Abstract
Rabbits and monkeys immunized with HIV type 1 (HIV-1) native-like BG505 SOSIP.664 (BG505s) glycoprotein trimers are known to induce antibodies that can neutralize the autologous tier-2 virus. Here, we assessed the induction of HIV-1 trimer binding and neutralizing antibody (nAb) titres when BG505s trimers were also delivered by non-replicating simian (chimpanzee) adenovirus and non-replicating poxvirus modified vaccinia virus Ankara (MVA) vaccine vectors. First, we showed that approximately two-thirds and one-third of the trimers secreted from the ChAdOx1.BG505s (C) and MVA.BG505s (M) vaccine-infected cells, respectively, were cleaved and in a native-like conformation. Rabbits were immunized intramuscularly with these vaccine vectors and in some cases boosted with ISCOMATRIX™–adjuvanted BG505s protein trimer (P), using CCC, MMM, PPP, CPP, MPP and CMP vaccine regimens. We found that the peak trimer-binding antibody and tier-1A and autologous tier-2 nAb responses induced by the CC, CM, PPP, CPP, MPP and CMP regimens were comparable, although only PPP induced autologous tier-2 nAbs in all the immunized animals. Three animals developed weak heterologous tier-2 nAbs. These results demonstrate that ChAdOx1 and MVA vectors are useful delivery modalities for not only T-cell, but also antibody vaccine development.
Introduction
Vaccines remain the best solution to ending of the HIV-1/AIDS epidemic. Ideally, a vaccine would induce protective levels of broadly neutralizing antibodies (bnAbs) against multiple HIV-1 variants [1, 2]. At least initially in an immunization regimen, while bnAb induction is sub-optimal, antibody responses may need to be supplemented by effective cytotoxic T-lymphocyte responses [3, 4]. Elicitation of either of these arms of immunity continues to pose major scientific challenges.
The envelope glycoprotein (Env) trimer is the only relevant target for antibodies on HIV-1 virions. bnAbs emerge in 20–30% of HIV-1-infected individuals after two or more years of infection, and multiple lineages have now been isolated [1, 5–19]. In these individuals, bnAbs do not provide clinical benefit because they are slow to mature and once they appear, the viruses can still escape [20–22]. Passive transfer of bnAbs can achieve sterilizing immunity in rhesus macaques, if sufficient concentrations are present at the time of or immediately after the virus challenge [23–30]. For active immunization by Env-based vaccines, the key obstacle is the need to induce bnAbs against tier-2 viruses, which dominate human transmission and are relatively neutralization-insensitive. Nevertheless, increasing understanding of the Env structure and its glycan shield, and the ability to follow virus and antibody co-evolution during infection and evolutionary convergence of bnAb specificities among some patients [31, 32] are cumulative, if gradual, steps towards the induction of bnAbs.
One approach to the problem of inducing bnAbs is the design platform represented by recombinant native-like soluble trimers, for which the prototype is the BG505 SOSIP.664 construct [33]. These trimers, from hereon referred to as BG505s, consist of furin-cleaved gp120 and gp41 ectodomains, are stabilized by an inter-subunit disulfide bond and a point substitution in gp41, and display multiple human bnAb epitopes [33–36]. In rabbit and macaque immunizations, BG505s trimers adjuvanted with ISCOMATRIX™ induced cross-reactive nAbs against tier-1 viruses and nAbs against the autologous BG505.T332N tier-2 virus, although nAbs against heterologous tier-2 viruses were only elicited sporadically and at low titres [37–39]. The BG505s trimer design is capable of improvement from the perspectives of trimer engineering and delivery [40].
Strategies combining simian (chimpanzee) adenoviruses and poxvirus modified vaccinia virus Ankara (MVA) into heterologous prime-boost regimens have induced both high-titre antibody [41–46] and T-cell [47–50] responses against the shared transgene products. Here, we assessed the in vivo expression of the BG505s trimers from the viral vector-infected cells, and immunogenicity of the viral vaccines alone and with boosting by adjuvanted BG505s trimers. We measured the titres of trimer-binding and virus-neutralizing Abs in the various regimens. We report that vector-expressed BG505s trimers are able to induce nAb titres that can be increased by boosting with soluble protein trimers.
Materials and methods
Construction of ChAdOx1.BG505s
The E1/E2-deleted vaccine vector ChAdOx1 is derived from chimpanzee adenovirus isolate Y25 of group E [51]. For the generation of recombinant ChAdOx1, the BG505 SOSIP.664 gene [33] was inserted into the E1 locus of the adenovirus genome (integrated in bacterial artificial chromosome) under the control of human cytomegalovirus immediate early promoter. The ChAdOx1.BG505s vaccine was rescued by transfection of HEK 293A T-Rex cells with the PmeI-excised ChAdOx1.BG505s genome. The transgene presence and absence of contaminating empty parental adenovirus in the virus stock was confirmed by PCR, the vaccine was titred and stored at –80°C until use.
Construction of MVA.BG505s
To generate MVA.BG505s, the BG505 SOSIP.664 gene [33] was subcloned under control of the modified H5 promoter. Through homologous recombination in chicken embryo fibroblast cells, the expression cassette was directed into the thymidine kinase locus of the MVA genome. Initially co-inserted green fluorescent protein marker was removed by trans-dominant recombination to generate markerless vaccine MVA.BG505s [52]. The virus was plaque-purified, expanded, purified on a 36% sucrose cushion, titred and stored at –80°C until use.
Production of BG505s trimers
BG505 SOSIP.664 trimers were produced and purified as reported previously [33, 36, 53]. Proteins were expressed in HEK 293T cells (i.e. have a human cell glycosylation profile) by transient transfection and purified using a 2G12 mAb affinity column followed by size-exclusion chromatography and stored at -80°C until use.
In vitro expression of BG505s trimers from vaccine-infected cells
HEK 293T cells were grown in 6-well plates to 70% confluence and infected with either ChAdOx1.BG505s or MVA.BG505s at a multiplicity of infection (MOI) of 5 or 50 for 1 h. The cells were then washed and grown for 24 or 48 h, and the culture supernatants were harvested and stored in 10% FBS at 4°C until use. To monitor the release of trimers into the cell culture supernatants, we developed a two-stage capture ELISA protocol.
In the first stage, high-binding ELISA plates (Greiner Bio-One) were coated with 4 μg/ml of capture mAb 2G12 in PBS at 4°C overnight. After washing 3x with PBS+0.05% Tween 20 (PBST), the wells were blocked using 200 μl/well of 2% BSA (Sigma) in PBST at room temperature (RT) for 1 h and washed 1x with PBST. A titration curve was prepared by diluting purified BG505s trimers of a known concentration in culture supernatant from untransfected cells. The Env proteins present in the various culture supernatants were captured onto the ELISA plates via the 2G12 mAb at RT for 2 h and the plates were washed 3x in PBST. Bound trimer was detected with biotinylated mAb VRC01, washed 3x with PBST, and labelled with horseradish peroxidase (HRP)-conjugated streptavidin (1:2000; GE Healthcare). Following 3 PBST washes, the colorimetric endpoint was obtained using the 1-Step ultra tetramethylbenzidine substrate (Thermo Scientific) until a signal of ~1.5 OD450 units was generated. Color development was stopped with 0.5 M sulfuric acid, and the 450-nm and 570-nm absorptions were recorded. After background subtraction, a “one-site-specific-binding-with-hill-slope” curve was fitted to the control titration curve using Prism v6 (GraphPad Software) and the concentrations of Env proteins present in the various culture supernatants were extrapolated.
In the second stage, the capture ELISA involved the same protocol, but with either 2G12 or PGT151 as the capture mAb and biotinylated PGT145 as the detection mAb. A series of samples with the same overall Env concentration were prepared, which were made up of purified trimeric and monomeric BG505 species mixed at different ratios from monomer only (0% correctly folded) to trimer only (100% correctly folded). Data analysis was performed as in the first stage.
Immunization of rabbits by modified HIV-1 Env
Rabbit immunization and blood sampling was carried out at MediTox, s.r.o. (The Czech Republic). The rabbits (10- to 14-week-old randomized male/female HYLA; http://www.eurolap.fr) were given either 5x1010 virus particles of ChAdOx1.BG505s, 108 plaque-forming units of MVA.BG505s or 30 μg of BG505s trimer protein adjuvanted in 75 units of ISCOMATRIX™, a saponin-based adjuvant obtained from CSL (Kankakee, Illinois, USA), per dose. All doses were administered intramuscularly. Serum was isolated from blood collected biweekly between weeks 0 and 38, and stored at -80°C until use. All animals were observed for clinical signs, morbidity or mortality once a day during the acclimatization, immunization and wash-out periods. Nine animals died during the study (S1 Table) for vaccine-unrelated causes as judged by the study vet. The study was carried out in full compliance with EMA, The European Agency for the Evaluation of Medicinal Products, Human Medicines Evaluation Unit, CPMP/SWP/465/95 and relevant Standard Operating Procedures of MediTox s.r.o.
Capture ELISA for quantifying binding Abs in rabbit sera
The 2G12 mAb (2.0 μg/ml) was coated onto ELISA plates (Greiner Bio-One) at 4°C during an overnight incubation. The wells were washed 3x in PBS-T and blocked with 2% BSA in PBS plus 0.05% Tween. After washing 1x in PBS-T, BG505s (0.2 μg/ml) was added to all wells at RT for 2 h. After washing 3x in PBS-T, serial 5-fold dilutions of sera starting from 1:100 were added to wells at RT for 2 h, the plates were washed 5x in PBS-T, incubated with 50 μl/well of secondary mouse anti-rabbit-HRP antibody (Millipore) diluted 1:15000 in 1% BSA in PBS at RT for 1 h, and washed 3x with PBS-T. The colorimetric reaction was carried out as described above. Note that this ELISA format was not identical to the one used in previous rabbit immunization studies [33–36]. Hence, the absolute titres may not be comparable across experiments.
HIV-1-neutralization assay
A validated TZM-bl neutralization assay using Env-pseudotyped viruses was described previously [54]. The test panel used here included the tier-1 viruses MN.3 and MW965.26, the autologous tier-2 virus BG505ΔCT/T332N and a global panel of heterologous tier-2 viruses (25710–2.43, TRO.11, BJOX0020_00.03.2, X1632-S2-B10, Ce1176_A3, 246-F3_C10_2, CH119.10, Ce70301_0217_B6 and CNE55) [55]. The glycan knock-in mutant viruses BG505ΔCT/T332N.S241N, BG505ΔCT/T332N.P291T and BG505ΔCT/T332N.S241N.P291T were used to assess the importance of glycan holes for neutralization [38]. For additional information on the assay and related protocols, see www.hiv.lanl.gov/content/nab-reference-strains/html/home.htm. No signal was detected when a murine leukemia virus-pseudotyped virus was used to estimate non-specific neutralization.
Statistical analysis
Statistical analyses were performed using Prism v6 (GraphPad Software). Two-group comparisons were performed with Mann-Whitney U test. Multi-group comparisons were performed with Kruskal-Wallis test and Dunn’s multiple comparison post-test. Correlation between neutralization of different viruses was analysed by using the Spearman nonparametric method. A P value (two-tailed) <0.05 was considered significant.
Results
Construction of the ChAdOx1.BG505s and MVA.BG505s vaccines
Two non-replicating virus-vectored vaccines ChAdOx1.BG505a and MVA.BG505s were constructed as described previously [52, 56]. The non-replicating ChAdOx1 vector is derived from a group E adenovirus Y25 with low human seroprevalence [51]. The parental non-replicating MVA originates directly from Professor Mayr, passage 575 dated 14-12-1983. Both vaccines express BG505 SOSIP.664 gp140 (ref. [33]) with the natural HIV-1 leader sequence replaced by that of the human tissue plasminogen activator; they do not include a gene expressing the furin protease, but instead rely on endogenous furin-family proteases for trimer maturation [57].
In vitro secreted BG505s trimers from vaccine-infected human cells maintain their integrity
To assess whether native-like BG505s trimers could be expressed from the virus vectors, we used cultured human HEK 293T cells as a model system. The cells were infected with each virus at multiplicity of infection (MOI) of 0 (no infection), 10, 33, 100, 333 and 1000 for 48 h. We first analysed the BG505 Env proteins released into the culture supernatants by non-denaturing gel electrophoresis and Western blotting using the glycan-specific 2G12 mAb as the detection antibody (Fig 1A). A 2G12-reactive protein band of relative molecular mass of ~600 kDa that corresponds to an Env trimer was detectable in the supernatants of cells infected with the ChAdOx1.BG505s virus at MOIs >100 (Fig 1A), but we were unable to detect any Env protein expression by Western blot at any MOI and at any time point in the MVA.BG505s-infected cell supernatants. This is probably because the poxvirus promoter is much weaker than the cytomegalovirus immediate early promoter employed in the recombinant adenovirus.
Fig 1. Expression of BG505s trimers in tissue culture.
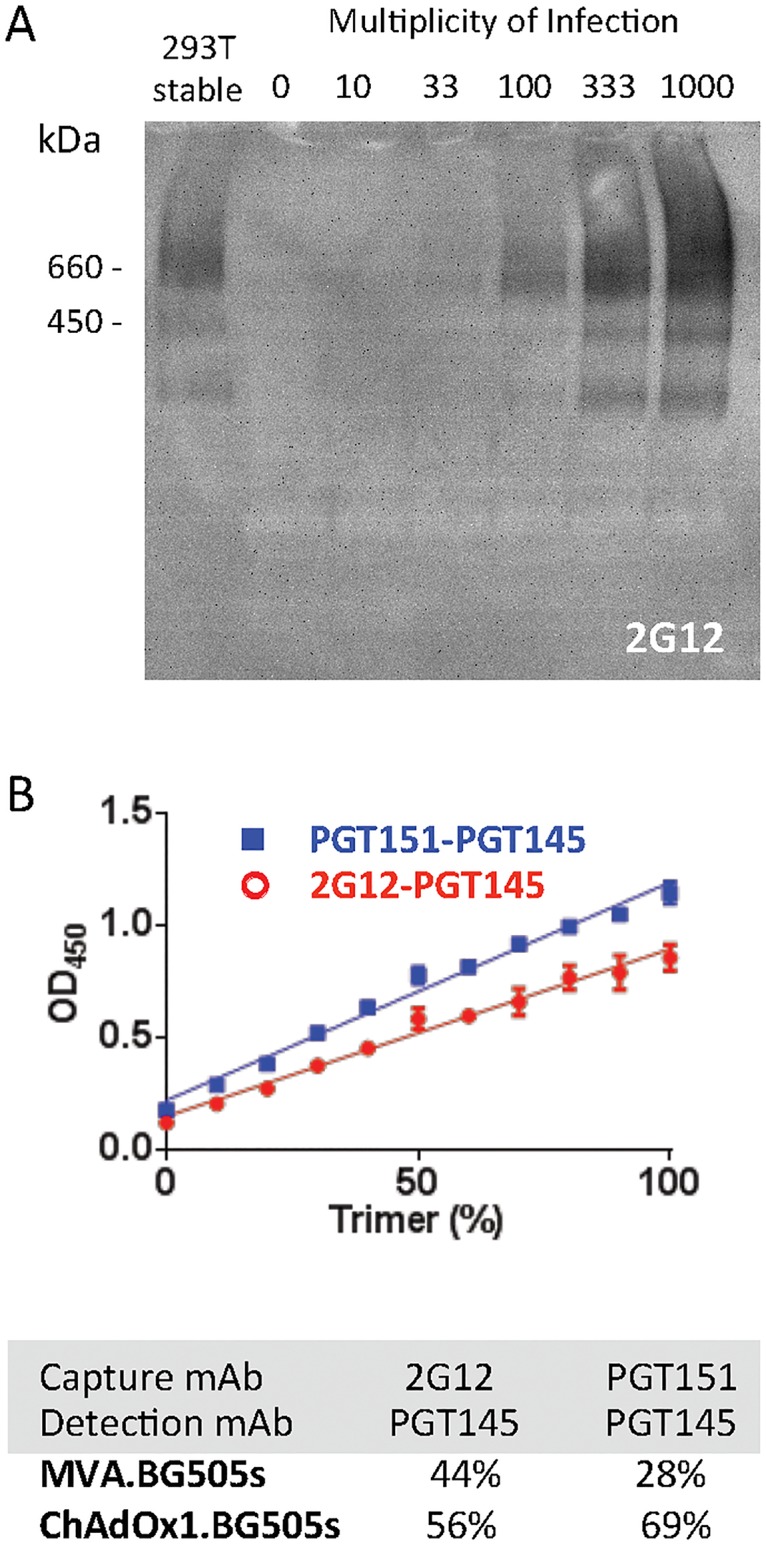
(A) Monolayers of HEK 293T cells were infected with ChAdOx1.BG505s at the indicated MOIs for 48 h. The proteins in the tissue culture supernatants were separated by non-denaturing polyacrylamide gel electrophoresis and transferred onto a nitrocellulose membrane. BG505 Env proteins were detected using the glycan-specific 2G12 mAb as the primary antibody. As a positive control, supernatant from a HEK 293T stable cell line that expresses fully cleaved BG505s trimers was analysed in the first lane. The relative molecular masses of 600 and 450 kDa are indicated. (B) To assess the percentage of correctly folded trimers in the ChAdOx1.BG505s- and MVA.BG505s-infected cell culture supernatants, the ELISA plates were first coated with either 2G12 or PGT151 Env-specific bnAbs and the captured native-like, furin-cleaved trimers were detected using PGT145 as the secondary mAb. To generate the standard curve used for trimer quantification (top), gp140 monomers and trimers purified from stably transfected cells were mixed at different ratios, but at a constant total concentration. The latter values were determined using a different, non-trimer specific capture ELISA (see Methods section). The percentages of native-like BG505s trimer present in the virus-infected cell culture supernatants are shown for the indicated antibody combinations (bottom).
Next, we assessed whether appropriately folded trimers were present in the culture supernatants by using a capture ELISA based on the properties of the PGT145 and PGT151 bnAbs. PGT145 recognizes an apex-specific quaternary epitope and PGT151 a gp120-gp41-specific quaternary epitope, and both bind only to native-like, furin-cleaved trimers. In this experiment, HEK 293T cells were infected with either vaccine at MOIs of 5 and 50 for 24 and 48 h. Using a 2G12-capture, biotinylated-VRC01 detection ELISA format, which does not distinguish between native-like trimers and other forms of Env, we estimated that the highest overall level of Env expression measured in the ChAdOx1.BG505s cultures was ~360 ng/ml at 48 h after infection at an MOI of 50, whereas for the MVA.BG505s cultures, the maximum expression at the same 48-h time-point was only ~4.5 ng/ml at an MOI of 5. The ~100-fold greater expression in the ChAdOx1.BG505s culture compared to MVA.BG505s concurs with the Western blotting data described above. Next, we used ELISA to estimate the fraction of native-like trimers in the culture supernatants by combining either 2G12- or PGT151-capture with biotin-PGT145 detection, thereby making use of the quaternary epitope specificity of these bnAbs. In a control experiment, we mixed purified “non-native” monomeric BG505 SOSIP.664 proteins with purified native-like trimers at different ratios, but a constant overall Env concentration. The use of the mixtures showed that there was a linear relationship between the proportion of native-like trimers in the sample and the ELISA signal (OD450). We used the resulting titration curve to estimate that approximately two-thirds and one-third of the total Env proteins in the ChAdOx1.BG505s and MVA.BG505s supernatants, respectively, were appropriately folded trimers (Fig 1B).
ChAdOx1.BG505s delivers a stronger prime than an adjuvanted protein trimer
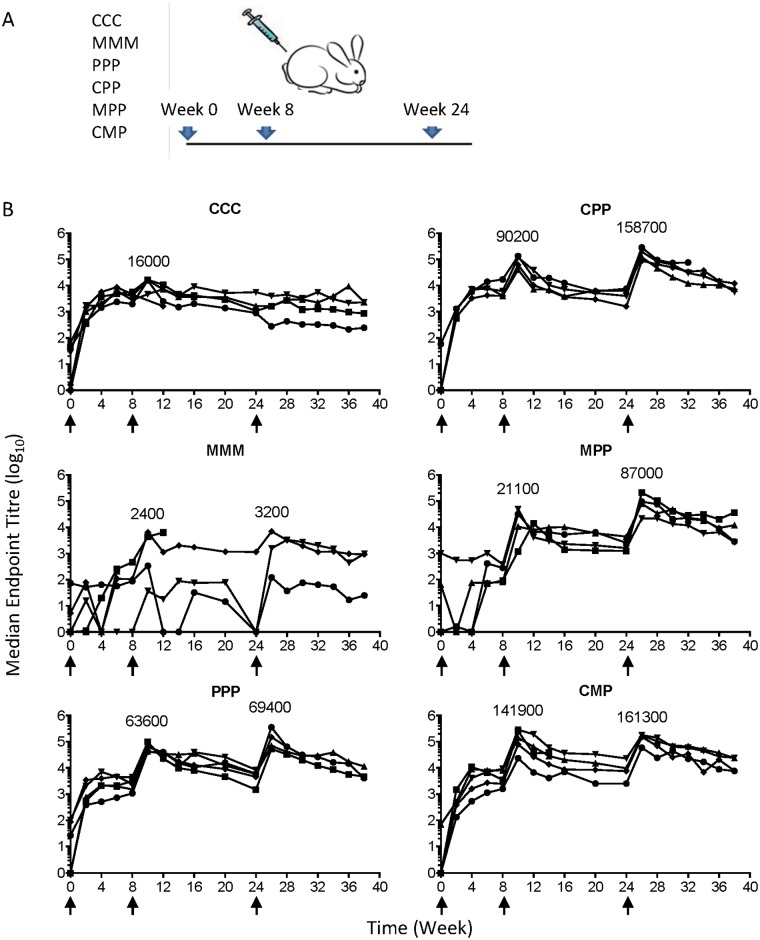
To assess immunogenicity, six groups of rabbits were immunized with the ChAdOx1.BG505s (C), MVA.BG505s (M) or HEK 293T-produced, ISCOMATRIX™-adjuvanted BG505s protein trimer (P) vaccines in either homologous or heterologous regimens of CCC, MMM, PPP, CPP, MPP and PPP (Fig 2A). For the PPP group, the 0-8-24 week immunization protocol, the 30-μg amount of trimer per dose, its source, and the adjuvant were all similar to those used in previous rabbit studies [37–39]. Serum samples were drawn at 2- to 4-week intervals until week 38, which was 14 weeks after the last immunization. In the first instance, we used a capture ELISA to monitor the induction kinetics of antibodies reactive with the BG505s trimer (Fig 2B, S1 Table). Comparing the peak trimer-binding titres to gauge the antibody response to a single C, M or P dose, the vaccine modalities ranked in the order of C > P > M with median reciprocal titres of 6300 (n = 13), 3100 (n = 5) and 100 (n = 9), respectively (Table 1 and S1 Table). The differences in peak responses induced by a single C, M and P administration were highly significant (P < 0.0001) (Kruskal-Wallis test), as was C vs P (P < 0.0001) (Dunn’s multiple comparison test).
Fig 2. Kinetics of BG505s trimer-binding antibody responses induced in rabbits by various immunization regimens.
(A) Groups of 5 rabbits were immunized with ChAdOx1.BG505s (C), MVA.BG505s (P) and ISCOMATRIX™-adjuvanted BG505s protein trimer (P) candidate vaccines, as depicted. The animals were immunized at weeks 0, 8 and 24 (arrows), and bled regularly. (B) Reciprocal endpoint titres of the BG505s trimer-binding antibodies in rabbit sera were determined by capture ELISA. The trimer-binding antibody titres at each time point are shown for individual rabbits, and the median values for group-peak time points are shown above each peak. Note that the time points at which the peak titres were measured differed among the rabbits in each group, and that some rabbits died for protocol-unrelated reasons (S1 Table).
Table 1. Peak reciprocal anti-trimer Ab end-point titres in rabbit sera, ranked by immunization group.
| Rank | Regimen | Median | Range | n |
|---|---|---|---|---|
| 1 | CMP | 161300 | 58900–179400 | 4 |
| 2 | CPP | 158700 | 88400–285600 | 4 |
| 3 | CM | 141900 | 23700–287500 | 5 |
| 4 | CP | 90200 | 40300–133600 | 4 |
| 5 | MPP | 87000 | 21600–210200 | 4 |
| 6 | PPP | 69400 | 52300–287500 | 5 |
| 7 | PP | 63600 | 41600–98500 | 5 |
| 8 | MP | 22800 | 10500–49600 | 4 |
| 9 | CC | 16000 | 9200–16700 | 4 |
| 10 | C | 6300 | 1600–17300 | 13 |
| 11 | CCC | 4200 | 400–9400 | 4 |
| 12 | MM | 3400 | 100–6400 | 4 |
| 13 | MMM | 3300 | 100–7000 | 3 |
| 14 | P | 3100 | 1100–7000 | 5 |
| 15 | M | 100 | 0–1000 | 9 |
PPP is the most immunogenic homologous vaccination regimen
Examining first the single-modality regimens, the binding titre for repeated ChAdOx1.BG505s immunizations peaked after the second dose (CC) at a median reciprocal titre of 16000; the third dose was inefficient, likely due to the anti-vector immunity. For the MVA.BG505s-only regimen, the initial titres were very low, but continued to increase with boosting; thus, the third vaccine administration (MMM) boosted the titres to a median of 3300. The sequential delivery of adjuvanted protein trimer (PPP) kept boosting the Ab responses and peaked at 69400 (Table 1).
Heterologous-modality and protein-only regimens induce similar BG505s-binding antibody titres
The best regimen after two immunizations was CM, with a median endpoint reciprocal titre of 141900, ~2-fold higher than the value of 63600 for the PP regimen (Table 1). Based on the peak titres recorded after the third immunization, the rank order for the various regimens was CMP > CPP > MPP > PPP with respective median reciprocal titres of 161300, 158700, 87000 and 69400 (non-significant using Kruskal-Wallis test) (Fig 2B, Table 1). Thus, from the perspective of inducing ELISA-reactive anti-trimer antibodies, priming with one or more virus vector (i.e., CMP or CPP) increases the subsequent response to an adjuvanted-protein boost by over 2-fold compared to the protein-only regimen (PPP).
Broad tier-1A and narrow-specificity tier-2 nAb responses
To assess HIV-1 neutralization, we used a validated TZM-bl cell assay [54] with the tier-1A viruses MN.3 and MW965.26, the BG505ΔCT/T332N vaccine strain and a Global Virus Panel of tier-2 viruses (Table 2). For the MMM, PPP, CPP, MPP and CMP immunization groups, we measured nAb responses using sera from week-26, i.e. 2 weeks after the third/final immunization (Fig 2A). We also included sera from the CCC and CMP groups that were obtained at week 10, which is 2 weeks after the second vector immunization. At that time, these animals had not received a BG505s trimer (P) boost, so for clarity these week-10 sera are designated as CC and CM, respectively. The eight analytical groups included in the nAb comparison sub-study and the resulting titre values are outlined in Table 2. BG505s trimers are known to induce a wide range of autologous nAb titres (from 50 to >10000) in individual rabbits, and tier-2 nAb titres also vary considerably [36, 39]. Such variation limits the power of any comparison between immunization groups that involve 4–5 rabbits. We have therefore generally refrained from making statistical comparisons between the various analytical groups.
Table 2. Neutralization of tier-1 and tier-2 viruses in TZM-bl cell assaya.
| Tier 1A | Tier 2 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clade B | Clade C | Clade A | Clade C | Clade B | CRF07 BC | Clade B | Clade C | Clade AC | CRF07 BC | Clade A | CRF01 AB | ||||
| Regimen | Animal ID | Bleed Week | Negative Control | MN.3 | MW965.26 | BG505ΔCT/T332N | 25710–2.43 | TRO.11 | BJOX002000.03.2 | X1632-S2-B10 | Ce1176 A3 | 246-F3 C10 2 | CH119.10 | Ce703010217 B6 | CNE55 |
| CC | M17 | 10 | <20 | 558 | 2819 | 73 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
| M19 | 10 | <20 | 554 | 895 | 70 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| M23 | 10 | 34 | 584 | 1207 | 40 | 32 | 32 | 28 | 35 | 28 | 35 | 40 | 27 | 24 | |
| F2 | 10 | <20 | 714 | 802 | 33 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| F21 | 10 | <20 | 635 | 1690 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| median | <20 | 584 | 1207 | 40 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | ||
| CCC | M17 | 26 | <20 | <20 | 189 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
| M19 | 26 | <20 | <20 | 76 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| M23 | 26 | <20 | <20 | <20 | 36 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| F2 | 26 | <20 | <20 | 65 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| median | <20 | <20 | 71 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | ||
| MMM | M8 | 26 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
| F20 | 26 | <20 | <20 | 710 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| F31 | 26 | <20 | <20 | <20 | 311 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| median | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | ||
| PPP | M10 | 26 | <20 | 496 | 5296 | 559 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
| M12 | 26 | <20 | 112 | 2171 | 925 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| M27 | 26 | <20 | 61 | 2385 | 45 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| F4 | 26 | <20 | 121 | 13856 | 150 | 80 | 29 | <20 | <20 | 77 | <20 | <20 | <20 | <20 | |
| F13 | 26 | <20 | 1714 | 33189 | 41 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| median | <20 | 117 | 5296 | 150 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | ||
| CPP | M7 | 26 | <20 | 235 | 8618 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
| F5 | 26 | <20 | 879 | 1397 | 151 | 27 | 21 | <20 | <20 | 39 | <20 | <20 | <20 | <20 | |
| F6 | 26 | <20 | 325 | 13309 | 553 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| F22 | 26 | <20 | 87 | 8532 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| median | <20 | 280 | 8575 | 76 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | ||
| MPP | M9 | 26 | <20 | 467 | 1770 | 125 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
| M15 | 26 | <20 | 521 | 8599 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| M18 | 26 | <20 | 1026 | 6008 | 400 | 42 | <20 | <20 | <20 | 45 | <20 | <20 | <20 | <20 | |
| F29 | 26 | <20 | 137 | 847 | 847 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| median | <20 | 494 | 3889 | 263 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | ||
| CM | M11 | 10 | <20 | 1130 | 940 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
| F1 | 10 | <20 | 20545 | 11454 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| F3 | 10 | <20 | 540 | 2770 | 90 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| F26 | 10 | <20 | 6558 | 2732 | 28 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| M16 | 10 | <20 | 2070 | 2071 | 245 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| median | <20 | 2070 | 2732 | 28 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | ||
| CMP | M11 | 26 | <20 | 468 | 1470 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
| F1 | 26 | <20 | 343 | 8624 | 37 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| F3 | 26 | <20 | 397 | 6355 | 2096 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| F26 | 26 | <20 | 6199 | 24627 | 1011 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| median | <20 | 432 | 7490 | 524 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | ||
a—Values are the serum dilution at which relative luminescence units (RLUs) were reduced 50% compared to virus control wells (no test sample). Highlighted bold values are considered positive for neutralization based on the criterion of signal ≤3x the signal of that sample against the MLV-pseudotyped negative-control virus.
The tier-1 nAb titres against the MN.3 and MW965.26 viruses were very low for the CCC and MMM regimens at week 26 (median titres <100). These minimal responses were markedly lower than those seen in the CC and CM groups at week 10 (584/1207 and 2070/2732, respectively, for the MN.3/MW965.26 viruses). The observations imply that the vector-only regimens induce early tier-1 nAb responses that are either not sustained or are not reboosted by a third immunization at week 24, perhaps due to anti-vector immunity. For the PPP, CPP, MPP, CMP regimens that involved one or more BG505s trimer immunizations the tier-1A nAb titres at week-26 were broadly comparable, the medians for the two test viruses differing by only ~2-fold. Thus, the CC, CM, CPP, MPP, CMP and PPP regimens induced similar levels of nAbs against the two tier-1 viruses.
The CC, CM (week 10) and CCC (week 26) regimens induced only sporadic and low (<75) autologous tier-2 nAb titres. Most of the rabbits receiving the PPP, CPP, MPP and CMP regimens developed autologous tier-2 nAb responses at week 26. Within the limitations imposed by the small group sizes, it seems reasonable to conclude that the CMP, MPP and PPP regimens were comparably effective at inducing nAbs against the autologous tier-2 virus.
Finally, sera from three rabbits, one from the PPP regimen (animal F4), one from CPP (animal F5) and one from MPP (animal M18), weakly neutralized (titres <100) 2 or 3 viruses in the tier-2 Global Panel. The viruses neutralized were TRO.11 (clade B), 25710–2.43 (clade C) and Ce1176_A3 (clade C). It is not possible to identify any pattern in these occasional low-level responses, although we note that all the rabbits had received a trimer boost.
Autologous tier-2 nAb responses are to epitopes involving a glycan hole at residues 241 and 289
We have reported that a PPP regimen induces autologous nAbs in rabbits that often, but not invariably, target a hole in the glycan shield created by the lack of a glycan at position 241 and/or 289 [38]. Thus, BG505.T332N virus mutants with glycans knocked in at these positions via S241N and P291T substitutions could have reduced or absent sensitivity to neutralization by rabbit sera [38]. Therefore, we tested 24 sera from the present study against the same BG505ΔCT/T332N.S241N, BG505ΔCT/T332N.P291T and BG505ΔCT/T332N.S241N.P291T glycan hole knock-in viruses used previously. No serum that was unable to neutralize the parental BG505.T332N virus could neutralize any of the mutant viruses, and both the single and double glycan-knock-in mutations resulted in markedly reduced autologous neutralization titres (Table 3). Thus, the nAb specificities induced in the present study are generally consistent with those we have described previously for the PPP regimen [38].
Table 3. Antibody recognition of glycan holesa.
| Regimen | Animal ID | Bleed Week | BG505ΔCT T332N | BG505ΔCT T332N.S241N | BG505ΔCT T332N.P291T | BG505ΔCT T332N.S241N.P291T |
|---|---|---|---|---|---|---|
| M17 | 26 | nd | nd | nd | nd | |
| CCC | M19 | 26 | <20 | nd | nd | nd |
| M23 | 26 | <20 | <20 | <20 | <20 | |
| F2 | 26 | <20 | <20 | <20 | <20 | |
| M8 | 26 | <20 | <20 | <20 | <20 | |
| MMM | F20 | 26 | <20 | <20 | <20 | <20 |
| F31 | 26 | 155 | 68 | 45 | 53 | |
| M10 | 26 | 247 | <20 | <20 | <20 | |
| PPP | M12 | 26 | 743 | 760 | 614 | 484 |
| M27 | 26 | <20 | <20 | <20 | <20 | |
| F4b | 26 | 39 | <20 | 23 | <20 | |
| F13 | 26 | <20 | <20 | <20 | <20 | |
| M7 | 26 | <20 | nd | nd | nd | |
| CPP | F5b | 26 | 74 | 51 | 44 | 58 |
| F6 | 26 | 304 | 261 | 184 | 128 | |
| F22 | 26 | <20 | <20 | <20 | <20 | |
| M9 | 26 | 102 | 68 | 62 | 54 | |
| MPP | M15 | 26 | <20 | <20 | <20 | <20 |
| M18b | 26 | 329 | <20 | <20 | <20 | |
| F29 | 26 | 405 | 30 | 29 | 26 | |
| M11 | 26 | <20 | <20 | <20 | <20 | |
| CMP | F1 | 26 | <20 | <20 | <20 | <20 |
| F3 | 26 | 2590 | 2381 | 1442 | 1237 | |
| F26 | 26 | 304 | <20 | <20 | <20 |
a—Values are the serum dilution at which relative luminescence units (RLUs) were reduced 50% compared to virus control wells (no test sample). Highlighted bold values are considered positive for neutralization based on the criterium of signal ≤3x the signal of that sample against the MLV-pseudotyped negative-control virus;
b—animals neutralized 2 to 3 tier-2 viruses (Table 2); nd—not done due to lack of sample.
Discussion
Developing a vaccine that induces Abs broadly neutralizing globally transmitted HIV-1 variants is a major goal of HIV/AIDS research, but remains a substantial scientific challenge. In the present work, we constructed novel vaccines vectored by chimpanzee adenovirus ChAdOx1 (C) and MVA (M) that express BG505 SOSIP.664 (BG505s) Env proteins and assessed systematically their immunogenicity in homologous and heterologous vaccine regimens that included ISCOMATRIX™-adjuvanted trimer proteins (P). The protein-only (PPP) regimen served as the comparator arm, as this immunization method has been tested previously [37–39]. Overall, viral vectors combined sequentially with adjuvanted BG505s trimers induced similar antibody responses to the PPP regimen.
Gp160 expressed within HIV-1-infected cells is almost always fully cleaved into the gp120 and gp41 subunits by the action of a furin-family protease [36]. Also SOSIP.664 trimers must be cleaved to adopt a stable and native-like structure. For in vitro SOSIP.664 trimer production by transient transfection or in stable CHO or HEK 293T cell lines, furin co-transfection is used to increase the extent of the recombinant gp140 cleavage. This compensates for the abnormally high Env expression levels driven by strong promoters relative to natural HIV-1 infection and a quite poor expression of furin-family proteases by the cells commonly used for trimer production. The extent to which gp140 is cleaved when expressed from virus vectors in vivo is unknown and the answer is not readily obtainable. Comparative immunogenicity experiments using various Env protein immunogens that in vitro do or do not require furin co-expression for the trimers native-like conformation, such as NFL, gp140-sc and UFO, would be required [40]. Here, we analyzed the Env proteins secreted from vaccine-infected HEK 293T cells. Even though neither of the ChAdOx1.BG505s or MVA.BG505s vaccines co-delivered a furin gene, two-thirds and one-third of the Env proteins in the supernatants, respectively, were native-like trimers as judged by their expression of quaternary (trimer-specific) epitopes. Furthermore, the immunogenicity data from our experiments clearly show that a substantial proportion of the expressed SOSIP.664 trimers must have been appropriately cleaved, and hence native-like, when expressed in the rabbits, because otherwise we would not have expected to induce an autologous nAb response. Thus, viral vectors can drive the production of native-like BG505 SOSIP.664 trimers in vitro, although other, non-native forms of Env are also present, and the resulting trimers are immunogenic in rabbits.
We and others have shown that viral vectors including simian adenoviruses and MVA can induce robust CD4+ and CD8+ T-cell responses particularly in heterologous prime-boost regimens [47–50]. Here, the rabbit data suggest equal trimer immunogenicity of the virus vector and protein-only regimens. This might be contributed by the likely strong induction of an Env-specific CD4+ T-cell help. For future regimens aiming to induce both effective CD8+ T cells and bnAbs in parallel and without mutual interference to achieve maximum benefit to the vaccine recipients [1, 3, 4], vaccination regimens may need to be rationally developed to optimize e.g. spatial or temporal separation of the T- and B-cell vaccine administration [58].
It seems unlikely that any single immunogen/Env shape can elicit bnAbs, because their broad specificity evolves from strain-specific Abs through multiple cycles of virus escape and Ab affinity maturation [16, 59, 60]. To mimic this process in the vaccine context may require more complex strategies such as the use of immunogens that engage the germline bnAb precursors [61, 62] followed by guided Ab affinity maturation by sequential isolates from infected individuals who developed bnAbs [31, 63–65]. As virus-vectored vaccines are cheaper to manufacture than protein trimers, they may play a role in delivering candidate Env immunogens in such strategies.
Supporting information
(PDF)
Data Availability
Data are all contained within the paper and/or Supporting Information.
Funding Statement
The work is jointly funded by the Medical Research Council (MRC) UK and the UK Department for International Development (DFID) under the MRC/DFID Concordat agreements (G1001757 and MR/N023668/1); National Institutes of Health grant P01 AI100657; National Institutes of Health grant UM1-AI100645 to the Duke Center for HIV/AIDS Vaccine Immunology-Immunogen Discovery (CHAVI-ID) - subcontract 210782; National Institutes of Health contract HHSN27201100016C; European AIDS Vaccine Initiative (EAVI) 2020 (681137); T.H. and Q.S. are the Jenner Institute Investigators, and Q.S. is a James Martin Senior fellow. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337(6091):183–6. Epub 2012/07/17. doi: 10.1126/science.1225416 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Julien JP, Lee PS, Wilson IA. Structural insights into key sites of vulnerability on HIV-1 Env and influenza HA. Immunological reviews. 2012;250(1):180–98. Epub 2012/10/11. doi: 10.1111/imr.12005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanke T. Conserved immunogens in prime-boost strategies for the next-generation HIV-1 vaccines. Expert Opin Biol Ther. 2014;14(5):601–16. doi: 10.1517/14712598.2014.885946 . [DOI] [PubMed] [Google Scholar]
- 4.Haynes BF, Shaw GM, Korber B, Kelsoe G, Sodroski J, Hahn BH, et al. HIV-Host Interactions: Implications for Vaccine Design. Cell host & microbe. 2016;19(3):292–303. doi: 10.1016/j.chom.2016.02.002 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, de la Pena AT, et al. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 2014;40(5):669–80. doi: 10.1016/j.immuni.2014.04.008 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annual review of immunology. 2013;31:705–42. doi: 10.1146/annurev-immunol-032712-095916 . [DOI] [PubMed] [Google Scholar]
- 7.Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, et al. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity. 2014;40(5):657–68. doi: 10.1016/j.immuni.2014.04.009 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, et al. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature. 2014. doi: 10.1038/nature13601 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491(7424):406–12. doi: 10.1038/nature11544 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature. 2012. Epub 2012/10/30. doi: 10.1038/nature11604 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mouquet H, Klein F, Scheid JF, Warncke M, Pietzsch J, Oliveira TY, et al. Memory B cell antibodies to HIV-1 gp140 cloned from individuals infected with clade A and B viruses. PloS one. 2011;6(9):e24078 Epub 2011/09/21. doi: 10.1371/journal.pone.0024078 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333(6049):1633–7. Epub 2011/07/19. doi: 10.1126/science.1207227 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Gils MJ, van den Kerkhof TL, Ozorowski G, Cottrell CA, Sok D, Pauthner M, et al. An HIV-1 antibody from an elite neutralizer implicates the fusion peptide as a site of vulnerability. Nat Microbiol. 2016;2:16199 doi: 10.1038/nmicrobiol.2016.199 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477(7365):466–70. Epub 2011/08/19. doi: 10.1038/nature10373 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326(5950):285–9. Epub 2009/09/05. doi: 10.1126/science.1178746 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.West AP Jr., Scharf L, Scheid JF, Klein F, Bjorkman PJ, Nussenzweig MC. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell. 2014;156(4):633–48. doi: 10.1016/j.cell.2014.01.052 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333(6049):1593–602. Epub 2011/08/13. science.1207532 [pii] doi: 10.1126/science.1207532 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329(5993):811–7. Epub 2010/07/10. science.1192819 [pii] doi: 10.1126/science.1192819 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity. 2013;39(2):245–58. Epub 2013/08/06. doi: 10.1016/j.immuni.2013.04.012 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bar KJ, Tsao CY, Iyer SS, Decker JM, Yang Y, Bonsignori M, et al. Early low-titer neutralizing antibodies impede HIV-1 replication and select for virus escape. PLoS pathogens. 2012;8(5):e1002721 doi: 10.1371/journal.ppat.1002721 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Euler Z, van Gils MJ, Bunnik EM, Phung P, Schweighardt B, Wrin T, et al. Cross-reactive neutralizing humoral immunity does not protect from HIV type 1 disease progression. J Infect Dis. 2010;201(7):1045–53. Epub 2010/02/23. doi: 10.1086/651144 . [DOI] [PubMed] [Google Scholar]
- 22.Piantadosi A, Panteleeff D, Blish CA, Baeten JM, Jaoko W, McClelland RS, et al. Breadth of neutralizing antibody response to human immunodeficiency virus type 1 is affected by factors early in infection but does not influence disease progression. Journal of virology. 2009;83(19):10269–74. Epub 2009/07/31. doi: 10.1128/JVI.01149-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gauduin MC, Parren PW, Weir R, Barbas CF, Burton DR, Koup RA. Passive immunization with a human monoclonal antibody protects hu-PBL-SCID mice against challenge by primary isolates of HIV-1. Nature medicine. 1997;3(12):1389–93. . [DOI] [PubMed] [Google Scholar]
- 24.Hessell AJ, Rakasz EG, Poignard P, Hangartner L, Landucci G, Forthal DN, et al. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS pathogens. 2009;5(5):e1000433 doi: 10.1371/journal.ppat.1000433 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hofmann-Lehmann R, Vlasak J, Rasmussen RA, Smith BA, Baba TW, Liska V, et al. Postnatal passive immunization of neonatal macaques with a triple combination of human monoclonal antibodies against oral simian-human immunodeficiency virus challenge. Journal of virology. 2001;75(16):7470–80. doi: 10.1128/JVI.75.16.7470-7480.2001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nature medicine. 2000;6(2):207–10. doi: 10.1038/72318 . [DOI] [PubMed] [Google Scholar]
- 27.Mc Cann CM, Song RJ, Ruprecht RM. Antibodies: can they protect against HIV infection? Curr Drug Targets Infect Disord. 2005;5(2):95–111. . [DOI] [PubMed] [Google Scholar]
- 28.Moldt B, Rakasz EG, Schultz N, Chan-Hui PY, Swiderek K, Weisgrau KL, et al. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(46):18921–5. doi: 10.1073/pnas.1214785109 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shingai M, Donau OK, Plishka RJ, Buckler-White A, Mascola JR, Nabel GJ, et al. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. The Journal of experimental medicine. 2014;211(10):2061–74. doi: 10.1084/jem.20132494 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veazey RS, Shattock RJ, Pope M, Kirijan JC, Jones J, Hu Q, et al. Prevention of virus transmission to macaque monkeys by a vaginally applied monoclonal antibody to HIV-1 gp120. Nature medicine. 2003;9(3):343–6. doi: 10.1038/nm833 . [DOI] [PubMed] [Google Scholar]
- 31.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496(7446):469–76. doi: 10.1038/nature12053 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore JP. HIV-1 Env antibodies: are we in a bind or going blind? Nature medicine. 2012;18(3):346–7; author reply 7–8. doi: 10.1038/nm.2689 . [DOI] [PubMed] [Google Scholar]
- 33.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS pathogens. 2013;9(9):e1003618 doi: 10.1371/journal.ppat.1003618 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, et al. Crystal Structure of a Soluble Cleaved HIV-1 Envelope Trimer. Science. 2013. Epub 2013/11/02. doi: 10.1126/science.1245625 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342(6165):1484–90. doi: 10.1126/science.1245627 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ringe RP, Sanders RW, Yasmeen A, Kim HJ, Lee JH, Cupo A, et al. Cleavage strongly influences whether soluble HIV-1 envelope glycoprotein trimers adopt a native-like conformation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(45):18256–61. Epub 2013/10/23. doi: 10.1073/pnas.1314351110 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Taeye SW, Ozorowski G, Torrents de la Pena A, Guttman M, Julien JP, van den Kerkhof TL, et al. Immunogenicity of Stabilized HIV-1 Envelope Trimers with Reduced Exposure of Non-neutralizing Epitopes. Cell. 2015;163(7):1702–15. doi: 10.1016/j.cell.2015.11.056 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klasse PJ, LaBranche CC, Ketas TJ, Ozorowski G, Cupo A, Pugach P, et al. Sequential and Simultaneous Immunization of Rabbits with HIV-1 Envelope Glycoprotein SOSIP.664 Trimers from Clades A, B and C. PLoS pathogens. 2016;12(9):e1005864 doi: 10.1371/journal.ppat.1005864 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, et al. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349(6244):aac4223 doi: 10.1126/science.aac4223 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanders RW, Moore JP. Native-like Env trimers as a platform for HIV-1 vaccine design. Immunological reviews. 2017;275(1):161–82. doi: 10.1111/imr.12481 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biswas S, Choudhary P, Elias SC, Miura K, Milne KH, de Cassan SC, et al. Assessment of humoral immune responses to blood-stage malaria antigens following ChAd63-MVA immunization, controlled human malaria infection and natural exposure. PloS one. 2014;9(9):e107903 doi: 10.1371/journal.pone.0107903 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Douglas AD, de Cassan SC, Dicks MD, Gilbert SC, Hill AV, Draper SJ. Tailoring subunit vaccine immunogenicity: maximizing antibody and T cell responses by using combinations of adenovirus, poxvirus and protein-adjuvant vaccines against Plasmodium falciparum MSP1. Vaccine. 2010;28(44):7167–78. Epub 2010/10/13. doi: 10.1016/j.vaccine.2010.08.068 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Draper SJ, Biswas S, Spencer AJ, Remarque EJ, Capone S, Naddeo M, et al. Enhancing blood-stage malaria subunit vaccine immunogenicity in rhesus macaques by combining adenovirus, poxvirus, and protein-in-adjuvant vaccines. Journal of immunology. 2012;185(12):7583–95. Epub 2010/11/26. doi: 10.4049/jimmunol.1001760 . [DOI] [PubMed] [Google Scholar]
- 44.Elias SC, Choudhary P, de Cassan SC, Biswas S, Collins KA, Halstead FD, et al. Analysis of human B-cell responses following ChAd63-MVA MSP1 and AMA1 immunization and controlled malaria infection. Immunology. 2014;141(4):628–44. doi: 10.1111/imm.12226 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ewer K, Rampling T, Venkatraman N, Bowyer G, Wright D, Lambe T, et al. A Monovalent Chimpanzee Adenovirus Ebola Vaccine Boosted with MVA. N Engl J Med. 2016;374(17):1635–46. doi: 10.1056/NEJMoa1411627 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheehy SH, Duncan CJ, Elias SC, Choudhary P, Biswas S, Halstead FD, et al. ChAd63-MVA-vectored blood-stage malaria vaccines targeting MSP1 and AMA1: assessment of efficacy against mosquito bite challenge in humans. Mol Ther. 2013;20(12):2355–68. Epub 2012/10/24. doi: 10.1038/mt.2012.223 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borthwick N, Ahmed T, Ondondo B, Hayes P, Rose A, Ebrahimsa U, et al. Vaccine-elicited human T cells recognizing conserved protein regions inhibit HIV-1. Mol Ther. 2014;22:464–75. Epub 2013/10/30. doi: 10.1038/mt.2013.248 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ewer KJ, O'Hara GA, Duncan CJ, Collins KA, Sheehy SH, Reyes-Sandoval A, et al. Protective CD8(+) T-cell immunity to human malaria induced by chimpanzee adenovirus-MVA immunisation. Nat Commun. 2013;4:2836 Epub 2013/11/29. doi: 10.1038/ncomms3836 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mutua G, Farah B, Langat R, Indangasi J, Ogola S, Onsembe B, et al. Broad HIV-1 inhibition in vitro by vaccine-elicited CD8+ T cells in African adults. Mol Ther Methods Clin Dev. 2016;3:16061 doi: 10.1038/mtm.2016.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rampling T, Ewer KJ, Bowyer G, Bliss CM, Edwards NJ, Wright D, et al. Safety and High Level Efficacy of the Combination Malaria Vaccine Regimen of RTS,S/AS01B With Chimpanzee Adenovirus 63 and Modified Vaccinia Ankara Vectored Vaccines Expressing ME-TRAP. J Infect Dis. 2016;214(5):772–81. doi: 10.1093/infdis/jiw244 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dicks MDJ, Spencer AJ, Edwards NJ, Wadell G, Bojang K, Gilbert SC, et al. A novel chimpanzee adenovirus vector with low human seroprevalence: improved systems for vector derivation and comparative immunogenicity. PloS one. 2012;7:e40385 doi: 10.1371/journal.pone.0040385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ondondo B, Murakoshi H, Clutton G, Abdul-Jawad S, Wee EG, Gatanaga H, et al. Novel Conserved-region T-cell Mosaic Vaccine With High Global HIV-1 Coverage Is Recognized by Protective Responses in Untreated Infection. Mol Ther. 2016;24(4):832–42. doi: 10.1038/mt.2016.3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Julien JP, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(11):4351–6. Epub 2013/02/22. doi: 10.1073/pnas.1217537110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarzotti-Kelsoe M, Bailer RT, Turk E, Lin CL, Bilska M, Greene KM, et al. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J Immunol Methods. 2013. doi: 10.1016/j.jim.2013.11.022 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.deCamp A, Hraber P, Bailer RT, Seaman MS, Ochsenbauer C, Kappes J, et al. Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. Journal of virology. 2014;88(5):2489–507. doi: 10.1128/JVI.02853-13 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abdul-Jawad S, Ondondo B, van Hateren A, Gardner A, Elliott T, Korber B, et al. Increased Valency of Conserved-mosaic Vaccines Enhances the Breadth and Depth of Epitope Recognition. Mol Ther. 2016;24:375–84. doi: 10.1038/mt.2015.210 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X, Liang G. Dual aggregation-induced emission for enhanced fluorescence sensing of furin activity in vitro and in living cells. Chem Commun (Camb). 2016. doi: 10.1039/c6cc09106g . [DOI] [PubMed] [Google Scholar]
- 58.Clutton G, Carpov A, Parks CL, Dean HJ, Montefiori DC, Hanke T. Optimizing parallel induction of HIV type 1-specific antibody and T-cell responses by multicomponent subunit vaccines. Aids. 2014;28(17):2495–504. doi: 10.1097/QAD.0000000000000468 . [DOI] [PubMed] [Google Scholar]
- 59.Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol. 2015;16(6):571–6. doi: 10.1038/ni.3158 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Taeye SW, Moore JP, Sanders RW. HIV-1 Envelope Trimer Design and Immunization Strategies To Induce Broadly Neutralizing Antibodies. Trends Immunol. 2016;37(3):221–32. doi: 10.1016/j.it.2016.01.007 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jardine JG, Ota T, Sok D, Pauthner M, Kulp DW, Kalyuzhniy O, et al. HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science. 2015;349(6244):156–61. doi: 10.1126/science.aac5894 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McGuire AT, Dreyer AM, Carbonetti S, Lippy A, Glenn J, Scheid JF, et al. HIV antibodies. Antigen modification regulates competition of broad and narrow neutralizing HIV antibodies. Science. 2014;346(6215):1380–3. doi: 10.1126/science.1259206 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dosenovic P, von Boehmer L, Escolano A, Jardine J, Freund NT, Gitlin AD, et al. Immunization for HIV-1 Broadly Neutralizing Antibodies in Human Ig Knockin Mice. Cell. 2015;161(7):1505–15. doi: 10.1016/j.cell.2015.06.003 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol. 2012;30(5):423–33. Epub 2012/05/09. doi: 10.1038/nbt.2197 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sliepen K, Medina-Ramirez M, Yasmeen A, Moore JP, Klasse PJ, Sanders RW. Binding of inferred germline precursors of broadly neutralizing HIV-1 antibodies to native-like envelope trimers. Virology. 2015;486:116–20. doi: 10.1016/j.virol.2015.08.002 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Data Availability Statement
Data are all contained within the paper and/or Supporting Information.