Abstract
Rationale: Small airways are the primary site of pathologic changes in chronic obstructive pulmonary disease (COPD), the major smoking-induced lung disorder.
Objectives: On the basis of the concept of proximal–distal patterning that determines regional specialization of the airway epithelium during lung development, we hypothesized that a similar program operates in the adult human lung being altered by smoking, leading to decreased regional identity of the small airway epithelium (SAE).
Methods: The proximal and distal airway signatures were identified by comparing the transcriptomes of large and small airway epithelium samples obtained by bronchoscopy from healthy nonsmokers. The expression of these signatures was evaluated in the SAE of healthy smokers and smokers with COPD compared with that of healthy nonsmokers. The capacity of airway basal stem cells (BCs) to maintain region-associated phenotypes was evaluated using the air–liquid interface model.
Measurements and Main Results: The distal and proximal airway signatures, containing 134 and 233 genes, respectively, were identified. These signatures included known developmental regulators of airway patterning, as well as novel regulators such as epidermal growth factor receptor, which was associated with the proximal airway phenotype. In the SAE of smokers with COPD, there was a dramatic smoking-dependent loss of the regional transcriptome identity with concomitant proximalization. This repatterning phenotype was reproduced by stimulating SAE BCs with epidermal growth factor, which was up-regulated in the SAE of smokers, during differentiation of SAE BCs in vitro.
Conclusions: Smoking-induced global distal-to-proximal reprogramming of the SAE represents a novel pathologic feature of COPD and is mediated by exaggerated epidermal growth factor/epidermal growth factor receptor signaling in SAE BCs.
Keywords: transcriptome, basal cells, stem cells, differentiation, chronic obstructive pulmonary disease
At a Glance Commentary
Scientific Knowledge on the Subject
The airway epithelium is heterogeneous along the proximal–distal axis of the tracheobronchial tree. Recent evidence suggests that distal, or small, airways and the epithelium that covers this anatomic region represent the primary site of pathologic changes in chronic obstructive pulmonary disease, the major smoking-induced lung disorder. Although the mechanisms that control proximal–distal patterning in the murine lung have been characterized, the transcriptional program that defines region-specific phenotypes in the adult human airways and how it changes in lung disease remain unknown.
What This Study Adds to the Field
By analyzing the transcriptomic profiles of epithelial samples from various airway regions of the human lung, the transcriptional program that characterizes the proximal–distal patterning of the human adult airway epithelium was identified. In the small airway epithelium of healthy smokers, and more so in smokers with chronic obstructive pulmonary disease, there was broad suppression of the distal patterning program, typical for this region, accompanied by acquisition of the proximal airway phenotype. This distal-to-proximal repatterning occurred independent from classic smoking-associated remodeling phenotypes and was mediated, at least in part, by augmented epidermal growth factor receptor signaling in small airway basal cells, the stem cells of the airway epithelium.
The human airway epithelium is composed of ciliated and secretory cells that mediate host defense at the airway surface, as well as basal stem cells (BCs) that reside above the basement membrane and operate as stem cells that self-renew and differentiate into ciliated and secretory cells (1, 2). The proportion and biologic properties of these cells change along the proximal–distal (P–D) axis (3–7). Compared with its proximal counterpart, the small airway epithelium (SAE), which covers the distal airways, has no mucus-producing (“goblet”) cells, substituted in this region by secretoglobin (SCGB)-producing (“club”) cells. BC numbers progressively decrease with each subsequent airway branch, although compared with the mouse small airways, where BCs are absent, BCs are found in humans throughout the tracheobronchial tree (4, 7).
The heterogeneity of the lung epithelium across the P–D axis is initially determined by the developmental P–D patterning program that allows endoderm-derived progenitors to specialize into the respiratory (“distal”) and conductive airway (“proximal”) epithelial cell types during lung morphogenesis (8–11). Whether a similar program is maintained in the adult human airways and how it changes in human lung diseases is unknown.
Chronic obstructive pulmonary disease (COPD) is a smoking-induced lung disorder characterized by progressive, irreversible airflow obstruction caused by the remodeling of small airways (12, 13). Although smoking causes airway remodeling across the tracheobronchial tree, smoking-induced pathologic changes in the small airways, including increased thickness of the SAE and occlusion of small airways by mucus, have been associated with airway obstruction in COPD (12). Capitalizing on the P–D patterning concept and the knowledge that airway BCs are responsible for the maintenance of normally differentiated airway epithelium (4, 5), we hypothesized that the SAE in smokers with chronic obstructive pulmonary disease (COPD-S) loses its region-specific phenotype as a result of pathologic programming of SAE BCs by smoking-dependent factors.
By analyzing the transcriptomes of the airway epithelium from different regions of human airways, we identified molecular signatures that define the P–D axis in the adult human airway epithelium. Consistent with our hypothesis, there was a dramatic smoking-dependent loss of the SAE transcriptomic identity in COPD-S, with a shift toward the proximal phenotype, which was reproduced in vitro by exposing the differentiating SAE BCs to epidermal growth factor (EGF), a growth factor up-regulated in the SAE of smokers. Thus, smoking-dependent reversal of the physiologic P–D airway pattern is a novel feature of small airway disordering in COPD driven by aberrant differentiation of SAE BCs.
Methods
Study Population and Sample Collection
All individuals were evaluated at the Weill Cornell Medical Center National Institutes of Health Clinical and Translational Science Center and Department of Genetic Medicine Clinical Research Facility under protocols approved by the Weill Cornell Medical Center Institutional Review Board. Before enrollment, written informed consent was obtained from each individual. The epithelium was collected from the proximal airways (total, n = 48; including trachea [n = 27] and fourth- to sixth-generation bronchi [n = 21]) of healthy nonsmokers and distal airways (SAE; 10th- to 12th-generation bronchi) of healthy nonsmokers (n = 63), healthy smokers (n = 73), and COPD-S (n = 37) by bronchoscopic brushing as described elsewhere (14). Inclusion and exclusion criteria are detailed in the online supplement, and demographic data are provided in Table 1.
Table 1.
Study Population and Biologic Samples
| Parameter | TR: Healthy Nonsmokers | Fourth- to Sixth-Generation Bronchi: Healthy Nonsmokers | SAE* |
||
|---|---|---|---|---|---|
| Healthy Nonsmokers | Healthy Smokers | COPD-S | |||
| n | 27 | 21 | 63 | 73 | 37 |
| Sex, M/F | 17/10 | 15/6 | 40/23 | 53/20 | 28/9 |
| Ethnicity, B/W/O | 15/8/4 | 10/7/4 | 28/24/11 | 45/16/12 | 13/14/11 |
| Age, yr | 40 ± 8 | 40 ± 8 | 40 ± 12 | 43 ± 8 | 52 ± 7 |
| Smoking history | |||||
| Pack-years | — | — | — | 27 ± 16 | 38 ± 24 |
| Nicotine, ng/ml | — | — | — | 1,381 ± 1,624 | 1,680 ± 1,548 |
| Cotinine, ng/ml | — | — | — | 1,325 ± 1,034 | 1,469 ± 625 |
| Lung function† | |||||
| FEV1 | 107 ± 16 | 107 ± 17 | 106 ± 14 | 107 ± 14 | 82 ± 21 |
| FVC | 107 ± 14 | 106 ± 13 | 107 ± 13 | 110 ± 13 | 104 ± 21 |
| FEV1/FVC | 82 ± 5 | 82 ± 5 | 82 ± 6 | 80 ± 5 | 63 ± 7 |
| TLC | 97 ± 13 | 99 ± 13 | 100 ± 13 | 101 ± 12 | 105 ± 19 |
| DlCO | 98 ± 16 | 101 ± 16 | 99 ± 15 | 94 ± 11 | 76 ± 16 |
| Epithelial cells‡ | |||||
| Number recovered, ×106 | 2.2 ± 2.9 | 6.8 ± 2.9 | 6.6 ± 3.0 | 7.5 ± 3.3 | 6.3 ± 2.7 |
| Epithelial cells, % | 99.9 ± 0.2 | 99.7 ± 0.6 | 98.9 ± 1.5 | 98.9 ± 1.4 | 98.3 ± 1.6 |
| Inflammatory cells, % | 0.1 ± 0.2 | 0.3 ± 0.6 | 1.1 ± 1.5 | 1.1 ± 1.4 | 1.7 ± 1.6 |
| Differential cell count | |||||
| Ciliated, % | 48.5 ± 8.6 | 53.6 ± 6.6 | 71.8 ± 8.9 | 64.0 ± 12.0 | 60.1 ± 10.7 |
| Secretory, % | 6.3 ± 3.6 | 9.7 ± 4.0 | 6.8 ± 3.6 | 8.4 ± 4.4 | 11.6 ± 5.7 |
| Basal, % | 27.3 ± 9.2 | 22.6 ± 3.4 | 12.5 ± 6.5 | 14.3 ± 8.0 | 14.7 ± 8.2 |
| Undifferentiated, % | 17.7 ± 6.9 | 14.3 ± 5.0 | 7.8 ± 3.5 | 12.3 ± 6.7 | 11.9 ± 3.8 |
Definition of abbreviations: B = black; COPD-S = smokers with chronic obstructive pulmonary disease; DlCO = diffusing capacity of the lung for carbon monoxide; O = other; SAE = small airway epithelium; TLC = total lung capacity; TR = trachea; W = white.
Data are presented as mean ± SD. For nine of the healthy nonsmokers, TR, bronchial, and SAE samples were from the same subjects; in an additional one, TR and bronchial samples were from the same subject; in an additional nine, TR and SAE samples were from the same subject; and in an additional eight, bronchial and SAE samples were from the same subject.
Nonsmokers (NS) were comparable to healthy smokers (S) and COPD-S with regard to sex (P > 0.3). There were more black subjects among S than in the NS or COPD-S groups (P < 0.04). NS were comparable to S in age and all lung function parameters (P > 0.05 for all comparisons), except for DlCO, which was lower in S (P < 0.03). The number of epithelial cells recovered, percentage of epithelial cells, percentage of inflammatory cells, and percentage of basal cells (BCs) were comparable in S versus NS (P > 0.1). The percentage of ciliated cells was lower, and the percentages of secretory and undifferentiated cells were higher, in S than in NS (P < 0.03). COPD-S were older than NS and S (P < 10−5) and had lower FEV1, FEV1/FVC, and DlCO than NS and S (P < 10−9). TLC and FVC were comparable (P > 0.07). Pack-years of smoking was lower in COPD-S than in S (P < 0.003). The number of epithelial cells recovered, percentage of epithelial cells, percentage of inflammatory cells, and percentage of BCs were comparable in COPD-S versus NS and S (P > 0.05). The percentage of ciliated cells was lower, and the percentages of secretory and undifferentiated cells were higher, in COPD-S (P < 10−5 vs. NS; P > 0.05 vs. S). The percentage of secretory cells was lower in COPD-S than in S (P < 0.003).
Pulmonary function testing parameters are given as percent predicted values, with the exception of FEV1/FVC, which is reported as percentage observed. Values were measured prebronchodilator use in NS and S and post-bronchodilator use in COPD-S.
As a percentage of the TR, bronchial (large airway epithelium [LAE]) or SAE cells recovered. The percentage of inflammatory cells was higher in SAE (all groups) than in TR-NS (P < 0.001) and LAE-NS (P < 0.05). The percentage of ciliated cells was higher in the SAE of nonsmokers (SAE-NS) than in LAE-NS and TR-NS (P < 10−11 for both comparisons) and in LAE-NS than in TR-NS (P < 0.03). The percentage of secretory cells in LAE-NS was higher than in SAE-NS (P < 0.003) and TR-NS (P < 0.004). The percentage of basal cells was higher in TR-NS than in LAE-NS (P < 0.03) and SAE-NS (P < 10−12) and in LAE-NS than in SAE-NS (P < 10−11). The percentage of undifferentiated cells was higher in TR-NS than in SAE-NS and in LAE-NS than in SAE-NS (P < 10−4 for both comparisons).
Gene Expression Analysis
Transcriptomic analysis of the airway epithelial samples was performed using the Human Genome U133 Plus 2.0 array (Affymetrix, Santa Clara, CA) and GeneSpring 7.3.1 software (Agilent Technologies, Palo Alto, CA) as described previously (14). Proximal and distal airway epithelial signatures were defined as sets of genes expressed significantly higher in one anatomic region than in another using the following criteria: (1) present call greater than or equal to 80% in the signature region, (2) fold change greater than or equal to 2, and (3) false discovery rate (Benjamini-Hochberg–corrected P value) less than 0.01. Ingenuity Pathway Analysis (QIAGEN Bioinformatics, Redwood City, CA) was used to identify enriched biologic functions and upstream regulators. Protein–protein interaction (PPI) networks were constructed using STRING9.1 (15) as described elsewhere (16). Enriched Gene Ontology categories and protein families were identified using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (17) and Gene Annotation Tool to Help Explain Relationships (GATHER) (18). Principal component and clustering analyses were performed using GeneSpring 7.3.1 software as previously described (19).
Expression of identified signatures in the SAE of different groups was compared using a P value less than 0.05 with the Benjamini-Hochberg correction as the criterion for differentially expressed genes. As a cumulative measure of signature expression, indices were calculated using previously described methodology as the percentage of genes with expression levels outside the range of 2 SD of the mean in healthy nonsmokers (14). Significant differences in indices between the groups were determined using the Mann-Whitney U test. The raw data are publicly available at the Gene Expression Omnibus website (www.ncbi.nlm.nih.gov/geo/) under accession number GSE64614. TaqMan (Life Technologies, Carlsbad, CA) real-time polymerase chain reaction (PCR) analysis was performed as previously described (16, 20).
Human Airway BC Studies
BCs were isolated from the airway epithelium of healthy nonsmokers as described previously (21, 22). BC differentiation was studied using the air–liquid interface (ALI) model as described elsewhere (23). The ALI cultures were grown with defined media (for proximal airway BCs, 2% Ultroser G; BioSerpa S.A., Cergy-Saint-Christophe, France; for SAE BCs, S-ALI media; Lonza, Basel, Switzerland) with or without recombinant human EGF (10 ng/ml; Sigma-Aldrich, St. Louis, MO) or cigarette smoke extract (3% CSE; a concentration known to be noncytotoxic in this model [16]). They were prepared using previously established protocols (24; see also Methods in the online supplement for details) and added from the basolateral ALI side every other day as described elsewhere (20, 25). In selected experiments, epidermal growth factor receptor (EGFR)-selective tyrosine kinase inhibitor AG1478 (10 µM; Calbiochem, San Diego, CA) was applied from the basolateral side for 1 hour prior to addition of media with or without CSE as described previously (25). See Methods in the online supplement for detailed description of BC studies.
Morphology
Immunohistochemistry and immunofluorescence of airway biopsy samples (obtained by bronchoscopy), lung tissue samples (US Biomax, Rockville, MD; and Lung Tissue Research Consortium, NHLBI, National Institutes of Health, Bethesda, MD) containing small airways (<2 mm in diameter), SAE brushing samples, ALI sections, and control tissues (US Biomax) were performed using standard protocols (20). Primary antibody information is provided in Table E6 in the online supplement. Quantification analysis of the immunofluorescence staining data was performed by counting at least 100 cells per sample in at least three samples per group for tissue sections and at least five samples per group for SAE brushings as previously described (19, 25).
Results
Identification of P–D Patterning Program in the Adult Human Airway Epithelium
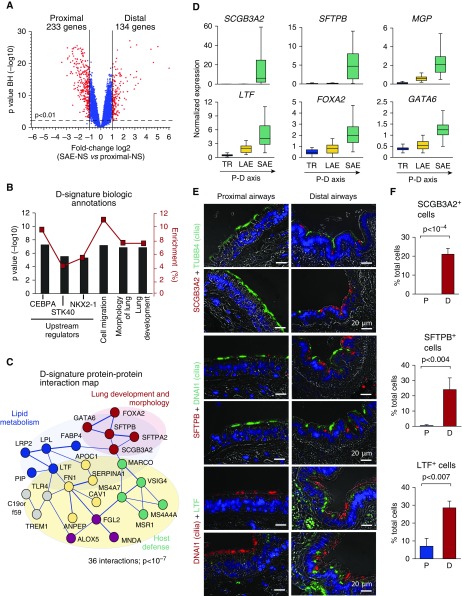
By genome-wide transcriptomic analysis, we identified 134 and 233 genes with significantly higher expression in the distal (“distal signature”) and proximal (“proximal signature”) airway epithelium, respectively (Figure 1A, Tables E1 and E2).
Figure 1.
Distal airway epithelial signature. (A) Volcano plot depicting differential gene expression between the distal (D; n = 63) and proximal (P; total n = 48, including trachea [TR; n = 27] and fourth- to sixth-generation bronchi [n = 21]) small airway epithelium of healthy nonsmokers (SAE-NS). D and P signatures were identified as sets of genes significantly up-regulated (red dots) in corresponding regions. Blue dots represent nonsignificant gene probe sets. (B) Biologic annotation categories enriched in the D signature on the basis of Ingenuity Pathway Analysis (IPA). Shown are the top three IPA-predicted upstream regulators and biologic functions based on the P value (bars, left y-axis) and enrichment score (red boxes, right y-axis). (C) STRING9.1-based protein–protein interaction networks in the D signature. Each circle corresponds to an individual gene; colors distinguish clusters determined using a Markov clustering algorithm and annotated using IPA and/or Gene Annotation Tool to Help Explain Relationships (GATHER). The structure of lines reflects the confidence of association, from thick (most confident) to thin (least confident). (D) Microarray-based normalized expression of selected D-signature genes (see Table E2 for full gene names and Figure E1 for more examples, statistics, and polymerase chain reaction validation) in the epithelium of TR (n = 27), large airway epithelium (LAE; fourth- to sixth-generation bronchi [n = 21]) and SAE (10th- to 12th-generation bronchi [n = 63]) of healthy nonsmokers. The direction of the proximal–distal (P–D) axis is shown. (E) Representative images of the P (bronchus) and D airway samples analyzed using immunofluorescence for D-signature markers secretoglobin family 3A member 2 (SCGB3A2), surfactant protein B (SFTPB), and lactotransferrin (LTF) in combination with cilia markers tubulin β4 chain (TUBB4) or dynein axonemal intermediate chain 1 (DNAI1). Nuclei are stained with 4′,6-diamidino-2-phenylindole (blue); scale bar = 20 μm. See Figure E2 for more examples. (F) Frequency of cells expressing SCGB3A2, SFTPB, and LTF in the P and D airway epithelium (n ≥ 3 samples; total, ≥500 cells/group). BH = Benjamini-Hochberg correction; CEBPA = CCAAT/enhancer-binding protein-α; FOXA2 = forkhead box protein A2; GATA6 = GATA-binding protein 6; MGP = matrix Gla protein; NKX2-1 = NK2 homeobox 1; STK40 = serine/threonine kinase 40.
Distal Airway Signature
The top enriched distal signature (D-signature) genes included secretoglobin family 3A member 2 (SCGB3A2), surfactant protein B (SFTPB), surfactant protein A2, LIM domain only 3 (LMO3), and matrix Gla protein (MGP) (Table 2). The top enriched D-signature transcription factors (TFs) were forkhead box protein A2 (FOXA2), SOX9, GATA6, MKX, and ARX (see Table 2 for full gene names). By pathway analysis, we identified CCAAT/enhancer-binding protein-α (CEBPA), STK40, and NK2 homeobox 1 (NKX2-1) as the top three upstream regulators of the D signature (Figure 1B, Table 2). The PPI cluster related to lung development dominated within the 36-PPI network identified in the D signature (P < 10−7 vs. random) (Figure 1C). Although the D signature was dominated by molecular features of epithelial differentiation (Figure 1B, Table E1), consistent with the epithelial nature of samples obtained by bronchoscopy (>98% epithelial cells) (Table 1), a number of genes coding for scavenger receptors (e.g., MARCO and MSR1) were found in the D signature to be enriched in the PPI cluster related to host defense (Figure 1C).
Table 2.
Region-Specific Signatures of Human Airway Epithelium
| Category | Gene Symbol, GO Identifier, or UniProt Keyword | Fold Enrichment* | FDR (BH) |
|---|---|---|---|
| Distal | |||
| Top five enriched genes† | |||
| Secretoglobin family 3A member 2 | SCGB3A2 | 64.1 | <10−18 |
| Surfactant protein B | SFTPB | 32.1 | <10−25 |
| Surfactant protein A2 | SFTPA2 | 24.6 | <10−20 |
| LIM domain only 3 (rhombotin-like 2) | LMO3 | 11.1 | <10−14 |
| Matrix Gla protein | MGP | 9.6 | <10−13 |
| Transcription factor–related‡ | |||
| Forkhead box A2 | FOXA2 | 3.9 | <10−10 |
| Mohawk homeobox | MKX | 3.0 | <10−11 |
| Sex-determining region Y (SRY) box 9 | SOX9 | 3.0 | <10−16 |
| GATA-binding protein 6 | GATA6 | 2.7 | <10−24 |
| Aristaless related homeobox | ARX | 2.3 | <10−14 |
| Top annotation categories§ | |||
| Extracellular space | GO:0005615 | 4.1 | <10−4 |
| Extracellular region part | GO:0044421 | 3.1 | <10−3 |
| Secreted | KW-0964 | 2.6 | <10−3 |
| Disulfide bond | KW-01015 | 2.1 | <10−3 |
| Signal | KW-0732 | 2.0 | <10−2 |
| Proximal | |||
| Top five enriched genes† | |||
| Plakophilin 1 (ectodermal dysplasia/skin fragility syndrome) | PKP1 | 10.8 | <10−20 |
| ATPase H+ transporting lysosomal 42kDa V1 subunit C2 | ATP6V1C2 | 7.8 | <10−16 |
| Tumor protein p63 | TP63 | 7.7 | <10−24 |
| Dystonin | DST | 7.6 | <10−21 |
| Tenascin C | TNC | 7.4 | <10−21 |
| Transcription factor–related‡ | |||
| Tumor protein p63 | TP63 | 7.7 | <10−24 |
| HOXB cluster antisense RNA 1 | HOXB-AS1 | 5.6 | <10−20 |
| Snail family zinc finger 2 | SNAI2 | 5.1 | <10−24 |
| Homeobox A1 | HOXA1 | 4.7 | <10−12 |
| Homeobox B3 | HOXB3 | 4.0 | <10−09 |
| Top annotation categories§ | |||
| Epithelial cell differentiation | GO:0030855 | 6.4 | <10−3 |
| Epidermal development | GO:0008544 | 5.9 | <10−3 |
| Ectodermal development | GO:0007398 | 5.5 | <10−3 |
| Skeletal system development | GO:0001501 | 3.9 | <10−2 |
| Extracellular matrix | GO:0031012 | 3.7 | <10−2 |
Definition of abbreviations: BH = Benjamini-Hochberg correction; FDR = false discovery rate; GO = Gene Ontology database; KW = keyword; UniProt = Universal Protein Resource.
Distal and proximal signatures are listed (see Tables E1 and E2 for complete gene lists).
Fold enrichment (for genes, fold change in the expression of a given gene in the signature region compared with its expression in the compared anatomic region; for categories, defined by Database for Annotation, Visualization and Integrated Discovery [DAVID]).
Top five genes (ranked on basis of fold enrichment) are listed.
Top five genes coding for transcription factors or transcription factor–related regulatory transcripts (ranked on basis of fold enrichment).
The two top D-signature genes, SCGB3A2 and SFTPB, were expressed exclusively in the SAE, whereas other D-signature genes were also detected in the proximal airway epithelium but progressively up-regulated along the P–D axis (Figure 1D; see Figure E1 for more examples and PCR validation). Consistent with the transcriptomic data, SCGB3A2+ cells were detected in the distal (frequency, 21.0 ± 3.2%) but not the proximal airway epithelium; SFTPB+ cells were found almost exclusively in the distal airway epithelium (24.0 ± 7.7% vs. 0.5 ± 0.6% in the proximal airway epithelium; P < 0.004); and cells expressing another D-signature marker, lactotransferrin (LTF), were more frequent in the distal airway epithelium (28.5 ± 3.8%) than in the proximal airway epithelium (7.0 ± 4.4%; P < 0.007) (Figures 1E and 1F). Nonciliated secretory cells expressed these proteins (Figures 1E and E2), consistent with the dominant secretory pattern of the D signature (Table 2).
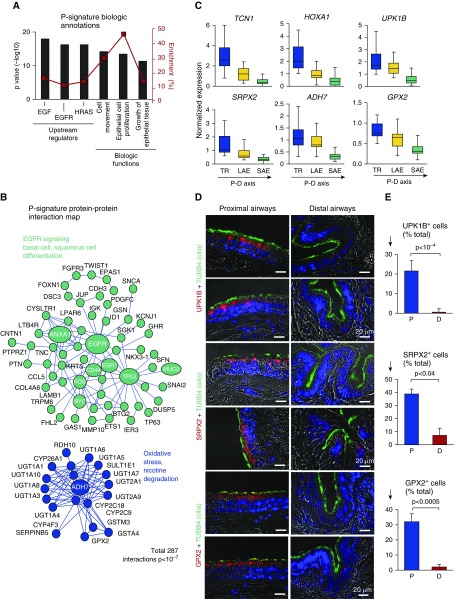
Proximal Airway Signature
The top enriched proximal (P)-signature genes were plakophilin 1, ATP6V1C2, TP63, dystonin, and tenascin C (Table 2). The top enriched P-signature TF-related genes included TP63, SNAI2, HOXA1, HOXB3, and HOXB cluster antisense RNA (Table 2). By pathway analysis, we identified EGF, EGFR, and HRAS as the top upstream regulators of the P signature (Figure 2A). Among the 287 PPI (P < 10−7 vs. random) (Figure 2B) identified in the P signature, the largest cluster was organized around the EGFR gene and included BC-related (KRT5, TP63, CD44), extracellular matrix–related (COL4A6, TNC), and epithelial-to-mesenchymal transition–related (SNAI2, TWIST) genes. The second-largest PPI cluster included genes related to oxidative stress (alcohol dehydrogenase [ADH7], glutathione peroxidase [GPX2]), xenobiotic metabolism, and nicotine degradation (Figure 2B). Gene categories related to the epidermis, ectoderm, skeletal system development, and extracellular matrix were enriched in the P signature (Table 2).
Figure 2.
Proximal airway epithelial signature. (A) Ingenuity Pathway Analysis–based biologic annotation categories enriched in the proximal signature. Shown are the top three Ingenuity Pathway Analysis–predicted upstream regulators and biologic functions based on the P value (bars, left y-axis) and enrichment score (red boxes, right y-axis). (B) STRING9.1-based protein–protein interaction networks in the P signature. Each circle corresponds to an individual gene (see Figure 1C legend for description). (C) Microarray-based normalized expression of selected P signature genes (see Table E3 for full gene names and Figure E3 for more examples, statistics, and TaqMan validation) in the epithelium from the trachea (TR), large airway epithelium (LAE; n = 21), and small airway epithelium (SAE; n = 63) of healthy nonsmokers. Direction of the proximal-to-distal (P–D) axis is shown. (D) Representative images of the P (bronchus) and D airway samples analyzed using immunofluorescence for P-signature genes uroplakin 1B (UPK1B), sushi repeat-containing protein X-linked 2 (SRPX2), and glutathione peroxidase 2 (GPX2) in combination with cilia marker tubulin β4 chain (TUBB4). Nuclei are stained with 4′,6-diamidino-2-phenylindole (blue); scale bar = 20 μm. See Figure E4 for more examples. (E) Frequency of cells expressing UPK1B, SRPX2, and GPX2 in the P and D airway epithelium (nc ≥ c3 samples; total, ≥500 cells per group). ADH7 = alcohol dehydrogenase 7 (class IV); EGF = epidermal growth factor; EGFR = epidermal growth factor receptor; HOXA1 = homeobox A1; HRAS = H-Ras; TCN1 = transcobalamin 1.
Expression of P-signature genes progressively decreased along the P–D axis (Figure 2C; see Figure E3 for more examples and PCR validation). Consistent with the transcriptomic data, cells expressing individual P-signature genes, including uroplakin 1B (UPK1B), sushi repeat-containing protein X-linked 2 (SRPX2), and GPX2, were found predominantly in the proximal airway epithelium (frequency, 21.5 ± 3.5%, 38.9 ± 3.7%, and 32.0 ± 5.3%, respectively) (Figures 2D and 2E). In the distal airway epithelium, only few cells expressed these markers (0.5 ± 0.75%, 7.2 ± 3.7%, and 2.1 ± 1.7%, respectively; P < 10−4, P < 0.05, and P < 10−3 vs. proximal, respectively) (Figure 2E). In the proximal airways, expression of these proteins was enriched in a subset of luminal, predominantly ciliated cells marked by the presence of cilia on the apical surface (Figures 2E and E4).
Smoking-Dependent Loss of SAE Transcriptome Identity in COPD
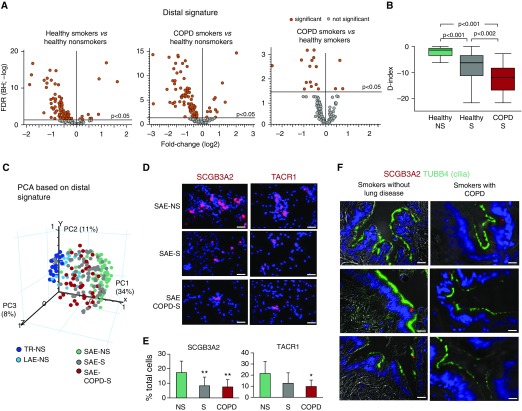
The transcriptomic analysis revealed that 47% of D-signature genes were down-regulated in the SAE of healthy smokers compared with nonsmokers (Figure 3A, left panel). In the SAE of COPD-S, the proportion (Figure 3A, middle panel) and identity (Table E3) of down-regulated D-signature genes were comparable to those in healthy smokers, with 10% of D-signature genes being further down-regulated compared with those of healthy smokers (Figure 3A, right panel). No significant difference was found between the study groups with regard to the distance traveled by the tip of the brush to reach distal and proximal airways during bronchoscopy (Table E4), suggesting that observed changes were not due to sampling.
Figure 3.
Down-regulation of distal (D)-signature expression in the small airway epithelium (SAE) in healthy smokers (S) and in smokers with chronic obstructive pulmonary disease (COPD-S). (A) Volcano plots illustrating differential expression of the D-signature gene probes in the SAE of healthy nonsmokers (NS; n = 63), healthy smokers (S; n = 73), COPD-S (n = 37), and significant probe sets (orange; Benjamini-Hochberg [BH]-corrected P < 0.05). (B) Mean D index in the SAE of indicated groups (described in A). Horizontal line within each bar represents the median for each group; P values are based on the Mann-Whitney U test. (C) Principal component analysis (PCA) of epithelial samples from the trachea (TR-NS; n = 27), large airway epithelium (LAE-NS; fourth- to sixth-generation bronchi; n = 21), and distal SAE (samples described in A) of nonsmokers based on the expression of the D-signature gene probes. (D) Representative images of SAE brushing samples from indicated groups analyzed using immunofluorescence for secretoglobin family 3A member 2 (SCGB3A2) and tachykinin receptor 1 (TACR1). Nuclei are stained with 4′,6-diamidino-2-phenylindole (blue); scale bar = 20 μm. See Figure E6 for more images and statistics. (E) SCGB3A2+ and TACR1+ cells (percentage of total) in the SAE brushing samples from NS (n = 9), S (n = 10), and COPD-S (n = 8). *P < 0.05, **P < 0.01. (F) Representative images of D airways of smokers without lung disease and COPD-S (n = 3 per group) analyzed using immunofluorescence for expression of SCGB3A1 (red) and cilia marker tubulin β4 chain (TUBB4) (green); nuclei are stained with 4′,6-diamidino-2-phenylindole (blue); scale bar = 20 μm. FDR = false discovery rate; PC = principal component.
Assessment of the SAE “D index” (see Methods) revealed remarkable heterogeneity of healthy smokers and COPD-S based on this parameter (Figure E5A). Overall, the SAE D index was dramatically lower in healthy smokers than in nonsmokers (P < 0.001) and further decreased in COPD-S (P < 0.002 vs. healthy smokers) (Figure 3B). Principal component analysis showed that, on the basis of D-signature expression, the SAE of healthy smokers, and more so COPD-S, shifted toward the transcriptomic pattern normally displayed by the epithelium of the trachea and large airways (Figure 3C).
Among the D-signature genes down-regulated in the SAE of smokers were secretory phenotype-related genes LTF, SCGB3A2, SFPTB, MGP, and WIF; receptor-coding genes tachykinin receptor 1 (TACR1) and folate receptor 1 (FOLR1); TF genes SOX9, FOXA2, and ARX; and others (see Table E3 for complete gene list). Expression of individual D-signature genes in the SAE of healthy smokers and COPD-S decreased to the levels expressed by these genes in the proximal airways (Figure E2). Consistent with the gene expression data, there was a decreased frequency of cells expressing D-signature markers SCGB3A2 and TACR1 in the SAE samples from healthy smokers and COPD-S compared with nonsmokers (Figures 3D, 3E, and E6), with loss of SCGB3A2+ secretory cells in the SAE luminal compartment (Figure 3F).
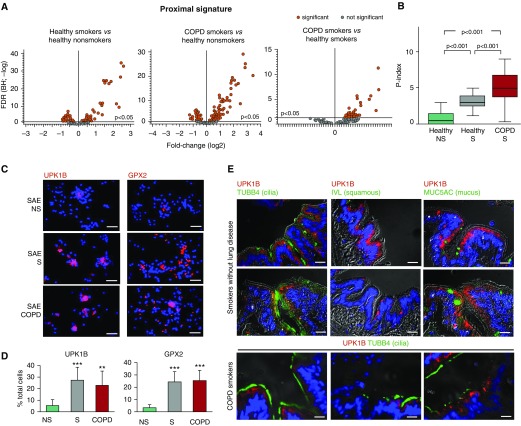
Proximalization of the SAE Transcriptome in COPD-S
By contrast to the D signature, which was suppressed in the SAE of smokers, 40 (about 17%) of 233 P-signature genes were up-regulated in the SAE of healthy smokers (Figure 4A, left panel), indicative of SAE “proximalization.” This phenomenon was even more dramatic in the SAE of COPD-S, where 70 (30%) of the P-signature genes were up-regulated compared with healthy nonsmokers (Figure 4A, middle panel), and 40 (about 17%) P-signature genes were further up-regulated compared with healthy smokers (Figure 4A, right panel).
Figure 4.
Proximalization of small airway epithelium (SAE) in smokers and in smokers with chronic obstructive pulmonary disease (COPD-S). (A) Volcano plots depicting differential expression of the proximal (P)-signature gene probes in the SAE of healthy nonsmokers (NS; n = 63), healthy smokers (S; n = 73), COPD-S (n = 37), and significant probe sets (orange; Benjamini-Hochberg [BH]-corrected P < 0.05). (B) P index in the SAE of indicated groups (study population as in A). Horizontal line within each bar represents the median value for each group. P values are based on the Mann-Whitney U test. (C) Representative images of SAE brushing samples from indicated groups analyzed using immunofluorescence for proximal signature genes uroplakin 1B (UPK1B) and glutathione peroxidase 2 (GPX2). Nuclei are stained with 4′,6-diamidino-2-phenylindole (blue); scale bar = 20 μm. See Figure E6 for more images and statistics. (D) UPK1B+ and GPX2+ cells (percentage of total) in the SAE brushing samples from healthy NS (n = 9), S (n = 10), and COPD-S (n = 8). **P < 0.01, ***P < 0.001. (E) Representative images of immunofluorescence analysis of the distal airways of S without lung disease and COPD-S (n = 3 per group) for expression of UPK1B and cilia marker tubulin β4 chain (TUBB4), as well as (for S without lung disease) UPK1B and involucrin (IVL) or mucin 5AC (MUC5AC). Scale bar = 20 μm. FDR = false discovery rate.
Assessment of the proximal index (P index) (see Methods) revealed that more than 90% of healthy nonsmokers, but less than 80% of healthy smokers and, strikingly, less than 30% of COPD-S, had an SAE P index within the ±2% of the ideal (“0”) range (Figure E5B). The average P index was markedly higher in the SAE of healthy smokers than in nonsmokers (P < 0.001) and further increased in COPD-S (P < 0.001 vs. healthy smokers) (Figure 4B). On the basis of expression of both P- and D-signature genes, the SAE of healthy smokers and more so that of COPD-S shifted toward the transcriptomic phenotype of the epithelium of trachea and large bronchi (Figures E5C and E5D).
Healthy smokers were particularly heterogeneous with regard to dysregulation of the SAE region–associated phenotype with nonsmoker-like, COPD-like, and intermediate subsets (Figures E5A–E5D). To determine possible factors that might contribute to this heterogeneity among healthy nonsmokers, we analyzed the relationship between the SAE D–P repatterning index, measured as a sum of D and P indices, and available subject characteristics. The SAE D–P repatterning index was significantly higher in healthy smokers older than 44 years of age (median age; P < 0.05) (Figure E5E) and with an FEV1/FVC ratio less than or equal to 80% of the predicted value (median ratio; P < 0.01) (Figure E5F).
Among the top up-regulated P-signature genes in the SAE of COPD-S were CEACAM5, GPX2, TCN1, SRPX2, UPK1B, IL1R2, and HOXA1 (see Table E5 for complete gene list). Expression of individual P-signature genes in the SAE of healthy smokers and COPD-S increased to levels expressed by these genes in the proximal airway epithelium (Figure E4). There was an increased proportion of cells expressing P-signature genes UPK1B and GPX2 in the SAE samples from healthy smokers and COPD-S compared with nonsmokers (Figures 4C, 4D, and E7).
To understand the relationship between smoking-induced SAE proximalization and histology, we analyzed the expression pattern of smoking-induced P-signature gene UPK1B. Similar to its expression pattern in the proximal airways, in the SAE of smokers, UPK1B was detected in the cytoplasm of ciliated cells (Figure 4E). The apical expression pattern of UPK1B resembled that in the urothelium and skin (FigureE8). In the SAE of smokers, UPK1B was detected in the areas free from squamous metaplasia, and its expression did not increase in association with mucous hyperplasia (Figure 4E), suggesting that molecular proximalization of the SAE in smokers may precede the development of classic smoking-induced histologic lesions.
EGF/EGFR Signaling in SAE BCs Promotes Smoking-associated D–P Repatterning
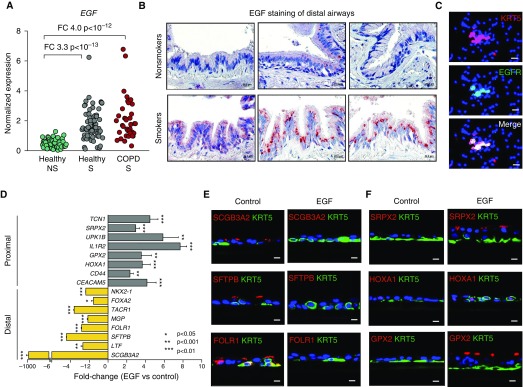
Because the EGF/EGFR pathway was predicted as the top upstream regulator of the P signature (Figure 2A), we focused on this signaling mechanism previously implicated in smoking-induced dysregulation of airway BCs (20). Application of EGFR inhibitor AG1478 to proximal airway BCs during differentiation of these cells in ALI (Figure E9A) decreased expression of the P-signature genes that were upregulated in the SAE of smokers, and it up-regulated D-signature genes in the epithelium derived from these cells (Figure E9B). Thus, EGFR signaling in proximal airway BCs promotes the proximal and suppresses the distal phenotype. This also suggests a possibility that smoking-associated D–P repatterning of the SAE might potentially be mediated by augmented EGF/EGFR signaling in this airway region. Indeed, EGF was up-regulated in the SAE of healthy smokers and COPD-S (Figure 5A), where it was detected in ciliated and secretory cells (Figure 5B). Similarly to proximal airways where EGFR was expressed largely in BCs (20, 26), EGFR was detected in a subset of BCs in the SAE (Figure 5C), suggesting the possibility that EGF/EGFR signaling might promote SAE proximalization by acting through SAE BCs.
Figure 5.
Epidermal growth factor (EGF)/epidermal growth factor receptor (EGFR) signaling in small airway epithelium (SAE) basal stem cells (BCs) promotes smoking-associated distal (D)-to-proximal (P) repatterning phenotype. (A) Microarray-based EGF gene expression in the SAE of healthy nonsmokers (NS; n = 63), healthy smokers (S; n = 73), and smokers with chronic obstructive pulmonary disease (COPD-S; n = 37). Fold changes (FCs) and Benjamini-Hochberg–corrected P values are shown. (B) Representative images showing EGF expression in the D airways of NS and S (n = 3 per group) using immunohistochemistry. (C) Immunofluorescence analysis of SAE brushing samples from healthy NS for BC marker keratin 5 (KRT5) and EGFR. In B and C, scale bar = 20 µm. (D) FC in expression of selected P- and D-signature genes in the epithelium derived from SAE BCs during 28 days of air–liquid interface culture in the presence of EGF (10 ng/ml) versus control BCs (n = 3 experiments). See Figures E9C and E9D for air–liquid interface time-course data and more examples. (E and F) Airway epithelium derived from control and EGF-treated SAE BCs as described in D analyzed using immunofluorescence for expression of indicated (E) D and (F) P signature genes (red), BC marker KRT5 (green), and nuclei (4′,6-diamidino-2-phenylindole [blue]). In E and F, scale bar = 10 µm. FOLR1 = folate receptor 1; GPX2 = glutathione peroxidase 2; HOXA1 = homeobox A1; SCGB3A2 = secretoglobin family 3A member 2; SFTPB = surfactant protein B; SRPX2 = sushi repeat-containing protein X-linked 2.
Indeed, SAE BCs treated with EGF during their differentiation in ALI generated the airway epithelium with markedly decreased expression of D-signature genes (Figures 5D and E9C), accompanied by a decreased proportion of cells expressing D-signature markers (Figure 5E), mimicking the phenotype observed in the SAE of smokers. Conversely, expression of the P-signature genes was up-regulated in the airway epithelium derived from EGF-stimulated SAE BCs (Figures 5D and E9D), accompanied by an increased proportion of cells expressing P-signature genes (Figure 5F). Hierarchical clustering analysis of airway brushing samples based on the expression of P- and D-signature features found to be reciprocally regulated by EGF/EGFR signaling in vitro revealed transition of the SAE from smokers toward the proximal-like airway pattern (Figure E9E). Similar to the effect of EGF, exposure of SAE BCs to CSE during their differentiation in ALI resulted in decreased expression of D-signature genes and up-regulation of P-signature genes (Figure E9F). The D–P repatterning phenotype induced by CSE was EGFR dependent because it was suppressed when SAE BCs were pretreated with the EGFR-selective inhibitor AG1478 prior to CSE exposure (Figure E9F). Together, these observations suggest that smoking-induced EGF shifts the SAE BC fate toward the proximal-like airway epithelium, mimicking the D–P repatterning phenotype observed in the SAE of smokers in vivo.
Discussion
Although the histologic heterogeneity of the human airway epithelium along the P–D axis is well established (3, 4, 6, 7), the transcriptional program that defines region-specific phenotypes in the adult human airway epithelium remained largely unknown. This study identifies the P–D patterning program in the adult human airways and demonstrates that this program is altered in the SAE of healthy smokers and COPD-S so that it loses its unique molecular identity and acquires the transcriptomic phenotype of the proximal airway epithelium.
The P–D patterning program identified in the present study included a number of molecular features implicated in lung development, including the genes coding for critical regulators of distal lung morphogenesis, such as SOX9, FOXA2, and GATA6 (27–31), which were enriched in the distal airway epithelium of the adult human lung. Further, NKX2-1, a TF that marks early lung endoderm progenitors (10, 32–35), was identified as an upstream regulator of the distal airway epithelial signature. This suggests that postnatal maintenance of the human SAE may involve mechanisms that control distal lung morphogenesis.
In addition to molecular features of lung epithelial development and differentiation, the D signature included a number genes coding for scavenger receptors, such as MARCO and MSR1, known to be expressed by macrophages, including alveolar macrophages (AMs) residing in the most distal alveolar compartment of the lung (36, 37). Although the origin of macrophage-related signals in the D signature identified in our study remains unclear, because more than 98% of cells in the analyzed samples were airway epithelial cells, it is possible that a small number of AMs may be present at the luminal SAE surface during their clearance from the distal lung region (38). Recently, expression of scavenger receptors has also been shown for airway epithelial cells (39), suggesting that both AMs and SAE cells may potentially contribute to this host defense mechanism in the distal airways.
By contrast, the proximal airway epithelium was enriched in molecular features related to ectodermal development and squamous differentiation. One explanation for this could be a higher proportion of BCs in this region (4, 7), a cell population that contributes to pseudostratified organization of the proximal airway epithelium and exhibits a transcriptomic pattern remarkably similar to that of ectoderm-derived stratified epithelia (21, 40). Consistent with this explanation, TP63, a BC-related TF associated with differentiation of stratified epithelia (41, 42), was identified as the top enriched TF in the proximal airway epithelium, followed by HOXA1 and HOXB3, implicated in embryonic patterning of the hindbrain, another ectoderm-derived tissue (43).
Although identified D and P signatures included known features of cell types enriched in corresponding anatomic regions, such as secretory cells and BCs more abundant in the distal and proximal airways, respectively (4, 7), this was not the case for a number of other signature genes. For example, the P-signature genes UPK1B, SRPX2, and GPX2 were expressed in a subset of ciliated cells, an epithelial cell type dominating in both airway regions of the human lung (3). Thus, in addition to being a transcriptomic “mirror” of differences in cellular composition along the P–D axis, the identified signatures contained information about the unique molecular features of individual cell types in different airway regions.
Another characteristic of the proximal airway epithelium was enrichment of molecular features related to oxidative stress, xenobiotic metabolism, and nicotine degradation, suggesting that this anatomic compartment could be better equipped than the SAE for defense against environmental oxidants and toxins. Lower expression levels of antioxidant genes in the SAE in steady state might explain, at least in part, why this anatomic region is particularly susceptible to smoking-induced changes relevant to COPD pathogenesis (12, 13). Alternatively, SAE may be naturally protected from the oxidative stress response, and its induction by smoking could drive COPD-relevant pathologic changes in this region, as previously proposed (14).
Whereas, as described above, the normal human SAE maintains its physiologic “distal” molecular pattern, in healthy smokers, and more so in COPD-S, it loses its region-associated transcriptomic identity and acquires the molecular phenotype of the proximal airway epithelium. We called this process distal-to-proximal (D–P) repatterning because it represents a reversal of the physiologic P–D patterning. Given that COPD is a smoking-induced lung disease in which airway obstruction results primarily from the remodeling of distal airways (12, 13), loss of the SAE transcriptomic identity as a result of smoking-induced D–P repatterning could be relevant to the pathogenesis of small airway disordering in this disease.
Similarly to other smoking-associated transcriptome changes, such as activation of the oxidative stress program (14) or suppression of the apical junctional complex program (16), healthy smokers were heterogeneous with respect to the degree to which the SAE transcriptome was repatterned. Although specific factors that contribute to this heterogeneity and their prognostic relevance require further investigation, age (≥45 yr) and FEV1/FVC ratio (≤80%) were associated with higher degree of D–P repatterning of SAE in healthy smokers in our study. This suggests that SAE transcriptomic repatterning may represent a subclinical phenotype associated with aging and COPD development in susceptible smokers.
It has been shown that the SAE of smokers loses SCGB-expressing secretory cells (44) and takes on some features of the proximal airways, such as increased numbers of BCs and mucus-producing cells (45). Although, as mentioned above, the P–D signatures identified in the present study capture a number of features that reflect regional differences in cellular composition, and smoking-suppressed distal signature genes are indeed enriched in secretory cells, detailed morphologic examination revealed that proximalization of SAE in smokers occurs independent from classic smoking-associated histologic changes (i.e., mucous cell hyperplasia and squamous metaplasia). This suggests the possibility that D–P repatterning of the SAE, particularly when observed in healthy smokers, may precede the development of smoking-associated, COPD-relevant histologic lesions in this anatomic region.
To explore the mechanism of smoking-associated D–P repatterning, we focused on SAE BCs, the stem cells that maintain the unique phenotype of this anatomic region (5), and the EGF/EGFR pathway identified in the present study as the top upstream regulator of the proximal airway signature. In a previous study, we showed that smoking up-regulates EGF in the large airway epithelium and that stimulation of large airway BCs with EGF shifts BC differentiation toward the epithelial-to-mesenchymal transition–like squamous phenotype (20). In the present study, we found that EGF is also up-regulated in the SAE of smokers and that stimulation of SAE BCs with EGF induced the D–P repatterning similar to that observed in the SAE of smokers in vivo. Inhibition of EGFR signaling in large airway BCs switched their differentiation from the proximal to the distal phenotype, whereas EGFR inhibition in SAE BCs suppressed smoking-induced D–P repatterning. These data suggest that EGFR signaling promotes the proximal airway epithelial pattern, and, when activated in the distal airways by smoking-associated factors such as EGF, shifts the fate of SAE BCs toward the proximal differentiation phenotype.
Together, the results of the present study describe the P–D patterning program in the adult human airway epithelium, which is reversed in the SAE of COPD-S in a smoking-dependent manner via a process that involves exaggerated EGF/EGFR signaling in SAE BCs. The latter observation further supports the concept that SAE BCs modified by the stress of cigarette smoking may serve as the cell of origin of small airway remodeling in COPD (46). Targeting augmented EGF/EGFR signaling in SAE BCs may represent a novel therapeutic approach to prevent small airway disordering in COPD via restoration of the physiologic P–D patterning.
Acknowledgments
Acknowledgment
The authors thank N. Mohamed for help with manuscript preparation.
Footnotes
Supported by National Institutes of Health grants R01 HL107882 and R01 HL1189541 (R.G.C.); R01 HL123544 and R01 HL127393 (R.S.); and UL1 TR000457 and UL1 RR0244143. R.S. was also supported in part by the Parker B. Francis Foundation and Qatar National Research Fund (NPRP 5-400-3-107).
Author Contributions: R.S. and R.G.C.: conceived of the study; R.S., J.Y., and R.G.C.: designed the experiments; R.S., J.Y., W.-L.Z., T.F., I.W.C., K.G., and B.L.: performed the experiments; R.S., M.R.S., R.J.K., Y.S.-B., J.S., and R.G.C.: contributed to collection and analysis of clinical data; R.S., J.Y., W.-L.Z., T.F., M.R.S., Y.S.-B., and R.G.C.: analyzed the data; R.S. and R.G.C.: wrote the manuscript; all authors: approved the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201608-1672OC on March 27, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Hajj R, Baranek T, Le Naour R, Lesimple P, Puchelle E, Coraux C. Basal cells of the human adult airway surface epithelium retain transit-amplifying cell properties. Stem Cells. 2007;25:139–148. doi: 10.1634/stemcells.2006-0288. [DOI] [PubMed] [Google Scholar]
- 2.Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, Hogan BL. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crystal RG, Randell SH, Engelhardt JF, Voynow J, Sunday ME. Airway epithelial cells: current concepts and challenges. Proc Am Thorac Soc. 2008;5:772–777. doi: 10.1513/pats.200805-041HR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rock JR, Randell SH, Hogan BL. Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis Model Mech. 2010;3:545–556. doi: 10.1242/dmm.006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar PA, Hu Y, Yamamoto Y, Hoe NB, Wei TS, Mu D, Sun Y, Joo LS, Dagher R, Zielonka EM, et al. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell. 2011;147:525–538. doi: 10.1016/j.cell.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercer RR, Russell ML, Roggli VL, Crapo JD. Cell number and distribution in human and rat airways. Am J Respir Cell Mol Biol. 1994;10:613–624. doi: 10.1165/ajrcmb.10.6.8003339. [DOI] [PubMed] [Google Scholar]
- 7.Boers JE, Ambergen AW, Thunnissen FB. Number and proliferation of basal and parabasal cells in normal human airway epithelium. Am J Respir Crit Care Med. 1998;157:2000–2006. doi: 10.1164/ajrccm.157.6.9707011. [DOI] [PubMed] [Google Scholar]
- 8.Morrisey EE, Hogan BL. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev Cell. 2010;18:8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domyan ET, Sun X. Patterning and plasticity in development of the respiratory lineage. Dev Dyn. 2011;240:477–485. doi: 10.1002/dvdy.22504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herriges M, Morrisey EE. Lung development: orchestrating the generation and regeneration of a complex organ. Development. 2014;141:502–513. doi: 10.1242/dev.098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardoso WV. Lung morphogenesis revisited: old facts, current ideas. Dev Dyn. 2000;219:121–130. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1053>3.3.co;2-8. [DOI] [PubMed] [Google Scholar]
- 12.Hogg JC, Chu F, Utokaparch S, Woods R, Elliott WM, Buzatu L, Cherniack RM, Rogers RM, Sciurba FC, Coxson HO, et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N Engl J Med. 2004;350:2645–2653. doi: 10.1056/NEJMoa032158. [DOI] [PubMed] [Google Scholar]
- 13.McDonough JE, Yuan R, Suzuki M, Seyednejad N, Elliott WM, Sanchez PG, Wright AC, Gefter WB, Litzky L, Coxson HO, et al. Small-airway obstruction and emphysema in chronic obstructive pulmonary disease. N Engl J Med. 2011;365:1567–1575. doi: 10.1056/NEJMoa1106955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tilley AE, O’Connor TP, Hackett NR, Strulovici-Barel Y, Salit J, Amoroso N, Zhou XK, Raman T, Omberg L, Clark A, et al. Biologic phenotyping of the human small airway epithelial response to cigarette smoking. PLoS One. 2011;6:e22798. doi: 10.1371/journal.pone.0022798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaykhiev R, Otaki F, Bonsu P, Dang DT, Teater M, Strulovici-Barel Y, Salit J, Harvey BG, Crystal RG. Cigarette smoking reprograms apical junctional complex molecular architecture in the human airway epithelium in vivo. Cell Mol Life Sci. 2011;68:877–892. doi: 10.1007/s00018-010-0500-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–75. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang JT, Nevins JR. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 2006;22:2926–2933. doi: 10.1093/bioinformatics/btl483. [DOI] [PubMed] [Google Scholar]
- 19.Shaykhiev R, Wang R, Zwick RK, Hackett NR, Leung R, Moore MA, Sima CS, Chao IW, Downey RJ, Strulovici-Barel Y, et al. Airway basal cells of healthy smokers express an embryonic stem cell signature relevant to lung cancer. Stem Cells. 2013;31:1992–2002. doi: 10.1002/stem.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaykhiev R, Zuo WL, Chao I, Fukui T, Witover B, Brekman A, Crystal RG. EGF shifts human airway basal cell fate toward a smoking-associated airway epithelial phenotype. Proc Natl Acad Sci USA. 2013;110:12102–12107. doi: 10.1073/pnas.1303058110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hackett NR, Shaykhiev R, Walters MS, Wang R, Zwick RK, Ferris B, Witover B, Salit J, Crystal RG. The human airway epithelial basal cell transcriptome. PLoS One. 2011;6:e18378. doi: 10.1371/journal.pone.0018378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Staudt MR, Buro-Auriemma LJ, Walters MS, Salit J, Vincent T, Shaykhiev R, Mezey JG, Tilley AE, Kaner RJ, Ho MW, et al. Airway basal stem/progenitor cells have diminished capacity to regenerate airway epithelium in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2014;190:955–958. doi: 10.1164/rccm.201406-1167LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fulcher ML, Randell SH. Human nasal and tracheo-bronchial respiratory epithelial cell culture. Methods Mol Biol. 2013;945:109–121. doi: 10.1007/978-1-62703-125-7_8. [DOI] [PubMed] [Google Scholar]
- 24.Brekman A, Walters MS, Tilley AE, Crystal RG. FOXJ1 prevents cilia growth inhibition by cigarette smoke in human airway epithelium in vitro. Am J Respir Cell Mol Biol. 2014;51:688–700. doi: 10.1165/rcmb.2013-0363OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuo WL, Yang J, Gomi K, Chao I, Crystal RG, Shaykhiev R. EGF-amphiregulin interplay in airway stem/progenitor cells links the pathogenesis of smoking-induced lesions in the human airway epithelium. Stem Cells. 2017;35:824–837. doi: 10.1002/stem.2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voynow JA, Fischer BM, Roberts BC, Proia AD. Basal-like cells constitute the proliferating cell population in cystic fibrosis airways. Am J Respir Crit Care Med. 2005;172:1013–1018. doi: 10.1164/rccm.200410-1398OC. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Hogan BL. Differential gene expression in the distal tip endoderm of the embryonic mouse lung. Gene Expr Patterns. 2002;2:229–233. doi: 10.1016/s1567-133x(02)00057-1. [DOI] [PubMed] [Google Scholar]
- 28.Rockich BE, Hrycaj SM, Shih HP, Nagy MS, Ferguson MA, Kopp JL, Sander M, Wellik DM, Spence JR. Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proc Natl Acad Sci USA. 2013;110:E4456–E4464. doi: 10.1073/pnas.1311847110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wan H, Kaestner KH, Ang SL, Ikegami M, Finkelman FD, Stahlman MT, Fulkerson PC, Rothenberg ME, Whitsett JA. Foxa2 regulates alveolarization and goblet cell hyperplasia. Development. 2004;131:953–964. doi: 10.1242/dev.00966. [DOI] [PubMed] [Google Scholar]
- 30.Yang H, Lu MM, Zhang L, Whitsett JA, Morrisey EE. GATA6 regulates differentiation of distal lung epithelium. Development. 2002;129:2233–2246. doi: 10.1242/dev.129.9.2233. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Rath N, Hannenhalli S, Wang Z, Cappola T, Kimura S, Atochina-Vasserman E, Lu MM, Beers MF, Morrisey EE. GATA and Nkx factors synergistically regulate tissue-specific gene expression and development in vivo. Development. 2007;134:189–198. doi: 10.1242/dev.02720. [DOI] [PubMed] [Google Scholar]
- 32.Martis PC, Whitsett JA, Xu Y, Perl AK, Wan H, Ikegami M. C/EBPα is required for lung maturation at birth. Development. 2006;133:1155–1164. doi: 10.1242/dev.02273. [DOI] [PubMed] [Google Scholar]
- 33.Roos AB, Berg T, Barton JL, Didon L, Nord M. Airway epithelial cell differentiation during lung organogenesis requires C/EBPα and C/EBPβ. Dev Dyn. 2012;241:911–923. doi: 10.1002/dvdy.23773. [DOI] [PubMed] [Google Scholar]
- 34.Yuan B, Li C, Kimura S, Engelhardt RT, Smith BR, Minoo P. Inhibition of distal lung morphogenesis in Nkx2.1(−/−) embryos. Dev Dyn. 2000;217:180–190. doi: 10.1002/(SICI)1097-0177(200002)217:2<180::AID-DVDY5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 35.Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox CH, Ward JM, Gonzalez FJ. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- 36.Gordon S, Plüddemann A, Martinez Estrada F. Macrophage heterogeneity in tissues: phenotypic diversity and functions. Immunol Rev. 2014;262:36–55. doi: 10.1111/imr.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desch AN, Gibbings SL, Goyal R, Kolde R, Bednarek J, Bruno T, Slansky JE, Jacobelli J, Mason R, Ito Y, et al. Flow cytometric analysis of mononuclear phagocytes in nondiseased human lung and lung-draining lymph nodes. Am J Respir Crit Care Med. 2016;193:614–626. doi: 10.1164/rccm.201507-1376OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowden DH. The alveolar macrophage. Environ Health Perspect. 1984;55:327–341. doi: 10.1289/ehp.8455327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dieudonné A, Torres D, Blanchard S, Taront S, Jeannin P, Delneste Y, Pichavant M, Trottein F, Gosset P. Scavenger receptors in human airway epithelial cells: role in response to double-stranded RNA. PLoS One. 2012;7:e41952. doi: 10.1371/journal.pone.0041952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fuchs E. Scratching the surface of skin development. Nature. 2007;445:834–842. doi: 10.1038/nature05659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Senoo M, Pinto F, Crum CP, McKeon F. p63 is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 42.Truong AB, Khavari PA. Control of keratinocyte proliferation and differentiation by p63. Cell Cycle. 2007;6:295–299. doi: 10.4161/cc.6.3.3753. [DOI] [PubMed] [Google Scholar]
- 43.Alexander T, Nolte C, Krumlauf R. Hox genes and segmentation of the hindbrain and axial skeleton. Annu Rev Cell Dev Biol. 2009;25:431–456. doi: 10.1146/annurev.cellbio.042308.113423. [DOI] [PubMed] [Google Scholar]
- 44.Lumsden AB, McLean A, Lamb D. Goblet and Clara cells of human distal airways: evidence for smoking induced changes in their numbers. Thorax. 1984;39:844–849. doi: 10.1136/thx.39.11.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baraldo S, Turato G, Saetta M. Pathophysiology of the small airways in chronic obstructive pulmonary disease. Respiration. 2012;84:89–97. doi: 10.1159/000341382. [DOI] [PubMed] [Google Scholar]
- 46.Shaykhiev R, Crystal RG. Early events in the pathogenesis of chronic obstructive pulmonary disease: smoking-induced reprogramming of airway epithelial basal progenitor cells. Ann Am Thorac Soc. 2014;11(Suppl 5):S252–S258. doi: 10.1513/AnnalsATS.201402-049AW. [DOI] [PMC free article] [PubMed] [Google Scholar]