Abstract
Fibronectin (FN) is an extracellular matrix (ECM) glycoprotein that plays an important role in a wide range of biological processes including embryonic development, wound healing, and fibrosis. Recent evidence has demonstrated that FN is mechanosensitive, where the application of force induces conformational changes within the FN molecule to expose otherwise cryptic binding domains. However, it has proven technically challenging to dynamically monitor how the nanostructure of FN fibers changes as a result of force-induced extension, due in part to the inherent complexity of FN networks within tissue and cell-generated extracellular matrix (ECM). This has limited our understanding of FN matrix mechanobiology and the complex bi-directional signaling between cells and the ECM, and de novo FN fiber fabrication strategies have only partially addressed this. Towards addressing this need, we have developed a modified surface initiated assembly (SIA) technique to engineer FN nanofibers that we can uniaxially stretch to >7-fold extensions and subsequently immobilize them in the stretched state for high resolution atomic force microscopy (AFM) imaging. Using this approach, we analyzed how the nanostructure of FN molecules within the nanofibers changed with stretch. In fully contracted FN nanofibers, we observed large, densely packed, isotropically-oriented nodules. With intermediate extension, uniaxially-aligned fibrillar regions developed and nodules became progressively smaller. At high extension, the nanostructure consisted of highly aligned fibrils with small nodules in a beads-on-a-string arrangement. In summary, we have established a methodology to uniaxially stretch FN fibers and monitor changes in nanostructure using AFM. Our results provide new insight into how FN fiber extension can affect the morphology of the constituent FN molecules.
Introduction
Fibronectin (FN) is an important extracellular matrix (ECM) glycoprotein that plays a major role in many biological processes including development, wound healing, blood clotting and fibrosis. In addition to providing binding sites for mammalian and bacterial cells,1, 2 FN contains binding sites for collagen, fibrin, heparin, growth factors, and self-assembly.3–5 Some of these sites have been identified as cryptic and require conformational changes in the tertiary and/or secondary protein structure to become exposed. While these sites may become accessible by enzymatic cleavage at specific sites, it is also believed that cells are capable of using contractile forces to unfold FN and expose these cryptic sites.6, 7 Understanding how cells can manipulate and mechanically deform FN fibers to modulate the exposure of these cryptic sites is critical for developing a mechanistic model of ECM mechanobiology, and how this underlies biological processes such as tissue morphogenesis and ECM remodeling in cancer, fibrosis, and other diseases.
Due to the complex structure and composition of the native ECM, a number of researchers have developed synthetic approaches to de novo assemble FN fibers in a cell-free environment to study FN’s biological, structural, and mechanical properties. Current strategies include the use of denaturants,8 disulfide reducing agents,9 protein-surface interactions,10–12 and surface tension at the air-liquid interface2, 13, 14 to control the unfolding of FN dimers from a compact, globular conformation and into a fibrillar conformation. This exposes cryptic self-assembly sites to enable the formation of synthetically assembled FN fibers in a cell-free environment. For example, Vogel and coworkers have used force-induced assembly at an air-liquid interface to create microscale FN fibers that can be deposited onto stretchable2, 13, 15 or microfabricated substrates.16, 17 They found that these microscale FN fibers can support upwards of 8-fold extension17 and that strain can modulate the exposure of cryptic cysteine residues.17 Stretching these microscale FN fibers also influenced the availability of bacterial adhesion sites,2 formation of FN-collagen interactions,16 and the unfolding of FN type III modules as quantified through FRET imaging.13, 14 Other researchers have used material-driven fibrillogenesis, which relies on protein-surface interactions, to assemble nanometer-scale FN fibrils on poly(ethylacrylate) (PEA),10, 18 poly(hydroxyethyl acrylate)19, and polysulfonated11 substrates. By using scanning electron microscopy (SEM) and/or atomic force microscopy (AFM) to directly image these smaller fibrils, researchers showed that surface chemistry can modulate the conformation of FN fibrils and affect cell adhesion12, 19 and differentiation.10 However, these FN fibrils were permanently bound to the surface, prohibiting a dynamic structural and/or mechanical characterization of the FN fibrils. While these and related techniques have provided important insight into FN mechanobiology, it has proven difficult to combine mechanical and structural characterization in order to directly correlate force-induced stretching with changes in molecular conformation and biological activity.
Here we directly addressed this challenge by using a surface-initiated assembly (SIA) technique20, 21 to engineer FN nanofibers that can be uniaxially stretched and then immobilized for nanoscale structural analysis. The SIA process uses thermally-driven dissolution of a poly(N-isopropylacrylamide) (PIPAAm) substrate to drive the fibrillogenesis of micropatterned ECM proteins, such as FN, into insoluble protein networks.21 Previous work has shown that FN nanofibers ~10 nm thick, 50 µm wide and over 1 cm long can be engineered and stretched to 8-fold extension using micromanipulators before failure.22 Further, using laminin nanofibers, we have shown that differences in microscale mechanical behavior can be correlated directly to differences in nanoscale molecular structure of the constituent molecules.23 In the current study, we combined uniaxial tensile deformation together with AFM imaging to directly study changes in nanoscale structure during stretch of FN nanofibers. To do this we developed a method to label the FN nanofibers with FN-based fiducial marks to accurately measure extension during uniaxial stretching, by using an adaptation of our previously published patterning-on-topography (PoT) technique.24 The FN nanofibers were then stretched in defined amounts from 1- to 8-fold extension and immobilized in the stretched state onto glass coverslips for AFM analysis. Using this approach, we were able to track how the structure of the FN molecules at the nanometer scale changes as a function of extension. Importantly, we quantified how FN molecules in the nanofibers change from a globular, folded structure to a fibrillar, unfolded structure and also determined how the network of FN molecules begins to fail at higher extensions. This provides unique insight into how the morphology of FN molecules assembled into fibers can be modulated by extension and ultimately towards our long-term goal to understand how extension can regulate FN biological activity through changes in secondary and/or tertiary structure.
Experimental
PDMS Stamp Fabrication
FN nanofibers were prepared by adapting the SIA approach described previously.20, 21 Briefly, poly(dimethylsiloxane) (PDMS) stamps for microcontact printing, patterned with either 1 cm long, 50 µm wide raised lines or 1 cm long, 10 µm wide raised lines were fabricated by first spincoating glass wafers with SPR 220.3 positive photoresist (Microchem). The photoresist was then exposed to UV light through a photomask and developed using MF-319 developer (Microchem). A negative of the patterned photoresist wafer was formed by casting PDMS prepolymer (Sylgard 184, Dow Corning) over it and curing it at 65° C for 4 hours. The cured PDMS was then peeled off the glass wafer and 1 cm2 stamps for microcontact printing were cut out.
Engineering Fibronectin Nanofibers with Fibronectin-Based Fiducial Marks
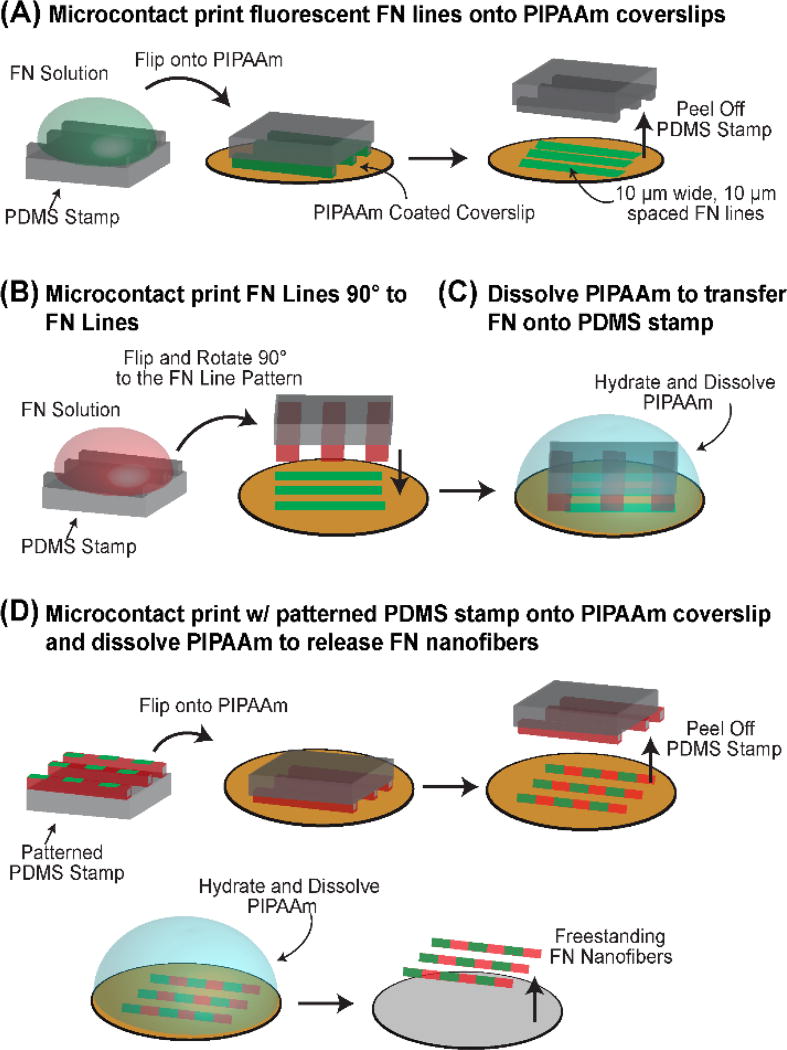
FN nanofibers were tagged with fluorescently labeled, FN-based fiducial marks by combining SIA20, 21 with Patterning-on-Topography (PoT)24 (Fig. 1). First, PDMS stamps with 10 µm wide, 10 µm spaced, 1 cm long ridges were cleaned by sonication in a 50% ethanol solution for 30 minutes. Following sonication, the stamps were dried under a stream of nitrogen, and then incubated with FN from human plasma (BD Biosciences) for 60 minutes at a concentration of 50 µg/ml in distilled water, where 40% of the FN was conjugated to an Alexa Fluor 488 dye (Life Technologies). After incubation, the stamps were washed, dried under a stream of nitrogen and then brought into conformal contact with PIPAAm coated coverslips for 15 minutes (Fig. 1A). The PIPAAm coated coverslips were prepared by spincoating a 10% PIPAAm (Polysciences Inc.) in 1-butanol (w/v) solution. Upon removal of the stamps, the fidelity of the transfer of the 10 µm wide FN lines to the PIPAAm coated coverslips was inspected using phase contrast microscopy.
Fig. 1.
Preparation of FN nanofibers with FN-based fiducial marks. (A) 10 µm wide, fluorescently conjugated FN lines were first microcontact printed onto a PIPAAm coated coverslip. (B) A second PDMS stamp was coated with FN and brought orthogonally into conformal contact with the 10 µm wide FN lines on PIPAAm. (C) The 10 µm wide lines were then released onto the FN coated stamp through the dissolution of PIPAAm. (D) The FN coated PDMS stamp with 10 µm wide FN fiducial marks was then microcontact printed onto a new PIPAAm coated coverslip and the FN nanofibers could be released by subsequent thermally triggered dissolution of PIPAAm.
Following microcontact printing, a second PDMS stamp with 50 µm wide and 1 cm long raised features was coated with a 50 µg/ml solution of FN dissolved in distilled water. The stamp was then washed to remove excess FN, dried under a stream of nitrogen and subsequently brought into conformal contact orthogonal to the PIPAAm coated coverslip containing the 10 µm wide FN lines (Fig. 1B). With the stamp still in conformal contact, the PIPAAm was hydrated with warm, 40° C, distilled water and allowed to cool below the LCST of PIPAAm thereby releasing the fluorescently labeled, 10 µm wide FN lines off of the PIPAAm onto the PDMS stamp, which was pre coated with FN (Fig. 1C). The stamp was then removed, washed in distilled water, dried under a stream of nitrogen and then brought into conformal contact with a new PIPAAm coated coverslip for 10 minutes to create arrays of 1 cm × 50 µm FN lines with 10 µm wide and 10 µm spaced fiducial marks (Fig. 1D).
Fibronectin Nanofiber Stretching Experiments
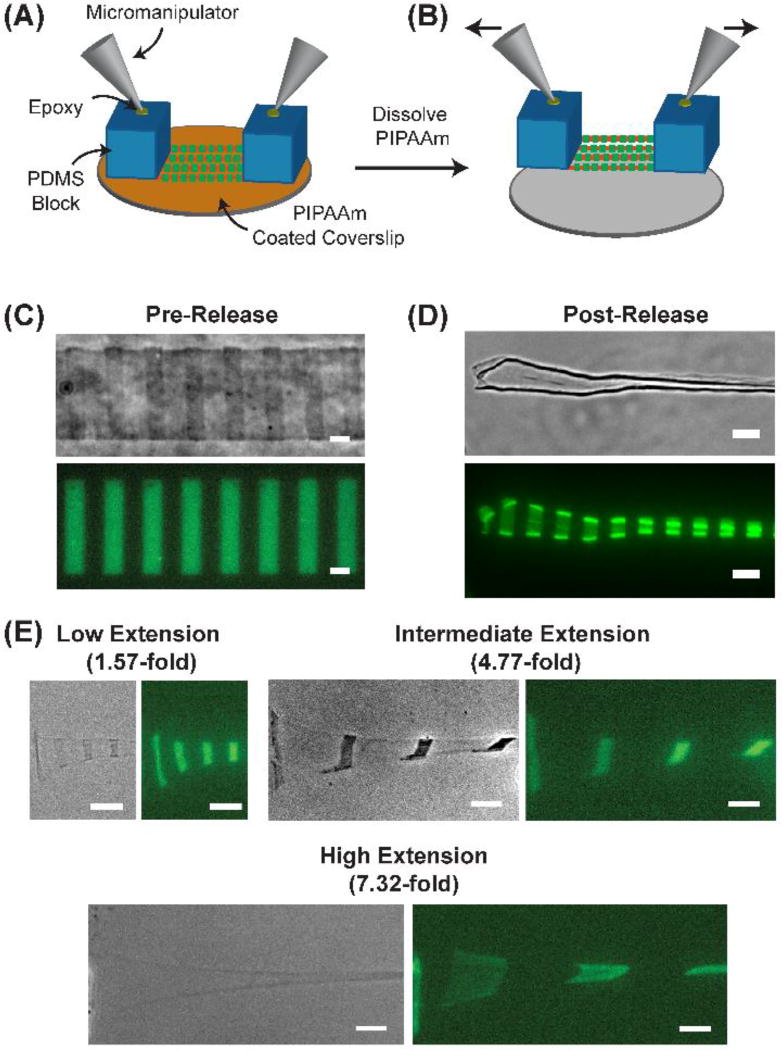
To stretch FN nanofibers, we first engineered 1 cm long, 50 µm wide FN nanofibers labeled with fiducial marks, as described above. Next, we prepared PDMS pads by casting PDMS prepolymer over a glass slide to a thickness of ~5 mm and curing it in an oven set to 65° C for 4 hours. Once the PDMS cured, we cut out 2 PDMS rectangles that were ~1 cm long and ~5mm wide and placed them at opposite ends on top of the FN nanofibers on PIPAAm. To interface PDMS pads to a pair of micromanipulators, glass capillary tubes attached to micromanipulators were then lowered and centered on top of the PDMS blocks and cured in place using a small drop of 5-minute epoxy (Devcon), applied to the top of the PDMS pads (Fig. 2A). Once the epoxy hardened for 30 minutes, we triggered the thermal dissolution of PIPAAm by hydration and subsequent cooling below the lower critical solution temperature (LCST) of PIPAAm, which led to the assembly and release of the nanofibers off of the PIPAAm and onto the PDMS pads. This resulted in FN fibers that were tethered at each end to a PDMS pad and freely suspended in between. First, the micromanipulators were brought towards each other until the FN nanofibers reached a fully contracted state with no slack. To apply uniaxial, tensile deformation the micromanipulators were moved in opposite directions to a defined distance (Fig. 2B), and the FN nanofibers were subsequently immobilized back onto the coverslip to maintain this extension for AFM imaging. To calculate extension, we used the center-to-center fiducial mark distance of a released and fully contracted FN nanofiber as the initial length (referred to as no stretch) and the center-to-center fiducial mark distance of the stretched FN nanofibers after extension as the final length. Stretching experiments were performed on top of an inverted epifluorescent microscope (Eclipse Ti, Nikon Instruments) with mounted micromanipulators (Transferman NK2, Eppendorf).
Fig. 2.
Using fiducial marks to measure FN stretch. (A) Schematic of the stretching apparatus where PDMS supports are placed on opposite ends of 1 cm long FN nanofibers. Micromanipulators are then attached to the PDMS supports using high strength epoxy. (B) After hydration and thermally triggered dissolution of PIPAAm, the FN nanofibers release onto the PDMS supports where they are freely suspended in between. (C) Phase contrast and fluorescent image of a FN nanofiber pre-release with 10 µm wide, 10 µm spaced fiducial marks. The center-to-center distance of the fiducial marks was 20.2 ± 0.6 µm. (D) Phase contrast and fluorescent image of a FN nanofiber post-release where the center-to-center distance of the fiducial marks decreased to 10.8 ± 0.4 µm. (E) FN nanofibers were subjected to low, intermediate, and high extensions as indicated by the separation of the fiducial marks. Scale bars are (C and D) 10 µm and (E) 20 µm.
Atomic Force Microscopy of Fibronectin Nanofiber Morphology
The AFM (MFP3D–Bio, Asylum Research) was used to analyze the nanostructure and quantify the morphology of FN nanofibers at low, intermediate, and high extensions. The FN nanofibers were scanned in air using AC mode with AC160TS cantilevers (Olympus Corporation). After each fiber was stretched, it was immobilized back onto the coverslip, washed three times with distilled water to remove any potential residual PIPAAm, and then dried in a 40°C oven. The FN nanofibers were initially imaged with a scan area of 20 µm to 30 µm. Areas of interest were then imaged with a scan size of 3 µm × 3 µm and used for quantitative analysis. The nodule size and the FN surface coverage were quantified using the IGOR Pro software environment (WaveMetrics) using the AFM height channel. To quantify surface coverage, we applied a mask to the fibrils within the nanostructure and measured the area occupied by the masked regions divided by the total area for the image. This was only performed on the high resolution, 3 µm × 3 µm scans to allow us to include the smallest fibrils within the mask. Nodule diameter and nodule-to-nodule spacing was performed also using the IGOR Pro software environment. For nodule diameter, a line was drawn over each nodule to generate a cross-sectional profile based on the height information of each pixel along the line. In these cross-sectional profiles, each nodule typically produced a peak. The width of each peak was measured to produce the diameter for each nodule. To measure the nodule spacing, we drew a line between consecutive nodules and measured the distance between the two points where the height was the highest. To measure regions under stretch that transitioned from nodular to fibrillar, a line was drawn along the fibril length to generate a height profile. The transverse widths of the nodular and fibrillar regions were measured orthogonally.
Results
Engineering Fibronectin Nanofibers with Fiducial Marks to Track Uniaxial Extension
To precisely track extension of the FN nanofibers, we developed a new technique to create fluorescent fiducial marks at defined positions. To do this, we combined SIA20, 21 with a modified PoT technique24 in order to engineer FN nanofibers that were initially ~10 nm thick, 50 µm wide and 1 cm long. This ribbon-like morphology was designed to ensure a single layer was maintained without folding over during high extensions, in order to resolve the nanostructure with high resolution AFM. The FN nanofibers were labeled with fluorescent, FN-based fiducial marks by (i) microcontact printing 10 µm wide, 10 µm spaced lines of fluorescently labeled FN onto PIPAAm (Fig. 1A); (ii) microcontact printing 50 µm wide lines of unlabeled FN orthogonal to the 10 µm wide FN lines, but keeping the PDMS stamp in conformal contact with the PIPAAm (Fig. 1B); (iii) dissolving the PIPAAm by hydration in 40° C water and cooling through the LCST of PIPAAm to transfer the FN pattern onto the PDMS stamp (Fig. 1C); and (iv) using this PDMS stamp to microcontact print the FN lines with fiducial marks onto a new PIPAAm coated coverslip and dissolving the PIPAAm to create the freestanding FN nanofibers (Fig. 1D). This process produces FN nanofibers that can be handled using micromanipulators, and similar nanofibers without the fiducial marks were previously shown to undergo large, 8-fold extensions before failure when uniaxially stretched with calibrated microneedles.22
Next, we developed a system to apply controlled uniaxial stretch to the FN nanofibers and then immobilize them onto glass coverslips. To do this, we took the array of FN nanofibers still adhered to the PIPAAm coated coverslip prior to release (as shown in Fig. 1D) and placed 10×5×5 mm PDMS blocks on top of the nanofibers at each end, in conformal contact. Glass capillary tubes attached to micromanipulators were then lowered and centered on top of the PDMS blocks and cured in place using 5-minute epoxy (Fig. 2A). Next, we submerged the coverslip and PDMS blocks in 40°C DI water and allowed it to cool to room temperature to dissolve the PIPAAm layer and release the FN nanofibers. The micromanipulators were then raised 100 µm above the surface of the coverslip, with the FN nanofibers freely suspended between the PDMS blocks (Fig. 2B). Note that pre-release nanofibers refer to the nanofibers when they are patterned on the PIPAAm, however, formation of freestanding nanofibers is not completed until PIPAAm dissolution occurs and the nanofibers are released from the substrate.
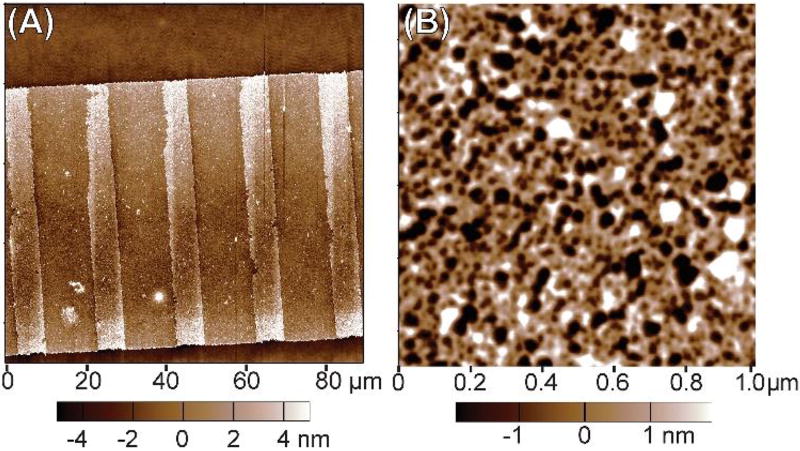
As an initial analysis, we performed AFM imaging of pre-release FN nanofibers at large (Fig. 3A) and small (Fig. 3B) scan areas. The large area AFM scan of an entire nanofiber width confirmed that we successfully formed the fiducial marks as a second patterned layer of FN stripes on top of a continuous underlying nanofiber (Fig. 3A). The small area AFM scan provided a higher resolution image of the FN molecules between the fiducial marks, and revealed an interconnected, fibrillar network (Fig. 3B). This suggests that the FN molecules have already formed fibrils on the PIPAAm layer prior to release. Importantly, this interconnected, fibrillar network morphology is consistent with that observed by AFM previously for pre-release FN and laminin nanofibers.21, 23
Fig. 3.
AFM images of as-patterned FN nanofibers pre-release while still attached to the PIPAAm-coated coverslip. (A) A large area AFM scan showing the FN nanofiber pre-release with the clearly defined fiducial marks of FN as raised features spaced at regular intervals. (B) A small area AFM scan showing the FN nanofiber molecular morphology in the region between fiducial marks. Note that the FN molecules appear to form an interconnected, fibrillar network with the fibrils forming interconnection points (nodes) where generally 3–5 fibrils meet.
Accurate quantification of extension requires the ability to measure fiducial mark spacing over a wide range of deformations. First, we measured the center-to-center distance of the fiducial marks on FN nanofibers pre-release on the PIPAAm surface (Fig. 2C). The distance of 20.2 ± 0.6 µm agreed well with the 20 µm center-to-center spacing of the original micropattern on the photomask and confirmed that we were achieving the intended fidelity with our nanofiber fabrication process. Next, we tethered one end of the FN nanofibers to a PDMS block and left the other end free, and then released the nanofibers through dissolution of the PIPAAm. This resulted in both lateral and longitudinal contraction of the nanofibers, with a decrease in fiducial mark center-to-center distance to 10.8 ± 0.4 µm, an ~0.5-fold change relative to pre-release (Fig. 2D). ECM protein nanofibers are known to be under a pre-stress and undergo dimensional change (contract) upon release as the constituent molecules undergo conformational changes, as previously reported for FN and laminin nanofibers.20–23 We used this post-release state as the starting point for calculating extension, and thus all nanofiber extensions were determined based on an initial center-to-center distance of 10.8 µm. We refer to this as the no stretch state. Representative examples of uniaxially stretched FN nanofibers showed center-to-center fiducial mark distances of 16.8 ± 1.6 µm, 51.4 ± 3.8 µm and 78.8 ± 2.6 µm, corresponding to 1.57-fold, 4.77-fold and 7.32-fold extensions, respectively (Fig. 2E). This large extensibility is comparable to what has been previously reported for uniaxial tensile testing of FN micro- and nanoscale fibers, confirming similar extensibility for our FN nanofibers engineered with fiducial marks.22.17
Nanostructure of Fibronectin Nanofibers Changes During Uniaxial Extension
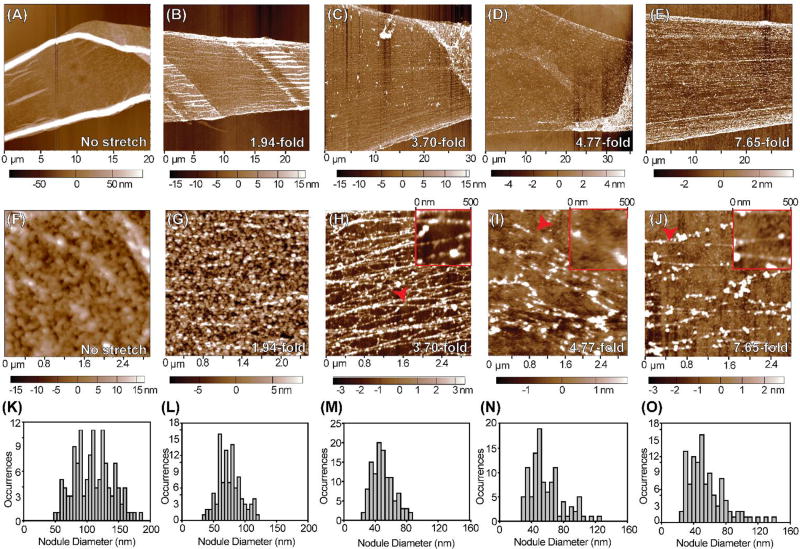
FN nanofibers were stretched up to ~7.5-fold and then immobilized in the stretched state by dehydrating back on to the coverslip for AFM analysis. We imaged FN nanofibers at 1- to 7.65-fold extensions at lower (Fig. 4A–E) and higher (Fig. 4F–J) scan sizes, and performed analysis of nodule diameter for each extension (Fig. 4K–O). We first imaged FN nanofibers that had fully contracted in solution (defined as no stretch) (Fig. 4A). At higher resolution FN molecules formed a dense array of nodules in an overall isotropic arrangement (Fig. 4F). Quantitative analysis showed nodule diameter varied from 50 to 200 nm, with a mean diameter of 107 ± 29 nm (Fig. 4K). Next, we scanned FN nanofibers that had a fiducial mark center-to-center distance of 20.8 ± 1.1 µm, a ~1.94-fold extension relative to the no stretch state (Fig. 4B). This was accomplished by releasing the nanofibers onto the PDMS supports and immediately lowering them back down onto the coverslip. Thus, these nanofibers had approximately the same extension as the pre-release nanofibers patterned on the coverslip prior to PIPAAm dissolution. At high resolution (Fig. 4G), it was clear that the nanostructure was still dominated by the presence of nodules, although they were markedly smaller with a mean diameter of 73 ± 19 nm (Fig. 4L).
Fig. 4.
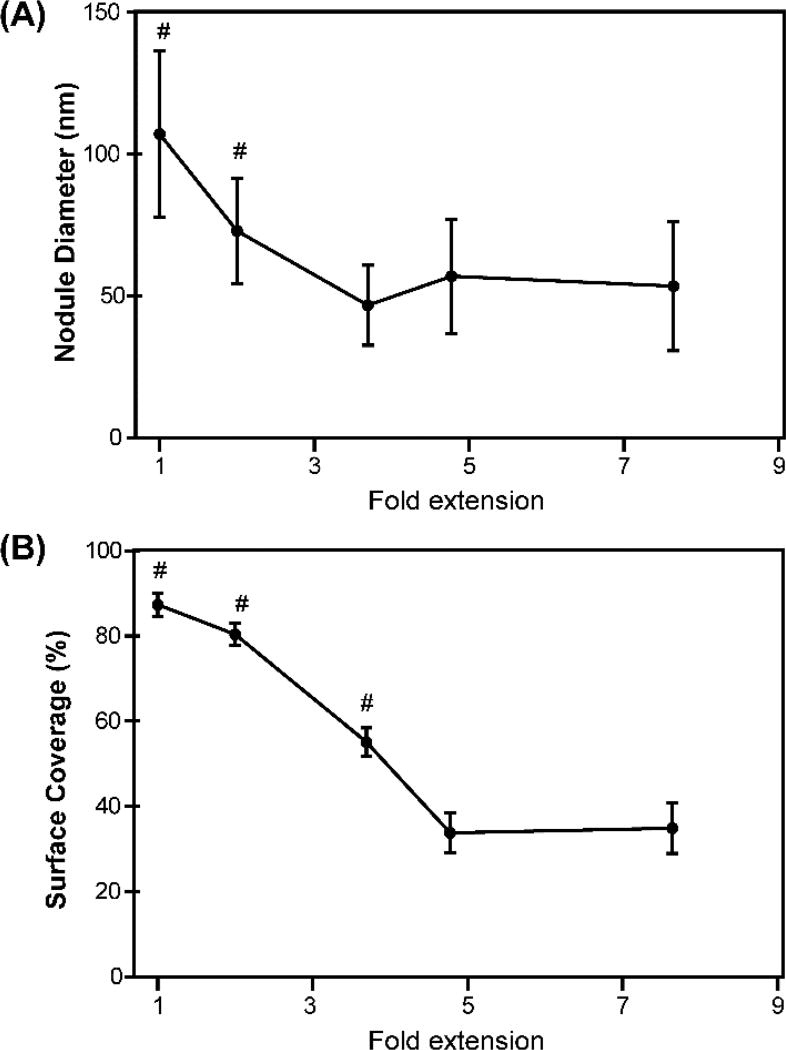
Investigating the nanostructure of fully contracted to highly stretched FN nanofibers with AFM. (A–E) Representative low resolution AFM scans of FN nanofibers that were (A) allowed to fully contract in solution prior to imaging, (B) stretched 1.94-fold, (C) stretched 3.70-fold, (D) stretched 4.77-fold, or (E) stretched 7.65-fold relative to the fully contracted state and immobilized prior to imaging. (F–J) Representative high resolution AFM scans of the same FN nanofibers from (A–E). (F) Fully contracted nanofibers had a nanostructure consisting mainly of large, isotropic nodules. (G) FN nanofibers stretched 1.94-fold had an isotropic, nodular network nanostructure with smaller nodules than fully contracted nanofibers. (H) FN nanofibers stretched 3.70-fold had a nodular nanostructure containing FN fibrils that align in the direction of stretch. (I) FN nanofibers stretched 4.77-fold also had a nodular nanostructure containing FN fibrils that align in the direction of stretch. (J) FN nanofibers stretched 7.65-fold had a nanostructure comprised of sparse, aligned FN fibrils that maintain a nodular structure. (K–O) Histograms showing nodule diameters for each amount of extension. (K) Nodules in fully contracted FN nanofibers had a mean diameter of 107 ± 29 nm, consistent with previous findings using smaller FN nanofibers. (L) Nodules in FN nanofibers stretched 1.94-fold had a mean diameter of 73 ± 19 nm. (M) Nodules in FN nanofibers stretched 3.70-fold had a mean diameter of 47 ± 14 nm. (N) Nodules in FN nanofibers stretched 4.77-fold had a mean diameter of 57 ± 20 nm. (O) Nodules in FN nanofibers stretched 7.65-fold had a mean diameter of 53 ± 23 nm.
Further increasing the extension to center-to-center fiducial mark distances of 39.8 ± 1.2 µm (Fig. 4C, ~3.70-fold extension), 51.4 ± 3.8 µm (Fig. 4D, 4.77-fold extension), and 117.4 ± 6.0 µm (Fig. 4E, 7.65-fold extension) revealed the presence of an overall fibrous nanostructure where the fibrils were anisotropically aligned in the direction of stretch. It is important to note that the fiducial marks for Fig. 4E were prepared using a different stamp and had a center-to-center distance of 29.4 ± 1.6 µm. The fiducial mark center-to-center distance of these FN nanofibers in a no stretch state was 15.4 ± 0.4 µm and was therefore used as the initial center-to-center distance.
In order to resolve the structural details of the aligned fibrils, we increased the resolution of the AFM by decreasing the scan area to ~3 µm × 3 µm. At this scale, it was clear that FN nanofibers stretched 3.70-fold were comprised of thick fibrils with an apparent nodular structure (Fig. 4H). Similarly, for FN nanofibers stretched 4.77-fold and 7.65-fold, there was also an apparent nodular nanostructure (Fig. 4I and 4J). FN nanofibers stretched 3.70-fold were found to have a mean nodule diameter of 47 ± 14 nm (Fig. 4M). In contrast, FN nanofibers that underwent 4.77-fold and 7.65-fold extensions were found to have mean nodule diameters of 57 ± 20 nm (Fig. 4N) and 53 ± 23 nm (Fig. 4O), respectively, which was larger than what was measured for nanofibers that underwent 3.70-fold extensions. We attributed this increase in nodule diameter to the constituent fibrils breaking during large extensions causing them to adopt a more globular conformation. Collectively, when comparing the mean nodule diameters for each amount of extension, we found that the mean nodule diameter significantly decreased from 107 ± 29 nm in fully contracted FN nanofibers to 47 ± 14 nm for FN nanofibers that underwent 3.70-fold extensions, about a 2.3-fold decrease in the mean nodule diameter (Fig. 5A).
Fig. 5.
Measuring nodule diameter and FN surface coverage as a function of extension. (A) Mean nodule diameter. Error bars are standard deviation and the symbol # indicates a statistically significant difference with P < 0.05 using an ANOVA rank sum test with a Dunn’s pairwise comparison. (B) Percent of FN molecule surface coverage as a function of extension. The symbol # indicates a statistically significant difference from the other points with P<0.05 using a one-way ANOVA rank sum test with a Dunn’s pairwise comparison.
During the transition from fully contracted to highly stretched, we also measured the surface coverage of the constituent FN fibrils. We found that as the nanofibers were stretched, there was a decrease in the surface coverage of the constituent fibrils (Fig. 5B). For example, fully contracted FN nanofibers had a mean surface coverage of 87.39% while after stretching the FN nanofibers ~2-fold, the surface coverage of the constituent FN molecules decreased to 80.4%. At higher extensions, we observed a sharper decline in the FN surface coverage from 55.1% for nanofibers stretched to 3.70-fold to 33.77% for nanofibers stretched to 4.77-fold. Finally, at high (7.65-fold) extension, the surface coverage was 34.86%. Altogether, it is clear that with increasing extension, the nodule size decreased until the extension became high enough that the constituent fibril strands began to break and reform larger nodules. With increasing extension there was also a measurable decrease in the FN surface coverage.
Fibronectin Nanofibers Undergo a Nodular to Fibrillar Transition in Nanostructure During Extension
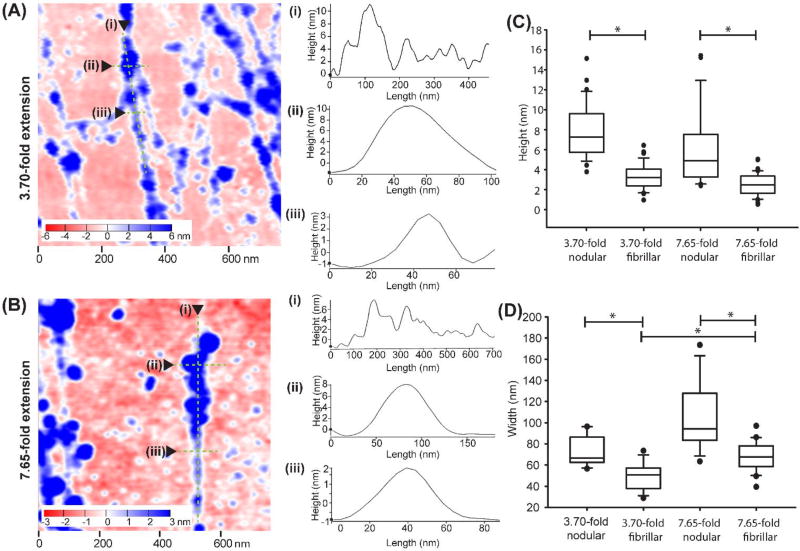
During higher stretch ratios, we observed that the nanostructure consisted of thicker fibrils with a nodular morphology that transitioned into thinner fibrils, as shown in the insets of Fig. 4H–J. This is similar to the nanostructure of fibronectin assembled in both cellular and cell-free environments, which contains thicker, nodular regions that adjoin thinner, fibrillar, or smooth regions.25, 26 To characterize this further, we compared FN nanofibers stretched 3.70-fold (Fig. 6A) and 7.65-fold (Fig. 6B). Representative height profiles along these fibrils showed that fibril height was ~6–10 nm in the nodular, sub-fibril regions and transitioned to ~1–5 nm for the narrower, fibrillar sub-fibril regions (Fig. 6A–i and Fig. 6B–i). For nanofibers stretched 3.70-fold, the mean nodular region height was 8.0 ± 2.7 nm and decreased to 3.3 ± 1.3 nm for the adjoining fibrillar regions (Fig. 6C). Similarly, for nanofibers stretched 7.65-fold, the mean nodular region height was 5.8 ± 3.7 nm and decreased to 2.5 ± 1.1 nm for the adjoining fibrillar regions. The transverse widths showed a similar decrease in size from nodular (Fig. 6A–ii and 6B–ii) to fibrillar (Fig 6A–iii and 6B–iii) regions. For nanofibers stretched 3.70-fold, the mean nodular sub-fibril region width was 74 ± 14 nm and decreased to 49 ± 12 nm for the adjoining fibrillar regions (Fig. 6D). Similarly, for nanofibers stretched 7.65-fold, the mean width of the nodular sub-fibril region was 103 ± 31 nm and decreased to 68 ± 14 nm for the adjoining fibrillar regions. While there was variability in the height and width of the nodular and fibrillar sub-regions across scan areas and samples, the general morphology of this transition from nodular to fibrillar along a fibril was consistent across the higher stretch ratios.
Fig. 6.
Analyzing the transitions between nodular and fibrillar regions in FN nanofiber fibrils. (A) A representative region from a 3 µm × 3 µm scan of a FN nanofiber stretched 3.70-fold. (A–i) Representative cross-sectional profiles of height along a fibril in an area of nodular to fibrillar transition at 3.70-fold extension. (A-ii and A-iii) Representative cross sectional profiles perpendicular through fibers at close nodular (A-ii) and fibrillar (A-iii) regions along the same fibril for 3.70-fold extension. (B) A representative region from a 3 µm×3 µm scan of a FN nanofiber stretched 7.65-fold. (B–i) Representative cross-sectional profiles of height along a fibril in an area of nodular to fibrillar transition at 7.65-fold extension. (B-ii and -iii) Representative cross sectional profiles perpendicular through fibers at close nodular (B-ii) and fibrillar (B-iii) regions along the same fibril for 7.65-fold extension. (C) Box plots of fibril heights in nodular to fibrillar transition areas of FN nanofibers stretched 3.70-fold and 7.65-fold. (D) Box plots of fibril widths in nodular to fibrillar transition areas of FN nanofibers stretched 3.70-fold and 7.65-fold. * indicates conditions are statistically different from each other with p<0.05 using one way ANOVA on ranks and Dunn’s pairwise comparison.
Highly Stretched Fibronectin Nanofibers Have a Bead-on-a-String Nanostructure
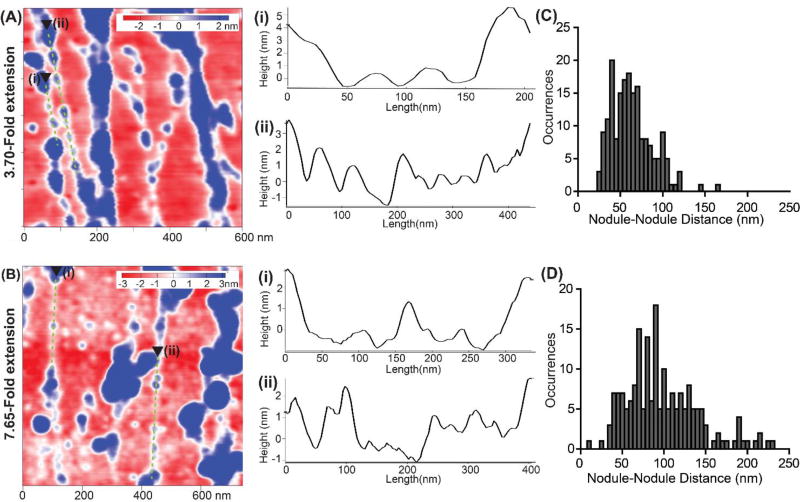
At higher stretch ratios, we observed a morphology consisting of small nodular structures that appeared to be interconnected in a beads-on-a-string configuration (Fig. 7A and 7B). We observed these small nodules linearly arranged between larger nodules or clusters of nodules. Recent reports investigating cell-generated FN fibers27 and FN fibers assembled on polysulfonate surfaces11 reported the presence of highly periodic, sub-fibril nodules. However, we found that the periodicity and nodule diameter varied even along single fibrils. While in some segments, the nodules were similar in size and spacing (Fig. 7A–i), there were also segments in which larger nodules transitioned into sections in which the nodule diameter and spacing decreased (Fig. 7A–ii).
Fig. 7.
Investigating the sub-fibril features of FN nanofibers stretched 3.70-fold and 7.65-fold. (A) A representative region from a 3 µm×3 µm scan of a FN nanofiber stretched 3.70-fold. (A-i and -ii) representative cross-sectional profiles along beads-on-a-string sub-fibril features for 3.70-fold extension. (B) A representative region from a 3 µm×3 µm scan of a FN nanofiber stretched 7.65-fold. (B-i and -ii) representative cross-sectional profiles along beads-on-a-string sub-fibril features for 7.65-fold extension. (C) Histogram showing nodule to nodule distance of FN nodules along beads on a string regions similar to those represented in (A-i and -ii) for 3.70-fold stretched nanofibers. (D) Histogram showing nodule to nodule distance of FN nodules along beads on a string regions similar to those represented in (B-i and -ii) for the 7.65-fold stretched nanofibers.
Height profiles along these linear, beads-on-a-string segments in FN nanofibers stretched 3.70-fold showed that the small nodules in the interior of these beads-on-a-string regions had height of ~2–3 nm. Similarly, FN nanofibers that were stretched 7.65-fold were also comprised of constituent fibrils with a beads-on-a-string nodular nanostructure (Fig. 7B). These nodules were also highly variable in size without a long-range periodicity. The nanostructure also contained regions in which larger nodules transitioned into segments with a sparse arrangement of nodules that were ~1–3 nm in height (Fig. 7B–i and 7B–ii). We observed that both sets of FN nanofibers (stretched 3.70-fold and 7.65-fold) had small nodules occurring between larger nodular features within linear beads-on-a-string segments (Fig. 7A–i, 7A–ii, 7B–i and 7B–ii).
In order to quantify how the nanostructure changed between FN fibers subjected to 3.70-fold and 7.65-fold extensions, we measured the nodule-to-nodule spacing of the small linear segments that spanned between larger nodular features (Fig. 7C and 7D). Nodule-to-nodule spacing was overall much larger in the FN nanofibers stretched 7.65-fold (Fig. 7C and 7D). Quantitatively, the distance increased from 60 ± 25 nm to 94 ± 41 nm for FN nanofibers that were stretched 3.70-fold and 7.65-fold, respectively. The nodule-to-nodule spacing in FN nanofibers stretched 7.65-fold was significantly higher than in FN nanofibers stretched 3.70-fold as determined by Mann-Whitney Rank Sum test with P<0.001. This suggests that within the beads-on-a-string structure, there are FN molecules in between the nodules that continue to unfold during increased extension.
Discussion
Both in vivo and in vitro, FN assembled into a matrix is able to undergo large deformations, with evidence suggesting this is due to folding and unfolding of the molecules. Our group and others have identified that both FN molecules and FN fibers assembled with and without cells contain a nodular nanostructure.11, 24, 25, 28–30 For example, FN fibrils formed on negatively charged, polysulfonate surfaces formed 1 µm long fibrils where each fibril contained nodules in a beads-on-a-string arrangement.11 These nodules were highly periodic, with a nodule-nodule spacing of 60 nm leading to the conclusion that these nodules were the result of FN dimers aligning in a staggered manner with an overlap between the first seven FN type III modules of each FN dimer. The globular FN nodules in this study were 12–22 nm diameter and ~2nm high, while the interspersing filaments ranged from ~0.2 nm to ~2 nm high. This conformation was determined to be initially driven by electrostatic interactions between FN and the negatively charged surface and eventually further stabilized by FN-FN interactions. A separate study used STORM super-resolution microscopy to measure the short range periodicity of cell-generated FN fibers.27 This study showed that FN fibers assembled by cells have a periodicity of ~95 nm, which arises from a 30–40 nm overlap between the first five FN type I modules on the N terminus of adjacent FN dimers. This study also showed that thicker FN fibrils formed from joining of thinner FN fibrils,27 consistent with other suggestions that thin fibrils become thicker by fibril bundling and lateral associations of FN molecules.26, 31 Another study used AFM on FN assembled by living cells, which revealed a beads-on-a-string nanostructure having bead-like regions 3–6 nm high and 12–25 nm wide, with interspersing filaments that were 1–2 nm high.26 Finally, SEM of cell generated FN fibers revealed the presence of both nodular and smooth regions along the same fibrils, where the smooth regions represent regions of the FN fiber that were under tension.25 Notably, immunolabeling suggested FN’s heparin II binding domain was denatured in highly smooth, fibrillar regions but accessible in nodular regions.25 Further study hypothesized the nodules, which had ~15 nm diameter, were clusters of 3–4 FN type III repeats.32 From these studies, it is clear that FN can form different molecular conformations that depend on FN-FN interactions, and presumably tension.
The nanofibers we engineered via SIA provide some insight into how FN molecular conformations are changing as a function of stretch. In the patterned state on the PIPAAm-coated coverslip pre-release, the FN in the nanofibers had a fibrillar, interconnected morphology (Fig. 3B). This structure is consistent with that previously reported for FN in pre-release nanofibers formed by SIA21 and similar to that in pre-release laminin nanofibers formed by SIA.23 This interconnected network suggests that the FN molecules have already started to assemble into fibrils in the pre-release state on the PIPAAm. The fact that the nanofibers release as an intact structure when the PIPAAm layer dissolves during SIA demonstrates that at least some degree of fibrillogenesis has occurred.
In the current study we developed the unique ability to stretch and subsequently immobilize FN fibers for AFM analysis to determine how the nanostructure changed as a function of extension. FN nanofibers that were released from the PIPAAm and allowed to fully contract in solution defined a “no stretch” state. AFM analysis revealed the formation of large nodules with a mean diameter of 107 ± 29 nm (Fig. 4F and 4K). Thus, we reasoned that FN exists in an interconnected fibrillar network in the pre-release state and that FN nanofiber release into water enables folding of the constituent FN molecules into compact conformations, potentially similar to the globular conformation of FN dimers in solution. This is further supported by previous work that showed an increase in FRET intensity between pre-release and post-release FN nanofibers, indicating that the constituent molecules folded into a more compact conformation.22 The diameter of nodules in no stretch FN nanofibers was appreciably larger than the diameter of nodules observed in cell-generated FN fibers,25 FN fibers formed on polysulfonate surfaces,11 and microfabricated PDMS surfaces.33 Since this diameter was also larger than the diameter of single FN dimers in a compact, globular conformation,28, 34 we reasoned that these nodules are comprised of several compact FN dimers clustered together.
The change in nodule size as a function of stretch provides insight into how the FN molecules are interacting. Nanofibers stretched 1.94-fold showed a decrease in nodule diameter to 71 ± 20 nm (Fig. 4G and 4L). By further stretching the FN nanofibers, we found that the mean nodule diameter decreased until about 3.70-fold extension. Nodule diameters for FN nanofibers stretched 3.70-fold and higher were relatively consistent, although there was a slight increase in nodule diameter above 3.70-fold stretch (Fig. 5A). At 3.70-fold stretch, nodules had a mean diameter of 47 ± 14 nm, the smallest mean diameter among the different stretch states, which is similar to the dimensions of globular, compact FN dimers.28, 34 Our AFM analysis approach cannot determine the molecular composition of the nodules as a function of stretch, but our data on mean diameter (Fig. 5A) and the distribution of diameters (Fig. 4K to 4O) provides a potential explanation. We propose that the smallest nodules observed at the 3.70-fold stretch are primarily folded FN dimers, with some portion unfolded and connected to adjacent, folded FN dimers. The larger nodules observed for no stretch and 1.94-fold stretch are multiple, folded FN dimers clustered together into larger nodules.
We reasoned that when tension was applied beyond 3.70-fold stretch, some of the nodules observed in the 3.70-fold stretched nanofibers unfold, lose their nodular tertiary structure, and become completely fibrillar. At stretches of 3.70-fold and higher, nodule diameter was relatively consistent. However the nodule-to-nodule distance increased, supporting the interpretation that unfolded FN fibrils were connecting the nodules. We also reasoned that as tension was applied beyond 3.70-fold stretch, some fibrils within the nanofibers may break and refold into nodules. In 3.70-fold stretch nanofibers, the nodules and fibrils had an interconnected appearance (Fig 4H). Qualitatively, in 4.77-fold and 7.65-fold stretch nanofibers, the fibrils and nodules appeared more anisotropic and less interconnected, which may be partially due to breaking of interconnecting fibrils. There was also a slight increase in average nodule diameter at 4.77-fold and 7.65-fold stretch relative to 3.70-fold stretch. From the histograms of nodule diameter for these conditions, (Fig. 4M to 4O), it is apparent that this shift is due to an increase in large nodules, which we attributed to FN fibrils breaking during extension and contracting back into an existing nodule, forming an overall larger nodule. Similarly, previous work has shown in FN matrices assembled by cells, that broken FN fibrils contract to less than one quarter their previous length.35 Notably, the width of fibrillar areas of FN fibrils was higher for 7.65-fold stretched fibers than for 3.70-fold stretched fibers. This may also be due to breakage of very thing fibrils, where the broken fibrils that likely occur more frequently at higher stretch refold and retract back onto the existing nodular and fibrillary regions, increasing the width. Altogether, our results indicate that while the FN nanofibers can withstand extensions upwards of 7.65-fold, constituent fibrils can begin to break within the nanostructure at much lower extensions.
Our results demonstrate that our engineered FN nanofibers have a nodular structure at no stretch and low extension, which then transitions to a more fibrillar structure at higher extension. While the nodules appear isotropic at no stretch, even at the 1.94-fold stretch, the nodules appear to start to align in the direction of extension (Fig. 4B and 4G). At 3.70-fold stretch and beyond, the large nodules have clearly transitioned to fibrils that are aligned in the direction of extension (Fig. 4C–F and 4H–J). AFM images of the FN nanofibers that underwent 3.70-fold (Fig. 7A) and 7.65-fold (Fig. 7B) extensions revealed that the nanostructure contains fibrils with smaller nodules in a beads-on-a-string arrangement. Qualitatively, periodicity and nodule diameter varied even along single fibrils. The small, sparse nodules located between larger nodular features (Fig. 7A and 7B) had diameters of ~30–40 nm. These nodule diameters are comparable to those observed on fibers assembled on polysulfonate surfaces,11 are comparable to the size of single FN dimers,28, 34 and are larger than ~15 nm diameter nodules that have been observed with SEM.32 Prior work has suggested similar nodules arise from the overlap between adjacent FN dimers in a stretched state,11 and that smaller, ~15 nm diameter nodules arise from globular conformations of 3–4 FN type III repeats.32
The beads-on-a-string structure in the highly stretched FN nanofibers suggests there is significant unfolding of the FN molecules in between the nodules (beads). As noted, the nodule-to-nodule distance increased from 60 ± 25 nm to 94 ± 41 nm for FN nanofibers that were stretched 3.70-fold and 7.65-fold, respectively (Fig. 7C and 7D). A single extended FN dimer, which has lost tertiary folding but maintains folding of FN type III domains, has a length of ~130 nm.28, 36 Each FN type III domain can extend from ~2.3 nm in the folded state to ~28.5 nm in the unfolded, extended state.37 Each FN dimer has 30 FN type III domains7 and can therefore theoretically extend ~700 nm with domain unfolding. Thus, the nodule-to-nodule distances from ~25 to ~200 nm that we observed are consistent with unfolded FN dimers that have lost tertiary structure, but minimal secondary structure of the type III domains. Similarly, the small nodules likely represent FN dimers that still retain some tertiary, globular structure, or alternately, overlapping regions of FN dimers that form a globular structure. It is difficult to achieve further insight with our existing AFM, which due to signal-to-noise, cannot resolve the fibrillar FN dimers that we hypothesize are connecting the small nodules in the beads-on-a-string structures.
Conclusions
We found that when the FN nanofibers were uniaxially stretched, large nodules in fully contracted FN fibers began to decrease in size, presumably as individual FN dimers became separated from large clusters of dimers. With further extensions, the nodules continued to decrease in size and align in the direction of stretch. We found that the FN nanofibers’ nanostructure did not homogenously change with stretch. Rather, within FN nanofibers extended 3.70-fold or higher, there are regions of large nodules and regions where these nodules transition to short, sparse sequences of small nodules in a beads-on-a-string arrangement. We attribute the large nodules to clusters of multiple FN dimers and the small nodules to globular portions of single molecules, or overlapping sections of multiple FN molecules. There are also regions where large nodular features transition to smaller fibrillar regions, suggesting conformational differences within adjoining regions along the fibronectin fibrils. Our results provide insight into how morphology of the nanostructure changes as a function of extension. Future work will investigate how conformational changes within the nanostructure of FN fibers, particularly at high extensions, influence the biological properties of FN, specifically the exposure of cryptic binding sites.
Acknowledgments
We acknowledge financial support from the Biomechanics in Regenerative medicine NIH T32 training program (T32EB003392) and Bertucci Graduate Fellowship to J.M.S. and the NIH Director’s New Innovator Award (DP2HL117750) and the Human Frontier Science Program Young Investigator Grant (RGY0071) to A.W.F. We acknowledge Rachelle N. Palchesko for helpful discussions and assistance with AFM data analysis.
References
- 1.Geiger B, Bershadsky A, Pankov R, Yamada KM. Nat Rev Mol Cell Biol. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 2.Chabria M, Hertig S, Smith ML, Vogel V. Nature communications. 2010;1:135. doi: 10.1038/ncomms1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwarzbauer JE, Sechler JL. Current opinion in cell biology. 1999;11:622–627. doi: 10.1016/s0955-0674(99)00017-4. [DOI] [PubMed] [Google Scholar]
- 4.Martino MM, Hubbell JA. The FASEB Journal. 2010;24:4711–4721. doi: 10.1096/fj.09-151282. [DOI] [PubMed] [Google Scholar]
- 5.Matsuka YV, Medved LV, Brew SA, Ingham KC. Journal of Biological Chemistry. 1994;269:9539–9546. [PubMed] [Google Scholar]
- 6.Zhong C, Chrzanowska-Wodnicka M, Brown J, Shaub A, Belkin AM, Burridge K. The Journal of Cell Biology. 1998;141:539–551. doi: 10.1083/jcb.141.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogel V. Annu. Rev. Biophys. Biomol. Struct. 2006;35:459–488. doi: 10.1146/annurev.biophys.35.040405.102013. [DOI] [PubMed] [Google Scholar]
- 8.Mosher D, Johnson R. Journal of Biological Chemistry. 1983;258:6595–6601. [PubMed] [Google Scholar]
- 9.Sakai K, Fujii T, Hayashi T. Journal of biochemistry. 1994;115:415–421. doi: 10.1093/oxfordjournals.jbchem.a124353. [DOI] [PubMed] [Google Scholar]
- 10.Salmerón-Sánchez M, Rico P, Moratal D, Lee TT, Schwarzbauer JE, García AJ. Biomaterials. 2011;32:2099–2105. doi: 10.1016/j.biomaterials.2010.11.057. [DOI] [PubMed] [Google Scholar]
- 11.Nelea V, Kaartinen MT. Journal Of Structural Biology. 2010;170:50–59. doi: 10.1016/j.jsb.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 12.Keselowsky BG, Collard DM, García AJ. Journal of Biomedical Materials Research Part A. 2003;66:247–259. doi: 10.1002/jbm.a.10537. [DOI] [PubMed] [Google Scholar]
- 13.Little WC, Smith ML, Ebneter U, Vogel V. Matrix Biology. 2008;27:451–461. doi: 10.1016/j.matbio.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith ML, Gourdon D, Little WC, Kubow KE, Eguiluz RA, Luna-Morris S, Vogel V. PLoS Biol. 2007;5:e268. doi: 10.1371/journal.pbio.0050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitsi M, Handschin S, Gerber I, Schwartländer R, Klotzsch E, Wepf R, Vogel V. Biomaterials. 2015;36:66–79. doi: 10.1016/j.biomaterials.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubow KE, Vukmirovic R, Zhe L, Klotzsch E, Smith ML, Gourdon D, Luna S, Vogel V. Nat Commun. 2015:6. doi: 10.1038/ncomms9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klotzsch E, Smith ML, Kubow KE, Muntwyler S, Little WC, Beyeler F, Gourdon D, Nelson BJ, Vogel V. Proceedings of the National Academy of Sciences. 2009;106:18267–18272. doi: 10.1073/pnas.0907518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mnatsakanyan H, Rico P, Grigoriu E, Maturana Candelas A, Rodrigo-Navarro A, Salmeron-Sanchez M, Sabater i, Serra R. ACS applied materials & interfaces. 2015 doi: 10.1021/acsami.5b05466. [DOI] [PubMed] [Google Scholar]
- 19.Rico P, Hernández JCR, Moratal D, Altankov G, Pradas MM, Salmerón-Sánchez M. Tissue Engineering Part A. 2009;15:3271–3281. doi: 10.1089/ten.TEA.2009.0141. [DOI] [PubMed] [Google Scholar]
- 20.Szymanski JM, Jallerat Q, Feinberg AW. Journal of visualized experiments: JoVE. 2014 doi: 10.3791/51176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feinberg AW, Parker KK. Nano letters. 2010;10:2184–2191. doi: 10.1021/nl100998p. [DOI] [PubMed] [Google Scholar]
- 22.Deravi LF, Su T, Paten JA, Ruberti JW, Bertoldi K, Parker KK. Nano Letters. 2012;12:5587–5592. doi: 10.1021/nl302643g. [DOI] [PubMed] [Google Scholar]
- 23.Szymanski JM, Ba M, Feinberg AW. Journal of Materials Chemistry B. 2015;3:7993–8000. doi: 10.1039/C5TB01003A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Y, Jallerat Q, Szymanski JM, Feinberg AW. Nature methods. 2015;12:134–136. doi: 10.1038/nmeth.3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, Zardi L, Peters DM. Scanning. 1997;19:349–355. doi: 10.1002/sca.4950190505. [DOI] [PubMed] [Google Scholar]
- 26.Gudzenko T, Franz CM. Molecular biology of the cell. 2015;26:3190–3204. doi: 10.1091/mbc.E15-06-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fruh SM, Schoen I, Ries J, Vogel V. Nat Commun. 2015:6. doi: 10.1038/ncomms8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erickson HP, Carrell N. Journal of Biological Chemistry. 1983;258:14539–14544. [PubMed] [Google Scholar]
- 29.Erickson HP, Carrell N, McDonagh J. The Journal of cell biology. 1981;91:673–678. doi: 10.1083/jcb.91.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dzamba BJ, Peters DM. Journal of cell science. 1991;100(Pt 3):605–612. doi: 10.1242/jcs.100.3.605. [DOI] [PubMed] [Google Scholar]
- 31.Ohashi T, Kiehart DP, Erickson HP. Journal of cell science. 2002;115:1221–1229. doi: 10.1242/jcs.115.6.1221. [DOI] [PubMed] [Google Scholar]
- 32.Peters DMP, Chen Y, Zardi L, Brummel S. Microscopy and microanalysis : the official journal of Microscopy Society of America, Microbeam Analysis Society, Microscopical Society of Canada. 1998;4:385–396. doi: 10.1017/s1431927698980369. [DOI] [PubMed] [Google Scholar]
- 33.Ulmer J, Geiger B, Spatz JP. Soft Matter. 2008;4:1998–2007. [Google Scholar]
- 34.Price TM, Rudee ML, Pierschbacher M, Ruoslahti E. European journal of biochemistry. 1982;129:359–363. doi: 10.1111/j.1432-1033.1982.tb07058.x. [DOI] [PubMed] [Google Scholar]
- 35.Ohashi T, Kiehart DP, Erickson HP. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:2153–2158. doi: 10.1073/pnas.96.5.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engel J, Odermatt E, Engel A, Madri JA, Furthmayr H, Rohde H, Timpl R. Journal of molecular biology. 1981;150:97–120. doi: 10.1016/0022-2836(81)90326-0. [DOI] [PubMed] [Google Scholar]
- 37.Oberhauser AF, Badilla-Fernandez C, Carrion-Vazquez M, Fernandez JM. Journal of molecular biology. 2002;319:433–447. doi: 10.1016/S0022-2836(02)00306-6. [DOI] [PubMed] [Google Scholar]