Abstract
Background
Orofacial clefts (OFCs), including nonsyndromic cleft lip with or without cleft palate (NSCL/P), are common birth defects. NSCL/P is highly heterogeneous with multiple phenotypic presentations. Two common subtypes of NSCL/P are cleft lip (CL) and cleft lip with cleft palate (CLP) which have different population prevalence. Similarly, NSCL/P can be divided into bilateral and unilateral clefts, with unilateral being the most common. Individuals with unilateral NSCL/P are more likely to be affected on the left side of the upper lip, but right side affection also occurs. Moreover, NSCL/P is twice as common in males as in females. The goal of this study is to discover genetic variants that have different effects in case subgroups.
Methods
We conducted both common variant and rare variant analyses in 1,034 individuals of Asian ancestry with NSCL/P, examining four sources of heterogeneity within CL/P: cleft type, sex, laterality, and side.
Results
We identified several regions associated with subtype differentiation – cleft type differences in 8q24 (p=1.00×10−4), laterality differences in IRF6, a gene previously implicated with wound healing (p=2.166×10−4), sex differences and side of unilateral CL differences in FGFR2 (p=3.00×10−4, p=6.00×10−4), and sex differences in VAX1 (p<1.00×10−4) among others.
Conclusions
Many of the regions associated with phenotypic modification were either adjacent to or overlapping functional elements based on ENCODE chromatin marks and published craniofacial enhancers. We have identified multiple common and rare variants as potential phenotypic modifiers of NSCL/P, and suggest plausible elements responsible for phenotypic heterogeneity, further elucidating the complex genetic architecture of OFCs.
Keywords: orofacial cleft, complex trait, genetic epidemiology
INTRODUCTION
Orofacial clefts (OFCs) are common birth defects, affecting approximately 1 in 800 births worldwide (Leslie and Marazita, 2013). Approximately 30% of OFCs are syndromic, occurring in combination with some other structural, cognitive, or developmental anomalies. The remaining 70% of OFCs occur as isolated (i.e. nonsyndromic) defects. Nonsyndromic OFCs have complex etiology with multiple genetic and environmental factors interacting to influence risk.
Nonsyndromic OFCs are highly heterogeneous with multiple phenotypic presentations (Dixon et al., 2011). OFCs are most commonly divided into three major subtypes: cleft lip (CL), cleft palate (CP), and cleft lip with cleft palate (CLP). CL and CLP share a defect of the lip and are commonly combined for analyses as cleft lip with or without cleft palate (CL/P) (Fogh-Andersen, 1942; Fraser, 1955). CL/P and CP have historically been considered distinct disorders with separate etiologies because of the different developmental origins of the lip and palate and markedly different prevalence rates in males and females (CP is twice as common in females as in males, while the opposite is true for CL/P (Mossey et al., 2009)). However, they occasionally occur within the same family, an even known as “mixed clefting” commonly observed in syndromic OFCs, including Van der Woude syndrome (Leslie and Marazita, 2013).
The CL/P subgroup itself is quite heterogeneous and can be further subdivided into bilateral and unilateral clefts, affecting either the left or right side of the upper lip. Of these, left sided unilateral clefts are the most common and bilateral clefts are the least common (Gundlach and Maus, 2006). The causes of variability in phenotype are largely unknown, and may arise due to underlying genetic factors, different environmental exposures, or other unknown factors. There have been many studies investigating the genetic architecture of NSCL/P, most collapsing cleft subtypes into one larger group (primarily CL/P) for analysis (Dixon et al., 2011; Leslie and Marazita, 2013). While this approach is powerful to identify sources of genetic variation that contribute to overall NSCL/P, any signal from genetic variation specific to only one subtype or that differentiates subtypes will be masked. Very few studies have explored genetic associations for clefting phenotypes beyond CL and CLP. There is some evidence that the 13q31 locus near SPRY2 has a stronger effect in CLP (Jia et al., 2015; Ludwig et al., 2012). Similarly, variants in IRF6 are more strongly associated with CL than CLP (Marazita et al., 2009; Rahimov et al., 2008). Recent evidence suggests that GREM1 is associated with clefts in the lip and soft palate (Ludwig et al., 2016). Furthermore, variants in GRHL3 are associated with CP and not with CL/P (Leslie et al., 2016b; Mangold et al., 2016; Wang et al., 2016). Examining CL/P subtypes may elucidate more of the complex genetic architecture of OFCs by identifying genetic mechanisms that modify cleft subtype.
We hypothesized that genetic components of phenotypic heterogeneity, including any contribution of rare variants, can be found for recognized clefting loci. We performed association tests for four sources of phenotypic heterogeneity within CL/P: cleft type (CL vs. CLP), sex (male vs. female), laterality (unilateral vs. bilateral), and side (right unilateral vs. left unilateral) in targeted sequencing from the CleftSeq study (Leslie et al., 2015).
METHODS
Sample
We compared subtypes within clefting cases from the CleftSeq study to investigate the potential genetic contribution to clefting heterogeneity. CleftSeq is a targeted sequencing study of 13 previously reported loci associated with NSCL/P (Leslie et al., 2015). These 13 regions, totaling 6.3 Mb, were comprised of 9 “high-priority” candidates from previous GWAS and/or genome-wide linkage studies and 4 regions containing candidate genes with prior evidence of rare variants contributing to NSCL/P (Table 1). Sequencing was performed on 1,498 case-parent trios from Europe, the United States, China and the Philippines.
Table 1.
CleftSeq regions.
| Region | Candidate Gene in Region |
Target Region (GRCh37) | Size (kb) |
|
|---|---|---|---|---|
| previous GWAS hits | 1p36 | PAX7 | chr1: 18,772,300 – 19,208,054 | 435.8 |
|
| ||||
| 1p22 | ARHGAP29 | chr1: 94,324,660 – 95,013,109 | 688.4 | |
|
| ||||
| 1q32 | IRF6 | chr1: 209,837,199 – 210,468,406 | 631.2 | |
|
| ||||
| 8q24 | -- | chr8: 129,295,896 – 130,354,946 | 1059.1 | |
|
| ||||
| 10q25 | VAX1 | chr10: 118,421,625 – 119,167,424 | 745.8 | |
|
| ||||
| 17p13 | NTN1 | chr17: 8,755,114 – 9,266,060 | 510.9 | |
|
| ||||
| 17p22 | NOG | chr17: 54,402,837 – 54,957,390 | 554.6 | |
|
| ||||
| 20q12 | MAFB | chr20: 38,902,646 – 39,614,513 | 711.9 | |
|
| ||||
| previous linkage hit | 9q22 | FOXE1 | chr9: 100,357,692 – 100,876,841 | 519.1 |
|
| ||||
|
candidate gene regions
(evidence for rare variants) |
4p16 | MSX1 | chr4: 4,825,126 – 4,901,385 | 76.3 |
|
| ||||
| 14q22 | BMP4 | chr14: 54,382,690 – 54,445,053 | 62.4 | |
|
| ||||
| 10q26 | FGFR2 | chr10: 123,096,374 – 123,498,771 | 402.4 | |
|
| ||||
| 9q22 | PTCH1 | chr9: 98,133,647 – 98,413,162 | 279.5 | |
From the 1,489 trios, we extracted 1,034 probands with NSCL/P of Asian (i.e. Chinese or Filipino) ancestry for analysis and cross-classified them using the four clefting subtype definitions (Table 2). Among the 1,034 cases, 33 with unknown laterality were excluded from the analysis of laterality and side of cleft lip groups.
Table 2.
Sample used for modifier analyses by population.
| Cleft Type | Sex | Laterality | Side of CL | |||||
|---|---|---|---|---|---|---|---|---|
| CL | CLP | Female | Male | Unilateral | Bilateral |
Right
unilateral |
Left
unilateral |
|
| China | 117 | 284 | 126 | 275 | 278 | 101 | 112 | 166 |
| Philippines | 171 | 462 | 219 | 414 | 440 | 182 | 147 | 293 |
|
| ||||||||
| Total | 288 | 746 | 345 | 689 | 718 | 282 | 259 | 459 |
Common Variant Analysis
For each factor (i.e. cleft type, sex, laterality, and side), we performed a case vs. case analysis, directly comparing allele frequencies at each SNP between the two groups (e.g. CL vs. CLP, male vs. female, etc.). This type of analysis has very high power to find genetic risk factors that differ between the two groups, but it has no power to find factors that are important in both groups. Thus this design is strictly a test for heterogeneity in the genotype/phenotype relationship, not an overall test of genetic effect. Ideally, this test will discover new loci for which there is an effect in only one subgroup; such loci may be masked in an overall scan when groups are combined.
We analyzed the association between the four cleft subtype phenotypes and 19,982 – 20,089 common SNPs (MAF > 0.01) in the thirteen candidate regions by directly comparing the two case subtypes using traditional Chi-Square tests for association. Each Asian population (Chinese and Filipino) was analyzed separately to account for any population stratification. Low-quality SNPs (missing genotypes > 5% or HWE p < 0.0001) were excluded from analyses.
Inverse-variance effects-based meta-analysis of the two population-specific scans was performed on 13,183 – 13,427 SNPs to detect any signal common to Asian populations. SNPs were excluded from the meta-analyses if they were flagged as low-quality in at least one population-specific analysis, or if effects were heterogeneous between populations (Cochran’s Q p < 0.05). Statistical significance was determined using a Bonferroni threshold adjusting for four scans of thirteen regions of 9.615 × 10−4 (i.e. 0.05/52). This threshold allows for the generation of hypotheses regarding the genetic mechanisms of clefting subtypes and thus is not as strictly conservative as a Bonferroni correction for the number of markers tested (5200 tests, p-value threshold of 1 × 10−5 (Leslie et al., 2015)). Thus, the suggestive associations found in this study should be followed up rigorously. Common variant analyses were performed using PLINK software (Purcell et al., 2007).
Rare Variant Analysis
Rare variants (MAF < 0.01) were also interrogated for association with subtype differentiation using the same phenotype definitions as in the common variant analysis.
First, variants within exons of canonical transcripts of each gene were examined using gene-based versions of the Collapsed Multivariate and Combining (CMC) test (Li and Leal, 2008) and the Sequence Kernel Association Test (SKAT) (Wu et al., 2011).
Secondly, two window-based approaches were used to investigate burdens of all rare variants. SNPs were combined into regions using two window-based methods – 2,662 windows using a fixed window size of 5Kb with 2.5Kb overlap between windows, and 14,232 windows using exactly 20 SNPs per window with 10 SNP overlap between windows (windows at the end of each region contained at least 14 SNPs). Each window was comprised of SNPs from only one of the candidate regions. Windows are highly correlated within each candidate region, so statistical significance was again determined using a Bonferroni threshold of 9.615 × 10−4. Rare variants were analyzed with the SKAT option in RVTESTS software (Zhan et al., 2016).
Functional Annotation of Rare Variant Windows
The CleftSeq project sequenced 6.3Mb of largely noncoding DNA around these GWAS and OFC candidate genes. We failed to identify significant associations in analyses of coding variants (results not shown), so we hypothesized that functional variants would be regulatory. We examined intervals containing overlapping windows for functional elements based on ENCODE chromatin marks (Consortium et al., 2011; Rosenbloom et al., 2013) and published craniofacial enhancers (Attanasio et al., 2013; Brinkley et al., 2016; Rada-Iglesias et al., 2012).
RESULTS
Cleft Type
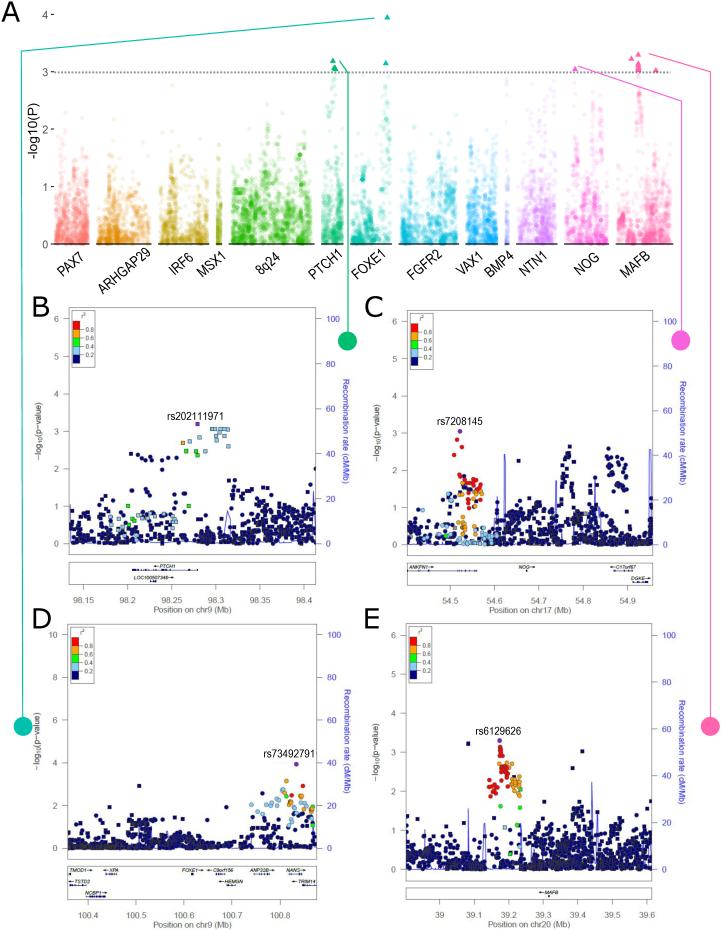
In the common variant meta-analysis, 20 SNPs from 4 loci were significantly associated with CL v. CLP differentiation (Figure 1A). These associations were seen in SNPs on 9q22 near PTCH1 and FOXE1, on 17p22 near NOG, and on 20q12 near MAFB. Specifically, a set of variants in and near PTCH1 were more strongly associated with CL than with CLP (lead SNP: rs202111971 p = 6.484 × 10−4, Figure 1B). A neighboring set of variants did not show formally significant differences by cleft type, but tended to be more strongly associated with CLP (Figure 1B). In the 9q22 region, a set of SNPs downstream of the FOXE1 transcription start site were more strongly associated with CLP than with CL (lead SNP: rs73492791 p = 1.138 × 10−4, Figure 1C). Moreover, minor alleles in the 17p22 regions and 20q12 regions were more strongly associated with CLP (lead SNPs: rs7208145 p = 9.041 × 10−4, rs6129626 p = 5.039 × 10−4, Figure 1D-E). Notably, none of these SNPs associated with cleft type differentiation (CL vs. CLP) was significantly associated with risk of OFC overall (Leslie et al., 2015).
Figure 1. CL vs. CLP cleft type modifiers.
A) Cleft type (CL vs. CLP) association results from the common-variant meta-analysis of Filipino and Chinese populations. (B) – (E) Regional association plots for 9q22 (x2), 17q22, and 20q12 showing −log10(P-values) for SNPs with stronger association with CL (squares) and stronger association with CLP (circles) based on the direction of the odds ratio. Plots were generated using LocusZoom (Pruim et al., 2010). The recombination overlay (blue line, right y-axis) indicates the boundaries of the LD-block. Points are color coded according to pairwise linkage disequilibrium (r2) with the index SNP.
Twenty-five windows of rare variants in the PAX7, ARHGAP29, 8q24, FOXE1, VAX1, NTN1, and NOG sequencing regions were significantly associated with cleft type differentiation (CL vs. CLP) (Supplementary Material, Table S1). Of these, two sets of three overlapping windows (8:129790677-129795772 [min p = 4.50 × 10−4] and 8:130298273-130305772 [min p = 1.00 × 10−4]) on 8q24 are particularly interesting because they contain SNPs that individually show strong association with NSCL/P in Europeans. Furthermore, one of these intervals (8:129790677-129795772) consisting of three overlapping windows was located adjacent to a putative regulatory element as defined by H3K27Ac marks in multiple cell types from ENCODE (Figure 4A).
Figure 4. Significant rare variant windows with potential regulatory effects.
(A) 8q24 for cleft type, (B) VAX1 for sex, and (C) FGFR2 for sex and side.
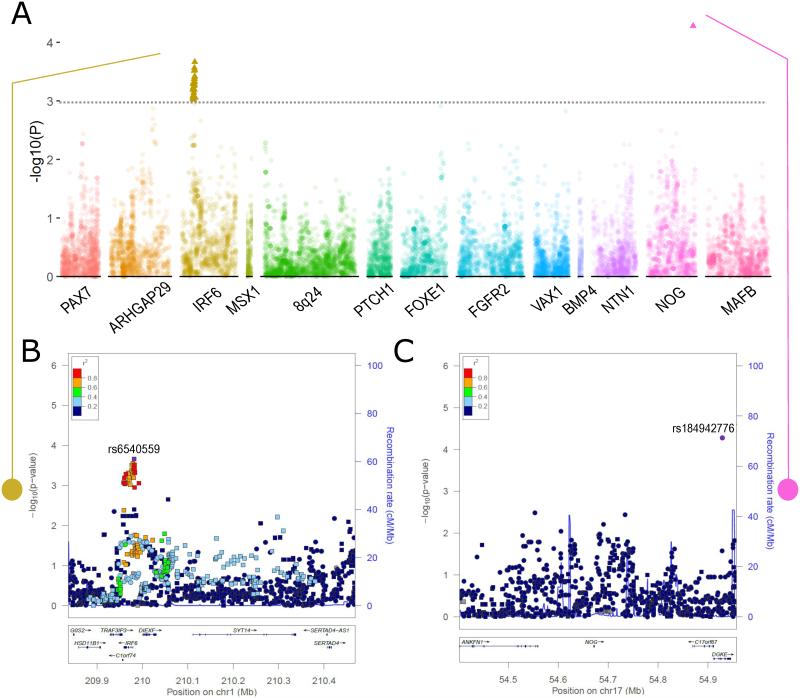
Laterality
In the common variant meta-analysis, 27 SNPs from 2 loci were significantly associated with laterality differences (Figure 2A). These associations were seen for 26 SNPs on 1q32 near IRF6 (lead SNP: rs6540559 p = 2.166 × 10−4, Figure 2B) and a single SNP on 17p22 near NOG (rs184942776 p = 5.262 × 10−5, Figure 2C). SNPs in IRF6 were associated with differentiation between bilateral and unilateral CL/P. Specifically, minor alleles of SNPs in IRF6 were associated with unilateral CL/P. The minor alleles at these SNPs also are significantly protective against overall OFC risk (Supplementary Material, Table S5).
Figure 2. Unilateral vs. bilateral CL/P modifiers.
(A) Laterality (unilateral vs. bilateral) association results from the common-variant meta-analysis of Filipino and Chinese populations. (B) – (C) Regional association plots for 1q32 and 17q22 showing −log10(P-values) for SNPs with stronger association with unilateral CL/P (squares) and stronger association with bilateral CL/P (circles) based on the direction of the odds ratio. Plots were generated using LocusZoom (Pruim et al., 2010). The recombination overlay (blue line, right y-axis) indicates the boundaries of the LD-block. Points are color coded according to pairwise linkage disequilibrium (r2) with the index SNP.
Differences in CL/P laterality were observed in 17 windows of rare variants (Supplementary Material, Table S2). Despite having many overlapping windows of rare variants, there was no evidence of known regulatory or enhancer elements within these intervals.
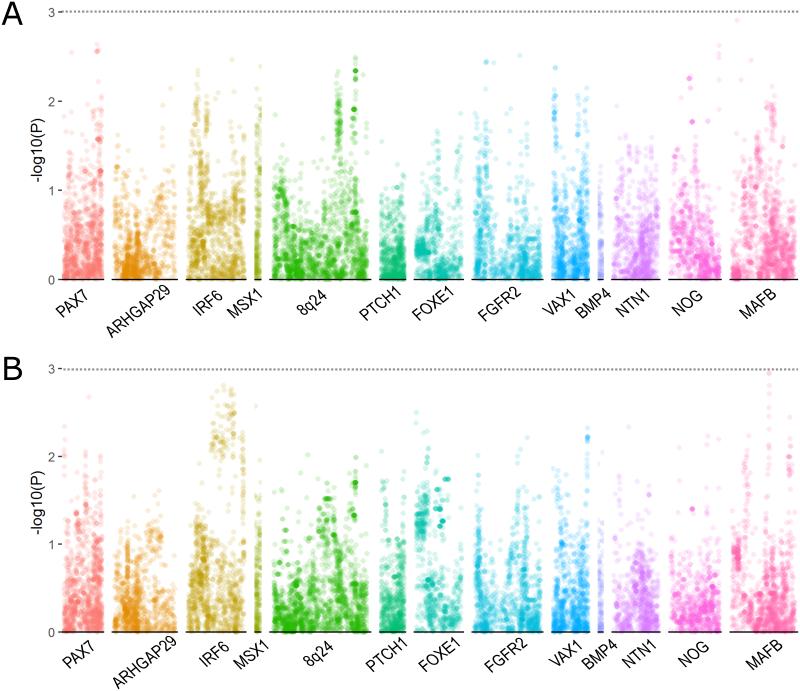
Sex
While no significant associations for sex differences were observed in the common variant analysis (Figure 3A), 28 windows of rare variants were significantly associated with sex differences (Supplementary Material, Table S3).
Figure 3. Sex-specific and side modifiers of CL/P.
(A) Sex (male vs. female) association results from the common-variant meta-analysis of Filipino and Chinese populations. (B) Side (right unilateral vs. left unilateral) association results from the common-variant meta-analysis of Filipino and Chinese populations.
Eight windows defining three larger intervals (10:118624030-118629029 [min p = 5.00 × 10−4], 10:118638519-118644029 [min p < 1.00 × 10−4], and 10:118851530-11885725 [min p < 1.00 × 10−4]) near VAX1 were significantly associated with sex differences in Filipinos. One of these intervals (10:118851530-11885725), comprised of three windows near VAX1, overlapped a craniofacial regulatory element identified from p300 ChIP-Seq in craniofacial tissue in mouse embryos (Attanasio et al., 2013; Visel et al., 2009) (Figure 4B). It is unclear what gene is regulated by this element, as the activity pattern of the enhancer resembles the endogenous expression of both adjacent genes VAX1 and SHTN1 (Armit et al., 2012; Diez-Roux et al., 2011). Interestingly, other significant windows in this region occurred immediately downstream of SHTN1.
Two non-overlapping windows near FGFR2 (10:123368869-123373868 [p = 3.00 × 10−4] and 10:123479803-123483275 [p = 8.00 × 10−4]) were also significantly associated with sex differences in Filipinos. The first of these windows overlapped multiple regulatory annotations including a binding site for p63, a transcription factor known to regulate FGFR2 (Ferone et al., 2012; Fomenkov et al., 2003) (Figure 4C). The second window overlaps more regulatory annotations characteristic of epithelial enhancers (Figure 4C).
Side of Lip
We did not observe any significant associations with right unilateral vs. left unilateral CL/P in the common variant analysis (Figure 3B). However, 13 windows of rare variants were significantly associated with side of cleft lip differentiation (Supplementary Material, Table S4). Interestingly, one window near FGFR2 (10:123431369-123436368 [p = 6.00 × 10−4]) was significantly associated with side of cleft lip differentiation in Filipinos and was adjacent to active enhancers from human neural crest cell lines and a putative palate enhancer from p300 ChIP-seq of mouse palatal tissue (Figure 4C).
DISCUSSION
NSCL/P is a complex disorder with many different anatomical forms. GWASs have identified dozens of genetic associations with NSCL/P (Beaty et al., 2010; Leslie et al., 2016a; Ludwig et al., 2012; Mangold et al., 2010); however, a small number of studies have identified cleft subtype specific associations, most of which are reflect differences between CL and CLP (Ludwig et al., 2012; Marazita et al., 2009; Rahimov et al., 2008). The current study adds to these findings by identifying both common and rare variants that are associated with subtype differentiation in cleft type, laterality, sex, and side of unilateral CL. We performed common and rare variant association testing with four cleft subtypes (cleft type: CL vs. CLP; laterality: unilateral vs. bilateral; sex; and side: right vs. left CL/P) to further interrogate OFC-associated regions from the CleftSeq targeting sequencing study. We identified several regions associated with cleft subtype differentiation – common variants in IRF6 and rare variants in 8q24, FGFR2, and VAX1 among others. Notably, these associations are found with both previously known clefting-associated variants and variants that were not significantly associated with overall clefting (CL/P). Multiple associations with regulatory (non-coding) elements and differences in clefting subtypes, contributing to the evidence that non-coding variants have a significant role in the genetic causes of NSCL/P (Leslie and Marazita, 2013; Leslie et al., 2015; Rahimov et al., 2008). However, it is not clear from the association results which alleles are relevant to these phenotypes; systematic studies in model systems will likely be required to identify functional SNPs and a possible mechanism.
We identified 26 SNPs within IRF6 associated with differences between unilateral and bilateral CL/P. Specifically, individuals with unilateral CL/P had higher frequencies of minor alleles in these 26 variants than did bilateral CL/P individuals. IRF6 has been previously implicated in wound healing (Biggs et al., 2014; Biggs et al., 2012; Jones et al., 2010), so these cleft laterality differences are particularly interesting. The same alleles showing a protective effect for overall cleft risk were more strongly associated with unilateral CL/P than bilateral. If we consider unilateral CL/P as a less severe presentation of clefting than bilateral CL/P, our finding that OFC-protective variants are associated more strongly with unilateral CL/P and previous evidence that IRF6 is associated with CL (Rahimov et al., 2008) together suggest that the IRF6 locus is associated with decreased risk of severe clefting.
Rare variants on 8q24 were found to significantly differ between CL and CLP, including an interval adjacent to a putative regulatory element. This provides strong evidence for a regulatory role of variants within 8q24 on the presentation of NSCL/P. Furthermore, SNPs on 8q24 have previously shown very strong association with cleft risk in European GWAS (Beaty et al., 2011; Birnbaum et al., 2009; Grant et al., 2009; Murray et al., 2012), but are not associated with cleft risk in Asian GWAS(i.e. in common variant analyses). This may be due to population-specific differences in SNP informativeness within 8q24, which reflects haplotype diversity (Murray et al., 2012). SNPs within 8q24 have markedly higher heterozygosity in Europeans than Asians, making common-variant associations within this region far more powerful among Europeans. We hypothesize that this region also is associated with clefting risk in other populations although the statistical evidence from analyses of common variants is lacking. The association with cleft type differentiation within windows of 8q24 rare variants observed in the Filipino population here may be evidence that some SNPs within 8q24 have with clefting risk in Asian populations.
Additionally, rare variant associations with potential regulatory elements were observed when examining sex differences and markers near VAX1 and FGFR2 and those near FGFR2 and the left vs. right side of unilateral CL/P. While it is not immediately clear how VAX1 and FGFR2 specifically contribute to sex differences in NSCL/P, biological hypotheses regarding sex differences in other disorders (e.g. autism) involve a multiple-threshold multifactorial liability model in which females have a higher threshold than males. In other words, affected females are hypothesized to carry a higher mutational burden than affected males. The same would hold for NSCL/P, where there are more affected males than females. Under this hypothesis, relatives of affected females are at increased risk for CL/P, which is supported by population-based recurrence risk estimates from Denmark (Grosen et al., 2010). A similar threshold model may also pertain to differences in laterality and severity of NSCL/P.
Contrary to the common disease-common variant hypothesis, we observed clear contributions from both common and rare variants in this study of the genetic underpinnings of NSCL/P and the potential differences within NSCL/P subtypes. This work adds to a growing body of evidence implicating rare variants in risk of NSCL/P (Al Chawa et al., 2014; Leslie and Murray, 2012; Leslie et al., 2015). Importantly, this work highlights the impact of rare variants as potential phenotypic modifiers, an area that needs larger studies in additional populations that are expanded to the entire genome. As costs of whole genome sequencing decrease, these studies will be more feasible for NSCL/P and will continue to improve our understanding of the genetic architecture of NSCL/P.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the families who participated in this study, recruitment staff, and the Operations Group and the Bioinformatics Group at the McDonnell Genome Institute. This work was supported by grants from the NIH (HG005925 [J.C.M., M.L.M.], DE008559 [J.C.M., M.L.M.], DE009886 [M.L.M.], DE016930 [M.L.M.], DE016148 [M.L.M.], DE014581 [T.H.B.], DE018993 [T.H.B.], and DE025060 [E.J.L].
LITERATURE CITED
- Al Chawa T, Ludwig KU, Fier H, Potzsch B, Reich RH, Schmidt G, Braumann B, Daratsianos N, Bohmer AC, Schuencke H, Alblas M, Fricker N, Hoffmann P, Knapp M, Lange C, Nothen MM, Mangold E. Nonsyndromic cleft lip with or without cleft palate: Increased burden of rare variants within Gremlin-1, a component of the bone morphogenetic protein 4 pathway. Birth defects research Part A, Clinical and molecular teratology. 2014 doi: 10.1002/bdra.23244. [DOI] [PubMed] [Google Scholar]
- Armit C, Venkataraman S, Richardson L, Stevenson P, Moss J, Graham L, Ross A, Yang Y, Burton N, Rao J, Hill B, Rannie D, Wicks M, Davidson D, Baldock R. eMouseAtlas, EMAGE, and the spatial dimension of the transcriptome. Mammalian genome : official journal of the International Mammalian Genome Society. 2012;23(9-10):514–524. doi: 10.1007/s00335-012-9407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attanasio C, Nord AS, Zhu Y, Blow MJ, Li Z, Liberton DK, Morrison H, Plajzer-Frick I, Holt A, Hosseini R, Phouanenavong S, Akiyama JA, Shoukry M, Afzal V, Rubin EM, FitzPatrick DR, Ren B, Hallgrimsson B, Pennacchio LA, Visel A. Fine tuning of craniofacial morphology by distant-acting enhancers. Science. 2013;342(6157):1241006. doi: 10.1126/science.1241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, Liang KY, Wu T, Murray T, Fallin MD, Redett RA, Raymond G, Schwender H, Jin SC, Cooper ME, Dunnwald M, Mansilla MA, Leslie E, Bullard S, Lidral AC, Moreno LM, Menezes R, Vieira AR, Petrin A, Wilcox AJ, Lie RT, Jabs EW, Wu-Chou YH, Chen PK, Wang H, Ye X, Huang S, Yeow V, Chong SS, Jee SH, Shi B, Christensen K, Melbye M, Doheny KF, Pugh EW, Ling H, Castilla EE, Czeizel AE, Ma L, Field LL, Brody L, Pangilinan F, Mills JL, Molloy AM, Kirke PN, Scott JM, Arcos-Burgos M, Scott AF. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nature genetics. 2010;42(6):525–529. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Ruczinski I, Murray JC, Marazita ML, Munger RG, Hetmanski JB, Murray T, Redett RJ, Fallin MD, Liang KY, Wu T, Patel PJ, Jin SC, Zhang TX, Schwender H, Wu-Chou YH, Chen PK, Chong SS, Cheah F, Yeow V, Ye X, Wang H, Huang S, Jabs EW, Shi B, Wilcox AJ, Lie RT, Jee SH, Christensen K, Doheny KF, Pugh EW, Ling H, Scott AF. Evidence for gene-environment interaction in a genome wide study of nonsyndromic cleft palate. Genetic epidemiology. 2011;35(6):469–478. doi: 10.1002/gepi.20595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs LC, Naridze RL, DeMali KA, Lusche DF, Kuhl S, Soll DR, Schutte BC, Dunnwald M. Interferon regulatory factor 6 regulates keratinocyte migration. J Cell Sci. 2014;127:2840–2848. doi: 10.1242/jcs.139246. Pt 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs LC, Rhea L, Schutte BC, Dunnwald M. Interferon regulatory factor 6 is necessary, but not sufficient, for keratinocyte differentiation. J Invest Dermatol. 2012;132(1):50–58. doi: 10.1038/jid.2011.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, Baluardo C, Ferrian M, Almeida de Assis N, Alblas MA, Barth S, Freudenberg J, Lauster C, Schmidt G, Scheer M, Braumann B, Berge SJ, Reich RH, Schiefke F, Hemprich A, Potzsch S, Steegers-Theunissen RP, Potzsch B, Moebus S, Horsthemke B, Kramer FJ, Wienker TF, Mossey PA, Propping P, Cichon S, Hoffmann P, Knapp M, Nothen MM, Mangold E. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nature genetics. 2009;41(4):473–477. doi: 10.1038/ng.333. [DOI] [PubMed] [Google Scholar]
- Brinkley JF, Fisher S, Harris MP, Holmes G, Hooper JE, Jabs EW, Jones KL, Kesselman C, Klein OD, Maas RL, Marazita ML, Selleri L, Spritz RA, van Bakel H, Visel A, Williams TJ, Wysocka J, FaceBase C, Chai Y. The FaceBase Consortium: a comprehensive resource for craniofacial researchers. Development. 2016;143(14):2677–2688. doi: 10.1242/dev.135434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP, Myers RM, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison RC, Bernstein BE, Gingeras TR, Kent WJ, Birney E. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS biology. 2011;9(4):e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, Lin-Marq N, Koch M, Bilio M, Cantiello I, Verde R, De Masi C, Bianchi SA, Cicchini J, Perroud E, Mehmeti S, Dagand E, Schrinner S, Nurnberger A, Schmidt K, Metz K, Zwingmann C, Brieske N, Springer C, Hernandez AM, Herzog S, Grabbe F, Sieverding C, Fischer B, Schrader K, Brockmeyer M, Dettmer S, Helbig C, Alunni V, Battaini MA, Mura C, Henrichsen CN, Garcia-Lopez R, Echevarria D, Puelles E, Garcia-Calero E, Kruse S, Uhr M, Kauck C, Feng G, Milyaev N, Ong CK, Kumar L, Lam M, Semple CA, Gyenesei A, Mundlos S, Radelof U, Lehrach H, Sarmientos P, Reymond A, Davidson DR, Dolle P, Antonarakis SE, Yaspo ML, Martinez S, Baldock RA, Eichele G, Ballabio A. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS biology. 2011;9(1):e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nature reviews Genetics. 2011;12(3):167–178. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferone G, Thomason HA, Antonini D, De Rosa L, Hu B, Gemei M, Zhou H, Ambrosio R, Rice DP, Acampora D, van Bokhoven H, Del Vecchio L, Koster MI, Tadini G, Spencer-Dene B, Dixon M, Dixon J, Missero C. Mutant p63 causes defective expansion of ectodermal progenitor cells and impaired FGF signalling in AEC syndrome. EMBO Mol Med. 2012;4(3):192–205. doi: 10.1002/emmm.201100199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogh-Andersen P. Inheritance of Harelip and Cleft Palate. Munksgaard; Copenhagen: 1942. [Google Scholar]
- Fomenkov A, Huang YP, Topaloglu O, Brechman A, Osada M, Fomenkova T, Yuriditsky E, Trink B, Sidransky D, Ratovitski E. P63 alpha mutations lead to aberrant splicing of keratinocyte growth factor receptor in the Hay-Wells syndrome. J Biol Chem. 2003;278(26):23906–23914. doi: 10.1074/jbc.M300746200. [DOI] [PubMed] [Google Scholar]
- Fraser FC. Thoughts on the etiology of clefts of the palate and lip. Acta genetica et statistica medica. 1955;5(4):358–369. doi: 10.1159/000150783. [DOI] [PubMed] [Google Scholar]
- Grant SF, Wang K, Zhang H, Glaberson W, Annaiah K, Kim CE, Bradfield JP, Glessner JT, Thomas KA, Garris M, Frackelton EC, Otieno FG, Chiavacci RM, Nah HD, Kirschner RE, Hakonarson H. A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24. The Journal of pediatrics. 2009;155(6):909–913. doi: 10.1016/j.jpeds.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Grosen D, Chevrier C, Skytthe A, Bille C, Molsted K, Sivertsen A, Murray JC, Christensen K. A cohort study of recurrence patterns among more than 54,000 relatives of oral cleft cases in Denmark: support for the multifactorial threshold model of inheritance. Journal of Medical Genetics. 2010;47(3):162–168. doi: 10.1136/jmg.2009.069385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundlach KK, Maus C. Epidemiological studies on the frequency of clefts in Europe and world-wide. Journal of cranio-maxillo-facial surgery : official publication of the European Association for Cranio-Maxillo-Facial Surgery. 2006;34(Suppl 2):1–2. doi: 10.1016/S1010-5182(06)60001-2. [DOI] [PubMed] [Google Scholar]
- Jia Z, Leslie EJ, Cooper ME, Butali A, Standley J, Rigdon J, Suzuki S, Gongorjav A, Shonkhuuz TE, Natsume N, Shi B, Marazita ML, Murray JC. Replication of 13q31.1 association in nonsyndromic cleft lip with cleft palate in Europeans. American journal of medical genetics Part A. 2015;167(5):1054–1060. doi: 10.1002/ajmg.a.36912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JL, Canady JW, Brookes JT, Wehby GL, L'Heureux J, Schutte BC, Murray JC, Dunnwald M. Wound complications after cleft repair in children with Van der Woude syndrome. The Journal of craniofacial surgery. 2010;21(5):1350–1353. doi: 10.1097/SCS.0b013e3181ec6aad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie E, Murray J. Evaluating rare coding variants as contributing causes to non-syndromic cleft lip and palate. Clinical genetics:n/a-n/a. 2012 doi: 10.1111/cge.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Carlson JC, Shaffer JR, Feingold E, Wehby G, Laurie CA, Jain D, Laurie CC, Doheny KF, McHenry T, Resick J, Sanchez C, Jacobs J, Emanuele B, Vieira AR, Neiswanger K, Lidral AC, Valencia-Ramirez LC, Lopez-Palacio AM, Valencia DR, Arcos-Burgos M, Czeizel AE, Field LL, Padilla CD, Cutiongco-de la Paz EM, Deleyiannis F, Christensen K, Munger RG, Lie RT, Wilcox A, Romitti PA, Castilla EE, Mereb JC, Poletta FA, Orioli IM, Carvalho FM, Hecht JT, Blanton SH, Buxo CJ, Butali A, Mossey PA, Adeyemo WL, James O, Braimah RO, Aregbesola BS, Eshete MA, Abate F, Koruyucu M, Seymen F, Ma L, de Salamanca JE, Weinberg SM, Moreno L, Murray JC, Marazita ML. A multi-ethnic genome-wide association study identifies novel loci for non-syndromic cleft lip with or without cleft palate on 2p24.2, 17q23 and 19q13. Hum Mol Genet. 2016a doi: 10.1093/hmg/ddw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Liu H, Carlson JC, Shaffer JR, Feingold E, Wehby G, Laurie CA, Jain D, Laurie CC, Doheny KF, McHenry T, Resick J, Sanchez C, Jacobs J, Emanuele B, Vieira AR, Neiswanger K, Standley J, Czeizel AE, Deleyiannis F, Christensen K, Munger RG, Lie RT, Wilcox A, Romitti PA, Field LL, Padilla CD, Cutiongco-de la Paz EM, Lidral AC, Valencia-Ramirez LC, Lopez-Palacio AM, Valencia DR, Arcos-Burgos M, Castilla EE, Mereb JC, Poletta FA, Orioli IM, Carvalho FM, Hecht JT, Blanton SH, Buxo CJ, Butali A, Mossey PA, Adeyemo WL, James O, Braimah RO, Aregbesola BS, Eshete MA, Deribew M, Koruyucu M, Seymen F, Ma L, de Salamanca JE, Weinberg SM, Moreno L, Cornell RA, Murray JC, Marazita ML. A Genome-wide Association Study of Nonsyndromic Cleft Palate Identifies an Etiologic Missense Variant in GRHL3. American journal of human genetics. 2016b;98(4):744–754. doi: 10.1016/j.ajhg.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Marazita ML. Genetics of cleft lip and cleft palate. American journal of medical genetics Part C, Seminars in medical genetics. 2013;163C(4):246–258. doi: 10.1002/ajmg.c.31381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Taub MA, Liu H, Steinberg KM, Koboldt DC, Zhang Q, Carlson JC, Hetmanski JB, Wang H, Larson DE, Fulton RS, Kousa YA, Fakhouri WD, Naji A, Ruczinski I, Begum F, Parker MM, Busch T, Standley J, Rigdon J, Hecht JT, Scott AF, Wehby GL, Christensen K, Czeizel AE, Deleyiannis FW, Schutte BC, Wilson RK, Cornell RA, Lidral AC, Weinstock GM, Beaty TH, Marazita ML, Murray JC. Identification of Functional Variants for Cleft Lip with or without Cleft Palate in or near PAX7, FGFR2, and NOG by Targeted Sequencing of GWAS Loci. American journal of human genetics. 2015 doi: 10.1016/j.ajhg.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. American journal of human genetics. 2008;83(3):311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig KU, Ahmed ST, Bohmer AC, Sangani NB, Varghese S, Klamt J, Schuenke H, Gultepe P, Hofmann A, Rubini M, Aldhorae KA, Steegers-Theunissen RP, Rojas-Martinez A, Reiter R, Borck G, Knapp M, Nakatomi M, Graf D, Mangold E, Peters H. Meta-analysis Reveals Genome-Wide Significance at 15q13 for Nonsyndromic Clefting of Both the Lip and the Palate, and Functional Analyses Implicate GREM1 As a Plausible Causative Gene. PLoS Genet. 2016;12(3):e1005914. doi: 10.1371/journal.pgen.1005914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig KU, Mangold E, Herms S, Nowak S, Reutter H, Paul A, Becker J, Herberz R, AlChawa T, Nasser E, Bohmer AC, Mattheisen M, Alblas MA, Barth S, Kluck N, Lauster C, Braumann B, Reich RH, Hemprich A, Potzsch S, Blaumeiser B, Daratsianos N, Kreusch T, Murray JC, Marazita ML, Ruczinski I, Scott AF, Beaty TH, Kramer FJ, Wienker TF, Steegers-Theunissen RP, Rubini M, Mossey PA, Hoffmann P, Lange C, Cichon S, Propping P, Knapp M, Nothen MM. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nature genetics. 2012;44(9):968–971. doi: 10.1038/ng.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold E, Bohmer AC, Ishorst N, Hoebel AK, Gultepe P, Schuenke H, Klamt J, Hofmann A, Golz L, Raff R, Tessmann P, Nowak S, Reutter H, Hemprich A, Kreusch T, Kramer FJ, Braumann B, Reich R, Schmidt G, Jager A, Reiter R, Brosch S, Stavusis J, Ishida M, Seselgyte R, Moore GE, Nothen MM, Borck G, Aldhorae KA, Lace B, Stanier P, Knapp M, Ludwig KU. Sequencing the GRHL3 Coding Region Reveals Rare Truncating Mutations and a Common Susceptibility Variant for Nonsyndromic Cleft Palate. American journal of human genetics. 2016;98(4):755–762. doi: 10.1016/j.ajhg.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold E, Ludwig KU, Birnbaum S, Baluardo C, Ferrian M, Herms S, Reutter H, de Assis NA, Chawa TA, Mattheisen M, Steffens M, Barth S, Kluck N, Paul A, Becker J, Lauster C, Schmidt G, Braumann B, Scheer M, Reich RH, Hemprich A, Potzsch S, Blaumeiser B, Moebus S, Krawczak M, Schreiber S, Meitinger T, Wichmann HE, Steegers-Theunissen RP, Kramer FJ, Cichon S, Propping P, Wienker TF, Knapp M, Rubini M, Mossey PA, Hoffmann P, Nothen MM. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nature genetics. 2010;42(1):24–26. doi: 10.1038/ng.506. [DOI] [PubMed] [Google Scholar]
- Marazita ML, Lidral AC, Murray JC, Field LL, Maher BS, Goldstein McHenry T, Cooper ME, Govil M, Daack-Hirsch S, Riley B, Jugessur A, Felix T, Morene L, Mansilla MA, Vieira AR, Doheny K, Pugh E, Valencia-Ramirez C, Arcos-Burgos M. Genome scan, fine-mapping, and candidate gene analysis of non-syndromic cleft lip with or without cleft palate reveals phenotype-specific differences in linkage and association results. Human heredity. 2009;68(3):151–170. doi: 10.1159/000224636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossey PA, Little J, Munger RG, Dixon MJ, Shaw WC. Cleft lip and palate. Lancet. 2009;374(9703):1773–1785. doi: 10.1016/S0140-6736(09)60695-4. [DOI] [PubMed] [Google Scholar]
- Murray T, Taub MA, Ruczinski I, Scott AF, Hetmanski JB, Schwender H, Patel P, Zhang TX, Munger RG, Wilcox AJ, Ye X, Wang H, Wu T, Wu-Chou YH, Shi B, Jee SH, Chong S, Yeow V, Murray JC, Marazita ML, Beaty TH. Examining markers in 8q24 to explain differences in evidence for association with cleft lip with/without cleft palate between Asians and Europeans. Genetic epidemiology. 2012;36(4):392–399. doi: 10.1002/gepi.21633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Prescott S, Brugmann SA, Swigut T, Wysocka J. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell stem cell. 2012;11(5):633–648. doi: 10.1016/j.stem.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M, Domann FE, Govil M, Christensen K, Bille C, Melbye M, Jugessur A, Lie RT, Wilcox AJ, Fitzpatrick DR, Green ED, Mossey PA, Little J, Steegers-Theunissen RP, Pennacchio LA, Schutte BC, Murray JC. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nature genetics. 2008;40(11):1341–1347. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, Lee BT, Barber GP, Harte RA, Diekhans M, Long JC, Wilder SP, Zweig AS, Karolchik D, Kuhn RM, Haussler D, Kent WJ. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic acids research. 2013;41:D56–63. doi: 10.1093/nar/gks1172. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461(7261):199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sun Y, Huang Y, Pan Y, Jia Z, Ma L, Ma L, Lan F, Zhou Y, Shi J, Yang X, Zhang L, Jiang H, Jiang M, Yin A, Cheng J, Wang L, Yang Y, Shi B. Association study between Van der Woude Syndrome causative gene GRHL3 and nonsyndromic cleft lip with or without cleft palate in a Chinese cohort. Gene. 2016;588(1):69–73. doi: 10.1016/j.gene.2016.04.045. [DOI] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. American journal of human genetics. 2011;89(1):82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Hu Y, Li B, Abecasis GR, Liu DJ. RVTESTS: An Efficient and Comprehensive Tool for Rare Variant Association Analysis Using Sequence Data. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.