang X., Wang H., Dragnev S., Thurber T., Seed B. A comprehensive collection of experimentally validated primers for polymerase chain reaction quantitation of murine transcript abundance. BMC Genomics. 2008;9:633. doi: 10.1186/1471-2164-9-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandidos A., Wang X., Wang H., Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010;38(Database issue):D792–D799. doi: 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefková K., Procházková J., Pachernik J. Alkaline phosphatase in stem cells. Stem Cells Int. 2015;2015:628368. doi: 10.1155/2015/628368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga K., Saitoh N., Goldberg I.G., Sakamoto C., Yasuda Y., Yoshida Y., Yamanaka S., Nakao M. Computational image analysis of colony and nuclear morphology to evaluate human induced pluripotent stem cells. Sci. Rep. 2014;4:6996. doi: 10.1038/srep06996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31:e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou K.H., Warfield S.K., Bharatha A., Tempany C.M., Kaus M.R., Haker S.J., Wells W.M., 3rd, Jolesz F.A., Kikinis R. Statistical validation of image segmentation quality based on a spatial overlap index. Acad. Radiol. 2004;11:178–189. doi: 10.1016/S1076-6332(03)00671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.